Abstract
Drug resistance is a major problem in health care, undermining therapy outcomes and necessitating novel approaches to drug design. Extensive studies on resistance to viral protease inhibitors, particularly those of HIV-1 and hepatitis C virus (HCV) protease, revealed a plethora of information on the structural and molecular mechanisms underlying resistance. These insights led to several strategies to improve viral protease inhibitors to counter resistance, such as exploiting the essential biological function and leveraging evolutionary constraints. Incorporation of these strategies into structure-based drug design can minimize vulnerability to resistance, not only for viral proteases but for other quickly evolving drug targets as well, toward designing inhibitors one step ahead of evolution to counter resistance with more intelligent and rational design.
Keywords: drug resistance, protease inhibitors, HIV-1 protease, substrate envelope, structure based drug design, resistance mutations
Drug Resistance and Viral Proteases as Drug Targets
Drug resistance is a major health burden in a wide range of diseases from cancer to bacterial and viral infections, causing treatment failure as well as severe economic impact on the healthcare system. Drug resistance can be conferred via various mechanisms including decreased intracellular levels of drug such as due to efflux pumps, altered gene expression, and changes in drug target [1–4]. The most common mechanism of resistance to drugs against quickly evolving targets involves mutation of the targeted protein, including resistance to small molecule inhibitors of viral proteases.
Viral proteases are ideal drug targets as they are essential in the viral life cycle, and inhibiting the viral protease prevents the generation of new infectious viral particles. Most viruses that infect humans and cause disease encode at least one viral protease [5, 6]. These proteases are responsible for cleaving the viral polyprotein precursors at specific sites to release individual functional proteins, including cis cleavage of the protease itself. Certain viral proteases have also been reported to cleave host cell proteins such as translation initiation factors (eIF4 and eIF3d) in HIV to inhibit host protein translation [7, 8], and transcription factors in hepatitis C virus (HCV) to confound the innate immune response [9]. Although most viral proteases share general backbone folds and catalytic mechanisms with host cellular proteases, they are generally more compact likely due to evolutionary pressure to maintain a small genome [6]. Nevertheless, viral proteases are able to recognize and cleave diverse substrate sequences with distinct specificities.
HIV-1 and HCV Protease Inhibitors
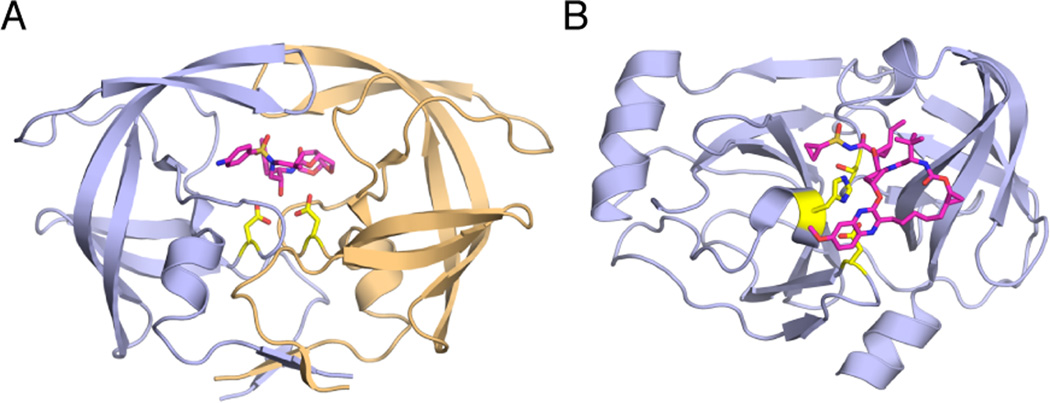
Among medically relevant viruses, the viral protease most extensively investigated is inarguably the aspartyl protease encoded by HIV-1, with hundreds of sequences in the Stanford Database and crystal structures in the Protein Data Bank [10, 11]. HIV-1 protease is comprised of two identical chains of 99 amino acids each, with the active site located at the dimer interface and each monomer contributing a catalytic Asp to the active site (Figure 1a). In the unliganded state, HIV-1 protease is symmetric with highly flexible flaps that allow access to the active site. In the liganded state, these flaps close upon the bound substrate or inhibitor at the active site and become much more rigid. HIV-1 protease has been the target of extensive drug discovery and development efforts for decades and had a major role in launching the field of structure-based drug design. These efforts resulted in 9 FDA-approved HIV-1 protease inhibitors (PIs). All HIV-1 PIs are competitive inhibitors that bind at the protease active site. Although these PIs are very effective in inhibiting the wild-type protease and have significantly contributed to clinical treatment outcomes in combination therapy [12–14], resistance has emerged to all HIV-1 PIs.
Figure 1.
HIV-1 and HCV Protease Structures. (A) HIV-1 protease bound to darunavir (PDB: 1T3R). The two monomers of HIV-1 protease are in light purple and gold. (B) HCV NS3/4A protease bound to MK-5172 (PDB: 3SUD). The inhibitors are in magenta and the catalytic residues in yellow sticks.
HCV, which infects millions of people worldwide and causes chronic liver disease, liver failure and liver cancer [15, 16], encodes a chymotrypsin-like serine protease, NS3/4A (Figure 1b). HCV NS3/4A protease is a prime therapeutic target for direct-acting antivirals, with four FDA-approved inhibitors [17–21] and several in various stages of clinical development. However, even before the drugs were approved for the clinic, resistant viral variants have emerged [22–24]. Rapid emergence of resistance and low efficacy against genotypes other than HCV Genotype 1 has mandated combination therapies, which also decreased treatment duration and increased cure rates especially for Genotype 1 [25–27].
