Summary
Defects in circadian rhythm influence physiology and behavior with implications for the treatment of sleep disorders, metabolic disease and cancer. Although core regulatory components of clock rhythmicity have been defined, insight into the mechanisms underpinning amplitude is limited. We show here that REV-ERBα, a core inhibitory component of clock transcription, is targeted for ubiquitination and subsequent degradation by the F-box protein FBXW7. By relieving REV-ERBα-dependent repression, FBXW7 provides an unrecognized mechanism for enhancing the amplitude of clock gene transcription. Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of REV-ERBα is necessary for FBXW7 recognition. Moreover, targeted hepatic disruption of FBXW7 alters circadian expression of core clock genes and perturbs whole body lipid and glucose levels. This CDK1-FBXW7 pathway controlling REV-ERBα repression defines an unexpected molecular mechanism for re-engaging the positive transcriptional arm of the clock, as well as a potential route to manipulate clock amplitude via small molecule CDK1 inhibition.
Introduction
Circadian rhythms align physiology and behavior with the daily light-dark cycle, enabling prediction of diurnal environmental changes (Dunlap, 1999). In mammals, while the central circadian clock is located in the suprachiasmatic nucleus (SCN) in the brain, peripheral cell-autonomous circadian systems function in all tissues (Dibner et al., 2010; Schibler and Sassone-Corsi, 2002). Disruption of normal circadian rhythmicity is associated with many disease conditions such as metabolic disorders and cancer (Bass and Takahashi, 2010; Gery and Koeffler, 2010; Turek et al., 2005), highlighting the importance of the circadian clock in maintaining homeostasis.
On the molecular level, circadian rhythms are generated and maintained by interlocked transcriptional-translational feedback loops (Reppert and Weaver, 2002). The core loop has been thought to be driven by the basic helix-loop-helix (bHLH) and PAS (Per-ARNT-Sim) domain-containing transcription factors BMAL1 (brain and muscle ARNT-like protein 1) and CLOCK (circadian locomotor output cycles kaput), which heterodimerize on E-box enhancers and activate the expression of repressive cofactors such as Cryptochrome (Cry1 and Cry2) and Period (Per1 and Per2) (Reppert and Weaver, 2002). Bmal and BMAL target genes, including Cry and Per, have been shown to be directly controlled by REV-ERBα, suggesting a role of REV-ERB cycling in rhythm generation (Bugge et al., 2012; Cho et al., 2012). REV-ERB represses transcription through binding to genomic response elements (termed ROREs) that are co-targeted by the positively-acting orphan nuclear receptors RORα and γ (Sato et al., 2004; Takeda et al., 2012). Coordination of the opposing activities of REV-ERB and ROR is key to understanding the mechanisms controlling the period and amplitude of the clock (Le Martelot et al., 2009; Preitner et al., 2002). Genome-wide cistromic analyses of REVERBα and β in murine liver revealed that their DNA binding sites localize to regulatory regions of all core circadian clock genes and overlap extensively with those of BMAL1 (Bugge et al., 2012; Cho et al., 2012). The genetic deletions of both REV-ERBα and β were shown to disrupt the circadian expression of core clock genes and induce circadian behavioral changes, establishing REV-ERBs as essential core clock components (Bugge et al., 2012; Cho et al., 2012). Moreover, the demonstration that small molecule REVERB agonists can affect circadian behavior as well as lipid and glucose metabolism establishes REV-ERBs as therapeutic targets for the treatment of metabolic disorders (Solt et al., 2012).
Post-translational modifications are essential strategies to generate and regulate the dynamics of circadian clocks by introducing delays in the transcriptional-translational feedback loops (Gallego and Virshup, 2007; Lee et al., 2001). For example, phosphorylation of PER by Casein Kinase I (CKI) and subsequent recruitment of the F-box protein β-Trcp modulate its stability and subcellular localization (Eide et al., 2005; Yagita et al., 2002). These are critical mechanisms underlying the regulation of the period length and phase of the circadian clock (Gallego and Virshup, 2007). Post-translational signaling allows the circadian clock machinery to be entrained by environmental cues and to coordinate with other physiological processes (Gallego and Virshup, 2007). For instance, AMPK-mediated phosphorylation of CRY and the subsequent recruitment of the F-box protein FBXL3 ubiquitin ligase complex for CRY degradation are important mechanisms for the entrainment of the circadian clock in peripheral tissues by food availability (Lamia et al., 2009; Siepka et al., 2007). Recently, FBXL21, a paralog of FBXL3, has been implicated in the regulation of circadian clock period length via control of CRY stability in a cellular compartmentalization-dependent manner (Hirano et al., 2013; Yoo et al., 2013). These observations implicate F-box protein family members as transducers of signals to the core clock components that regulate the period of circadian rhythms. However, as an intrinsic property of circadian oscillation, the mechanism that regulates the amplitude of rhythmicity is not known. Despite the central roles of REVERBs as transcriptional regulators in circadian rhythm, the post-translational events that modulate their activity and/or stability are poorly understood.
Here we identify a REV-ERBα post-translational regulatory circuit in which cyclin-dependent kinase 1 (CDK1) phosphorylation of REV-ERBα is recognized by the F-box protein, FBXW7α to direct REV-ERBα degradation via the proteasome. Disruption of this CDK1-FBXW7-mediated REV-ERBα degradation pathway in mouse liver alters circadian rhythmicity, in particular amplitude, and whole body lipid/glucose homeostasis. This work uncovers an important module of the cell-autonomous circadian rhythm regulatory network, and suggests that control of nuclear hormone receptor stability is an important mechanism that contributes to normal circadian clock amplitude and whole body energy homeostasis.
