Abstract
Disruption of the gene encoding Protein Tyrosine Kinase 6 (Ptk6) delayed differentiation and increased growth in the mouse intestine. However, Ptk6 null mice were also resistant to azoxymethane-induced colon tumorigenesis. To further explore functions of PTK6 in colon cancer, expression of epithelial and mesenchymal markers, as well as proliferation, migration, and xenograft tumor growth were examined in human colon tumor cell lines with knockdown or overexpression of PTK6. PTK6 protein, transcript, and activation were also examined in a human colon tumor tissue array, using immunohistochemistry (IHC) and qRT-PCR. Knockdown of PTK6 led to the epithelial-mesenchymal transition (EMT) in SW480 and HCT116 cells, while overexpression of PTK6 in SW620 cells restored an epithelial phenotype in a kinase-independent manner. PTK6 knockdown also increased xenograft tumor growth of SW480 cells, suggesting tumor suppressor functions. In clinical specimens, PTK6 expression was highest in normal differentiated epithelial cells and reduced in tumors. In contrast, overexpression of constitutively active PTK6 promoted STAT3 and ERK5 activation in colon cancer cells, and endogenous PTK6 promoted cell survival and oncogenic signaling in response to DNA damaging treatments. These data indicate that PTK6 has complex, context specific functions in colon cancer; PTK6 promotes the epithelial phenotype to antagonize the EMT in a kinase-independent manner, while activation of PTK6 promotes oncogenic signaling.
INTRODUCTION
Protein Tyrosine Kinase 6 (PTK6, also referred to as BRK) is an intracellular tyrosine kinase expressed in differentiated epithelial cells of the skin, gastrointestinal tract, and prostate (1). PTK6 is distantly related to the SRC-family and contains SRC-homology SH2 and SH3 domains, but lacks an amino terminal SH4 domain that directs lipid modification and membrane targeting. Mice with systemic disruption of Ptk6 were viable, but exhibited increased growth and delayed epithelial differentiation in the small intestine (2). Although PTK6 is normally expressed in nondividing differentiated cells in the gastrointestinal tract, PTK6 expression was induced in proliferating progenitor cells following DNA damage where it promoted DNA damage-induced apoptosis (3). While functions elucidated for PTK6 in normal epithelia suggested that it might have tumor suppressor functions, PTK6 promoted azoxymethane induced colon tumorigenesis in mouse models by promoting activation of STAT3 and tumor initiation (4). These data illustrate the context dependent functions of PTK6 and highlight opposing roles for PTK6 in normal epithelia and cancer.
PTK6 has been most widely studied in breast cancer. It is induced and activated in a majority of human breast tumors, where it promotes oncogenic signaling and breast tumor growth (5). In prostate cancers, PTK6 is activated and associated with the plasma membrane, where it promotes tumorigenesis and the epithelial to mesenchymal transition (EMT) (6) [reviewed in (7)]. The PTK6 gene is amplified and PTK6 is overexpressed in high-grade serous ovarian carcinomas and may promote the development and growth of ovarian tumors (8). PTK6 has been identified as a potential therapeutic target in non-small cell lung carcinoma, where its expression is increased (9) and associated with decreased survival (10). It was shown that PTK6 is expressed in pancreatic cancer and pancreatic cancer cell lines, and knockdown of PTK6 reduced migration and invasion of pancreatic cancer cells (11).
Interestingly, recent studies suggest that PTK6 may also have tumor suppressor roles in some epithelial cancers. It is down-regulated in human esophageal squamous cell carcinomas (ESCC) (12, 13) and knockdown of PTK6 in human ESCC cells enhanced xenograft tumor growth (12). Low PTK6 expression correlates with poor prognosis in patients with laryngeal squamous cell carcinoma (14). PTK6 expression is also reduced with increasing malignancy in squamous cell carcinomas of the skin (15, 16) and oral mucosa (17).
Dedifferentiation, loss of adhesive constraints, and enhanced motility are hallmarks of the EMT, and are characteristic of metastatic cancer cells. In contrast to findings in breast cancer and prostate cancer, we found that knockdown of PTK6 in colon cancer cell lines led to increased growth and cell migration, as well as increased xenograft tumor growth. In patient colon tumor samples, we demonstrate decreased PTK6 expression in less differentiated, more metastatic tumors. Our findings indicate that although PTK6 plays a prominent role in promoting colon tumor initiation in a mouse model (4), this kinase also has tumor suppressor functions in established tumors where it contributes to the maintenance of the epithelial phenotype. Our data underscore the importance of understanding the context-dependent functions of potential therapeutic targets.
MATERIALS & METHODS
Cell Lines
SW480, SW620, and HCT116 cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and cultured as per their instructions. ATCC authenticates human cell lines using a variety of techniques including morphology, karyotyping, and PCR based approaches.
PTK6 Expression and Knockdown
Myc-tagged full-length human wild-type (WT), constitutively active (YF), and kinase-defective (KM) PTK6 recombinant constructs in the pBABE-puro vector (Cell Biolabs, Inc., San Diego, CA) have been described (18). Empty vector was used as a control. Lentiviruses expressing TCRN0000021549 (shRNA49), TCRN0000021552 (shRNA52), from the PTK6 Mission TCR shRNA Target Set (Sigma-Aldrich) and scrambled control (shSCR) were used to generate SW480 and HCT116 cells with stable knockdown of PTK6. For both overexpression and knockdown cells, stable pools were selected in growth medium containing 2 μg/mL puromycin (Sigma-Aldrich, St. Louis, MO) for 2 weeks, stable lines were maintained in selection media.
Immunoblotting, immunohistochemistry, and antibodies
Protein preparation, immunoblotting, and immunohistochemistry were performed as described (19) (20). For immunofluorescence, cells grown on coverslips were fixed with Carnoys solution (6:3:1 ethanol:chloroform:acetic acid), and biotinylated secondary anti-mouse or anti-rabbit antibody (Vector Laboratories; Burlingame, CA) binding was detected with fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories) or AlexaFluor 594-conjugated avidin (Life Technologies Corp., Carlsbad, CA). Coverslips were mounted on slides with mounting medium containing DAPI (Vector Laboratories). Antibodies against human PTK6 (G6, sc-166171), E-Cadherin (sc-7870), and ZEB1 (sc-25388) were purchased from Santa Cruz Biotechnology. Antibodies against N-Cadherin (13116), ZO-1 (8193), Vimentin (5741), Claudin-1 (13255), and GAPDH (2118) were purchased from Cell Signaling Technology, and β-Actin (A5411) from Sigma-Aldrich. Antibodies against inactive PTK6 (PY447; ab138368), and Ki67 (ab16667) were purchased from Abcam (Cambridge, MA). Antibody against active PTK6 (PY342; 09-144) and was purchased from EMD Millipore (Temecula, CA). Densitometry analysis was done using ImageJ software (NIH) and values were normalized to loading control.
Cell Line quantitative real-time PCR (qRT-PCR) and primers
Total RNA was extracted from subconfluent cultures with TRIzol reagent (Invitrogen, Carlsbad, CA). 1 μg of RNA was used to generate cDNA using a cDNA synthesis kit (Roche, Indianapolis, IN). qPCR was performed using SYBR Green I reagent (Roche) on a Roche LightCycler 480 II instrument with the following primers: PTK6 (Fwd: 5′-CGGACCCGTGGTTCTTTG-3′, Rev: 5′-ACTCGGCTTCTCGCTGAC -3′); CDH1 (Fwd: 5′-ATGCTGATGCCCCCAATACC-3′, Rev: 5′-TCCAAGCCCTTTGCTGTTTTC -3′); ZEB1 (Fwd: 5′-AACGCTTTTCCCATTCTGGC -3′, Rev: 5′-GAGATGTCTTGAGTCCTGTTCTTGG -3′); SNAI1 (Fwd: 5′-CTGCGGGAAGGCCTTCTCT -3′, Rev: 5′-CGCCTGGCACTGGTACTTCTT -3′), SNAI2 (Fwd: 5′-GCTCAGAAAGCCCCATTAGTGATG-3′, Rev: 5′-GCCAGCCCAGAAAAAGTTGAATAG-3′), TWIST1 (Fwd: 5′-GTCCGCAGTCTTACGAGGAG -3′, Rev: 5′-CCAGCTTGAGGGTCTGAATC -3′), and VIMENTIN (Fwd: 5′-TTGACAATGCGTCTCTGGCAC-3′, Rev: 5′-CCTGGATTTCCTCTTCGTGGAG-3′), CYCLOPHILIN (Fwd: 5′-GCAGACAAGGTCCCAAAGACAG -3′, Rev: 5′-CACCCTGACACATAAACCCTGG -3′), GAPDH (Fwd: 5′-AATGAAGGGGTCATTGATGG-3′, Rev: 5′-AAGGTGAAGGTCGGAGTCAA-3′); andB2M (Fwd: 5′-TCTCTGCTGGATGACGTGAG-3′, Rev: 5′-TAGCTGTGCTCGCGCTACT -3′). Gene expression levels were calculated using the standard curve method and normalized to the average of the three reference genes.