Mutations Confer Resistance by Selectively Weakening Inhibitor Binding but Retaining Specific Substrate Recognition and Cleavage
For a virus to become resistant to a PI, the viral genome acquires mutations that allow the protease to thwart inhibition by the drug but still retain the ability to cleave the viral polyprotein substrate at the required specific sites to allow viral maturation. HIV-1 evolves rapidly due to a high viral replication rate (107–109 newly infected cells/day in a patient [28]) and the error-prone mechanism of the viral reverse transcriptase, which generates a diverse pool of viral variants. This rapid evolution enables the targeted viral protease to acquire mutations that abrogate the efficiency of inhibitor–protein binding. Many mutations already pre-exist at low levels even before the start of therapy in infected patients, and quickly become selected under drug pressure. Critically, these selected protease variants still retain their substrate recognition and cleavage activity and allow viral propagation. HIV-1 protease needs to process the Gag and Gag-Pro-Pol polyproteins at nine distinct sites, while HCV NS3/4A protease cleaves viral polyprotein precursors at four cleavage sites and cleaves two human immune proteins to confound the innate immune response. However, these cleavage sites are highly diverse in amino acid sequence, and unlike most known human proteases, the viral proteases do not have simple substrate sequence recognition motifs.
How is a viral protease able to recognize and cleave apparently dissimilar substrates with high specificity? And how can the targeted protease mutate to avoid inhibition but still process these substrates? The answer to these questions for HIV-1 protease led to major breakthroughs in elucidating molecular mechanisms underlying selection of primary mutations in drug resistance.
Substrate Envelope Explains Selection of Active Site Resistance Mutations
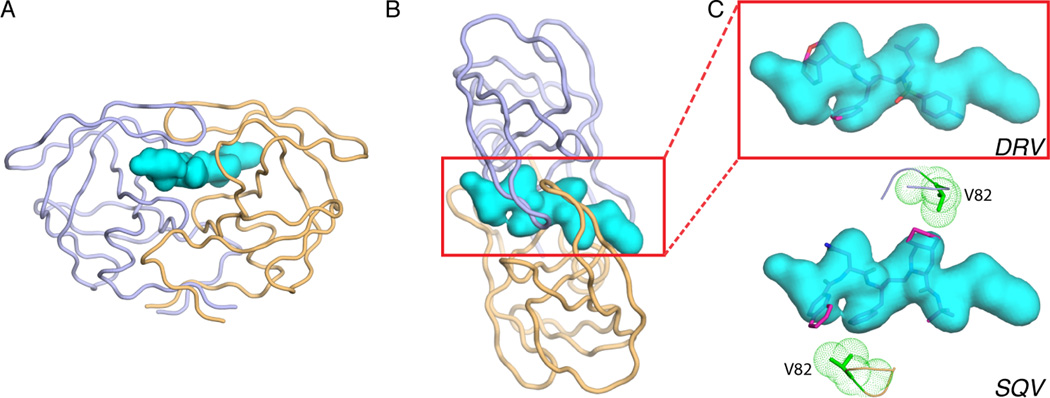
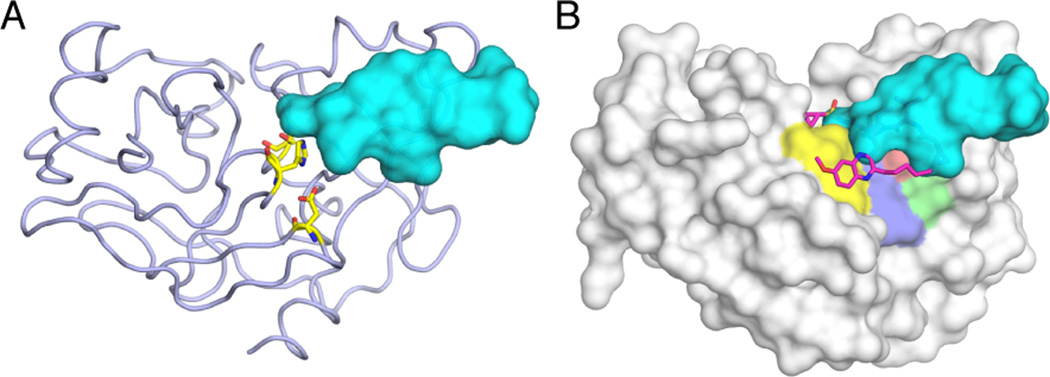
Although not similar in amino acid sequence, the substrates cleaved by HIV-1 protease all occupy a similar ‘shape’ or volume when bound at the active site, as revealed by high-resolution crystal structures of protease-bound peptides corresponding to cleavage sites in the viral polyprotein [29, 30]. Hence, the recognition motif for the viral protease is not a particular amino acid or sequence but a common shape assumed by the substrates, termed the substrate envelope (Figure 2). Similarly, the substrate envelope for HCV NS3/4A protease has also been determined as the overlapping volume shared by bound peptides in complex crystal structures [31] (Figure 3).
Figure 2.
Substrate Envelope of HIV-1 Protease and Fit of PIs in the Envelope. (A) The substrate envelope (teal volume) displayed at the active site of HIV-1 protease (light purple and gold ribbons), and (B) top view of this. (C) The fit of protease inhibitors darunavir (DRV, red sticks) and saquinavir (SQV, magenta sticks) in the substrate envelope. The side chain of protease residue V82, which mutates to A to confer resistance to SQV, is displayed as green sticks and mesh surface.
Figure 3.
Substrate Envelope of HCV NS3/4A Protease and the Fit of PI MK-5172 in the Envelope. (A) The substrate envelope (teal volume) displayed at the active site of HCV protease (light purple ribbon). The catalytic side chains are in yellow sticks. (B) The fit of protease inhibitor MK-5172 (magenta sticks) in the substrate envelope. The protease is displayed as a gray surface, with catalytic residues (yellow) and residues at the S2 subsite (purple, green, and red) that mutate to confer resistance colored on the surface. The large P2 extension moiety of MK-5172 protrudes beyond the envelope but avoids contacts with the S2 residues and instead packs against the catalytic residues.
In addition to explaining how the viral proteases recognize their substrates, the substrate envelope effectively explains the mechanism of resistance due to active site mutations. Active site residues that make essential and conserved interactions with the substrate, including the catalytic side chains, cannot mutate to confer resistance without compromising substrate binding. However, whenever an inhibitor protrudes beyond the substrate envelope to contact a protease residue, that residue can mutate to differentially weaken contact with the inhibitor relative to substrate binding resulting in drug resistance. Accordingly, specific resistance mutations near the viral protease active site that confer resistance to a given inhibitor occur at positions where that inhibitor protrudes beyond the substrate envelope to make contact with the enzyme (Figure 2) that are not necessary for substrate recognition but compromise inhibitor binding.
