SUMMARY
Throughout the bacterial domain, the alarmone ppGpp dramatically reprograms transcription following nutrient limitation. This “stringent response” is critical for survival and antibiotic-tolerance and is a model for transcriptional regulation by small ligands. We report that ppGpp binds to two distinct sites 60 angstroms apart on E. coli RNA polymerase (RNAP), one characterized previously (Site 1) and a second identified here at an interface of RNAP and the transcription factor DksA (Site 2). The location and unusual tripartite nature of Site 2 account for the DksA-ppGpp synergism and suggest mechanisms for ppGpp enhancement of DksA’s effects on RNAP. Site 2 binding results in the majority of ppGpp’s effects on transcription initiation in vitro and in vivo, and strains lacking Site 2 are severely impaired for growth following nutritional shifts. Filling of the two sites at different ppGpp concentrations would expand the dynamic range of cellular responses to changes in ppGpp levels.
Graphical Abstract

INTRODUCTION
The global regulators ppGpp and pppGpp (guanosine 5′-diphosphate 3′-diphosphate and guanosine 5′-triphosphate 3′-diphosphate), here referred to collectively as ppGpp, are the molecular effectors of the bacterial stringent response, an extensive reprogramming of transcription and metabolism in response to nutrient deprivation (Potrykus and Cashel, 2008). The enzyme(s) that synthesize ppGpp, the RelA/RSH family, are almost universal in bacteria and are also found in archaea and in plant chloroplasts (Atkinson et al., 2011). ppGpp plays an essential role in pathogenesis and persistence (Dalebroux et al., 2010; Hauryliuk et al. 2015) and regulates evolutionarily conserved as well as lineage-specific pathways (Boutte and Crosson, 2013).
In E. coli, changes in ppGpp levels alter expression of ~700 genes including those encoding ribosomal RNA and other parts of the translational machinery, flagella, fatty acids, amino acids, and many other central cellular components (Durfee et al., 2008; Traxler et al., 2008; Haugen et al., 2008; Ross et al., 2013; and references therein). Gene expression is regulated by direct ppGpp binding to RNAP, decreasing or increasing transcription initiation depending on the kinetic properties of the specific promoter (Haugen et al. 2008), and effects of ppGpp on transcription elongation have been reported as well (Zhang et al., 2014; Roghanian et al., 2015).
ppGpp usually works in conjunction with the small protein DksA, a transcription factor that modifies RNAP by binding in the RNAP secondary channel (Paul et al., 2004; Perederina et al., 2004; Lennon et al., 2012). DksA inhibits rRNA transcription initiation in vitro in the absence of ppGpp, but DksA and ppGpp together have much larger effects than either alone, and they are both required for direct positive control of promoters (Paul et al., 2005). In bacteria distantly related to proteobacteria, including B. subtilis, ppGpp inhibits rRNA promoter function indirectly by binding to enzymes required for synthesis of GTP, the initiating nucleotide for promoters negatively regulated by ppGpp in these species, rather than by binding directly to RNAP (Krasny and Gourse, 2004; Liu et al., 2015).
Although it is often assumed that RNAP is the major target of ppGpp in E. coli, ppGpp also binds directly to, and regulates, additional protein targets, complicating assignment of its effects to specific mechanisms. Some of these targets are GTP binding-proteins, central players in replication and translation, where ppGpp inhibits enzyme function by competing with GTP (e.g., Liu et al., 2015; Hauryliuk et al., 2015). In other cases, competition with GTP is not involved and ppGpp works allosterically (Kanjee et al., 2012).
Multiple genetic and biochemical attempts have been made to locate the binding site for ppGpp on RNAP, but a specific site that fully accounts for its profound effects in vivo has not been found previously (reviewed in Potrykus and Cashel, 2008; Ross et al., 2013; Hauryliuk et al., 2015). A ppGpp binding site was reported in a T. thermophilus RNAP co-crystal (Artsimovitch et al., 2004), but its biological significance has been challenged (Vrentas et al., 2008). A 32P-6-thio-ppGpp crosslink to the E. coli RNAP β’ subunit (Toulokhonov et al. (2001) was later mapped to the interface of β’ and ω (Ross et al., 2013), and ppGpp binding to this site was observed in E. coli RNAP co-crystals (Zuo et al., 2013; Mechold et al., 2013). Substitutions in this site, here referred to as Site 1, eliminated inhibition of transcription by ppGpp in vitro in the absence of DksA, but effects of chromosomal mutations on growth were relatively mild, seemingly too small to account for the severity of the stringent response (Ross et al., 2013).
We now identify an additional ppGpp binding site on E. coli RNAP by analysis of ppGpp function, crosslinking, and binding in the presence of the transcription factor DksA using RNAP lacking Site 1. The newly-identified site (Site 2) is created by the interaction of DksA with RNAP and is located at the interface of DksA and the β’ subunit rim-helices, far from the site identified in the absence of DksA (Site 1) or the site identified in the T. thermophilus cocrystal. Substitutions in Site 2 eliminate the DksA-ppGpp synergy, and ppGpp (with or without DksA) has no effect on transcription initiation in vitro when RNAP lacks both sites. Chromosomal mutations that eliminate Site 2 alone or both Sites 1 and 2, created by CRISPR-Cas9 and recombineering approaches, have dramatic effects on growth and transcription initiation in vivo, similar (but not identical) to deletions that eliminate synthesis of ppGpp or DksA. The location of Site 2 on RNAP suggests a mechanism for ppGpp/DksA action, an explanation for the ppGpp/DksA synergism, and a potential target for small molecule inhibitors of RNAP. Furthermore, the strain lacking the ppGpp binding sites on RNAP should facilitate investigation of other ppGpp targets, thus allowing examination of the full-breadth of the stringent response.
RESULTS
RNAPs Lacking the Previously-Identified ppGpp Binding Site Retain Sensitivity to ppGpp When DksA is Present
The previously-identified binding site for ppGpp on E. coli RNAP (Site 1) is required for inhibition of rRNA transcription by ppGpp in vitro in the absence of the transcription factor DksA (Ross et al., 2013). However, this site is at least 30Å from the secondary channel where the transcription factor DksA binds to RNAP, providing no straightforward explanation for the synergistic effects of DksA and ppGpp together.
We addressed directly whether this site was needed for the synergy with DksA by testing whether substitutions in Site 1 reduced inhibition of rRNA transcription by ppGpp when DksA was present (Figure 1). Consistent with our previous report (Ross et al., 2013), transcription from the rrnB P1 promoter by wild-type (WT) RNAP was inhibited 3 to 4-fold by ppGpp in vitro, whereas two mutant RNAPs lacking Site 1 (Δω RNAP or M8 RNAP) were not inhibited by ppGpp at all (Figures 1A, 1B, and S1A). Surprisingly, however, the Site 1 mutants did not prevent inhibition of transcription by ppGpp when DksA was included in the reaction (Figures 1B and 1C). Because these mutant RNAPs lack as many as 8 amino acids that contribute to binding Site 1 (Ross et al., 2013; Zuo et al., 2013), residual ppGpp binding to the mutant site seemed unlikely, suggesting that another binding site (Site 2) might function when DksA is present. This hypothesis would explain DksA’s previously-reported ability to “rescue” inhibition by ppGpp when RNAP lacks the ω subunit (Vrentas et al., 2005), which we now know forms part of Site 1 (Ross et al., 2013).