Proliferation, wound-healing, migration, and soft agar assays
For proliferation assays, 5 x 105 cells were seeded in triplicate in 6-well plates on day 0; at each timepoint, live cells were selected by trypan blue exclusion and quantified using the Countess Automated Cell Counter (Invitrogen). Wound healing and migration assays have been described (20). For soft agar assays, 1 x 104 cells were resuspended in 0.35% low-melting agarose and plated over a 0.7% agarose base in wells of 6-well plates. Cells were monitored and supplemented with normal growth media every two days.
DNA Damaging Cell Treatment
For apoptosis experiments cells were plated at 1 x 106 cells/plate on 10 cm dishes 16 hours before treatment. Fresh medium containing either DMSO (control cells) or 5 μM or 10 μM of doxorubicin (Sigma) was added to the cells and incubated for 24 hours at 37°C. Both floating and adherent cells were harvested for protein lysate.
Xenograft assays
Animal studies were approved by the UIC Institutional Animal Care and Use Committee. Female athymic nude Foxn1nu mice were purchased from Harlan laboratories. 5 x 106 cells were resuspended in 100 μl of Matrigel and injected subcutaneously into the flanks of 7-week old mice; SW480 cells expressing shScr on the left side and shRNA49 or shRNA52 on the right side. Five animals were used for each for shRNA49 and shRNA52 cell lines, for a total of ten animals. Palpable tumors were first detected after one week and animals were sacrificed after two weeks due to humane endpoint conditions.
Clinical samples and tissue microarrays (TMAs)
Studies involving patient-derived tissues were approved by the UIC Institutional Review Board. Formalin-fixed paraffin-embedded tissues were obtained from patients diagnosed with colorectal cancer at the University of Illinois Medical Center. One hundred thirty one (n=131) colorectal resection specimens for colonic adenocarcinoma TMAs were designed and constructed based on prescribed practices (21). A total of four TMA recipient blocks were prepared comprising duplicated cores representing normal colonic mucosa (NCM), hyperplastic colonic mucosa (HCM), dysplastic colonic mucosa (DCM) and colonic adenocarcinoma (CAC) tissue cores from all 131 colorectal resection specimens included in the study. Four-micron sections from the TMAs were placed on positively charged glass slides for immunohistochemical studies.
Quantitative real-time PCR (qPCR) and primers for patient samples
Tissue samples were collected from adenocarcinoma colorectal cancer patients and polyp-free, tumor free controls from the Chicago Colorectal Cancer Consortium (22). RNA was extracted using the Maxwell® 16 Total RNA Purification Kit from Promega (Madison, WI, USA) according to manufacture’s protocol. cDNA was prepared using the iScript cDNA synthesis kit (Biorad; Hercules, CA). qPCR was performed using the FASTSTART Universal Probe Master reagent (Roche) on the VIIA7 instrument (Applied Biosystems, Life Technologies). The following human taqman probes (cat. no. 4331182) were purchased from Applied Biosystems and used for gene expression: PTK6 (Hs00963386_m1) and E-cadherin (CDH1; Hs01023894_m1); β2 microglobulin (B2M; Hs00984230_m1) was used as a reference gene (23). Expression levels were calculated using the ddCt method and normalized to B2M.
Statistical analysis
For cell culture studies, one-tailed t-tests were performed to determine p-values (Microsoft Excel). For xenograft studies, paired t-tests were performed to determine p-values (Microsoft Excel). To evaluate whether the ordinal epithelial score and stroma score were statistically different among different colon cancer stages in the TMA cores, we used ordinal logistic regression models (Fig. 6C). To account for the correlation for scores from different cell types obtained from the same subject, we used the Generalized Estimation Equation (GEE) (24) (Fig. 6B), PROC GENMOD in SAS 9.4 (Cary, NC) was used to perform the analysis. For analysis of the patient-derived qPCR studies, Ct values for PTK6 were normalized to the B2M housekeeping gene for each sample; the resulting values were averaged for adenocarcinoma cases and polyp-free controls. A non-parametric Mann-Whitney U test was performed to determine the P-value comparing the two groups. A regression analysis (Graphpad PRISM; La Jolla, CA) was performed to calculate the R-square value for the correlation between PTK6 and CDH1 expression.
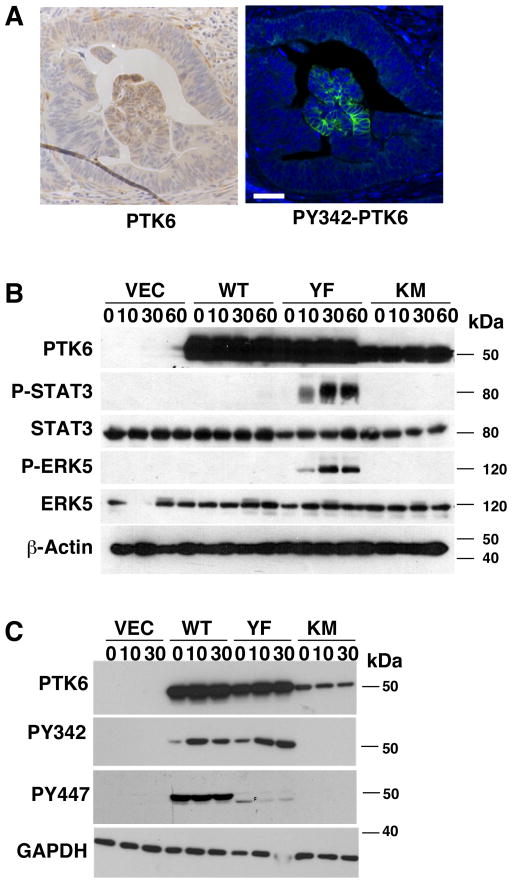
Figure 6. PTK6 Kinase Activity in Colon Tumors May Promote Oncogenic Signaling.
(A) Serial sections of patient tissue were stained for total PTK6 (left panel) and active PTK6 PY342 (right panel). Active PTK6 (PY342) was infrequently observed in colon cancer sections; an example of a rare patch of PY342 positive cells is shown. Size bar = 50 μm. (B, C) Activation of PTK6 and its downstream signaling were examined in serum starved and stimulated stable SW620 cell lines. SW620 cells expressing PTK6-WT, -YF, or -KM, empty vector (VEC) were serum starved (0 min) and stimulated with 20% FBS-containing media for indicated timepoints (minutes). Immunoblotting was performed with antibodies against PTK6, P-STAT3 (PY705), STAT3, P-ERK5, and ERK5 (B), and PTK6, PY342-PTK6 and PY447-PTK6 (C). (B) Phosphorylation of STAT3 at Y705, a substrate of PTK6, and activating phosphorylation of ERK5 were only detected in the PTK6-YF cells. (C) While both PTK-WT and PTK6-YF display activating phosphorylation (PY342), PTK6-WT is also subjected to inhibitory phosphorylation on Y447. β-Actin and GAPDH were used as loading controls.