Correlations between active site resistance mutations and protrusion of a specific inhibitor out of the substrate envelope have been demonstrated for a variety of HIV-1 and HCV PIs [29, 31–34]. HIV-1 protease signature resistance mutations (such as D30N to nelfinavir, I50V/L to amprenavir/atazanavir, G48V to saqunavir, and V82A to saquanavir/ritonavir) all occur where that particular inhibitor contacts the protease active site beyond the substrate envelope (Figure 2). Similarly, the major HCV NS3/4A protease resistance mutations occur in the S2 pocket residues (R155, A156, and D168) that are contacted by large P2 extension moieties protruding out the substrate envelope [24, 31]. Hence, a structural analysis of the fit of a given inhibitor within the viral protease substrate envelope is a good predictor of active site residues that may mutate to confer resistance to that inhibitor.
Strategies in Designing Against Resistance
Drug design typically focuses on improving affinity against a single target of interest, typically the wild-type variant of the most common genotype in the case of viral proteases. Indeed, all the HIV-1 PIs have been designed on wild-type HIV-1 subtype B, the dominant viral strain in Europe and the Americas. Inadvertently, this current drug design paradigm may actually facilitate the occurrence of drug resistance, since the standard process is to develop potent compounds that very specifically target the wild-type structure of a single subtype. Polymorphisms in other subtypes can severely impact inhibitor potency and lead to different pathways to resistance, as seen in HCV genotype 3 [35] and HIV-1 [36]. Disrupting the activity of a single wild-type target is necessary, but not sufficient, for developing a robust drug that avoids resistance. Instead of trying to combat resistance only after resistant viral variants arise in clinic, we need a paradigm shift to incorporate elucidation of drug resistance mechanisms to the design strategy.
Detailed analysis of molecular resistance mechanisms for HIV-1 and HCV viral PIs has led to several strategies that can be incorporated into the design strategy toward more robust inhibitors, which should extend to other rapidly evolving drug targets (Box 1).
Box 1. Strategies to Design Potent Viral PIs Robust Against Resistance.
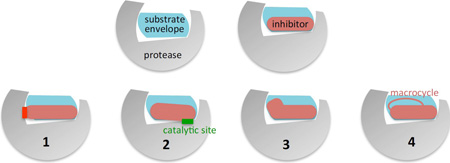
Figure I. Depiction of Strategies for Design of Inhibitors to Prevent Resistance
The substrate envelope of viral proteases can be incorporated into structure-based drug design to minimize the vulnerability of inhibitors to the emergence of resistance. The main strategies to design resistance-avoiding PIs include:
Stay within the substrate envelope: avoid contacts with protease residues (red bar) beyond those of the natural substrate(s).
Extend beyond the substrate envelope only if contacting invariant conserved residues such as the catalytic side chains (green bar), or toward the solvent.
Exploit untapped regions of the substrate envelope to further increase potency.
Add macrocycles to preposition the inhibitor in a binding-competent state, making sure the macrocycle itself obeys the rules above.
The first strategy is to design inhibitors that fit within the substrate envelope. This strategy originates from the realization that substrate envelope efficiently explains susceptibility to resistance due to active site mutations. As protrusions out the substrate envelope render an inhibitor vulnerable to resistance, avoiding such protrusions will minimize the chances of active site mutations that can confer resistance. Analogous pairs of inhibitors designed to stay within versus extend beyond the substrate envelope supported this strategy [37]. Darunavir (DRV) is the proof-of-concept for the viability and validity of this strategy and is the most potent and robust HIV-1 PI in the clinic [38–40]. Although not designed according to the substrate envelope, DRV fits very well within HIV-1 protease substrate envelope, avoiding unessential contacts that would render the inhibitor susceptible to resistance mutations. Accordingly, no single or even double mutation confers significant DRV resistance; protease needs to extensively evolve to accumulate at least ~7 mutations to escape DRV inhibition in vitro at therapeutic concentrations [38, 41]. In clinical trials, 11 mutations in protease were associated with resistance to DRV and the presence of at least 3 of these mutations was associated with a diminished virological response to this drug [42]. DRV also proves that achieving high potency (single digit picomolar) while staying within the substrate envelope is possible.
How widely applicable is the design strategy of using the substrate envelope to counter resistance due to active site mutations? Both HIV-1 and HCV proteases have diverse substrates that are recognized by shape, which defines the envelope. Similarly, most viral proteases also lack a sequence-based recognition motif and process diverse substrates that also likely bind to protease in a conserved shape. However, multiple or diverse substrates are not a prerequisite to define a substrate envelope for a given target. The key to this strategy is to define target-substrate contacts (even for a single natural substrate) that are essential for biological function, and avoid any contacts beyond those in inhibitor design.
A complementary strategy in identifying residues that are essential to the target’s biological function is exploiting knowledge on sequence conservation. Whether with or without selection pressure, the viral protease can evolve only if the enzyme maintains residues essential for function, including the catalytic residues. Designing inhibitors that pack on these strictly conserved residues minimizes chances of mutations that the enzyme can tolerate to confer resistance. The HCV NS3/4A PI MK-5172 is a good example where the extended P2 moiety protrudes out the substrate envelope but packs on the catalytic residues (rather than the mutation-prone S2 subsite) (Figure 3). In addition to the catalytic residues, other regions that cannot tolerate mutations can be identified by sequence alignment or by systematic probing of the mutational fitness landscape through saturating mutagenesis [43, 44]. In such an analysis, all possible point mutations can be introduced into the protease and the effect on enzymatic fitness measured. Residues that cannot tolerate mutations without compromising fitness may have less chance of mutating to confer resistance.
One strategy in designing against resistance is maximizing hydrogen bonds of the inhibitor with main chain atoms of active site residues. The idea here is that the protease backbone atoms are invariant regardless of the side chain, which can change due to a mutation [45, 46]. While less likely than a side chain hydrogen bond, main chain hydrogen bonds can still be readily lost due to steric effects or shifts in the inhibitor-binding mode due to side chain changes. Therefore, this strategy may be the most effective when the binding mode of natural substrates is taken into consideration, by either mimicking the hydrogen bonds of natural substrates or making sure the inhibitors do not violate the substrate envelope constraints.