Figure 1. DksA Restores a Response to ppGpp to RNAPs Lacking the Previously-Identified ppGpp Binding Site.
(A) Inhibition of rrnB P1 transcription by ppGpp in vitro using WT RNAP or two different RNAPs lacking Site 1, Δω RNAP and M8 RNAP (ωΔ 2–5, β’R362A, R417A, K615A, Y626A). Means and ranges shown here and in (C), (D), and (F) are from two independent experiments. See Figure S1A and Supplemental Experimental Procedures.
(B) Representative gel showing in vitro transcripts from rrnB P1 and the vector-encoded RNA I promoter in reactions with RNAP lacking Site 1 (Δω RNAP) and either 2 μM DksA and 0–160 μM ppGpp or ppGpp without DksA.
(C) Inhibition of rrnB P1 transcription by ppGpp as in (A) but with 2 μM DksA. 2 μM DksA alone, with no ppGpp, inhibited transcription similarly for each RNAP (transcript levels ~0.6 relative to no DksA).
(D) Relative rrnB P1 transcript levels in vitro, normalized to levels without DksA or ppGpp for each RNAP.
(E) IC50s for ppGpp for inhibition of rrnB P1 with WT or RNAP lacking Site 1 (Δω RNAP) and the indicated DksA concentrations. Values with 95% confidence intervals are shown. For WT RNAP without DksA (Site 1 only), n=10; WT RNAP with 2 μM DksA (Sites 1 and 2), n=8; Δω RNAP with 0.5 μM DksA (Site 2 only), n=2; Δω RNAP with 2 μM DksA (Site 2 only), n=16; Δω RNAP with 6 μM DksA (Site 2 only), n=2.
(F) Inhibition of rrnB P1 transcription by ppGpp using RNAP lacking Site 1 (Δω RNAP) and 0, 0.5, 2, or 6 μM DksA. Inhibition by DksA alone, with no ppGpp, varied with the DksA concentration; transcript levels relative to no DksA, were 1.07 ± 0.18 at 0.5 μM DksA; 0.60 ± 0.04 at 2 μM DksA; 0.29 ± 0.02 at 6 μM DksA. See Figure S1B for plot without normalization.
(G) Crosslinking of 32P-6-thio-ppGpp to DksA in reactions with DksA (2.5, 5 or 10 μM WT or 1.7, 3.3 or 6.6 μM N88I) and 0.6 μM RNAP lacking ppGpp Site 1 (Δω RNAP, lanes 1–8), or DksA alone (lanes 11–16). Reactions in lanes 1, 2, 9, 10, 17, and 18 used RNAP lacking Site 1 (Δω or M8) or WT RNAP but no DksA. The crosslink with WT RNAP is to β’. Both gels (4–12% SDS-PAGE) were from the same experiment.
(H) Crosslinking as in (G) with 2.5 μM N88I-DksA and 0.75 μM WT RNAP or RNAP lacking Site 1 (Δω) ± 1 mM unlabeled ppGpp competitor.
(I) Crosslinking as in (G) with 0.75 μM WT RNAP and 6 μM WT, N88I, or L15F-DksA, ± 1 mM unlabeled ppGpp competitor. See also Figure S1B.
At a DksA concentration that approximated its effective concentration in vivo (2 μM) (Rutherford et al., 2007), transcription with the Site 1 mutant RNAPs was inhibited ~5-fold by saturating concentrations of ppGpp, less than the 15 to 20-fold inhibition observed with WT RNAP (Figures 1C and 1D). The IC50 values for RNAP containing only the putative Site 2 (43 μM for mutant RNAP lacking Site 1 at 2 μM DksA) were ~2-fold higher than for RNAP containing only Site 1 (21 μM for WT RNAP in the absence of DksA) (Figure 1E). The combined effects of ppGpp binding to Site 1 (~3–4 fold) and Site 2 (~5-fold) account for the ~15 to 20-fold inhibition observed with WT RNAP and DksA (Figure 1D), similar to the inhibition of rrn P1 promoters observed during a stringent response in vivo (Paul et al., 2004). The extent of inhibition of Site 1 mutant RNAP (Δω RNAP) by ppGpp/DksA depended on the DksA concentration (Figures 1E and F), reflecting primarily the degree of saturation of RNAP with DksA rather than a weaker intrinsic affinity of ppGpp for Site 2 than Site 1 (see Discussion).
Identification and Characterization of Site 2 by Crosslinking with 32P-6-thio-ppGpp
Crosslinking of 32P-6-thio-ppGpp to RNAP lacking Site 1 (Δω RNAP) in the presence of DksA was used to search for the proposed Site 2. 32P-6-thio-ppGpp crosslinked to WT RNAP but not to the Site 1 mutant (Δω RNAP; Figure 1G-1I), as reported previously (Ross et al., 2013). Inclusion of DksA in the complex did not restore crosslinking of 32P-6-thio-ppGpp to the Site 1 mutant RNAP, but instead resulted in crosslinking to DksA itself (weakly to WT DksA, strongly to the higher affinity DksA variant, N88I; Figure 1G, lanes 3–8) (Blankschien et al., 2009). The level of ppGpp crosslinking to DksA was similar in complexes containing or lacking Site 1 (Figure 1H) and was not observed in the absence of RNAP (Figure 1G). DksA crosslinking was higher not only to N88I-DksA but also to another higher affinity variant, L15F-DksA (Figure 1I) (Blankschien et al., 2009). DksA crosslinks were specific: unlabeled competitor ppGpp eliminated crosslinking (Figures 1H and 1I), but GTP did not (Figure S1B). Taken together, the transcription inhibition and crosslinking data suggest there are two binding sites for ppGpp on RNAP, the previously-identified DksA-independent site at the interface of the β’ and ω subunits (Site 1) and a site that requires both DksA and RNAP (Site 2).
RNAP Secondary Channel Rim Residues are Required for ppGpp Binding and its Effects on Transcription in the Presence of DksA
To identify Site 2, we took a genetic approach, screening for substitutions in RNAP and DksA that eliminated ppGpp function in the complex lacking Site 1. The requirement for both DksA and RNAP for ppGpp crosslinking to DksA suggested that the second ppGpp binding site might be at the DksA-RNAP interface. Although there are no crystal structures of DksA-RNAP complexes, models based on genetic and biochemical studies suggest that DksA interacts with the β’ rim-helices at the entrance to the RNAP secondary channel, with its coiled-coil extending into the secondary channel and its coiled-coil tip in close proximity to the RNAP active site (Rutherford et al., 2009; Lennon et al., 2012; Lee et al., 2012; Furman et al., 2013; Parshin et al., 2015).
To define RNAP residues in Site 2, we initially screened RNAP variants that lacked Site 1 and contained single substitutions for one of seven arginine or lysine residues in β’ near the DksA-RNAP interface in models of the complex (Figure 2A and legend). These residues were chosen because previously-characterized ppGpp binding sites contain basic side chains (e.g., Kanjee et al., 2012; Zuo et al., 2013; Liu et al., 2015).
Figure 2. Separation of Function Substitutions in RNAP.