Results
FBXW7 regulates REV-ERBα stability
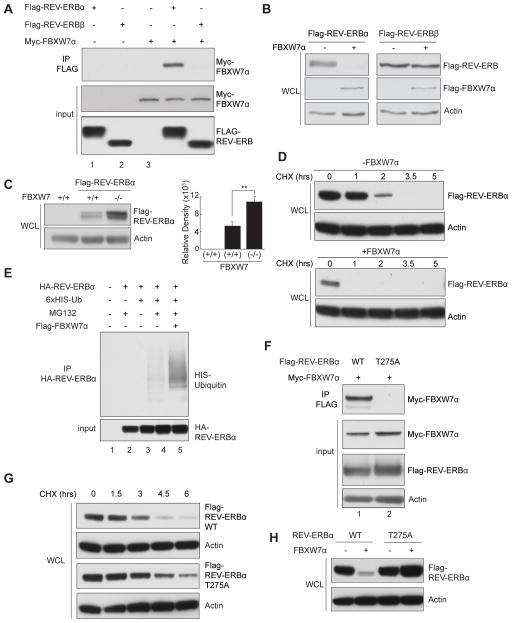
To identify proteins involved in regulating the stability and/or activity of REV-ERBs, REV-ERBα and β protein complexes were purified from HEK293T cells using a tandem affinity approach, and associating proteins identified via Multidimensional Protein Identification Technology (MudPIT) (Figure S1A). Notably, several components of the Skp1-Cullin-F-box (SCF) ubiquitin E3 ligase complex, including adaptor protein Skp1A and substrate recognition subunit F-box protein FBXW7, co-purify with REV-ERBα. We confirmed the specific interaction between REV-ERBα and FBXW7α in transiently transfected cells using reciprocal co-immunoprecipitations (Figures 1A and S1B). Interestingly, FBXW7α interacts with REV-ERBα, but not with REV-ERBβ (Figure 1A) or other circadian clock core components such as cryptochrome 1 (CRY1) (data not shown), suggestive of a specific interaction.
Figure 1. FBXW7α regulates REV-ERBα stability in a T275-dependent manner.
Western blots of (A) coimmunoprecipitated FBXW7α from cos-7 cells expressing FBXW7α and REV-ERBα or β. Cells were pretreated with MG132 for 4 hrs. (B) Total REV-ERBα and β levels with or without FBXW7α co-expression. (C) REV-ERBα in FBXW7 (+/+) or (−/−) cells; quantification shown on right. Three independent experiments were conducted. Error bars indicate SEM; statistical significance was determined by Student’s t-test (** P<0.01). (D) Time course of REV-ERBα levels in cycloheximide (CHX)-treated cells with or without FBXW7α. (E) REV-ERBα ubiquitination in cells co-expressing REV-ERBα and His-tagged ubiquitin with or without FBXW7α. Cells were pretreated with MG132 for 4 hrs. (F) Immunoprecipitated FBXW7α from cells co-expressing WT and T275A REV-ERBα. (G) Time course of WT and T275A REV-ERBα levels in CHX-treated cells. (H) WT and T275A REV-ERBα levels with and without FBXW7α. See also Figure S1, S2, and S3.
As FBXW7α is the substrate recognition subunit of the SCF(FBXW7) ubiquitin E3 ligase complex, we asked if FBXW7α might be a REV-ERBα stability modulating factor. Indeed, REV-ERBα levels were dramatically reduced upon co-expression of FBXW7α in the absence of MG132, which should allow for proteasomal degradation (Figure 1B). Other F-box proteins, including SKP2, β-TRCP and FBXL3, did not promote REV-ERBα degradation (Figure S1C). Interestingly, REV-ERBβ abundance is not altered by FBXW7α co-expression, consistent with the lack of interaction between these proteins (Figure 1B). Furthermore, both endogenous and ectopically expressed REV-ERBα protein levels are significantly elevated in FBXW7α null colorectal adenocarcinoma cells (Figures 1C and S1D) and after siRNA-mediated knockdown of FBXW7α in Hepa1-6 hepatocarcinoma cells (Figure S1E). Expression of a dominant-negative FBXW7α lacking the N-terminal F-box domain that renders it unable to recruit the SCF complex (FBXW7αDN) (Strohmaier et al., 2001) dramatically increased the levels of endogenous, as well as ectopically expressed REV-ERBα (Figures S1F and S1G), while having minimal effect on the abundance of REV-ERBβ (Figure S2A).
To further explore the role of FBXW7α in modulating REV-ERBα stability, we examined REV-ERBα half-life in cells treated with the protein synthesis inhibitor cycloheximide (CHX). Co-expression of FBXW7α decreased the REV-ERBα half-life to less than 1 hr (Figures 1D and S2B) while expression of FBXL3 known to destabilize CRY1/2, (Siepka et al., 2007) only had a minimal effect (Figure S2C). Notably, the ability of FBXW7α to reduce REV-ERBα half-life is completely blocked by MG132 (Figure S2C). As a component of the SCF ubiquitin E3 ligase complex, we asked if FBXW7α modulates REV-ERBα stability through catalyzing REV-ERBα ubiquitination. Indeed, ectopic expression of ubiquitin and co-expression of FBXW7α, increase REVERBα ubiquitination (Figure 1E), supporting the notion that an SCF(FBXW7α) complex regulates REV-ERBα stability.