Another strategy against resistance, proposed for both HIV-1 protease and reverse transcriptase and may be more generally applicable, is designing flexibility into the inhibitors so that they can slightly shift or change conformation in response to active site mutations [47–50]. One key caveat in applying this strategy may be having an ‘anchor’ moiety with extensive interactions that are invariant, which can help ensure sustained potency even when part of the inhibitor adapts in response to the mutation.
Increasing Potency to Avoid Resistance
Although common practice is to report fold-changes in affinity (Ki or IC50 values) with respect to wild-type protease, the absolute value of affinity against a given variant is what determines resistance at a given drug concentration. For instance, DRV has a Ki in the low picomolar range against wild-type HIV-1 protease. Even with a 10 or 100-fold change due to a mutation, DRV may still be more potent against a ‘resistant’ variant compared to other inhibitors with much lower affinity. Hence, increasing potency of a viral PI contributes to the barrier to resistance.
Potency of PIs can benefit from additional design strategies based on recent findings, in addition to standard structure based drug design. First, inhibitors can be modified to more optimally fill the substrate envelope, without compromising low susceptibility to resistance. For example, the P1’ moiety in DRV can be extended without violating the substrate envelope, a strategy that resulted in inhibitors even more potent and more robust than DRV [51–53]. Similarly, a comparison of HCV NS3/4A inhibitors with the substrates reveals a large portion of the substrate envelope in the P4-P5 region not utilized by any current inhibitor [54] (Figure 3). Extending inhibitors to this untapped region of the protease active site can enhance inhibitor potency. Another strategy that significantly enhanced HCV PI potency is adding a macrocycle to link either the P2–P4 or P1–P3 moieties [55–57]. The macrocycle is thought to stabilize the inhibitor in a conformation competent for protease binding, thus increasing the entropic contribution to the free energy of binding. However, macrocycle design may need to be optimized between the rigidity required for enhancing entropy versus the flexibility required for adapting to a mutated active site [55, 58].
Can Resistance-Proof PIs Be Designed?
PIs of HIV-1 have two unique advantages toward becoming resistance-proof: (i) PIs have highly cooperative dose-response curves. This implies that at clinical levels of drug concentration, which is typically much higher than IC50 values, very high levels of (or complete) enzyme inhibition can be achieved [59–61]. (ii) PIs are the only transition-state analogs among direct-acting antivirals (DAAs). As transition-state analogs, HIV-1 PIs have unprecedented potential to achieve exceptional potencies, as enzymes have evolved to bind tightest to the transition state.
In clinical practice, emergence of drug resistance by HIV-1 is successfully thwarted by multidrug therapy where the rapid addition of high levels of a combination of drugs presents a high enough genetic barrier so that resistance does not develop. This happens even though resistance would evolve to each of the drugs if they were given sequentially and additively. This clearly shows that there is a limited sequence space the virus can occupy during the time of therapy initiation, and that there is not enough time (i.e. rounds of replication) to explore greater space during the period when viral load is falling in the presence of multiple drugs. Even though there is no imminent effort to completely replace multidrug therapy with a single inhibitor, the possibility of a ‘resistance-proof protease inhibitor’ is a compelling concept, and may find clinical use in maintenance therapy.
As increasingly potent transition state analogs are developed, the threshold may be crossed where the potency is so high that the addition of one, two, or three resistance mutations does not confer a useful level of resistance and thus does not allow sustained replication. This approach could lead to a path where single drug therapy could be a viable option in a maintenance strategy for treatment-naïve patients, greatly reducing the cost of lifelong therapy. In fact, therapy failure with DRV in such individuals is rarely associated with drug resistance mutations [62], suggesting that failure is primarily due to compliance or extreme metabolism of the drug. In few patients where DRV-containing regimens as first line of therapy fail, in addition to no mutations in the protease, the reverse transcriptase resistance mutations were only rarely observed [63]. Hence, DRV may be close to an inhibitor where the appearance of the first resistance mutation is insufficient to confer enough of a growth advantage to allow evolution of subsequent resistance mutations, and further improvements in inhibitor potency could lead to resistance-avoiding PIs. A recent clinical study actually found DRV monotherapy to be acceptable for long-term management of HIV infection, with regular viral load monitoring and prompt reintroduction of combination treatment for those who rebound [64]. However, considering the scarcity of data on inflammation and clinical progression, long-term DRV monotherapy needs to be administered with caution and only in selected patients, and we may need to even further improve pharmacokinetics, potency and clinical outcomes for long-term PI monotherapy.
Remaining Challenges in Overcoming Resistance
While we have considerable knowledge on the molecular mechanisms underlying resistance conferred by various protease mutations, still multiple challenges remain in understanding and overcoming PI resistance (see Outstanding Questions). In many drug resistant protease variants, multiple mutations co-evolve to both decrease the affinity of a particular inhibitor, and increase the viability and fitness of the enzyme. In HCV, a single mutation is often enough to confer resistance to PIs, but variants with multiple mutations can be found in clinical samples, as well as multiple changes in amino acid sequence (polymorphisms) across different genotypes. In HIV-1, multi-drug resistant patient isolates contain multiple mutations with up to 25% of the amino acids in the enzyme able to mutate and contribute to resistance [65, 66]. The impact of such combinations of mutations on conferring drug resistance is not simply additive, but instead these mutations can have complex interdependent effects [67, 68]. One remaining challenge is to unravel the interdependency of multiple mutations in conferring drug resistance.