(A) E. coli RNAP (PDB 4JKR; Zuo et al., 2013) showing residues tested for DksA-dependent inhibition by ppGpp (single β’ substitutions N680A, K681A, R692A, K695A, R731A, R738A, or the double substitution K598A / K599A in blue spacefill or E677G in magenta spacefill). β’ secondary channel rim-helices: yellow. Active site Mg2+: red sphere. β’: pink. β: cyan. αII NTD: green. αI NTD: grey. ω: slate blue. σ70: light grey. ppGpp bound in PDB 4JKR (Site 1): red spacefill.
(B) Representative gel showing in vitro transcripts from rrnB P1 and the RNA I control promoter with 2 μM DksA and the indicated ppGpp concentrations, with RNAP lacking Site 1 (lanes 1–8) or lacking Site 1 and containing β’N680A / K681A (lanes 9–13). Intervening lanes between lanes 8 and 9 were removed. See also Figure S2.
(C) Transcription from rrnB P1 with RNAPs lacking Site 1 (Δω), or with either β’K681A or β’N680A and lacking Site 1. Reactions contained 2 μM WT DksA and indicated ppGpp concentrations. Values for each RNAP with both DksA and ppGpp are normalized to the values without ppGpp. Transcript levels with DksA alone and no ppGpp, were lower (~0.6) than without DksA (Figures 2F and 4D). Means and ranges shown here and in (D–F) are from two independent experiments.
(D) rrnB P1 transcription as in (C), but with Δω, Δω β’N680A/K681A, or Δω β’ E677G RNAP.
(E) rrnB P1 transcription with Δω, Δω β’N680A, or Δω β’K681 RNAP at the indicated DksA concentrations without ppGpp.
(F) rrnB P1 transcription without ppGpp as in (E), but with Δω Δω β’N680A/K681A, or Δω β’ E677G RNAP.
(G) Representative 4–12% SDS-PAGE gels from the same experiment showing DksA binding to Δω RNAP (left gel), N680A / K681A Δω RNAP (middle gel), or E677G Δω RNAP (right gel) using the Fe2+ cleavage assay (Lennon et al., 2009). Full length 32P-DksA (F–L) is cleaved by ·OH when Fe2+ is bound in the RNAP active site, producing the N-terminally labeled product DksA 1–73. RNAP concentrations: 0 to 1.5 μM. See Figure S2B for quantitation.
Of these residues, only rim-helix substitution β’K681A had the “separation of function” properties predicted for a Site 2 mutant, i.e., loss of inhibition of rrnB P1 transcription by ppGpp without loss of DksA binding to RNAP. A substitution for the adjacent β’ residue, N680A, had similar properties. Singly or in combination, these substitutions reduced or abolished DksA-dependent inhibition by ppGpp (Figures 2B–2D), but retained similar or greater sensitivity to inhibition by DksA alone in the absence of ppGpp (Figures 2E and 2F). Consistent with retention of DksA function, DksA binding to the double substitution mutant RNAP (β’K681A/N680A Δω) and the control Δω RNAP was observed in a direct DksA-RNAP binding assay (Figure 2G). Therefore, the rim-helix substitutions did not reduce ppGpp Site 2 function by disrupting DksA interactions with RNAP. Transcript levels from the rrnB P1 and RNA I promoters (Figure 2B) and intrinsic lifetimes of RNAP-promoter complexes (Figure S2) were similar for the mutant and WT RNAPs in the absence of ppGpp or DksA, supporting the conclusion that the loss of inhibition by ppGpp did not derive from altered kinetic properties of the mutant RNAPs. In contrast, an adjacent β’ substitution, E677G, eliminated both transcription inhibition and DksA binding, with or without ppGpp (Figures 2D, 2F, and 2G), consistent with its original selection as a mutant resistant to overexpression of N88I-DksA (Satory et al., 2013).
The properties of the rim-helix mutant RNAPs β’K681A Δω and β’K681A/N680A Δω in vitro were consistent with disruption of a ppGpp binding site, but the rrnB P1 inhibition defects could potentially have resulted from alteration of a ppGpp-induced conformational change and not from direct loss of ppGpp binding. Therefore, we used the differential radial capillary action of ligand assay (DRaCALA; Roelofs et al., 2011) to test directly whether the substitutions prevented ppGpp binding to the mutant RNAPs in vitro.
First, as a positive control, we established that specific 32P-ppGpp binding to Site 1 in WT RNAP was detected by the DRaCALA assay (Figure 3A, column 1, and Figure 3B). Substitutions inactivating Site 1 (Δω or M8; defined in Figure 1A legend) eliminated ppGpp binding to RNAP in the absence of DksA, and 32P-ppGpp did not bind to ω alone (Figure 3B).
Figure 3. Binding of 32P-ppGpp and Crosslinking of 32P-6-thio-ppGpp to Site 2.
(A) Nitrocellulose filter assay (DRaCALA) for 32P-ppGpp binding. Column 1: no DksA and the indicated RNAP (2 μM). WT RNAP without DksA (Site 1 only). Δω RNAP and M8 RNAP (described in Figure 1A legend) without DksA (no Site 1 or Site 2). Column 2: 20 μM WT DksA and 2 μM Δω RNAP (Site 2 only), or β’ K681A Δω or β’N680A/K681A Δω RNAP (no Site 1 or Site 2). Column 3: 20 μM DksA and 2 μM WT RNAP (has Sites 1 and 2). Unlabeled ppGpp competitor: 1 mM. DksA alone: 20 μM. Duplicate filters shown for each. All filters were from the same image in the same experiment.
(B) Effects of RNAP mutants or 1 mM unlabeled competitors on 32P-ppGpp binding to Sites 1 and/or 2, from DRaCALA experiments as in (A). Means and ranges from duplicate filters in each of two independent experiments are shown. Values were normalized to WT RNAP (12.9 ± 1.3 % of counts bound) for samples without DksA or to Δω RNAP with DksA (12.7 ± 1.3 % of counts bound) for samples with DksA. No binding above background was detected for ω alone, DksA alone, or with ppGpp competitor.
(C) Relative crosslinking of 32P-6-thio-ppGpp to N88I-DksA in complexes with RNAP lacking Site 1 (Δω RNAP), or with Δω RNAP and the indicated β’ substitutions, or with WT RNAP. Error bars indicate ranges from replicate experiments relative to Δω RNAP.
(D) Representative 4–12% SDS-PAGE gel showing crosslinking of 32P-6-thio-ppGpp to N88I-DksA or to the β’ subunit in reactions with various RNAPs (0.75 μM) and N88I or WT DksA (2.5 μM). Crosslink with β’N680A Δω RNAP is shifted from N88I-DksA to β’ (lane 7). See also Figure S3.
(E) 32P-6-thio-ppGpp crosslink with β’N680A Δω RNAP and N88I-DksA maps to the β’ rim-helices. Representative 16% SDS-Tricine gel showing radiolabeled products of thrombin digestion of crosslinked complexes. RNAPs in lanes 2–4 contained β’N680A Δω and engineered thrombin sites at β’ 648 and/or 709. Arrow indicates crosslink to the 61 residue rim-helix peptide (648–709) in lane 4 detected in 3 independent experiments. See Figure S3 for identification of other labeled β’ thrombin fragments.