We next sought to identify upstream signaling events mediating the interaction between REV-ERBα and the SCF(FBXW7α) ubiquitin E3 ligase complex. FBXW7α recognizes a short, phosphothreonine-containing motif known as the Cdc4 phosphodegron (CPD) (Welcker and Clurman, 2008). A highly conserved optimal CPD sequence is present in the REV-ERBα hinge region, centered on threonine 275 (T275) (Figure S2D). This motif is not conserved in REV-ERBβ, consistent with FBXW7α failing to interact with REV-ERBβ. Supporting the hypothesis that the T275 CPD is required for the REV-ERBα-FBXW7α interaction, mutating T275 to alanine (T275A) completely abolishes their co-precipitation (Figure 1F). In addition, mutating the conserved S279 in the CPD motif (Figure S2D) to alanine also eliminates the REV-ERBα-FBXW7α interaction (data not shown), suggesting the importance of an intact CPD in FBXW7 recruitment. To address whether the T275 residue is important for regulating REV-ERBα stability, we stably expressed REV-ERBα T275A in NIH3T3 cells. We observe ~1.6 fold increase in the abundance of the mutant protein compared to WT REV-ERBα (Figure S2E). Furthermore, the half-life of REV-ERBα T275A is increased compared to that of WT REV-ERBα in CHX-treated cells (Figure 1G) and T275A REV-ERBα is resistant to FBXW7α-induced protein degradation when co-expressed (Figure 1H), indicating that FBXW7α modulates REV-ERBα stability in a T275 CPD-dependent manner. Moreover, the CPD motif is sufficient for FBXW7-mediated degradation, as a chimeric protein in which the REV-ERBα CPD is introduced into REV-ERBβ (termed REV-ERBβ-CPD) acquires the ability to interact with and be targeted for degradation by FBXW7α (Figure S2F).
Cyclin-dependent kinase 1 (CDK1) phosphorylates REV-ERBα
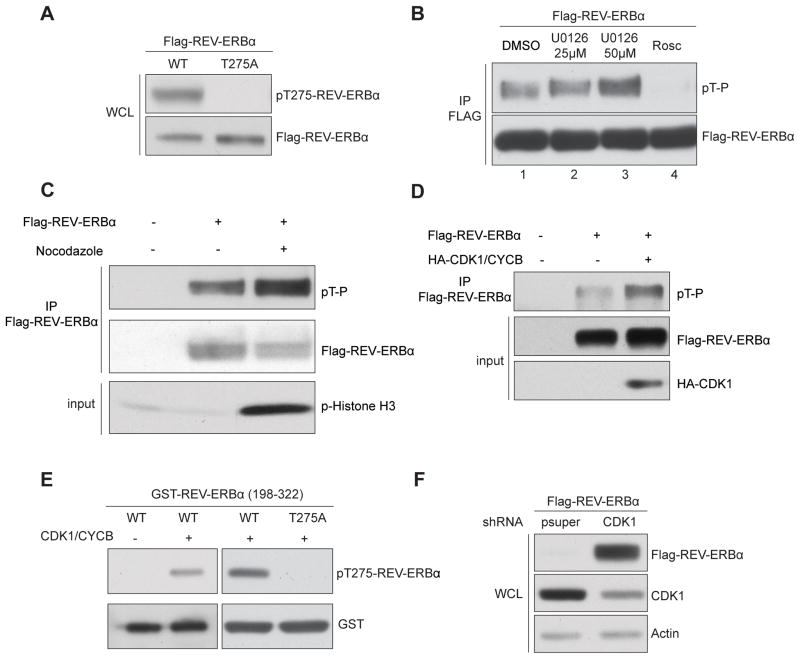
Next we generated a phospho-specific antibody that recognizes T275 phosphorylated REV-ERBα (anti-pT275 Ab). The phosphorylation of WT but not T275A REV-ERBα in cultured cells was confirmed using anti-pT275 Ab (Figure 2A), as well as via a phosphorylated CDK substrate antibody (pT-P Ab) (Figure S3A).
Figure 2. CDK1 phosphorylates REV-ERBα at T275.
Western blots of (A) total and T275 phosphorylated REV-ERBα in AD293 cells expressing WT or T275A REV-ERBα. (B) Phosphorylated REV-ERBα after 2h pretreatment with MEK1/2 inhibitor (U0126, 25μM and 50 μM) or CDK inhibitor (Roscovitine, 50μM). (C) Total and phosphorylated REV-ERBα in synchronized AD293 cells (with or without nocodazole, 100ng/ml for 16 hrs). Phospho-histone H3 is shown as a G2/M phase marker. (D) Total and phosphorylated REV-ERBα in cells with or without CDK1 and cyclin B co-expression. (E) In vitro phosphorylation of WT (left) or T275A REV-ERBα (198–322) (right) by CDK1/cyclin B. (F) Total REV-ERBα in AD293 cells with and without shRNA-mediated CDK1. See also Figure S4.