RESULTS
PTK6 Knockdown Results in an EMT Phenotype in Colon Cancer Cells
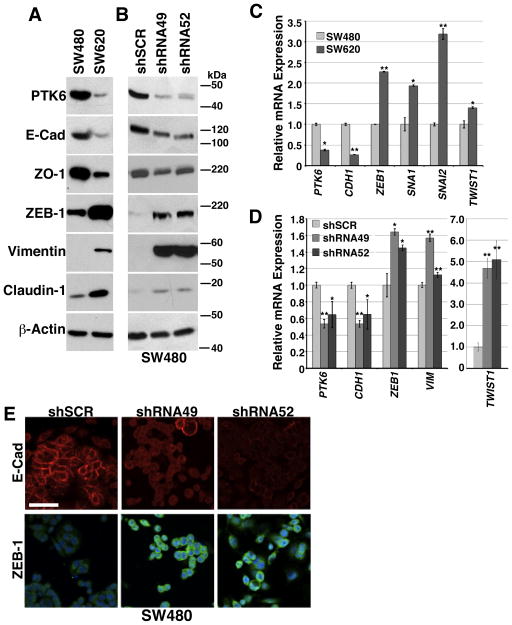
PTK6 is expressed in differentiated epithelial cells of the murine small intestine and colon. Disruption of the Ptk6 gene led to impaired differentiation and increased proliferation in the mouse small intestine (2). To determine if PTK6 plays a role in the EMT in these tissues, we used the human colon adenocarcinoma cell lines SW480 and SW620, which represent a primary colon tumor and a lymph node metastasis from the same patient, respectively (25). Using immunoblotting, we confirmed that SW480 cells have an epithelial phenotype, and are characterized by high levels of E-Cadherin and ZO-1 protein expression and relatively low levels of ZEB1, Vimentin, and Claudin-1 (Fig. 1A), while SW620 cells appear to have undergone an EMT and have an opposite protein expression profile. PTK6 protein (Fig. 1A) and mRNA (Fig. 1C) expression are reduced in the mesenchymal-like SW620 cell line compared with the epithelial-like SW480 cell line. Decreased expression of CDH1, the gene that encodes E-Cadherin, as well as increased expression of the mesenchymal markers ZEB1, SNAI1, SNAI2, and TWIST1 was detected in SW620 cells (Fig. 1C). In both the SW480 and SW620 cell lines, most PTK6 is localized in the cytoplasm, and basal activation measured by phosphorylation of tyrosine residue 342 could not be detected (Fig. S1 A).
Figure 1. PTK6 promotes the epithelial phenotype in human colon cancer cell lines.
Relative expression of PTK6, the epithelial markers E-Cadherin and ZO-1, and the mesenchymal markers N-Cadherin, ZEB-1, Claudin-1 and Vimentin was examined in SW480 and SW620 colon cancer cell lines, and SW480 cells stably expressing shRNA constructs targeting PTK6, shRNA49 and shRNA52, or a non-targeted shRNA control, shSCR. Immunoblotting was performed using total cell lysates from preconfluent cultures. (A) SW480 and SW620 parental cell lines and (B) SW480 PTK6 knockdown cell lines. Expression of RNAs encoding EMT markers was analyzed using qRT-PCR (CDH1 = E-Cadherin, SNAI1 = Snail, SNAI2 = Slug). (C) parental SW480 and SW620 cell lines and (D) SW480 PTK6 knockdown cell lines (*p<0.05 **p<0.005). (E) Immunofluorescent staining for protein expression and localization of E-Cadherin and ZEB-1 in SW480 PTK6 control and knockdown cells is shown (size bar = 100 μm).
To study the role of PTK6 in the EMT phenotype, two different shRNA sequences, shRNA49 and shRNA52 were used to stably knock down PTK6 expression in SW480 cells. These shRNAs target different regions of PTK6; shRNA49 targets the 3′ UTR, while shRNA52 targets coding sequence. A non-targeted scrambled shRNA, shSCR, was used as a control. Stable knockdown of PTK6 with both shRNAs results in reduced expression of E-Cadherin and ZO-1 and increased expression of ZEB-1, Vimentin and Claudin-1 in SW480 cells (Fig. 1B). We observed a similar result by qPCR analysis for mRNA expression levels; PTK6 knockdown results in decreased CDH1 (E-cadherin) and increased ZEB1, VIM, and dramatic increases in TWIST1 expression in both shRNA49 and shRNA52 expressing cell lines (Fig. 1D). Immunofluorescent staining demonstrates reduced membrane staining for E-Cadherin and increased nuclear staining for ZEB-1 in PTK6 knockdown SW480 cells (Fig. 1E). We also found that knockdown of PTK6 led to a more mesenchymal phenotype in HCT116 cells (Fig. S1 B), while epithelial differentiation in the Caco-2 cell line correlated with increased PTK6 expression (Fig. S1 C). These data support a functional role for PTK6 in promoting and maintaining the epithelial phenotype in colon cancer cells and that the loss of PTK6 results in an EMT phenotype.
PTK6 Plays a Tumor Suppressor Role in SW480 Colon Cancer Cells
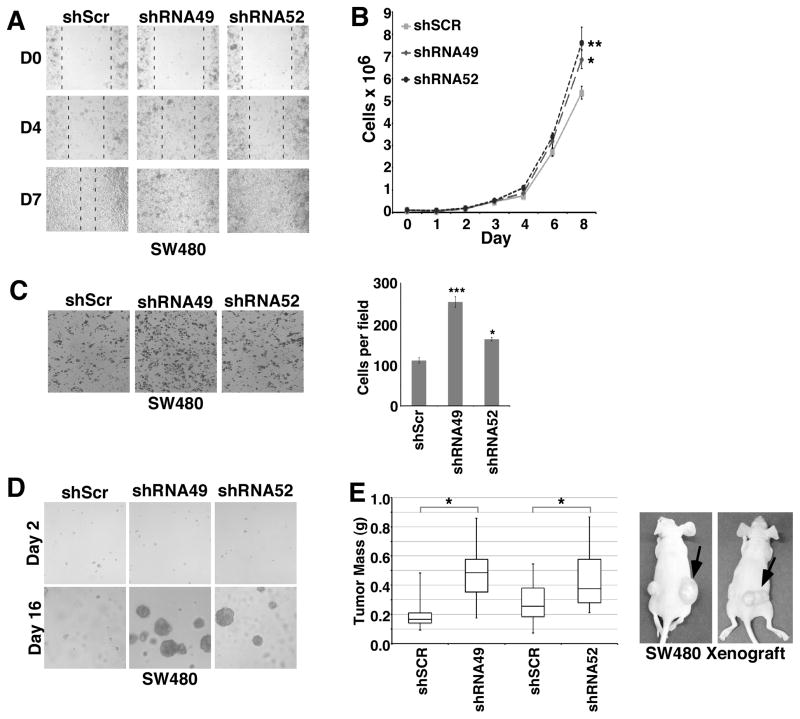
In addition to changes in gene and protein expression, tumor cells that have undergone the EMT are characterized by increased proliferative and migratory properties that promote their dissociation from primary tumors and metastasis to distant sites (26). Wound healing is a physiological process for which the EMT is required for epithelial cells to proliferate and migrate to fill in a wounded region. When SW480 cells were allowed to grow to confluence and then wounded, the PTK6 knockdown lines, shRNA49 and shRNA52, demonstrated increased wound healing capacity when compared with control cells (Fig. 2A). PTK6 knockdown confers a statistically significant proliferative advantage to SW480 cells compared with control cells (Fig. 2B, *p < 0.03, **p < 0.02). PTK6 knockdown also resulted in increased migration when cells were plated on transwells and stimulated with serum to migrate across a porous membrane (Fig. 2C, ***p<0.001, *p<0.02).
Figure 2. PTK6 plays a tumor suppressor role in SW480 cells.