32P-ppGpp binding to Site 2 in the RNAP-DksA complex lacking Site 1 (Δω RNAP) was observed, but no binding was seen with Δω RNAP containing the rim-helix substitutions β’ K681A or β’N680A/K681A (Figure 3A, column 2) or with DksA alone (Figure 3A, column 3, and Figure 3B). Binding was higher to RNAP containing both sites than to RNAPs with only one site, and unlabeled competitor ppGpp eliminated binding whereas GTP only competed weakly and ATP did not compete for binding to Sites 1 or 2 (Figure 3A and 3B).
RNAP substitutions β’ K681A or K681A/N680A also disrupted crosslinking of 32P-6-thio-ppGpp to DksA in DksA-RNAP complexes (Figure 3C), while other β’ substitutions tested in the screen (β’ K598A/K599A, R731A, R692A, and K695A; Figure 2A) or an RNAP deleted for the nearby lineage-specific insertion β’ SI3 (Figure S1C) retained both Site 2 function and crosslinking to DksA. We conclude that β’ K681 and N680 are in or near Site 2.
32P-6-thio-ppGpp Crosslink Mapping Localizes Site 2 to the β’ Rim-Helices
Although the β’N680A substitution prevented Site 2 function and reduced crosslinking to N88I-DksA, unexpectedly it generated a strong crosslink signal to RNAP (β or β’; Figure 3D, lane 7). This shift in crosslink target from DksA to β/β’ was observed only with this combination of mutant proteins, i.e. only with both N88I-DksA and β’N680A Δω RNAP and not with the other mutant RNAPs (β’K681A Δω RNAP, β’K681A/N680A Δω RNAP) or with WT DksA (Figure 3D). We suggest that β’N680A does not entirely prevent binding of ppGpp to Site 2, but it shifts the orientation of the bound ppGpp slightly, such that the crosslinkable 6-thio moiety at the interface of the two proteins is positioned closer to β’ than to DksA.
Mapping this shifted crosslink would provide physical evidence for localization of Site 2 to the β’ rim-helices. Therefore, we repeated the crosslinking reaction with β’N680A RNAP derivatives containing thrombin sites introduced at each end of the β’ secondary channel rim (at β’648 and/or β’709). Digestion of the complexes with thrombin produced unique β’ sub-fragments for each RNAP, including a 61 residue 32P-6-thio-ppGpp-labeled fragment consisting only of the rim-helices when the RNAP contained thrombin sites at both 648 and 709 (Figures 3E and S3; Ross et al., 2013; Figure 3 legend). These data provide direct evidence that when 6-thio-ppGpp is bound at Site 2, the 6-thio moiety is sandwiched between DksA and the β’ rim-helices.
Identification of DksA Residues Contributing to Site 2
Strains lacking DksA or lacking ppGpp fail to grow on minimal medium lacking amino acids (Paul et al., 2004), suggesting that Site 2 might be required for growth under these conditions. In an initial screen of 24 plasmid-encoded alanine substitutions in DksA, 8 variants complemented the ΔdksA strain poorly if at all (Table S3; Supplemental Experimental Procedures). Four DksA substitutions, L95A, K98A, R129A, and K139A, reduced or abolished the inhibition of transcription by ppGpp from rrnB P1 with RNAP lacking Site 1 in vitro (Figures 4A and C), but these DksA variants were able to inhibit rrnB P1 in the absence of ppGpp (Figures 4B and D). The 4 substitutions reduced or abolished ppGpp binding to RNAP-DksA complexes in the DRaCALA assay (Figures 4E and F).
Figure 4. DksA Residues in Site 2.
(A) Inhibition of rrnB P1 transcription by ppGpp in vitro with RNAP lacking Site 1 (Δω RNAP) and 2 μM WT, K98A, or L95A-DksA. See also Figure S4. Means and ranges shown here and in (B–D) are from two independent experiments.
(B) Inhibition of rrnB P1 transcription by WT, K98A, or L95A-DksA alone with Δω RNAP (no ppGpp).
(C) Inhibition of rrnB P1 transcription by ppGpp as in (A), but with 2 μM WT, R129A, or K139A-DksA.
(D) Inhibition of rrnB P1 transcription by DksA alone (no ppGpp) as in (B), but with WT, R129A, or K139A-DksA.
(E) DRaCALA assay for binding of 32P-ppGpp to Site 2 in complexes with 2.6 μM Δω RNAP lacking Site 1 and 20 μM WT or variant DksAs. Duplicate filters are shown in top 2 rows. Samples in bottom 2 rows contain DksA but no RNAP.
(F) Summary of 32P-ppGpp binding to Site 2 in complexes containing DksA variants and Δω RNAP in DRaCALA assays. Values were normalized to WT DksA (24 ± 0.9% counts bound). Means and ranges are from 2 independent experiments like that in (E).
(G) DksA structure (adapted from PDB 1TJL; Perederina et al., 2004) showing Site 2 residues (L95, K98, R129, K139) in blue, coiled-coil tip residues D74 and A76 in red, and N88 in yellow. (See also Figure S4).
(H) Flexibility of the DksA C-terminal helix. Two different DksA molecules (A in green and H in blue) from DksA crystal structure (PDB 1TJL) were aligned in Pymol to illustrate variable position of C-terminal helix (C1 and C2). Residues implicated in Site 2 function are in stick form.
L95, K98, R129, and K139 form a cluster in DksA near the junction of the globular domain with the C-terminal helix (Figures 4G and 4H). Substitutions in other nearby residues also altered DksA/ppGpp function. R91A strongly decreased ppGpp function, and therefore could also be part of Site 2, although its role in ppGpp-dependent inhibition could be indirect since R91A also reduced the effect of DksA on transcription in the absence of ppGpp (Figure S4).
Other nearby substitutions, L84A and E85A, caused much smaller defects (Figures 4F and S4). In contrast, N88I resulted in a DksA that not only bound RNAP better (Blankschien et al., 2009) but also formed an RNAP-DksA complex hypersensitive to ppGpp (Figures 4E, 4F, and S4), perhaps reflecting a local change in the conformation of the complex.
Taken together, our data suggest that RNAP rim-helix residues β’K681 and β’N680 and DksA residues L95, K98, R129, K139 (and perhaps R91) comprise Site 2.