We sought to identify the kinase responsible for REV-ERBα phosphorylation. Motif analysis (Obenauer et al., 2003) predicts T275 as a cyclin-dependent kinase (CDK) consensus site, consistent with the amino acid sequence recognized by pT-P Ab (Figure S3A), implicating a member of the CDK clade as the target kinase. In support of this, the CDK inhibitor, Roscovitine, significantly reduces REV-ERBα phosphorylation, whereas the ERK inhibitor U0126 does not (Figures 2B and S3B). Interestingly, Roscovitine perturbs circadian gene expression in human osteosarcoma U2OS cells (Hirota et al., 2008) and mouse liver (Iurisci et al., 2009). We found that REV-ERBα phosphorylation is also sensitive to Olomoucine (CDK1, 2 and 5 inhibitor), CGP74514A (selective CDK1 inhibitor), and a CDK1/5 inhibitor, whereas PNU112455A (Cdk2/5 inhibitor) and NSC625987 (Cdk4 inhibitor) have reduced impact (Figure S3C). In addition, GSK-3β inhibitors (SB216763 and LiCl (Yin et al., 2006)) had no effect (data not shown). These findings implicate CDK1 as the dominant kinase in REV-ERBα phosphorylation. Notably, CDK1 displays the highest activity for REV-ERBα phosphorylation in an in vitro assay (Figure S3D).
To explore the role of CDK1 in REV-ERBα phosphorylation, we activated endogenous CDK1 in cultured cells using a microtubule-destabilizing agent, nocodazole. Nocodazole treatment substantially increased REV-ERBα phosphorylation while concomitantly reducing total REV-ERBα levels compared with untreated cells (Figure 2C). Similarly, co-expression of CDK1 and CYCLIN B significantly increased REVERBα phosphorylation (Figure 2D). Conversely, expression of a dominant-negative form of CDK1 reduced REV-ERBα phosphorylation (Figure S3E). In in vitro kinase assays, purified CDK1/CYCLIN B phosphorylated T275 in both truncated (amino acids 198–322) and full length REV-ERBα (Figures 2E, S3F and S3G). Importantly, the T275A mutation completely abolished the phosphorylation induced by CDK1 (Figure 2E). This data implicates CDK1 as responsible for REV-ERBα T275 phosphorylation.
We next asked if CDK1 directly controls REV-ERBα stability. ShRNA-mediated knockdown of CDK1 dramatically increases REV-ERBα abundance (Figure 2F). Conversely, activation of the CDK1/CYCLIN B complex by nocodazole treatment markedly reduces endogenous REV-ERBα levels (Figure S4A). Expression of a dominant-negative CDK1 increased REV-ERBα stability in the presence of FBXW7α (Figure S4B), and the co-immunoprecipitation of CDK1 and REV-ERBα in AD293 cells further supports a physical association between these proteins (Figure S4C). Together these findings implicate CDK1 in directing FBXW7α-mediated REV-ERBα stability.
CDK1-mediated REV-ERBα T275 phosphorylation and FBXW7 modulate amplitude of circadian clock
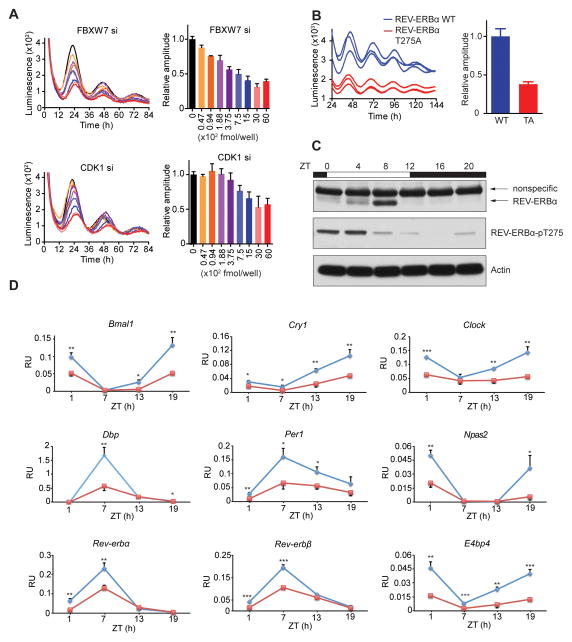
As REV-ERBα acts as a molecular brake on the circadian clock machinery by repressing the expression of Bmal1 and other clock genes (Bugge et al., 2012; Cho et al., 2012), we examined whether perturbation of CDK1-FBXW7-mediated REV-ERBα stability affects circadian oscillation. We utilized U2OS osteosarcoma cells expressing a destabilized luciferase reporter controlled by Bmal1 promoter (Bmal1-dLuc) to monitor circadian oscillation in real time. In wild type cells, resetting the circadian clock by switching the growth medium causes a robust and prolonged oscillation of luciferase reporter expression with a period length of approximately 24 hours (Hirota et al., 2010). Notably, siRNA-mediated knockdown of Fbxw7α in these cells reduces the amplitude of Bmal1 reporter expression in a dose-dependent manner (Figure 3A). Similarly, siRNA-mediated knockdown of CDK1 dose-dependently dampened the amplitude of Bmal1 reporter expression (amplitude is defined as half the oscillation; (peak value-trough value)/2). These results suggest that FBXW7α and CDK1 play pivotal roles in regulating the amplitude of Bmal1 oscillation. Next, we examined the activity of the T275A mutant in REV-ERBα null fibroblasts harboring a Bmal1 reporter (Liu et al., 2008). Consistent with the observed increased stability of the T275A mutant, the amplitude of Bmal1 reporter expression was reduced by 60–80% in T275A mutant expressing cells relative to WT REV-ERBα (Figures 3B and S5A). Endogenous Bmal1 expression is also significantly reduced in MEFs expressing the T275A mutant compared to WT REVERBα (Figure S5B), indicating that the CDK1-pT275-FBXW7 cascade plays a key role in establishing the amplitude set-point for Bmal1 cyclic expression.