Functional assays were performed with SW480 cells with stable knockdown of PTK6 (shRNA49 and shRNA52), or a non-targeted scrambled shRNA (shSCR) as a control to further examine the EMT phenotype. (A) Confluent cells were serum starved for 24 hours, wounded with a yellow pipette tip, stimulated with normal growth media and photographed daily to monitor wound healing. Dashed lines mark the boundaries of the wound in each panel. (B) An increase in cell proliferation was detected in stable cells with knockdown of PTK6. Equal cell numbers were plated in triplicate for each timepoint, then trypsinized and quantified to assess cell proliferation (*p < 0.03, **p < 0.02). (C) Increased cell migration is observed following knockdown of PTK6. Cells were plated in transwells, grown to confluence and serum starved for 24 hours. Growth media containing 20% serum was added to the lower chamber as a chemoattractant, and following their migration, cells were fixed and stained with crystal violet. Graph represents quantification of migrated cells (***p < 0.001 *p < 0.02). (D) Cells with knockdown of PTK6 form large colonies in soft agar. Cells were grown under anchorage-independent conditions in 0.35% agar and supplemented with normal growth media to assess resistance to anoikis. (E) Knockdown of PTK6 resulted in increased xenograft tumor growth (*p < 0.05). 5 x 106 SW480 shRNA49 or shRNA52 cells were resuspended in 100 μl matrigel and injected subcutaneously into the right flanks of nude mice, while equal numbers of control shSCR cells in matrigel were injected into the left flanks to serve as internal controls.
In order for tumor cells to metastasize, they must be able to evade anoikis and survive in the circulation. To assess contributions of PTK6 to anchorage-independent survival and growth, SW480 cell lines were grown in 3D soft agar, which does not contain any ECM structures or proteins. Knockdown of PTK6 in SW480 cells confers an anchorage-independent survival and growth advantage in both shRNA49 and shRNA52 cell lines, which formed large colonies when grown in soft agar, as compared with shSCR control cells (Fig. 2D).
To examine the role of PTK6 in tumorigenesis in vivo, we injected the SW480 cell lines subcutaneously into the flanks of nude mice. In each mouse, shSCR control cells were injected into the left flank and either shRNA49 or shRNA52 cells were injected into the right flank. Both PTK6 knockdown cell lines formed larger tumors than the scrambled control cells demonstrating by xenograft assay that PTK6 knockdown enhances SW480 tumorigenesis in nude mice (Fig. 2E, *p<0.05).
PTK6 Overexpression Reverses the EMT Phenotype in SW620 Cells
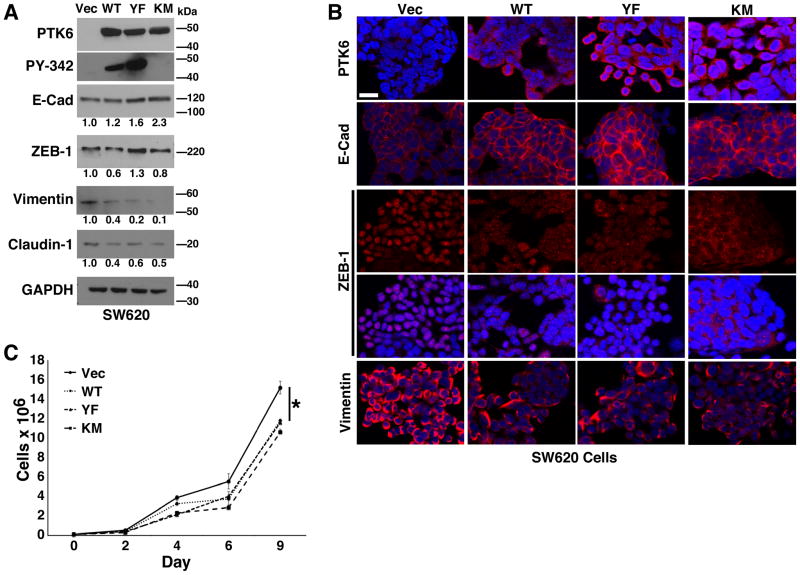
To determine contributions of PTK6 in promoting an epithelial phenotype, we stably overexpressed full length wild-type (PTK6-WT), constitutively active (PTK6-YF), and kinase-dead (PTK6-KM) PTK6, or empty vector as a control in the metastatic SW620 cell line (19). PTK6-YF has a substitution of the regulatory tyrosine, Y447, to phenylalanine, resulting in a constitutively active kinase that cannot be negatively regulated by tyrosine phosphorylation at its carboxy terminus. PTK6 KM has a substitution of a critical lysine (K219) in the ATP-binding site to methionine resulting in a kinase dead mutant. Immunoblot analysis of resulting stable cell lines demonstrates overexpression of PTK6, as well as kinase activity of ectopic PTK6-WT and PTK6-YF detected by an antibody specific for phosphorylation of PTK6 tyrosine residue 342 (PY342); PY342 PTK6 is not detectable in the vector control (VEC) or PTK6-KM cells (Fig. 3A). We also see a modest increase in E-Cadherin in cells expressing each of the three PTK6 isoforms accompanied by deceased expression of mesenchymal markers Vimentin and Claudin-1; the mesenchymal marker ZEB-1 is decreased in PTK6-WT and PTK6-KM expressing cells, but its expression is mildly upregulated in PTK6-YF cells (Fig. 3A). Immunofluorescent staining more clearly shows increased E-Cadherin at cell-cell contacts (Fig. 3B). Although total ZEB-1 levels are not decreased in PTK6-YF cells, as seen by immunoblot (Fig. 3A), immunofluorescent staining demonstrates a loss of nuclear expression of this transcriptional repressor of E-Cadherin in cells overexpressing each of the different PTK6 isoforms, PTK6-WT, -YF and -KM (Fig. 3B). In addition, reduced Vimentin expression was observed in all PTK6-expressing lines (Fig. 3B). To further assess the effect of PTK6 reintroduction in SW620 cells, we assayed the stable lines for proliferation rate and detected decreased proliferation of SW620 cells overexpressing PTK6-WT, -YF and -KM as compared with vector control cells (Fig. 3C). Together, these data demonstrate that reintroduction of PTK6 promotes a reversal of EMT, or a mesenchymal to epithelial transition (MET), of metastatic colon cancer cells in a kinase-independent manner.
Figure 3. Reintroduction of PTK6 promotes an epithelial phenotype in SW620 cells in a kinase-independent manner.
Recombinant wild type (WT), active (YF), and kinase dead PTK6 (KM), and empty vector (VEC) were stably introduced into SW620 cells. (A) Expression of PTK6 and EMT markers was determined by immunoblotting. Phosphorylation of PTK6 at PY342 was detected in PTK6-WT and -YF expressing cells but not -KM cells. (B) Immunofluorescent staining was performed to detect protein expression (size bar = 50 μM). Ectopic expression of all isoforms of PTK6 led to increased E-Cadherin as well as reduced Vimentin expression and ZEB-1 nuclear localization. (C) A significant decrease in proliferation of PTK6-expressing stable SW620 cell lines was detected compared with vector control cells at days 4, 6, and 9 days (*p < 0.05). Equal cell numbers were plated in triplicate for each timepoint, then trypsinized and quantified to assess cell proliferation.