The Site 2 Substitutions in β’ and DksA Reduce Positive Control of Transcription by ppGpp/DksA
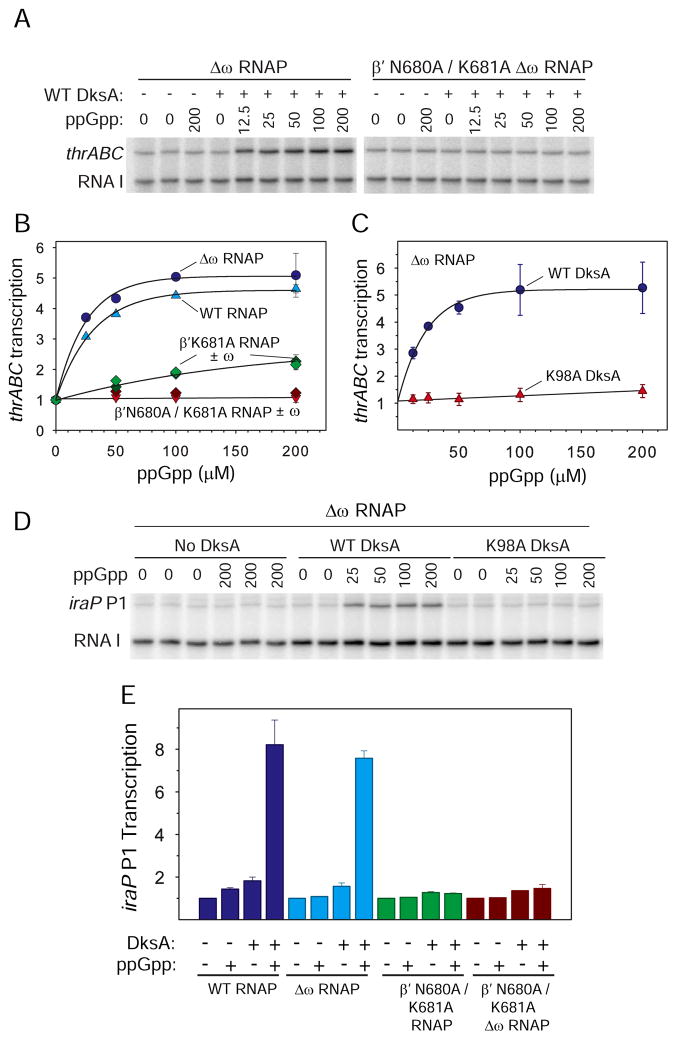
ppGpp and DksA together activate a relatively large number of promoters different from those inhibited by ppGpp and DksA (Paul et al., 2005), including thrABC, several other promoters that are needed for amino acid biosynthesis, and iraP, a promoter responsible for synthesizing the “anti-adapter” protein that is needed for stabilization of the stationary phase sigma factor σS (Bougdour and Gottesman, 2007).
thrABC promoter activity increased ~5-fold (Figures 5A–C), and iraP promoter activity increased 6 to 7-fold (Figures 5D and E) in the presence of ppGpp and WT DksA in vitro, whereas the control promoter RNA I was not activated. There was no activation by ppGpp or DksA alone (Paul et al., 2005; Figure 5D). The extent of activation of thrABC or iraP was similar with WT RNAP or RNAP containing only Site 2 (Δω RNAP) (Figures 5B and E), and there was little or no activation without Site 2 (i.e., with β’K681A/N680A RNAP or K98A DksA). We conclude that Site 2 is required for positive control of transcription by ppGpp.
Figure 5. Substitutions in Site 2 Decrease or Abolish Activation by ppGpp/DksA.
(A) Representative gel showing activation of thrABC transcription in vitro by WT DksA (2 μM) and ppGpp (0–200 μM) and RNAP lacking Site 1 (Δω RNAP) or lacking both Sites 1 and 2 (β’ N680A/K681A Δω RNAP). The RNA I transcript from the same plasmid was unaffected and serves as a control. All lanes were from the same gel image. Intervening lanes between the Δω RNAP and N680A / K681A Δω RNAP samples were removed.
(B) Activation of thrABC transcription by WT DksA and ppGpp, as in (A), with the indicated RNAPs: WT (both Sites 1 and 2, i.e., 1+2+); Δω (1−2+); β’K681A or β’N680A/K681A + ω (1+2−); β’K681A or β’N680A/K681A Δω (1−2−). Transcription is normalized to the level for each RNAP in the absence of DksA or ppGpp. Means and ranges here and in (C) and (E) are from 2 independent experiments.
(C) Activation of thrABC transcription, as in (B), but with RNAP lacking Site 1 (Δω), and either WT DksA or Site 2 mutant DksA K98A (2 μM). Transcription is normalized to the level in the absence of ppGpp.
(D) Representative gel showing activation of iraP P1 transcription by RNAP lacking Site 1 (Δω RNAP) and either no DksA, WT DksA or Site 2 mutant DksA K98A (2 μM) and the indicated ppGpp concentrations.
(E) Relative activation of iraP P1 transcription by WT DksA (2 μM) and ppGpp (200 μM) with RNAPs containing both Sites 1 and 2 (WT), only Site 2 (Δω), only Site 1 (β’N680A/K681A), or neither Site (β’N680A/K681A Δω). Values normalized to that without ppGpp or DksA for each RNAP.
Model for ppGpp Site 2 Based on Genetic and Biochemical Data
Effects of specific substitutions in RNAP and DksA on ppGpp function, binding, and crosslinking, together with localization of a ppGpp crosslink to the β’ rim-helices, suggested that DksA binding to the secondary channel rim creates a pocket for ppGpp, Site 2. These data, other genetic and biochemical data (Supplemental Information), and crystal structures of E. coli RNAP holoenzyme (Zuo et al., 2013) and DksA (Perederina et al., 2004) were used to construct a plausible model for Site 2. In this model, the DksA coiled-coil tip region (DxxDxA motif) and the second helix of the coiled-coil domain are positioned similarly to the analogous regions of Gfh1 in its cocrystal with T. thermophilus RNAP (Sekine et al., 2015). The position of DksA in this complex is similar to that in a recent DksA-RNAP model based on an independent set of genetic and biochemical constraints (Parshin et al., 2015).
Residues implicated by mutational analysis in Site 2 (L95, K98, R129, and K139 in DksA and N680 and K681 in β’) are in close proximity in our model of the DksA-RNAP complex, forming a pocket between the RNAP secondary channel rim and the adjacent surface at the junction of the DksA globular domain, C-terminal helix, and coiled-coil domain, with dimensions sufficient to accommodate ppGpp (or pppGpp) (Figures 6A and 6B). Our data do not allow orientation of ppGpp within the proposed pocket with certainty, but the model suggests that ppGpp could fit in this location without steric clash. The model also is consistent with the observed crosslink of the 6-thio moiety of 6-thio-ppGpp to DksA (with WT RNAP) or to the β’ rim (with β’N680A RNAP) (Figures 3D, 3E, and 6B). Site 2 in this model is ~60Å from Site 1 (Figure 6A), ~30Å from the site proposed in the T. thermophilus RNAP-ppGpp complex (Artsimovitch et al., 2004), and ~90Å from another recently proposed binding site (Syal and Chatterji, 2015).
Figure 6. Model for ppGpp Site 2 Based on Genetic and Biochemical Data.
(A) Model showing proposed location of ppGpp Site 2, constructed in Pymol using structures of E. coli RNAP with ppGpp bound at Site 1 (PDB 4JKR; Zuo et al., 2013) and DksA (chain A from PDB 1TJL; Perederina et al., 2004). See text and Supplemental Information for details about modeling positions of DksA and Site 2-bound ppGpp. ppGpp (red spacefill); secondary channel rim (yellow), β’ (pink), β (cyan), ω (slate blue), DksA (green), σ70 (gold), β’ bridge helix (BH; magenta), active site Mg2+ (grey sphere).
(B) Proposed location of Site 2 (rotated right ~90° from view in A), showing residues implicated in Site 2 binding and function from genetic and biochemical data in Figures 1–5. A possible orientation of ppGpp is shown, indicating that the pocket is large enough to accommodate it. ppGpp (red spacefill); guanine O6 in ppGpp (orange); β’ N680 (blue spacefill); DksA-R91, K98, R129, and K139 (green spacefill).