Figure 3. CDK1-FBXW7-REV-ERBα pathway modulates circadian amplitude.
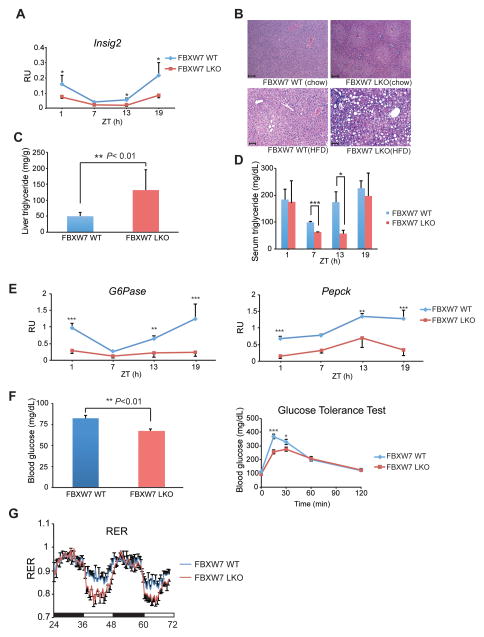
(A) Real-time bioluminescence imaging (rt-BLI) in synchronized U2OS cells stably expressing destabilized luciferase reporter controlled by Bmal1 promoter (Bmal1-luc) after transfection with indicated amount of FBXW7α (upper) or CDK1 (lower) siRNAs (left), and normalized amplitude (right, n=4). (B) rt-BLI in REV-ERBα null fibroblasts harboring Bmal1-luc after stable expression of WT or T275A REV-ERBα. Representative profiles from three individual assays (left) and normalized amplitude (right, mean +/− SEM, n=7). Error bars indicate standard error of means (SEM). (C) Western blot of total (top) and T275 phosphorylated (middle) REV-ERBα in liver lysates (β-actin loading control, lower). (D) Hepatic gene expression in WT (blue) and Fbxw7-LKO (red) mice at indicated times, determined by RT-PCR analysis. Error bars= SD, n=3. *P<0.05, **P<0.01, ***P<0.001 by Student’s t-test. See also Figure S5, Table S2.
To extend the in vitro observations in vivo, we measured REV-ERBα T275 phosphorylation changes in cell lysates from mouse livers over a 24-hour period (Figure 3C). REV-ERBα levels exhibited a circadian oscillatory pattern, with the peak at ZT8 and the trough at ZT20, as previously reported (Preitner et al., 2002). Notably, a robust circadian rhythmicity in endogenous REV-ERBα phosphorylation was revealed with the pT275-specific antibody. Interestingly, the levels of total and phosphorylated REV-ERBα are out of phase, suggesting that the oscillation in phosphorylation is not due to changes in protein levels but rather the reciprocal. Indeed, normalizing phosphorylated to total protein levels reveals that the highest ratio of T275 phosphorylation occurs at ZT20 when REV-ERBα levels are lowest (Figure S5C). This inverse relationship suggests that the pT275-mediated degradation program is, as expected, gating REV-ERBα levels. Furthermore, treatment of mice with a CDK inhibitor (Roscovitine, 4 hr) significantly reduced T275 phosphorylation. This reduction is accompanied by a modest increase of REV-ERBα protein levels, despite the negative regulation of its own expression (Figure S5D) (Adelmant et al., 1996).
As the FBXW7 whole body knockout is embryonic lethal, we created a liver-specific Fbxw7 knockout mouse (Fbxw7-LKO; Fbxw7-floxed crossed with albumin-Cre). As shown in Figure 3D, tissue-selective FBXW7 knockout reduces the cycling amplitude of Bmal1, as well as key circadian clock components such as Rev-erbβ, Cry1, Per1, Clock, Npas2, Dbp and E4bp4 (Bugge et al., 2012; Cho et al., 2012). This hints at a strategic logic for the CDK1-FBXW7α cascade in specifically targeting REV-ERBα and suggests that rhythmic cycling of transcriptional repression, achieved through temporally regulated expression and targeted degradation, may comprise the general molecular underpinning of clock amplitude.
To explore acute changes in hepatic diurnal gene expression, we retro-orbitally injected adenovirus expressing Cre recombinase into Fbxw7-floxed mice. As seen with albumin-Cre-induced deletion, acute Fbxw7 loss suppressed core clock gene expression including Bmal1, Clock and Npas2 at ZT20 (Figure S5E). Conversely, acute adenovirus-mediated Fbxw7a expression in mouse liver reduced endogenous REV-ERBα levels and increased Bmal1 mRNA levels, effects that were largely lost in adenovirus-mediated expression of an enzymatic-deficient FBXW7 (FBXW7 ED) (Figure S5F).