PTK6 expression is downregulated during colon tumor progression
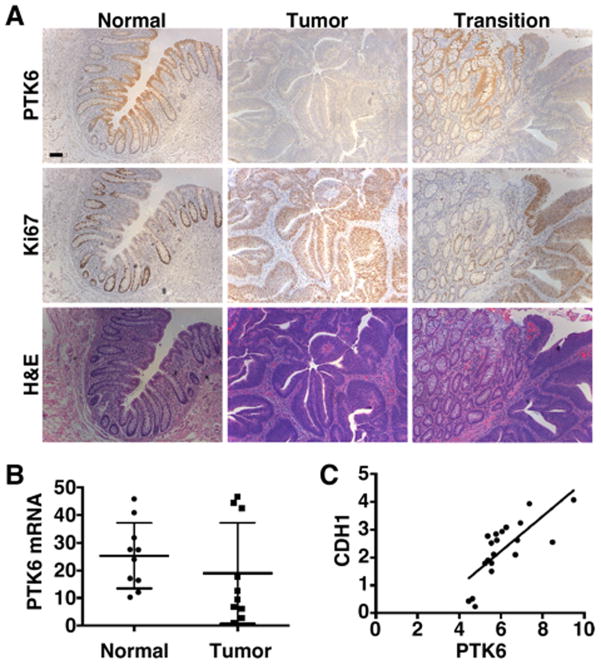
Our findings that PTK6 is important for maintaining an epithelial phenotype and inhibiting xenograft tumor growth in mice led us to examine PTK6 expression in human tissues. Serial sections of colon tissue from colon cancer patients were stained for PTK6 and Ki67, a marker for proliferation. As previously reported in mice (2), PTK6 expression is restricted to the differentiated surface epithelium of normal colon and is not detectable in the proliferative crypt regions, which stain positive for Ki67 (Fig. 4A, left panels). However, in carcinomas (Fig. 4A, center panel), PTK6 is barely detectable, while a strong signal for Ki67 is detected in the tumor epithelium. Expression patterns at the transition between normal epithelium and an adenoma highlight the inverse patterns of PTK6 and Ki67 expression and absence of PTK6 in the tumor epithelium (Fig. 4A, Transition, right panels).
Figure 4. PTK6 expression decreases in human colorectal cancer.
(A) Formalin fixed and paraffin embedded serial sections of patient tissue were immunostained for PTK6 (top row) and Ki67 expression (middle row), and stained with hematoxylin and eosin (H&E, bottom row) (size bar = 100 μ0). An inverse pattern of PTK6 and Ki67 expression was observed. (B) qRT-PCR analysis of mRNA from biopsy samples of human adenocarcinoma cases polyp-free controls for PTK6, *p = 2 X 10−7. (C) Regression analysis of PTK6 and E-cadherin (CDH1) expression R-square = 0.57
In addition to examining patterns of PTK6 protein expression, we analyzed PTK6 mRNA expression in a small cohort of normal (n = 10) and tumor (n = 10) patient biopsy samples using qRT-PCR. We detect a statistically significant decrease in PTK6 mRNA levels in tumors samples as compared with polyp-free control tissues (Fig. 4B). Additionally, qPCR analysis for E-cadherin (CDH1) in the same sample set finds a correlation between PTK6 and CDH1 expression, demonstrating an association of PTK6 with epithelial differentiation in patient tissue (Fig. 4C).
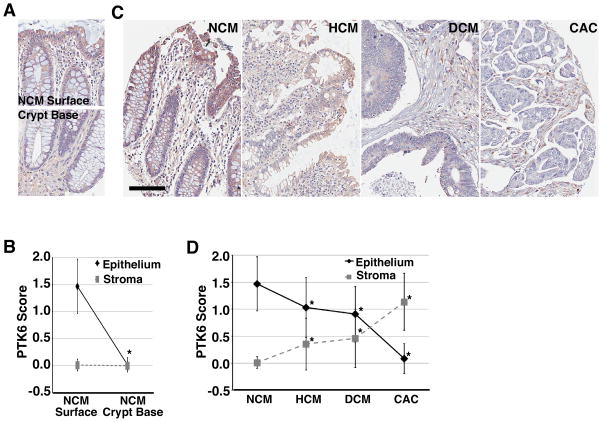
To study PTK6 expression in a larger cohort of patient samples, we stained a human colon tissue microarray (TMA) composed of 560 tissue samples from 130 patients. Based on histological scoring of staining intensity, we find that PTK6 expression is highest in normal (NCM) differentiated surface epithelium (Fig. 5A and C) and is reduced or absent toward the crypt base regions (Fig. 5A and B; *p<0.0001). Additionally, in the normal tissue sections, PTK6 staining is not detectable in stromal tissue (Fig. 5A and B). We observe that PTK6 expression decreases in tumors with increasing tumor stage. Highest levels of PTK6 protein expression are detected in the normal surface epithelium, with moderate expression in hyperplasia (HCM) and adenoma (DCM) tissues, and very low to undetectable expression in invasive carcinomas (CAC) (Fig. 5C and D; *p<0.0001). Interestingly, PTK6 staining in cells within the stromal tissue increases with tumor stage; it is not detectable in normal stromal tissue at either the surface or base but is highly expressed in the stromal tissue surrounding invasive carcinomas (Fig. 5A – C; *p<0.0001).
Figure 5. PTK6 expression decreases in epithelial cells during tumor progression.
Representative images from a tissue microarray with immunohistochemical staining for PTK6 in normal colonic mucosa (NCM) (A), and in NCM, hyperplastic (HCM), and dysplastic (DCM) colonic mucosa, and colonic adenocarcinoma (CAC) (C) (size bar = 100 μm). (B, D) PTK6 staining intensity was scored on a 0, 1+, 2+ scale for epithelial and stromal cells for all tissue samples.
Kinase-Active PTK6 Promotes Oncogenic Signaling
Membrane-association of active PTK6 promotes tumorigenesis and the EMT in prostate cancer cells (6)(reviewed in (7)). Activation of PTK6 at the plasma membrane has also been detected in human breast tumors of different subtypes (5). To determine if PTK6 is activated in colon tumors, we stained serial sections of patient tissue with the antibody against active PTK6 PY342. Active PTK6 was not appreciably detected in the normal tissue or in the majority of colon tumor tissue. However, rare pockets of membrane-associated active PTK6 were detected in invasive colon tumor samples (Fig. 6A), in these same regions, total PTK6 expression was also found to be mildly upregulated.
PTK6 has oncogenic substrates that are activated by tyrosine phosphorylation, including the transcription factor STAT3 (4, 27–29). We serum starved and stimulated SW620 cells stably expressing PTK6-WT, -YF and -KM to promote growth factor receptor-mediated PTK6 activation, and probed cell lysates for STAT3 activation (phosphorylation at tyrosine residue 705), as well as activation of the downstream effectors ERK1/2 (11) and ERK5 (20, 30). In SW620 cells expressing PTK6-YF, serum stimulation resulted in STAT3 and ERK5 activating phosphorylation, which was not detectable in vector control, PTK6-WT and PTK6-KM expressing cells (Fig. 6B).
While serum stimulation increased phosphorylation of tyrosine residue 342 in both PTK6-WT and PTK6-YF (Fig. 6C), indicating PTK6 activation, only the PTK6-YF cells exhibited increased downstream signaling (Fig. 6B). We probed the cell lysates with an antibody against PTK6 phosphorylated on regulatory tyrosine residue 447, which is present in PTK6-WT and PTK6-KM, but mutated in PTK6-YF. Only PTK6-WT was phosphorylated on tyrosine residue 447 (PY447), which leads to inhibition of its kinase activity (Fig. 6C). The absence of PY447 in cells expressing kinase-dead PTK6-KM suggests that PTK6 activity is required for Y447 phosphorylation, and overexpressed wild type PTK6 is capable of autophosphorylation and inhibition. Inhibitory phosphorylation of PTK6-WT prevents activation of STAT3 and ERK5 in cells overexpressing PTK6-WT (Fig. 6B).