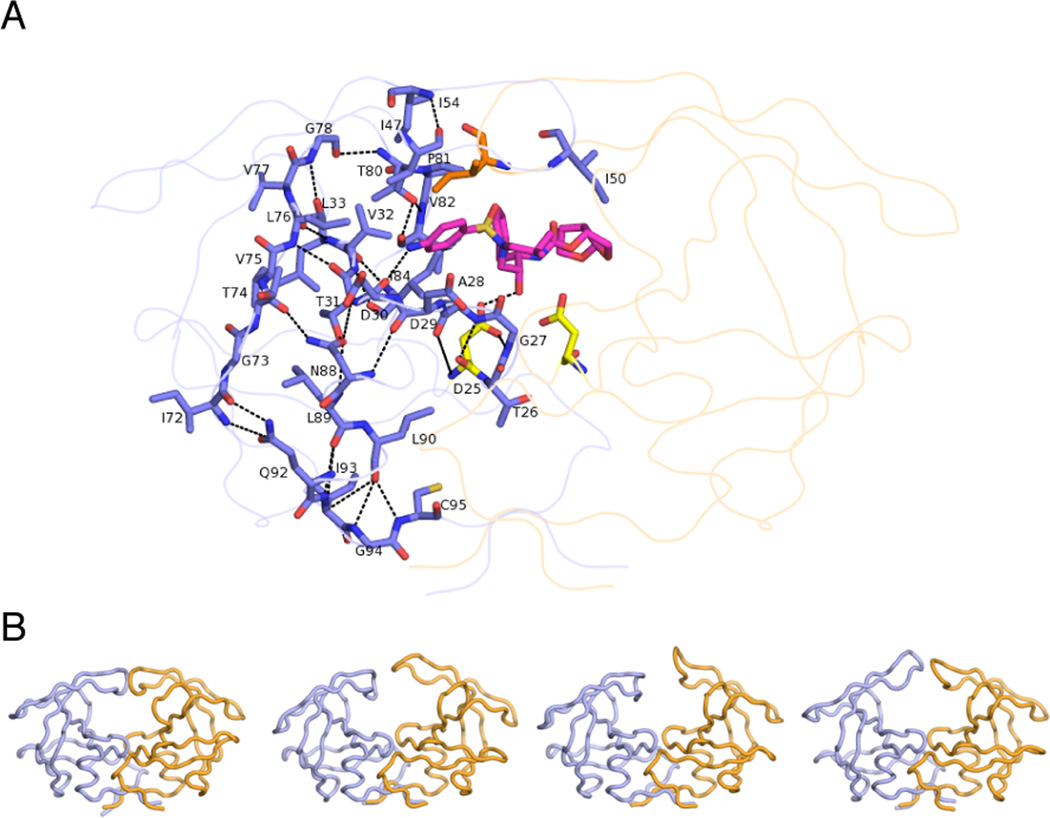
Another challenge is revealing the molecular mechanisms of resistance conferred by mutations at residues outside of the active site (Figure 4). While the substrate envelope efficiently explains the selection of active site mutations, the mechanism of resistance associated with mutations outside the active site still remain elusive. Mutations distal to the active site may compensate for the losses in enzymatic fitness or protein conformational stability due to the active site mutations [69, 70]. In HIV-1 protease, many of these mutations are known to compensate for enzymatic fitness, but these and others may also directly confer resistance. Residues outside the active site that mutate to confer resistance are in the flaps (M46, F53, and I54) and core of the enzyme (L24, L33, L76, N88, and L90). In HCV protease, the distal mutation V36M is commonly associated with resistance [71]. Incorporating the effect of such distal mutations into rational drug design remains a complete challenge. The effect of mutations away from the active site may be propagated to the active site through a network of interactions within the protease, similar to a domino effect, as suggested by a detailed analysis of changes in the interaction network of HIV-1 protease structure due to such mutations (Figure 4A) [72].
Figure 4.
Mutations Remote From the Active Site Can Contribute to Conferring Resistance. (A) The network hypothesis postulates that the impact of mutations away from the active site can be transmitted to the active site via the network of hydrogen-bonded interactions in the protein (dashed lines, displayed on one monomer of HIV-1 protease). (B) HIV-1 protease is highly dynamic, as revealed in snapshots from MD simulations, and mutations may modulate ligand binding to confer resistance by altering such conformational dynamics.
While explaining the mechanism by which such mutations outside the active site confer resistance is challenging, one likely mechanism is through alterations in protein dynamics (Figure 4B). HIV-1 protease is a flexible enzyme that undergoes extensive conformational changes to allow opening of the flaps and access to the active site [73–76]. These changes at the flaps are coupled to extensive side chain repacking at the hydrophobic core, or hydrophobic sliding [77, 78]. Mutations that cause changes in this dynamic process can confer resistance by altering the balance between substrate processing versus inhibitor binding at the active site. Altered dynamics differentially impact substrate processing versus inhibitor binding, as the inhibitor needs to stay bound at the active site for efficient inhibition, while the substrates need to be processed and released for efficient turnover. Such changes in dynamics have been revealed in molecular dynamics simulations as well as experimental NMR and electron paramagnetic resonance (EPR) dynamics of HIV-1 protease drug resistant variants [79–82]. This resistance mechanism through changes in conformational dynamics propagating to the active site [72] may be common to distal mutations in other proteases.
Mutations in other viral proteins beyond protease can contribute to drug resistance. Viral maturation is a complex interdependent process where additional levels of resistance can occur due to mutations in other viral proteins, and especially the protease cleavage sites in the Gag and Gag-Pro-Pol polyproteins. Mutations in the HIV-1 Gag polyprotein co-evolve with primary resistance mutations in the protease [83–87] and some mutations can even directly confer resistance in the absence of protease mutations [85, 88–90]. Thus drug resistance occurs within a complex system of interactions between the protease and the viral and host substrates; with the interdependent mechanisms by which changes outside the protease contribute to resistance remaining to be elucidated. Whether there are certain key signature mutations and/or common mechanisms through which resistance is conferred in highly mutated viral variants is still to be explored.
Concluding Remarks
To avoid resistance, the design of inhibitors against viral proteases needs to consider the essential biological function and evolutionary constraints on the protease. This entails design not just against the wild-type target but considering all potential mutant variants that can exist and still carry out the protease’s biological function. Combining strategies to minimize vulnerability to resistance while enhancing potency can lead toward designing inhibitors that are more robust against resistance. Regardless of whether ‘resistance-proof’ inhibitors are achievable or not, lessons from HIV-1 and HCV proteases can be incorporated into structure-based drug design. Reaching ‘resistance-avoiding’ inhibition is achievable given the constraints on viral evolution as the population size plummets with the initiation of potent therapy. Instead of lagging behind evolution and waiting for resistance to emerge, we can move one step ahead of evolution to counter resistance with more intelligent and rational design.