(C) Conservation of β’ secondary channel rim residues in proteobacteria from ConSurf analysis of representative species (consurf.tau.ac.il), shown in ConSurf color scale on surface representation of E. coli RNAP (PDB 4JKR). See Table S4 for species analyzed and Figure S5. Position of DksA (green cartoon) is as in (A). β’ (dark grey); β (light grey); α (white); ω (light blue); σ70 (pale yellow); active site (orange).
(D) Close-up of secondary channel rim from (C).
(E) As in (D), but showing rim helix conservation in the non-proteobacterial phyla listed in Table S4.
(F) Conservation of DksA residues in representative proteobacteria from ConSurf analysis. See also Figure S5.
Evolutionary Conservation of Site 2
Site 1 is highly conserved in proteobacteria but not in more distantly-related bacterial phyla (Ross et al., 2013), although the enzymes required for synthesis of ppGpp are found throughout the bacterial domain (Atkinson et al., 2011). In species evolutionarily distant from E. coli (e.g., B. subtilis), ppGpp binds to other targets instead of RNAP and regulates transcription by modulating GTP levels (Krasny and Gourse, 2004; Liu et al., 2015).
We examined the evolutionary conservation of Site 2 using the ConSurf Server. Site 2 residues and a short adjacent patch in β’ near the turn between the two rim-helices in E. coli RNAP are highly conserved in representative proteobacterial β’ sequences (magenta in Figures 6C and D; β’N680 100% and β’K681 93% conserved; see also Figure S5 and Table S4), consistent with a critical role for these residues in vivo. In contrast, other surface-exposed residues in the rim-helices are much less conserved. DksA residues in Site 2 are also highly conserved in the same set of proteobacteria (L95, 88% conserved; K98, 88%; R129, 93%; K139, 53%K and 43%Q; Figures 6F and S5). In the non-proteobacterial species analyzed, DksA homologs have not been verified experimentally, and the Site 2 residues in β’ are much less conserved than in the proteobacteria (Figures 6E and S5; Supplemental Experimental Procedures). Although there may be some non-proteobacterial species where only one of the ppGpp binding sites on RNAP is present, in general it appears that ppGpp binds to both Sites 1 and 2 on RNAP in most proteobacteria and it binds to non-RNAP targets in more evolutionarily-distant species.
Both Sites are Required in vivo for Responses to Nutritional Shifts and Amino Acid Starvation
To evaluate the physiological significance of ppGpp binding Sites 1 and 2 in vivo in E. coli, we constructed a set of isogenic strains containing wild-type or mutant ppGpp binding sites: 1+2+ (WT); 1+2− (Site 1 only); 1−2+ (Site 2 only); and 1−2− (neither site present). β’ and/or ω variants that eliminated responses to ppGpp in vitro were introduced into the bacterial chromosome by recombineering and CRISPR/Cas9 selection, followed by transduction with phage P1 into E. coli MG1655 (Supplemental Experimental Procedures). The entire genomes of the 4 strains were sequenced to confirm that no unintended mutations had occurred during strain construction.
The 4 strains grew similarly in LB, with steady-state doubling times of ~22 min for strains containing Site 2 (1+2+ and 1−2+) and ~27 min for strains lacking Site 2 (1+2− and 1−2−) (Figures 7A and C). However, when shifted from LB into a defined minimal medium, the three mutant strains displayed longer lag times than the WT strain before attaining exponential growth (Figure 7B). The 1−2+ strain had the briefest lag, similar to that reported previously (~6 h vs ~3 h for the WT strain; Ross et al., 2013) and formed relatively normal-sized colonies on minimal agar plates. However, the 1+2− and 1−2− strains had much longer lags (~15 h and 30 h, respectively) and formed tiny colonies on minimal agar. The growth of the 1−2− strain at 30 h after the shift did not reflect the presence of suppressor mutations, since cells from the population that started growing after 30 h retained a long-lag phenotype when shifted from rich to minimal medium again. Consistent with this conclusion, all cells in the 1−2− population formed tiny colonies on minimal agar after a long incubation period (efficiency of plating of ~1 relative to the WT strain) (Figure 7C).
Figure 7. Growth and Transcription Regulation of Strains Containing RNAP Substitutions in ppGpp Binding Sites 1 and/or 2.
(A) Growth of four isogenic E. coli strains in LB: 1+2+ (WT), 1−2+ (lacking Site 1), 1+2− (lacking Site 2), or 1−2− (lacking both Sites). The Site 1 mutant is β’ R362A / R417A / K615A / ω Δ(2–5). The Site 2 mutant is β’ K681A / N680A. Means and standard deviations here and in (B–E) are from 3 independent experiments.
(B) Growth of ΔdksA, ΔrelAΔspoT, or strains described in (A) after a shift from LB to glucose minimal medium lacking amino acids (MM).
(C) Cell doubling times in exponential growth and efficiency of plating on MM vs LB (EOP).
(D) qPCR analysis of unstable rrn P1 leader transcripts from chromosomal rRNA operons (rrn) at indicated times after induction of the stringent response with serine hydroxamate (SHX) in the indicated strains. See also Figure S6.
(E) qPCR analysis of iraP transcripts after SHX addition.
rRNA promoter activity in the mutant strains was measured by qPCR during a stringent response by monitoring the synthesis of the unstable leader region generated by the rrn P1 promoters (Figure 7D). ppGpp levels in all four strains increased after induction of amino acid starvation by serine hydroxamate (SHX) addition (Figure S6), and rRNA promoter activity in the 1+2+ strain declined by ~16-fold, as expected. In contrast, rRNA promoter activity in the 1−2− strain was not inhibited at all following SHX addition, a defect similar to that observed in strains lacking DksA or ppGpp (ΔdksA or ΔrelA ΔspoT) (Figure 7D). The 1+2− strain was almost as defective as the 1−2− strain, but partial function was observed in the 1−2+ strain. Thus, Site 1 contributed somewhat to rRNA promoter regulation under these conditions, but Site 2 accounted for the majority of the effect of ppGpp. Similar results were obtained when ppGpp synthesis was induced using a plasmid-encoded relA gene, indicating that the defects observed in the mutant strains were not an indirect effect of starvation (data not shown).
Effects of the ppGpp site mutants on positive control by ppGpp in vivo were determined by measuring iraP mRNA levels by qPCR following SHX addition (Figure 7E). iraP transcript levels increased dramatically after starvation of the strains containing Site 2 (1+2+ or 1−2+), but a much smaller increase was observed in strains lacking Site 2 (1−2− or 1+2−). We suspect that the residual increase in iraP transcription in the Site 2 mutant strains could derive from indirect effects of the reduction in ribosome synthesis on RNAP availability after SHX treatment. Thus, positive control by ppGpp and DksA depends on Site 2 at the high ppGpp concentrations produced by SHX treatment.
DISCUSSION
Our data show that there are two distinct binding sites for ppGpp on E. coli RNAP and that together they account for effects of ppGpp on transcription during the stringent response. Site 1 is at the β’ - ω subunit interface and does not require the participation of DksA, whereas Site 2 is at the DksA - β’ interface. Thus, Site 2 accounts for the previously-reported synergistic effects of DksA and ppGpp on transcription initiation (Paul et al., 2004, 2005), and under the in vivo conditions measured in our experiments (induction of a stringent response; Figure 7), Site 2 has much larger effects than Site 1.