FBXW7 affects the circadian transcriptome in the liver
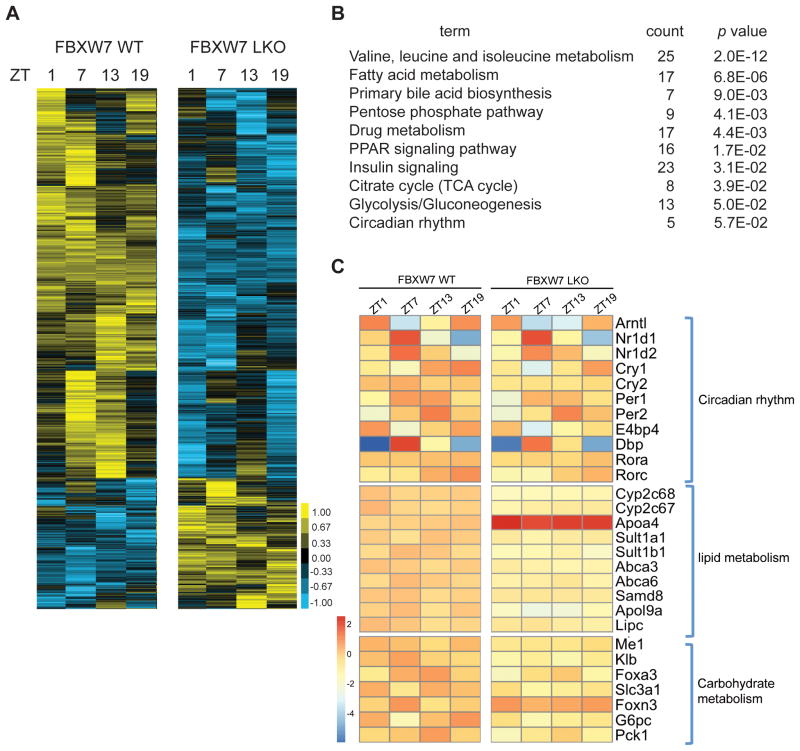
To further explore the functional interaction of FBXW7 and REV-ERBα in circadian clock regulation, we compared liver transcriptomes from WT and Fbxw7-LKO mice at different zeitgeber times (ZT), with a known gene set that contains all hepatic genes with circadian expression patterns (Circa DB, (Hughes et al., 2009)). Notably, over 30% (1559 out of 4881) of hepatic circadian expressed genes are disrupted in Fbxw7-LKO mice (Figure 4A). Of these 1559 genes, 75% show reduced amplitude, demonstrating that up-regulation of REV-ERBα suppresses hepatic circadian gene expression. Comparison of our Fbxw7-LKO gene sets with that of our previously described Rev-erbα KO (Cho et al., 2012) revealed extensive overlap (30–40%) between FBXW7- and REV-ERBα-dependent genes at each time point examined. Pathway analysis of FBXW7 and REV-ERBα co-regulated genes revealed that amino acid metabolism, lipid and bile acid metabolism, carbohydrate metabolism, PPAR signaling pathway and circadian rhythm pathways are all highly enriched (Figure 4B). The extent of dysregulation induced by Fbxw7-LKO is illustrated in Figure 4C, where the altered circadian expression patterns of selected REV-ERB target genes in the specified pathways are shown. Interestingly, the genes described in Fig 4C were virtually all shown by ChIP-Seq (Cho et al., 2012) to be direct binding targets of REV-ERB. These findings not only support the role of FBXW7 in regulating REV-ERBα stability, but also additionally implicate FBXW7 as a critical coordinator of clock amplitude with transcriptional output of circadian metabolic target genes in the liver.
Figure 4. Hepatic ablation of FBXW7 disrupts circadian transcriptome.
(A) Heatmap of hepatic genes with circadian expression pattern in WT and Fbxw7-LKO. (B) PANTHER pathway analysis of genes co-regulated by REV-ERB and FBXW7. (C) Heatmap of relative expression of a subset of genes co-regulated by REV-ERB and FBXW7 in WT and Fbxw7-LKO livers. See also Table S2.
FBXW7 liver-specific knockout disrupts whole body lipid and glucose homeostasis
The extensive overlap of REV-ERBα and FBXW7-regulated genes involved in lipid and carbohydrate metabolism is consistent with circadian rhythm being tightly linked to metabolism (Bass and Takahashi, 2010; Sancar and Brunner, 2014), and prompted us to explore the role of FBXW7 in metabolism. In Fbxw7-LKO mice, the amplitude of diurnal expression of Insig2, an important component of the SREBP pathway (Le Martelot et al., 2009) and direct REV-ERBα target gene (Cho et al., 2012), is reduced by about 66% (Figure 5A). Consistent with this, 14-week old Fbxw7-LKO mice develop hepatic centrilobular steatosis with elevated lipid content, as revealed by liver triglyceride measurements (Figures 5B and 5C). A more striking hepatic steatosis phenotype was observed when the animals were challenged with high fat diet for 4 weeks (Figure 5B).
Figure 5. Liver-specific depletion of FBXW7 affects glucose and whole body metabolism.
(A) Hepatic Insig 2 expression from WT and Fbxw7-LKO animals, measured by RT-PCR. Error bars= SD, n=3. *P<0.05 by Student’s t-test. (B) H&E staining of liver from WT and Fbxw7-LKO mice fed normal chow (top) and HFD (lower). Scale bar:100 μm. (C) Hepatic triglyceride (TGs) levels in WT and Fbxw7-LKO mice (n=3). (*P<0.05 by Student’s t-test) (D) Serum TGs in WT and Fbxw7-LKO mice at indicated times (n=3). (*P<0.05 by Student’s t-test). (E) Circadian expression patterns for hepatic PEPCK and G6Pase in WT (blue) and Fbxw7-LKO (red) mice, determined by RT-PCR. (**P<0.01, ***P<0.001 by Student’s t-test) (F) Fasting blood glucose levels (left) and glucose tolerance test (right) from WT and Fbxw7-LKO mice (n=9). (*P<0.05, **P<0.01, ***P<0.001 by Student’s t-test) (G) Respiratory exchange ratio (RER) of WT (blue) and Fbxw7-LKO (red) mice (n = 5). Error bars indicate standard error of means (SEM). See also Figure S6, Table S1, S2.