Chemotherapeutic Modulation of PTK6 Signaling and Kinase Activity
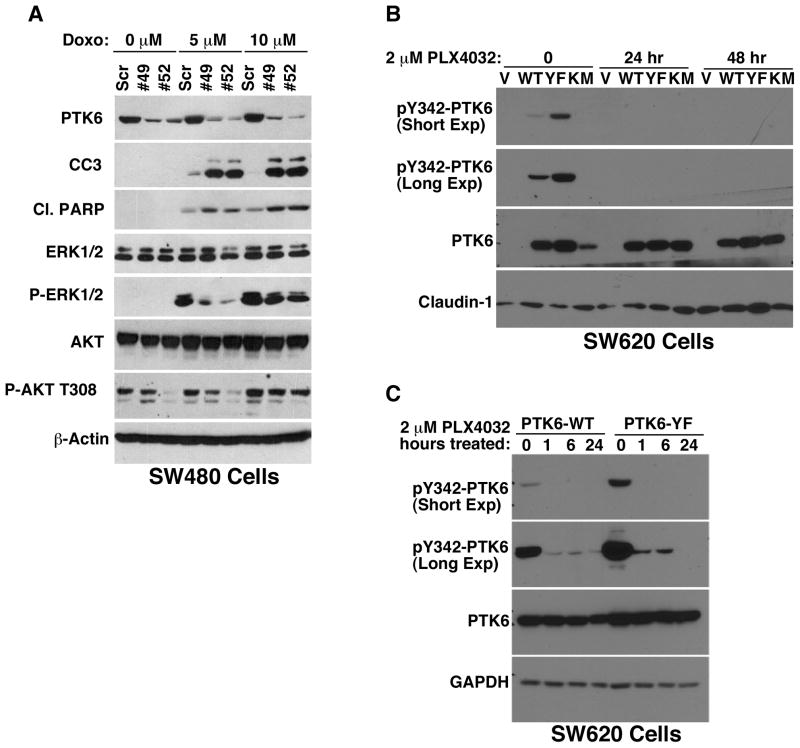
PTK6 has been shown to promote human colon cancer cell survival in response to DNA-damaging agents (28). Doxorubicin is a DNA-damaging anthracycline antibiotic used in chemotherapy treatment of a wide range of malignancies (31), which induces apoptosis in sensitive cancer cells. To explore the role of PTK6 in the response of SW480 cells to doxorubicin treatment, SW480 cells expressing PTK6 knockdown constructs shRNA49 and shRNA52 were treated with 0, 5 and 10 μM doxorubicin for 24 hours (Fig 7A). PTK6 knockdown resulted in increased apoptosis as assayed by cleaved caspase 3 (CC3) and cleaved PARP expression, indicating a role for PTK6 in resistance to DNA damage-induced apoptosis. DNA damage-mediated activation of ERK1/2 and AKT were also decreased in PTK6-knockdown cells. While heregulin stimulation activated PTK6 and p38 MAPK in breast cancer cell lines (30), activation of p38 was not detected in colon cancer cell lines (Fig. S2A). In fact, stable knockdown of PTK6 appeared to enhance p38 activation in response to heregulin stimulation in the SW480 (Fig. S2B) and HCT116 (Fig. S2C) colon cancer cell lines.
Figure 7. Chemotherapeutic Modulation of PTK6 Signaling and Activity.
(A) Immunoblotting was performed with lysates from doxorubicin treated SW480 cells with stable knockdown of PTK6 (shRNA49 and shRNA52), with a scrambled shRNA (Scr) as a control. Apoptotic cell death was assayed by examining expression of cleaved caspase 3 (CC3) and cleaved PARP (Cl. PARP). Activation of oncogenic targets ERK1/2, ERK5, STAT3 and AKT was detected with phospho-specific antibodies. (B) Immunoblotting of total cell lysates from SW620 cells transfected with vector control (V), wild type PTK6, PTK6-YF, or PTK6-KM, treated with 2 μM PLX4032 for 24 or 48 hours. Membranes were probed with antibodies against PY-342 PTK6, total PTK6, and Claudin-1. (C) PTK6-WT and -YF cells were treated with 2 μM PLX4032 for 1, 6, or 24 hours; membranes were probed for total PTK6 and PY342-PTK6, GAPDH was used as a loading control. For both experiments, DMSO was used as the 0 timepoint vehicle control.
Kinase inhibitors have shown promise in the treatment of various cancers. PLX4032 (vemurafenib) is approved for treatment of metastatic melanoma with activating mutations in the BRAF gene. Out of a panel of kinases tested, PTK6 was also inhibited in the nanomolar range by PLX4032 (32, 33). The mechanism for PTK6 inhibition is unclear, as this compound was developed to inhibit mutant B-Raf, which is a serine/threonine kinase. Treatment of SW620-PTK6 overexpressing cells with PLX4032 results in complete inhibition of PY342-PTK6 in both PTK6-WT and PTK6-YF cells that is sustained for at least 48 hours (Fig. 7B). Closer examination of SW620 PTK6-WT and -YF cells shows that PLX4032 treatment inhibits PTK6 PY342 phosphorylation as early as 1 hour post-treatment with complete inhibition after 24 hours (Fig. 7C). These data demonstrate that PLX4032 functions effectively as an inhibitor of PTK6 activation and may be useful in the treatment of PTK6-expressing colon tumors.
DISCUSSION
PTK6 was initially identified in cultured human melanocytes (34), breast cancer cells (35) and the mouse small intestine (36, 37). Several studies indicate that PTK6 plays oncogenic roles in breast cancer [reviewed in (38)], and we recently demonstrated that Ptk6 contributes to tumor initiation and metastasis in an MMTV-ERBB2 mouse model of breast cancer (29). Although PTK6 contributes to epithelial cell differentiation in the normal intestine, it can also promote STAT3 activation and tumor initiation in the AOM/DSS mouse colon cancer model (4). Differences in the intracellular localization of PTK6, which would regulate its access to specific substrates, may account for some of the opposing activities of PTK6 in normal cells and cancer. PTK6 is predominantly nuclear in normal prostate epithelial cells, but nuclear localization of PTK6 is lost and PTK6 is activated at the plasma membrane in prostate cancers (6, 7).
Although targeting PTK6 expression to the membrane or nucleus in colon cancer cell lines had opposing outcomes (19), comparison of PTK6 intracellular localization in colon cancer cells with epithelial and mesenchymal phenotypes did not reveal any notable differences in PTK6 intracellular localization (Fig. S1 A). In contrast to human prostate (5, 6), skin (16), and breast tumors (5), appreciable activation of PTK6 at the plasma membrane was not detected in colon cancer cell lines (Fig. S1 A) and in most human colon tumors. In addition, manipulation of PTK6 expression did not have an impact on the activating tyrosine phosphorylation of two of its substrates, FAK (20) and BCAR1 (20) (data not shown), as previously demonstrated in human prostate cell lines (20), and in tumors that formed in mouse models of skin (16), and breast (29) cancer.
Our studies suggest that the ability of PTK6 to promote an epithelial phenotype and oppose the EMT is not dependent upon its tyrosine kinase activity. Ectopic expression of PTK6-KM, which has a mutation that abolishes kinase activity, suppressed expression of mesenchymal markers and led to increased association of E-cadherin with the plasma membrane and decreased ZEB-1 expression in the nucleus (Fig. 3). PTK6-WT, which also promotes an epithelial phenotype in SW620 cells, is phosphorylated on tyrosine residue 447 (Fig. 6), which would render it inactive. PTK6 contains SH3 and SH2 domains that mediate protein-protein interactions, and PTK6 may have specific functions as an adaptor protein in colon cancer cells.
Protein tyrosine kinases contribute to oncogenic signaling in a variety of cancer types and have emerged as important therapeutic targets. A goal of our studies was to determine if PTK6 should be targeted in colon cancer. Systemic knockout of the Ptk6 gene conferred resistance to AOM and led to reduced initiation and formation of colon tumors in the mouse (4), suggesting that PTK6 is a prospective therapeutic target. Knockdown of PTK6 also enhanced apoptosis of colon cancer cell lines treated with chemotherapeutic agents and radiation (28) (Fig. 7A), further suggesting targeting PTK6 may be beneficial to patients. However, induction of PTK6 in mouse intestinal crypts in response to radiation-induced DNA damage led to increased apoptosis in vivo (3), and a recent paper demonstrated that PTK6 induction promotes gemcitabine-induced apoptosis of pancreatic caner cells (39), underscoring the need to consider different context specific roles for PTK6.