Outstanding Questions.
How do mutations outside the protease active site confer resistance? What is the molecular mechanism by which the impact of such mutations propagates to the active site?
How is protein conformational dynamics related to drug resistance? Can we predict resistance from changes in dynamics?
Are there key signature mutations that are pivotal in conferring resistance in highly mutated protease variants, and/or redundant mutations? Is there interdependency or synergy between coexisting mutations? Can we design inhibitors to avoid them?
Can predictive and quantitative models of drug resistance be developed based on the kinetic balance between inhibitor binding versus substrate processing? Can the effect of multiple mutations in the protease, substrates sites, and other viral components be incorporated in the models?
Is a ‘resistance-proof protease inhibitor possible? Is there a reachable potency threshold beyond which a protease inhibitor avoids clinical resistance?
Trends Box.
Viral proteases recognize substrates through a conserved shape, defining the substrate envelope.
A variant that does not bind inhibitors efficiently but still processes substrates is resistant.
Resistance mutations occur where inhibitors protrude outside of the substrate envelope.
High potency inhibitors that fit within the substrate envelope can avoid resistance.
Resistance-avoiding strategies need to be included in structure-based drug design.
Acknowledgments
We thank Shurong Hou for help with preparing the figures. This research was supported by National Institute of Health (NIH) Grant P01 GM109767.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Hughes D, Andersson DI. Evolutionary consequences of drug resistance: shared principles across diverse targets and organisms. Nat. Rev. Genet. 2015;16:459–471. doi: 10.1038/nrg3922. [DOI] [PubMed] [Google Scholar]
- 3.Poole K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 4.Szakacs G, et al. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 5.Krausslich HG, Wimmer E. Viral proteinases. Annu. Rev. Biochem. 1988;57:701–754. doi: 10.1146/annurev.bi.57.070188.003413. [DOI] [PubMed] [Google Scholar]
- 6.Tong L. Viral proteases. Chem. Rev. 2002;102:4609–4626. doi: 10.1021/cr010184f. [DOI] [PubMed] [Google Scholar]
- 7.Jager S, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ventoso I, et al. HIV-1 protease cleaves eukaryotic initiation factor 4G and inhibits cap-dependent translation. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foy E, et al. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145–1148. doi: 10.1126/science.1082604. [DOI] [PubMed] [Google Scholar]
- 10.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee SY, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogg RS, et al. Improved survival among HIV-infected individuals following initiation of antiretroviral therapy. Jama-J Am Med Assoc. 1998;279:450–454. doi: 10.1001/jama.279.6.450. [DOI] [PubMed] [Google Scholar]
- 13.Hogg RS, et al. Decline in deaths from AIDS due to new antiretrovirals. Lancet. 1997;349:1294–1294. doi: 10.1016/S0140-6736(05)62505-6. [DOI] [PubMed] [Google Scholar]
- 14.Palella FJ, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. New Engl. J. Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 15.Alter MJ. Epidemiology of hepatitis C virus infection. World journal of gastroenterology : WJG. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modi AA, Liang TJ. Hepatitis C: a clinical review. Oral Dis. 2008;14:10–14. doi: 10.1111/j.1601-0825.2007.01419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andreone P, et al. ABT-450, ritonavir, ombitasvir, and dasabuvir achieves 97% and 100% sustained virologic response with or without ribavirin in treatment-experienced patients with HCV genotype 1b infection. Gastroenterology. 2014;147:359–365. doi: 10.1053/j.gastro.2014.04.045. e351. [DOI] [PubMed] [Google Scholar]
- 18.Kwong AD, et al. Discovery and development of telaprevir: an NS3-4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat. Biotechnol. 2011;29:993–1003. doi: 10.1038/nbt.2020. [DOI] [PubMed] [Google Scholar]
- 19.Malcolm BA, et al. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob. Agents Chemother. 2006;50:1013–1020. doi: 10.1128/AAC.50.3.1013-1020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perni RB, et al. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 2006;50:899–909. doi: 10.1128/AAC.50.3.899-909.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenquist Å, et al. Discovery and Development of Simeprevir (TMC435), a HCV NS3/4A Protease Inhibitor. J. Med. Chem. 2014;57:1673–1693. doi: 10.1021/jm401507s. [DOI] [PubMed] [Google Scholar]
- 22.Halfon P, Locarnini S. Hepatitis C virus resistance to protease inhibitors. J. Hepatol. 2011;55:192–206. doi: 10.1016/j.jhep.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Pan D, et al. Understanding the drug resistance mechanism of hepatitis C virus NS3/4A to ITMN-191 due to R155K, A156V, D168A/E mutations: a computational study. Biochim. Biophys. Acta. 2012;1820:1526–1534. doi: 10.1016/j.bbagen.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Romano KP, et al. The molecular basis of drug resistance against hepatitis C virus NS3/4A protease inhibitors. PLoS Pathog. 2012;8:e1002832. doi: 10.1371/journal.ppat.1002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everson GT, et al. Efficacy of an interferon- and ribavirin-free regimen of daclatasvir, asunaprevir, and BMS-791325 in treatment-naive patients with HCV genotype 1 infection. Gastroenterology. 2014;146:420–429. doi: 10.1053/j.gastro.2013.10.057. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez JA, et al. Interferon-free, direct-acting antiviral therapy for chronic hepatitis C. J. Viral Hepat. 2015;22:861–870. doi: 10.1111/jvh.12422. [DOI] [PubMed] [Google Scholar]
- 27.Lawitz E, et al. Efficacy and safety of 12 weeks versus 18 weeks of treatment with grazoprevir (MK-5172) and elbasvir (MK-8742) with or without ribavirin for hepatitis C virus genotype 1 infection in previously untreated patients with cirrhosis and patients with previous null response with or without cirrhosis (C-WORTHY): a randomised, open-label phase 2 trial. Lancet. 2015;385:1075–1086. doi: 10.1016/S0140-6736(14)61795-5. [DOI] [PubMed] [Google Scholar]