FBXW7 has been previously implicated in hepatic triglyceride metabolism (Kumadaki et al., 2011; Onoyama et al., 2011), but the mechanism has been controversial. Our work, in conjunction with previous ChIP studies (Bugge et al., 2012; Cho et al., 2012) suggests that the post-translational control of REV-ERBα stability and thereby circadian clock output gene expression is a critical factor contributing to liver metabolic homeostasis. As REV-ERBα is also directly involved in the circadian SREBP pathway (Kornmann et al., 2007; Le Martelot et al., 2009), our data suggest that FBXW7-dependent control of REV-ERBα stability may represent another layer of regulation that coordinates the circadian clock with liver metabolism. Thus, FBXW7 controls SREBP function through at least two different pathways: transcription level control of Insig2 circadian expression by regulating REV-ERBα stability, and direct control of SREBP protein degradation (Onoyama et al., 2011). Supporting this idea, hepatic knockout of FBXW7 boosted diurnal variation of serum triglyceride (TG) levels, with significant reductions of TG levels at the trough phase (Figure 5D). Strikingly, Fbxw7-LKO and REV-ERBα knockout animals display diametric changes in multiple serum metabolic parameters, including HDL and LDL (Supplemental Table 1), whereas the transcriptional changes in hepatic circadian clock and metabolic gene expression in Fbxw7-LKO parallel those from REV-ERBα hepatic transgenic animals (Kornmann et al., 2007; Le Martelot et al., 2009) (Supplemental Table 2). These correlations support the notion that REV-ERBα is an FBXW7 target in vivo.
Hepatic glucose production is key to whole body glucose homeostasis and tightly controlled by circadian clocks (Lamia et al., 2011; Tong and Yin, 2013). REV-ERBα has been shown as an important factor mediating this process through repressing PEPCK and G6Pase, two genes encoding rate-limiting steps of liver gluconeogenesis (Yin et al., 2007). Notably, ablation of Fbxw7 in mouse liver disrupts circadian oscillation of both PEPCK and G6Pase (Figure 5E), further supporting an in vivo role for FBXW7 in REVERBα-regulated metabolic gene expression. Consistent with this notion, Fbxw7-LKO animals had lower fasting blood glucose levels and improved glucose tolerance compared to WT littermates (Figure 5F), whereas their insulin tolerance was not affected (data not shown). This phenotype resembles Bmal1 liver specific knockout animals where improved glucose tolerance was observed (Lamia et al., 2008). In addition, diurnal rhythms of circulating factors involved in whole body glucose metabolism including glucagon, insulin, PAI-1 and GLP-1 were altered in Fbxw7-LKO animals (Figure S6A). We also observed a marked reduction of hepatic glycogen storage in Fbxw7-LKO livers (Figure S6B), consistent with the dampened expression pattern of glycogen synthase 2 (Gys2), a diurnally regulated gene encoding the key enzyme for hepatic glycogen synthesis (Figure S6C). Furthermore, the reduced respiratory exchange ratio (RER) in Fbxw7-LKO animals (Figure 5G) indicates a shift in energy utilization towards fatty acid oxidation.
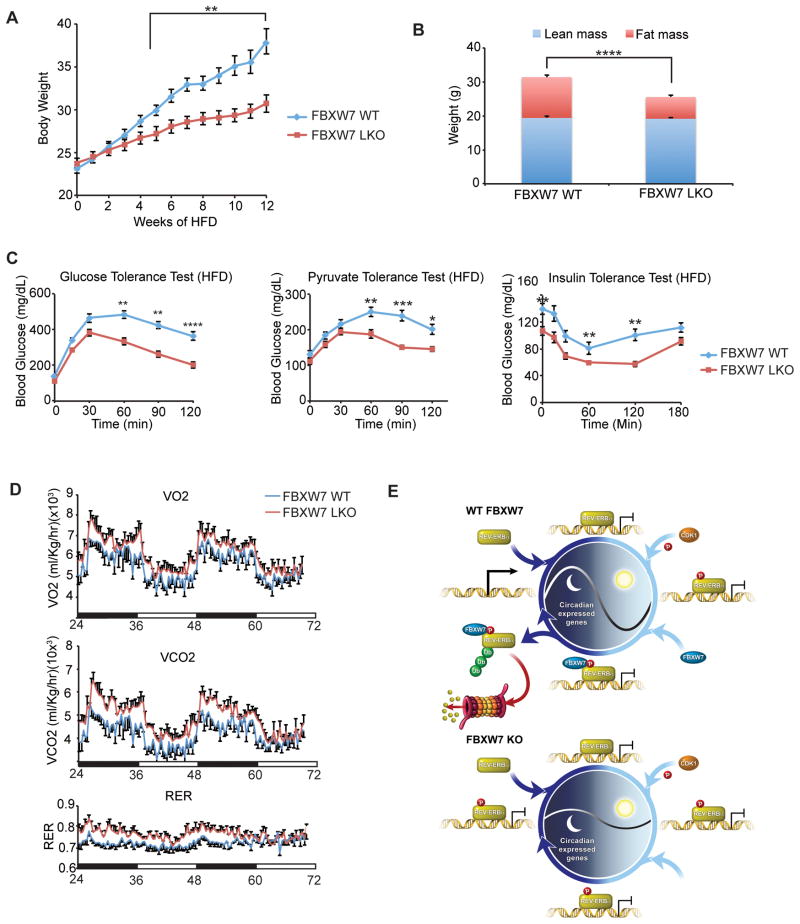
To further explore the role of FBXW7 in circadian clock and metabolic regulation, we challenged Fbxw7-LKO animals with a high-fat diet (HFD, 60% of calories from fat). Whereas high-fat feeding caused pronounced body weight increases in WT animals, Fbxw7-LKO mice were protected against HFD-induced obesity (weight increases in Fbxw7-LKO mice are comparable to normal chow-fed animals, Figure 6A). This weight differential is attributed to reduced fat mass in Fbxw7-LKO mice, as measured by MRI (Figure 6B). Paradoxically, Fbxw7-LKO animals have improved glucose management, as measured by glucose tolerance tests, despite a marked increase in hepatic steatosis (Figures 5B and 6C). This apparent disparity may be due to compromised hepatic gluconeogenesis in Fbxw7-LKO mice (suggested by pyruvate tolerance tests) as insulin sensitivity is not significantly affected (Figure 