Site 2 is an unusual ligand binding site in that it is formed at the interface of two proteins, RNAP and an RNAP-bound transcription factor, DksA. Signaling through this site therefore integrates input from both the concentration of ppGpp and the fractional occupancy of RNAP with DksA. Intracellular ppGpp concentrations vary widely, from ~10 to 100 μM in steady-state growth on different carbon sources (“basal” levels) to ~1 mM after starvation for amino acids (Ryals et al., 1982). Under the conditions examined, the DksA concentration in E. coli varies only ~2−fold (Rutherford et al., 2007) and is non-saturating, since DksA overexpression leads to increased effects on transcription (Potrykus et al., 2006). However, because other E. coli secondary channel-binding transcription factors, such as the elongation factors GreA and GreB, bind to an overlapping site on RNAP, changes in their concentrations could potentially affect DksA binding by competition. The Gre factors lack the residues in DksA that interact with ppGpp, do not act synergistically with ppGpp, and are normally expressed at levels that are too low to regulate rRNA transcription directly (Rutherford et al., 2007; Lee et al., 2012). However, they compete with DksA when overexpressed (Potrykus et al., 2006).
Because the phenotype of the strain lacking both sites (1−2−) was almost as severe as that of strains lacking DksA or ppGpp (Figure 7), RNAP appears to be the major ppGpp target in E. coli. Conservation of the ppGpp binding sites on RNAP suggests that this could be the case throughout the proteobacteria. However, ppGpp has also been reported to play roles in DNA replication, translation, central metabolism, and bacterial persistence by binding to and regulating the activities of other enzymes besides RNAP (Potrykus and Cashel, 2008; Kanjee et al., 2012; Liu et al., 2015; Hauryliuk et al., 2015). We expect that the strains lacking the ppGpp binding sites in RNAP will be an essential tool for distinguishing effects of ppGpp binding to non-RNAP targets from indirect effects resulting from direct binding to RNAP. For example, it has been proposed that ppGpp-binding proteins different from RNAP are responsible for bacterial persistence (Hauryliuk et al., 2015), predicting that persistence might still occur in the strain lacking RNAP Sites 1 and 2.
Why two binding sites for ppGpp?
Our data suggest that Sites 1 and 2 might fill at different concentrations of ppGpp. At a non-saturating DksA concentration in vitro (2 μM), a concentration that mimics its effects on transcription in vivo (Paul et al., 2004), IC50 values for ppGpp at Site 2 were ~2-fold higher than for Site 1 (Figure 1E). The degree of saturation of RNAP with DksA in vivo, the precise differences in affinity of ppGpp for Sites 1 and 2, and the potential for cooperativity in binding of ppGpp to the two sites remain to be determined. In any case, having two ppGpp sites with different affinities would expand the dynamic range for responses to ppGpp under different growth conditions.
Our in vitro data predict that Site 1 would be responsible for effects on transcription initiation in vivo when ppGpp concentrations are low, e.g. during growth in rich medium or early in a starvation response. However, when ppGpp concentrations are high enough to fill both sites, the larger effects of Site 2 on transcription observed in vitro predict that it would be responsible for the majority of the effects in vivo. Consistent with this prediction, Site 2 mutants were more defective than Site 1 mutants in recovering from severe nutritional shifts and in responding to amino acid starvation (Figure 7).
ppGpp binding to the two different sites could also affect RNAP-promoter interactions by different mechanisms, perhaps by favoring distinct conformational states in RNAP. Individual promoters could be differentially sensitive to effects of ppGpp binding at one site or the other, depending on the intrinsic kinetic properties of the particular promoter. Although most promoters that are regulated by ppGpp are also responsive to DksA, there have been reports of promoters regulated by only one or the other (e.g. Aberg et al., 2009). Our identification of two distinct ppGpp binding sites that fill under different nutritional or environmental conditions could provide an explanation for these differential responses.
Mechanism of ppGpp Action
Site 1 is located ~60Å from Site 2, at the primary interface connecting two rigid-body domains in RNAP, the core and shelf modules, raising the possibility that ppGpp binding to Site 1 might block hinge-like motions between the domains (Ross et al., 2013; Zuo et al., 2013; Sekine et al., 2015). In contrast, since Site 2 is partly composed of DksA, ppGpp might enhance the function proposed for DksA by itself. DksA is thought to act by an allosteric mechanism in which the DksA coiled-coil tip region interacts with the RNAP trigger loop and bridge helix to alter RNAP-promoter contacts, shifting the equilibrium occupancy of intermediates on the pathway to the transcription-competent complex (Rutherford et al., 2009; Lee et al., 2012; Lennon et al., 2012). For negatively-regulated promoters, RNAP interactions with promoter DNA downstream of the −10 hexamer are disfavored by DksA binding, enriching for an RPC-like closed complex (Rutherford et al., 2009; Mekler et al., 2014). Consistent with the idea that ppGpp at Site 2 might work by enhancing DksA function, DksA coiled-coil tip residues A76 and D74 are required for DksA function alone and for effects of ppGpp bound to Site 2 (Figure S5; Lee et al., 2012).
In theory, ppGpp could simply increase the occupancy of the DksA-RNAP complex by causing a conformational change in DksA that increases its binding to RNAP. However, the requirement for both ppGpp and DksA for positive control of transcription and the requirement for ppGpp for positive control even at high concentrations of the high affinity DksA variant N88I (Blankschien et al., 2009) suggest that simply increasing DksA binding to RNAP is insufficient to account for the full effect of ppGpp binding to Site 2.
The position of Site 2 at the junction of the three sub-domains of DksA (globular, coiled-coil, and C-terminal helix) suggests how it could cause a conformational change in DksA (Figure 4). The C-terminal helix is critical for DksA function (Furman et al., 2013; Parshin et al., 2015), and its position relative to the rest of the protein varies in the 10 monomers captured in the E. coli DksA crystal (Perederina et al. 2004; Figure 4H). We speculate that ppGpp binding could affect RNAP-promoter interactions by stabilizing a DksA conformation that involves the C-terminal helix interaction with nearby regions of RNAP, namely β SI1 and/or β’ SI3 (Parshin et al. 2015). Site 2-dependent ppGpp effects could thereby be mediated through altered or stabilized DksA coiled-coil tip region interactions with RNAP, but in addition through a second, uncharacterized mechanism involving DksA C-terminal helix interactions with RNAP β SI1 and/or β’ SI3.
Prospect
We have shown here that ppGpp binds to two sites on RNAP and that substitutions in these sites eliminate effects of ppGpp on transcription initiation in vitro and in vivo. Although the details of Site 2 will require structural information, the enzymes and strains lacking one or both sites provide tools for examining the allosteric transitions responsible for inhibition and activation of promoters by ppGpp, for studying effects of ppGpp on nutritional regulation of the transcription initiation and elongation machineries genome-wide, and for identifying molecular targets of ppGpp other than RNAP. Finally, definition of the ppGpp binding sites could facilitate design of antibiotics that could deprive pathogens of an essential survival mechanism during infection.