PTK6 expression was previously examined in a small set of human intestinal tumors (n = 5), and PTK6 mRNA levels were higher in 3/5 tumors compared with adjacent normal mucosa (40). Here we examined PTK6 expression in multiple samples from 130 patients and found that PTK6 protein levels are highest in normal differentiated epithelial cells and decrease in epithelial tumor cells as cancer progresses (Fig. 5). We also detected an increase in PTK6 positive stromal cells in colon adenocarcinomas, and our data suggest induction of PTK6 in stromal cells in human colon cancers may be a marker for advanced stage/grade in colon cancer. Recently, PTK6 mRNA was identified in stromal cells of normal endometrium and breast and bladder cancers, (41). PTK6 protein expression was detected in stromal cells that had epithelial characteristics in skin spindle cell tumors (16). PTK6 expression has also been reported in activated cultured T-cells (42). Further studies will be required to characterize the PTK6 positive stromal cells and understand their significance in colon cancer.
Earlier we reported that PTK6 expression levels decrease in human squamous cell carcinomas of the skin (16). Comparison of PTK6 mRNA expression in several datasets (43–50) suggests that PTK6 expression decreases in cancers that develop in tissues, which have appreciable levels of PTK6 in normal differentiated epithelial cells (Fig. S3). These include head and neck cancer (47), oral (48), and cervical squamous cell carcinomas (49) and esophageal squamous cell carcinomas (ESCC) (50). Similar to our findings with colon cancer, PTK6 expression was shown to decrease in human ESCC and knockdown of PTK6 enhanced ESCC xenograft tumor growth (12). While inhibiting PTK6 may be effective in breast and prostate tumors, targeting it alone could potentially have adverse effects in cancers that develop in tissues where it is normally associated with epithelial cell differentiation, such as the colon and esophagus. However, since knockdown of PTK6 can enhance the response of colon cancer cells to therapeutic DNA damaging agents (28) (Fig. 7A), PTK6 inhibitors may prove useful in the development of combination therapies to more effectively kill colon tumor cells.
Supplementary Material
IMPLICATIONS.
Understanding context specific functions of PTK6 is important, because although it promotes cell survival and oncogenic signaling after DNA damage, expression of PTK6 in established tumors may maintain the epithelial phenotype, preventing tumor progression.
Acknowledgments
Studies were supported by NIH grant RO1 DK044525 (ALT). We thank Dr. Dragana Kopanja (UIC) for her assistance with the xenograft studies, and Dr. Priti Marwaha for help obtaining tissue specimens.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors have no conflicts of interest to report.
This is an original manuscript; data have not been published elsewhere.
STATEMENT OF AUTHOR CONTRIBUTIONS
P.S.M.: Acquisition of data, analysis and interpretation of data, drafting the manuscript
J.J.G.: Acquisition of data and analysis and interpretation of data
G.G.: Acquisition of data and analysis and interpretation of data
H.X.: Statistical Analysis
R.M.X.: Acquisition of data and analysis and interpretation of data
X.L.: Acquisition of data and analysis and interpretation of data
M.C.: Analysis and interpretation of data
A.O.P.: Acquisition of data and analysis and interpretation of data
A.L.T.: Study concept and design, analysis and interpretation of data, drafting the manuscript
REFERENCES CITED
- 1.Brauer PM, Tyner AL. Building a better understanding of the intracellular tyrosine kinase PTK6 - BRK by BRK. Biochimica et biophysica acta. 2010;1806:66–73. doi: 10.1016/j.bbcan.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haegebarth A, Bie W, Yang R, Crawford SE, Vasioukhin V, Fuchs E, et al. Protein tyrosine kinase 6 negatively regulates growth and promotes enterocyte differentiation in the small intestine. Molecular and Cellular Biology. 2006;26:4949–57. doi: 10.1128/MCB.01901-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haegebarth A, Perekatt AO, Bie W, Gierut JJ, Tyner AL. Induction of protein tyrosine kinase 6 in mouse intestinal crypt epithelial cells promotes DNA damage-induced apoptosis. Gastroenterology. 2009;137:945–54. doi: 10.1053/j.gastro.2009.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gierut J, Zheng Y, Bie W, Carroll RE, Ball-Kell S, Haegebarth A, et al. Disruption of the Mouse Protein Tyrosine Kinase 6 Gene Prevents STAT3 Activation and Confers Resistance to Azoxymethane. Gastroenterology. 2011;141:1371–80. e2. doi: 10.1053/j.gastro.2011.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng M, Emmadi R, Wang Z, Wiley EL, Gann PH, Khan SA, et al. PTK6/BRK is expressed in the normal mammary gland and activated at the plasma membrane in breast tumors. Oncotarget. 2014;5:6038–48. doi: 10.18632/oncotarget.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y, Wang Z, Bie W, Brauer PM, Perez White BE, Li J, et al. PTK6 activation at the membrane regulates epithelial-mesenchymal transition in prostate cancer. Cancer Research. 2013;73:5426–37. doi: 10.1158/0008-5472.CAN-13-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, Tyner AL. Context-specific protein tyrosine kinase 6 (PTK6) signalling in prostate cancer. Eur J Clin Invest. 2013;43:397–404. doi: 10.1111/eci.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmandt RE, Bennett M, Clifford S, Thornton A, Jiang F, Broaddus RR, et al. The BRK Tyrosine Kinase is Expressed in High-Grade Serous Carcinoma of the Ovary. Cancer Biol Ther. 2006;5:1136–41. doi: 10.4161/cbt.5.9.2953. [DOI] [PubMed] [Google Scholar]
- 9.Fan C, Zhao Y, Liu D, Zhang X, Wang E. Detection of Brk expression in non-small cell lung cancer: clinicopathological relevance. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2011;32:873–80. doi: 10.1007/s13277-011-0188-z. [DOI] [PubMed] [Google Scholar]
- 10.Zhao C, Chen Y, Zhang W, Zhang J, Xu Y, Li W, et al. Expression of protein tyrosine kinase 6 (PTK6) in nonsmall cell lung cancer and their clinical and prognostic significance. Onco Targets Ther. 2013;6:183–8. doi: 10.2147/OTT.S41283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono H, Basson MD, Ito H. PTK6 promotes cancer migration and invasion in pancreatic cancer cells dependent on ERK signaling. PLoS ONE. 2014;9:e96060. doi: 10.1371/journal.pone.0096060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma S, Bao JY, Kwan PS, Chan YP, Tong CM, Fu L, et al. Identification of PTK6, via RNA sequencing analysis, as a suppressor of esophageal squamous cell carcinoma. Gastroenterology. 2012;143:675–86. e1–12. doi: 10.1053/j.gastro.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Chen YF, Ma G, Cao X, Huang ZL, Zeng MS, Wen ZS. Downregulated expression of PTK6 is correlated with poor survival in esophageal squamous cell carcinoma. Med Oncol. 2014;31:317. doi: 10.1007/s12032-014-0317-9. [DOI] [PubMed] [Google Scholar]
- 14.Liu XK, Zhang XR, Zhong Q, Li MZ, Liu ZM, Lin ZR, et al. Low expression of PTK6/Brk predicts poor prognosis in patients with laryngeal squamous cell carcinoma. J Transl Med. 2013;11:59. doi: 10.1186/1479-5876-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang TC, Jee SH, Tsai TF, Huang YL, Tsai WL, Chen RH. Role of breast tumour kinase in the in vitro differentiation of HaCaT cells. Br J Dermatol. 2005;153:282–9. doi: 10.1111/j.1365-2133.2005.06604.x. [DOI] [PubMed] [Google Scholar]