- 28.Coffin J, Swanstrom R. HIV pathogenesis: dynamics and genetics of viral populations and infected cells. Cold Spring Harb Perspect Med. 2013;3:a012526. doi: 10.1101/cshperspect.a012526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King NM, et al. Combating susceptibility to drug resistance: Lessons from HIV-1 protease. Chem. Biol. 2004;11:1333–1338. doi: 10.1016/j.chembiol.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Prabu-Jeyabalan MNEA, Schiffer CA. Substrate shape determines specificity of recognition for HIV-1 Protease: analysis of crystal structures of six substrate complexes. Structure. 2002;10:369–381. doi: 10.1016/s0969-2126(02)00720-7. [DOI] [PubMed] [Google Scholar]
- 31.Romano KP, et al. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc. Natl. Acad. Sci. U. S. A. 2010;107:20986–20991. doi: 10.1073/pnas.1006370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolli M, et al. HIV-1 Protease-Substrate Coevolution in Nelfinavir Resistance. J. Virol. 2014;88:7145–7154. doi: 10.1128/JVI.00266-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mittal S, et al. Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50. J. Virol. 2013;87:4176–4184. doi: 10.1128/JVI.03486-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soumana DI, et al. Structural analysis of asunaprevir resistance in HCV NS3/4A protease. ACS Chem Biol. 2014;9:2485–2490. doi: 10.1021/cb5006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu M, et al. In vitro efficacy of approved and experimental antivirals against novel genotype 3 hepatitis C virus subgenomic replicons. Antiviral Res. 2013;100:439–445. doi: 10.1016/j.antiviral.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 36.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS reviews. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 37.Shen Y, et al. Testing the substrate-envelope hypothesis with designed pairs of compounds. ACS Chem Biol. 2013;8:2433–2441. doi: 10.1021/cb400468c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Meyer S, et al. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 2005;49:2314–2321. doi: 10.1128/AAC.49.6.2314-2321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King NM, et al. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 2004;78:12012–12021. doi: 10.1128/JVI.78.21.12012-12021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surleraux DL, et al. Discovery and selection of TMC114, a next generation HIV-1 protease inhibitor. J. Med. Chem. 2005;48:1813–1822. doi: 10.1021/jm049560p. [DOI] [PubMed] [Google Scholar]
- 41.Lefebvre E, Schiffer CA. Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir. AIDS reviews. 2008;10:131–142. [PMC free article] [PubMed] [Google Scholar]
- 42.de Meyer S, et al. Resistance profile of darunavir: combined 24-week results from the POWER trials. AIDS Res. Hum. Retroviruses. 2008;24:379–388. doi: 10.1089/aid.2007.0173. [DOI] [PubMed] [Google Scholar]
- 43.Hietpas RT, et al. Experimental illumination of a fitness landscape. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7896–7901. doi: 10.1073/pnas.1016024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roscoe BP, Bolon DN. Systematic exploration of ubiquitin sequence, E1 activation efficiency, and experimental fitness in yeast. J. Mol. Biol. 2014;426:2854–2870. doi: 10.1016/j.jmb.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghosh AK, et al. Enhancing protein backbone binding--a fruitful concept for combating drug-resistant HIV. Angew. Chem. Int. Ed. Engl. 2012;51:1778–1802. doi: 10.1002/anie.201102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh AK, et al. Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance. Acc Chem Res. 2008;41:78–86. doi: 10.1021/ar7001232. [DOI] [PubMed] [Google Scholar]
- 47.Das K, Arnold E. HIV-1 reverse transcriptase and antiviral drug resistance. Part 1. Curr Opin Virol. 2013;3:111–118. doi: 10.1016/j.coviro.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das K, et al. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J. Med. Chem. 2004;47:2550–2560. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 49.Muzammil S, et al. Unique thermodynamic response of tipranavir to human immunodeficiency virus type 1 protease drug resistance mutations. J. Virol. 2007;81:5144–5154. doi: 10.1128/JVI.02706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velazquez-Campoy A, et al. The binding energetics of first- and second-generation HIV-1 protease inhibitors: implications for drug design. Arch. Biochem. Biophys. 2001;390:169–175. doi: 10.1006/abbi.2001.2333. [DOI] [PubMed] [Google Scholar]
- 51.Nalam MN, et al. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J. Virol. 2010;84:5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nalam MN, et al. Substrate envelope-designed potent HIV-1 protease inhibitors to avoid drug resistance. Chem. Biol. 2013;20:1116–1124. doi: 10.1016/j.chembiol.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nalam MN, Schiffer CA. New approaches to HIV protease inhibitor drug design II: testing the substrate envelope hypothesis to avoid drug resistance and discover robust inhibitors. Current opinion in HIV and AIDS. 2008;3:642–646. doi: 10.1097/COH.0b013e3283136cee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozen A, et al. Improving the Resistance Profile of Hepatitis C NS3/4A Inhibitors: Dynamic Substrate Envelope Guided Design. J Chem Theory Comput. 2013;9:5693–5705. doi: 10.1021/ct400603p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali A, et al. Evaluating the role of macrocycles in the susceptibility of hepatitis C virus NS3/4A protease inhibitors to drug resistance. ACS Chem Biol. 2013;8:1469–1478. doi: 10.1021/cb400100g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harper S, et al. Discovery of MK-5172, a Macrocyclic Hepatitis C Virus NS3/4a Protease Inhibitor. ACS Med Chem Lett. 2012;3:332–336. doi: 10.1021/ml300017p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vendeville S, et al. Discovery of novel, potent and bioavailable proline-urea based macrocyclic HCV NS3/4A protease inhibitors. Bioorg. Med. Chem. Lett. 2008;18:6189–6193. doi: 10.1016/j.bmcl.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Soumana D, et al. Structural and Thermodynamic Effects of Macrocyclization in HCV NS3/4A Inhibitor MK-5172. ACS Chem Biol. 2015 doi: 10.1021/acschembio.5b00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jilek BL, et al. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat. Med. 2012;18:446–451. doi: 10.1038/nm.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sampah MES, et al. Dose-response curve slope is a missing dimension in the analysis of HIV-1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7613–7618. doi: 10.1073/pnas.1018360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siliciano JD, Siliciano RF. Recent trends in HIV-1 drug resistance. Curr Opin Virol. 2013;3:487–494. doi: 10.1016/j.coviro.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Orkin C, et al. Final 192-week efficacy and safety of once-daily darunavir/ritonavir compared with lopinavir/ritonavir in HIV-1-infected treatment-naive patients in the ARTEMIS trial. HIV medicine. 2013;14:49–59. doi: 10.1111/j.1468-1293.2012.01060.x. [DOI] [PubMed] [Google Scholar]