EXPERIMENTAL PROCEDURES
Further experimental detail is supplied in Supplemental Information.
Plasmids, Strain Construction, and Protein Purification
Strains, plasmids, and oligonucleotides are listed in Tables S1 and S2. Site-directed mutations were introduced into multisubunit RNAP overexpression plasmids encoding β’ with a His-tag or DksA with a His-tag. E. coli strains with substitutions in ppGpp binding Sites 1 and/or 2 were constructed by recombineering methods together with CRISPR-Cas9 counterselection (see below). Δω RNAPs (Site 1-) were purified from a BL21DE3 ΔrpoZ strain (Ross et al., 2013). DksA was purified from BL21DE3 ΔdksA (Paul et al., 2004).
In Vitro Transcription
Multiple round transcription reactions contained 170 mM NaCl, 20 nM RNAP, with DksA and/or ppGpp as indicated. Promoter-specific transcripts were produced from plasmid DNA templates containing a cloned promoter followed the rrnB T1 terminator (Ross et al, 2013). These plasmids also encode the RNA I promoter.
Crosslinking
32P-6-thio-ppGpp was synthesized from 6-thio-GDP and γ-32P-ATP and was used at ~5 μM. Crosslinking was induced by UV treatment for 5 min (Ross et al., 2013).
ppGpp and DksA Binding Assays
32P-ppGpp binding to RNAP or to RNAP-DksA complexes was determined by the Differential Radial Capillary Action of Ligand Assay (DRaCALA; Roelofs et al., 2011). 32P-ppGpp was synthesized from GDP and γ-32P-ATP, purified, incubated with the indicated proteins. Reactions were spotted onto nitrocellulose filters and counts retained in the central protein spot were quantified by phosphorimaging. DksA binding was determined using the Fe2+ cleavage assay (Lennon et al., 2009).
Effects of ppGpp Binding Site Mutants in vivo
Mutant and wild-type strains were shifted from rich medium (LB) to minimal medium (MOPS-glucose), and cell growth was followed in a plate reader. Effects of amino acid starvation on promoter activity were measured by extraction of RNA from cultures at the indicated times after addition of serine hydroxamate (SHX) and qPCR of the rrn P1 leader transcript or the iraP mRNA.
Modeling of ppGpp Binding Site 2
Models for DksA binding to RNAP and for Site 2 were created in Pymol using existing structures of E. coli RNAP (Zuo et al., 2013; PDB 4JKR) and DksA (Perederina et al., 2004; chain A from PDB 1TJL).
Conservation of ppGpp Site 2 Residues
A 125 amino acid segment of the β’ subunit sequence and DksA sequences from 57 species representing all orders of proteobacteria and β’ subunit sequences from 26 species representing 6 non-proteobacterial phyla were analyzed for conservation of individual residues using the ConSurf Server (consurf.tau.ac.il).
Supplementary Material
Highlights.
ppGpp still regulates a DksA-RNAP complex lacking the known ppGpp binding site
A ppGpp binding site forms at the DksA-RNAP interface, ~60Å from the first site
ppGpp binding to this 2nd site strongly affects transcription in vitro and in vivo
Cells lacking both sites have dramatic defects in recovery from nutritional shifts
Acknowledgments
We thank C. Gross, S. Borukhov, and A. Parshin for communication of results prior to publication, L. Marraffini for providing pCas9, and S. Ades, K. Gerdes, R. Landick, M. Laub, and members of our lab for comments on the manuscript. This work was supported by NIH R37 GM37048 to R.L.G. Support for P.S.V. and H.L.B. was provided in part by an NIH Molecular Biosciences predoctoral training grant and fellowships from UW-Madison.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, writing, and editing the manuscript: W.R. and R.L.G.; Methodology and Investigation: W.R., P.S.V., H.B., A.C., and J.H.L.; Supervision: W.R. and R.L.G.; Funding Acquisition: R.L.G.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aberg A, Fernández-Vázquez J, Cabrer-Panes JD, Sánchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol. 2009;191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankschien MD, Lee JH, Grace ED, Lennon CW, Halliday JA, Ross W, Gourse RL, Herman C. Super DksAs: substitutions in DksA enhancing its effects on transcription initiation. EMBO J. 2009;28:1720–1731. doi: 10.1038/emboj.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA. 2007;104:12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013;21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalebroux SD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J Mol Biol. 2013a;425:82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjee U, Ogata K, Houry WA. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol. 2012;85:1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Lennon CW, Ross W, Gourse RL. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J Mol Biol. 2012;416:503–517. doi: 10.1016/j.jmb.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Gaal T, Ross W, Gourse RL. Escherichia coli DksA binds to free RNA polymerase with higher affinity than to RNA polymerase in an open complex. J Bacteriol. 2009;191:5854–5858. doi: 10.1128/JB.00621-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee JH, Butcher SE, Gourse RL. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Myers AR, Pisithkul T, Claas KR, Satyshur KA, Amador-Noguez D, Keck JL, Wang JD. Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol Cell. 2015;57:735–749. doi: 10.1016/j.molcel.2014.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 2013;41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V, Minakhin L, Borukhov S, Mustaev A, Severinov K. Coupling of downstream RNA polymerase-promoter interactions with formation of catalytically competent transcription initiation complex. J Mol Biol. 2014;426:3973–3984. doi: 10.1016/j.jmb.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parshin A, Shiver AL, Lee J, Ozerova M, Schneidman-Duhovny D, Gross CA, Borukhov S. DksA regulates RNA polymerase in Escherichia coli through a network of interactions in the secondary channel that includes Sequence Insertion 1. Proc Natl Acad Sci USA. 2015;112:e6862–6871. doi: 10.1073/pnas.1521365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel-structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Vinella D, Murphy H, Szalewska-Palasz A, D'Ari R, Cashel M. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J Biol Chem. 2006;281:15238–15248. doi: 10.1074/jbc.M601531200. [DOI] [PubMed] [Google Scholar]
- Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roghanian M, Zenkin N, Yuzenkova Y. Bacterial global regulators DksA/ppGpp increase fidelity of transcription. Nucleic Acids Res. 2015;43:1529–1536. doi: 10.1093/nar/gkv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Lemke JJ, Vrentas CE, Gaal T, Ross W, Gourse RL. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals J, Little R, Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol. 1982;151:1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–421. doi: 10.1016/j.molcel.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Satory D, Halliday JA, Sivaramakrishnan P, Lua RC, Herman C. Characterization of a novel RNA polymerase mutant that alters DksA activity. J Bacteriol. 2013;195:4187–4194. doi: 10.1128/JB.00382-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syal K, Chatterji D. Differential binding of ppGpp and pppGpp to E. coli RNA polymerase: photo-labeling and mass spectral studies. Genes Cells. 2015;20:1006–1016. doi: 10.1111/gtc.12304. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, Shulgina I, Hernandez VJ. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta’-subunit. J Biol Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–2387. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Mooney RA, Grass JA, Sivaramakrishnan P, Herman C, Landick R, Wang JD. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell. 2014;53:766–778. doi: 10.1016/j.molcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, Wang Y, Steitz TA. The structure of magic spot bound to RNA polymerase suggests how it is regulated by ppGpp. Mol Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.