- 16.Chastkofsky MI, Bie W, Ball-Kell SM, He YY, Tyner AL. Protein Tyrosine Kinase 6 Regulates UVB-Induced Signaling and Tumorigenesis in Mouse Skin. The Journal of investigative dermatology. 2015;135:2492–501. doi: 10.1038/jid.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petro BJ, Tan RC, Tyner AL, Lingen MW, Watanabe K. Differential expression of the non-receptor tyrosine kinase BRK in oral squamous cell carcinoma and normal oral epithelium. Oral Oncol. 2004;40:1040–7. doi: 10.1016/j.oraloncology.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Y, Peng M, Wang Z, Asara JM, Tyner AL. Protein tyrosine kinase 6 directly phosphorylates AKT and promotes AKT activation in response to epidermal growth factor. Molecular and Cellular Biology. 2010;30:4280–92. doi: 10.1128/MCB.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palka-Hamblin HL, Gierut JJ, Bie W, Brauer PM, Zheng Y, Asara JM, et al. Identification of beta-catenin as a target of the intracellular tyrosine kinase PTK6. J Cell Sci. 2010;123:236–45. doi: 10.1242/jcs.053264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng Y, Asara JM, Tyner AL. Protein-tyrosine Kinase 6 Promotes Peripheral Adhesion Complex Formation and Cell Migration by Phosphorylating p130 CRK-associated Substrate. The Journal of Biological Chemistry. 2012;287:148–58. doi: 10.1074/jbc.M111.298117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogel UF, Bueltmann BD. Simple, inexpensive, and precise paraffin tissue microarrays constructed with a conventional microcompound table and a drill grinder. American journal of clinical pathology. 2006;126:342–8. doi: 10.1309/F2Q38DXN1V1V4GQM. [DOI] [PubMed] [Google Scholar]
- 22.Xicola RM, Gagnon M, Clark JR, Carroll T, Gao W, Fernandez C, et al. Excess of proximal microsatellite-stable colorectal cancer in African Americans from a multiethnic study. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:4962–70. doi: 10.1158/1078-0432.CCR-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dydensborg AB, Herring E, Auclair J, Tremblay E, Beaulieu JF. Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. American journal of physiology Gastrointestinal and liver physiology. 2006;290:G1067–74. doi: 10.1152/ajpgi.00234.2005. [DOI] [PubMed] [Google Scholar]
- 24.Liang KY, Zeger SL. On the use of concordant pairs in matched case-control studies. Biometrics. 1988;44:1145–56. [PubMed] [Google Scholar]
- 25.Hewitt RE, McMarlin A, Kleiner D, Wersto R, Martin P, Tsokos M, et al. Validation of a model of colon cancer progression. The Journal of pathology. 2000;192:446–54. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH775>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L, Gao Y, Qiu H, Miller WT, Poli V, Reich NC. Identification of STAT3 as a specific substrate of breast tumor kinase. Oncogene. 2006;25:4904–12. doi: 10.1038/sj.onc.1209501. [DOI] [PubMed] [Google Scholar]
- 28.Gierut JJ, Mathur PS, Bie W, Han J, Tyner AL. Targeting protein tyrosine kinase 6 enhances apoptosis of colon cancer cells following DNA damage. Molecular cancer therapeutics. 2012;11:2311–20. doi: 10.1158/1535-7163.MCT-12-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng M, Ball-Kell SM, Tyner AL. Protein tyrosine kinase 6 promotes ERBB2-induced mammary gland tumorigenesis in the mouse. Cell death & disease. 2015;6:e1848. doi: 10.1038/cddis.2015.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ostrander JH, Daniel AR, Lofgren K, Kleer CG, Lange CA. Breast tumor kinase (protein tyrosine kinase 6) regulates heregulin-induced activation of ERK5 and p38 MAP kinases in breast cancer cells. Cancer Research. 2007;67:4199–209. doi: 10.1158/0008-5472.CAN-06-3409. [DOI] [PubMed] [Google Scholar]
- 31.Weiss RB. The anthracyclines: will we ever find a better doxorubicin? Seminars in oncology. 1992;19:670–86. [PubMed] [Google Scholar]
- 32.Sala E, Mologni L, Truffa S, Gaetano C, Bollag GE, Gambacorti-Passerini C. BRAF silencing by short hairpin RNA or chemical blockade by PLX4032 leads to different responses in melanoma and thyroid carcinoma cells. Molecular cancer research : MCR. 2008;6:751–9. doi: 10.1158/1541-7786.MCR-07-2001. [DOI] [PubMed] [Google Scholar]
- 33.Vin H, Ojeda SS, Ching G, Leung ML, Chitsazzadeh V, Dwyer DW, et al. BRAF inhibitors suppress apoptosis through off-target inhibition of JNK signaling. Elife. 2013;2:e00969. doi: 10.7554/eLife.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee ST, Strunk KM, Spritz RA. A survey of protein tyrosine kinase mRNAs expressed in normal human melanocytes. Oncogene. 1993;8:3403–10. [PubMed] [Google Scholar]
- 35.Mitchell PJ, Barker KT, Martindale JE, Kamalati T, Lowe PN, Page MJ, et al. Cloning and characterisation of cDNAs encoding a novel non-receptor tyrosine kinase, brk, expressed in human breast tumours. Oncogene. 1994;9:2383–90. [PubMed] [Google Scholar]
- 36.Siyanova EY, Serfas MS, Mazo IA, Tyner AL. Tyrosine kinase gene expression in the mouse small intestine. Oncogene. 1994;9:2053–7. [PubMed] [Google Scholar]
- 37.Vasioukhin V, Serfas MS, Siyanova EY, Polonskaia M, Costigan VJ, Liu B, et al. A novel intracellular epithelial cell tyrosine kinase is expressed in the skin and gastrointestinal tract. Oncogene. 1995;10:349–57. [PubMed] [Google Scholar]
- 38.Goel RK, Lukong KE. Tracing the footprints of the breast cancer oncogene BRK - Past till present. Biochimica et biophysica acta. 2015;1856:39–54. doi: 10.1016/j.bbcan.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Ono H, Basson MD, Ito H. PTK6 Potentiates Gemcitabine-Induced Apoptosis by Prolonging S-phase and Enhancing DNA Damage in Pancreatic Cancer. Molecular cancer research : MCR. 2015;13:1174–84. doi: 10.1158/1541-7786.MCR-15-0034. [DOI] [PubMed] [Google Scholar]
- 40.Llor X, Serfas MS, Bie W, Vasioukhin V, Polonskaia M, Derry J, et al. BRK/Sik expression in the gastrointestinal tract and in colon tumors. Clin Cancer Res. 1999;5:1767–77. [PubMed] [Google Scholar]
- 41.Kiflemariam S, Ljungstrom V, Ponten F, Sjoblom T. Tumor Vessel Up-Regulation of INSR Revealed by Single-Cell Expression Analysis of the Tyrosine Kinome and Phosphatome in Human Cancers. The American journal of pathology. 2015;185:1600–9. doi: 10.1016/j.ajpath.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Kasprzycka M, Majewski M, Wang ZJ, Ptasznik A, Wysocka M, Zhang Q, et al. Expression and Oncogenic Role of Brk (PTK6/Sik) Protein Tyrosine Kinase in Lymphocytes. Am J Pathol. 2006;168:1631–41. doi: 10.2353/ajpath.2006.050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Provenzani A, Fronza R, Loreni F, Pascale A, Amadio M, Quattrone A. Global alterations in mRNA polysomal recruitment in a cell model of colorectal cancer progression to metastasis. Carcinogenesis. 2006;27:1323–33. doi: 10.1093/carcin/bgi377. [DOI] [PubMed] [Google Scholar]
- 44.Hong Y, Ho KS, Eu KW, Cheah PY. A susceptibility gene set for early onset colorectal cancer that integrates diverse signaling pathways: implication for tumorigenesis. Clin Cancer Res. 2007;13:1107–14. doi: 10.1158/1078-0432.CCR-06-1633. [DOI] [PubMed] [Google Scholar]
- 45.Alhopuro P, Sammalkorpi H, Niittymaki I, Bistrom M, Raitila A, Saharinen J, et al. Candidate driver genes in microsatellite-unstable colorectal cancer. International journal of cancer Journal international du cancer. 2012;130:1558–66. doi: 10.1002/ijc.26167. [DOI] [PubMed] [Google Scholar]
- 46.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuriakose MA, Chen WT, He ZM, Sikora AG, Zhang P, Zhang ZY, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cellular and molecular life sciences : CMLS. 2004;61:1372–83. doi: 10.1007/s00018-004-4069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toruner GA, Ulger C, Alkan M, Galante AT, Rinaggio J, Wilk R, et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer genetics and cytogenetics. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 49.Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer research. 2007;67:10163–72. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 50.Hu N, Clifford RJ, Yang HH, Wang C, Goldstein AM, Ding T, et al. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.