- 63.Llibre JM, et al. Genetic Barrier to Resistance for Dolutegravir. AIDS reviews. 2015;17:56–64. [PubMed] [Google Scholar]
- 64.Paton NI, et al. Protease inhibitor monotherapy for long-term management of HIV infection: a randomised, controlled, open-label, non-inferiority trial. Lancet HIV. 2015;2:e417–e426. doi: 10.1016/S2352-3018(15)00176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhee SY, et al. HIV-1 protease mutations and protease inhibitor cross-resistance. Antimicrob. Agents Chemother. 2010;54:4253–4261. doi: 10.1128/AAC.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu TD, et al. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 2003;77:4836–4847. doi: 10.1128/JVI.77.8.4836-4847.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foulkes-Murzycki JE, et al. Cooperative effects of drug-resistance mutations in the flap region of HIV-1 protease. ACS Chem Biol. 2013;8:513–518. doi: 10.1021/cb3006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohtaka H, et al. Multidrug resistance to HIV-1 protease inhibition requires cooperative coupling between distal mutations. Biochemistry (Mosc) 2003;42:13659–13666. doi: 10.1021/bi0350405. [DOI] [PubMed] [Google Scholar]
- 69.Chang MW, Torbett BE. Accessory mutations maintain stability in drug-resistant HIV-1 protease. J. Mol. Biol. 2011;410:756–760. doi: 10.1016/j.jmb.2011.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, et al. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J. Mol. Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 71.Zhou Y, et al. Phenotypic characterization of resistant Val36 variants of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 2008;52:110–120. doi: 10.1128/AAC.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ragland DA, et al. Drug resistance conferred by mutations outside the active site through alterations in the dynamic and structural ensemble of HIV-1 protease. J. Am. Chem. Soc. 2014;136:11956–11963. doi: 10.1021/ja504096m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Freedberg DI, et al. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hornak V, et al. HIV-1 protease flaps spontaneously open and reclose in molecular dynamics simulations. Proc. Natl. Acad. Sci. U. S. A. 2006;103:915–920. doi: 10.1073/pnas.0508452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishima R, et al. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure. 1999;7:1047–1055. doi: 10.1016/s0969-2126(99)80172-5. [DOI] [PubMed] [Google Scholar]
- 76.Scott WR, Schiffer CA. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure. 2000;8:1259–1265. doi: 10.1016/s0969-2126(00)00537-2. [DOI] [PubMed] [Google Scholar]
- 77.Foulkes-Murzycki JE, et al. Hydrophobic sliding: a possible mechanism for drug resistance in human immunodeficiency virus type 1 protease. Structure. 2007;15:225–233. doi: 10.1016/j.str.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittal S, et al. Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease. J. Am. Chem. Soc. 2012;134:4163–4168. doi: 10.1021/ja2095766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai Y, et al. Differential Flap Dynamics in Wild-type and a Drug Resistant Variant of HIV-1 Protease Revealed by Molecular Dynamics and NMR Relaxation. J Chem Theory Comput. 2012;8:3452–3462. doi: 10.1021/ct300076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cai Y, et al. Drug Resistance Mutations Alter Dynamics of Inhibitor-Bound HIV-1 Protease. J Chem Theory Comput. 2014;10:3438–3448. doi: 10.1021/ct4010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Vera IMS, et al. Elucidating a Relationship between Conformational Sampling and Drug Resistance in HIV-1 Protease. Biochemistry (Mosc) 2013;52:3278–3288. doi: 10.1021/bi400109d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goldfarb NE, et al. Defective hydrophobic sliding mechanism and active site expansion in HIV-1 protease drug resistant variant Gly48Thr/Leu89Met: mechanisms for the loss of saquinavir binding potency. Biochemistry (Mosc) 2015;54:422–433. doi: 10.1021/bi501088e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fun A, et al. Human Immunodeficiency Virus Gag and protease: partners in resistance. Retrovirology. 2012;9:63. doi: 10.1186/1742-4690-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kolli M, et al. Co-evolution of nelfinavir-resistant HIV-1 protease and the p1-p6 substrate. Virology. 2006;347:405–409. doi: 10.1016/j.virol.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 85.Kolli M, et al. Human Immunodeficiency Virus Type 1 Protease-Correlated Cleavage Site Mutations Enhance Inhibitor Resistance. J. Virol. 2009;83:11027–11042. doi: 10.1128/JVI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ozen A, et al. Structural basis and distal effects of Gag substrate coevolution in drug resistance to HIV-1 protease. Proc. Natl. Acad. Sci. U. S. A. 2014;111:15993–15998. doi: 10.1073/pnas.1414063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Prabu-Jeyabalan M, et al. Structural basis for coevolution of a human immunodeficiency virus type 1 nucleocapsid-p1 cleavage site with a V82A drug-resistant mutation in viral protease. J. Virol. 2004;78:12446–12454. doi: 10.1128/JVI.78.22.12446-12454.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dam E, et al. Gag Mutations Strongly Contribute to HIV-1 Resistance to Protease Inhibitors in Highly Drug-Experienced Patients besides Compensating for Fitness Loss. PLoS Path. 2009;5:e1000345. doi: 10.1371/journal.ppat.1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maguire MF, et al. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to 150V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 2002;76:7398–7406. doi: 10.1128/JVI.76.15.7398-7406.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nijhuis M, et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS medicine. 2007;4:e36. doi: 10.1371/journal.pmed.0040036. [DOI] [PMC free article] [PubMed] [Google Scholar]