6C). In addition, HFD-fed Fbxw7-LKO mice have increased expression of Cyp7a1 (Figure S6D), a rate-limiting enzyme controlling primary bile acid synthesis, consistent with the role of REV-ERBα in regulating bile acid metabolism (Le Martelot et al., 2009). Notably, the selective loss of FBXW7 in the liver increases whole body energy expenditure of HFD-fed mice, with increases in both oxygen consumption and CO2 production (Figure 6D). Consistent with the reduced Bmal1 expression amplitude, Fbxw7-LKO mice phenocopy the Bmal1 knockout animal as shown by greater hypoketotic hypoglycemia under prolonged fasting condition (Figure S6E) and accumulation of medium/long chain free fatty acids in liver (data not shown) (Peek et al., 2013). Collectively, these findings implicate an FBXW7-regulated liver metabolic program in whole body energy homeostasis, and reinforce the importance of post-translational modifications in the circadian regulation of metabolism.
Figure 6. FBXW7 regulates glucose and whole body metabolism.
(A) Body weights of HFD-fed WT and FBXW7 LKO mice (n=11). (B) Body composition, (C) Glucose tolerance test, pyruvate tolerance test and insulin tolerance test and (D) Oxygen consumption, CO2 production and Respiratory Exchange Ratio (RER) for WT (blue) and Fbxw7-LKO (red) mice fed a HFD for 12 weeks (n=11). For all panels: Error bars indicate standard error of means (SEM). Statistical significance was determined by Student’s t-test (*P<0.05, **P<0.01, ***P<0.001). (E) Model of circadian amplitude regulation via FBXW7-targeted REV-ERBα degradation. See also Figure S6.
Discussion
The circadian oscillatory program requires not only the regulation of cycle time (period and phase) but also amplitude, with low amplitude molecular oscillations unable to drive strong rhythmic physiology. Circadian amplitude is coupled with physiology and disease, sleep disruption and aging dampen, whereas physical activities increase the amplitude of circadian rhythms (Ramkisoensing and Meijer, 2015). Notably, transplantation of SCN tissue from a young to an aged animal efficiently rescues the low-amplitude behavior and gene expression rhythms in old animals (Cai et al., 1997; Hurd et al., 1995), suggesting that amplitude control is intrinsic and cell autonomous. However, the mechanisms underpinning robustness or amplitude of clock rhythm remain poorly understood (Luo et al., 2012). Our findings that REV-ERBα is targeted for ubiquitination and subsequent degradation by the F-box protein FBXW7α provides a pathway for regulating the amplitude of clock transcription (Figure 6E).
Mechanistically, our data identifies that CDK1-dependent phosphorylation is required for REV-ERBα degradation. CDK1 is epistatic to FBXW7, and both FBXW7 and CDK1 knockout and inhibition affect the amplitude of cell autonomous circadian rhythms in cultured cells. In mouse liver, diurnal oscillation of REV-ERBα T275 phosphorylation contributes to the long-recognized rhythmic changes of REV-ERBα protein levels. Strikingly, hepatic FBXW7 knockout alters the amplitude of diurnal expression of many core clock genes, as well as large numbers of genes controlling liver metabolic pathways. Similarly, CDK1 inhibitor Roscovitine, modulates circadian gene expression in U2OS cells (Hirota et al., 2008) and mouse liver (Iurisci et al., 2009). As REV-ERBα is a critical regulator coordinating circadian rhythm and liver metabolism, the abnormal and muted amplitude of expression of genes involved in lipid and glucose metabolism, such as Insig2 and G6Pase, appears to directly contribute to liver steatosis and dysregulated gluconeogenesis in FBXW7-deficient animals. These observations underscore the importance of precise regulation of REV-ERBα stability and circadian amplitude in maintaining whole body energy homeostasis, and are consistent with recent studies linking REV-ERBα SNPs to obesity (Garaulet and Gomez-Abellan, 2014; Goumidi et al., 2013)
Of interest is the selectivity of the FBXW7 interaction with REV-ERBα, but not REV-ERBβ or CRY1, owing to the exclusive presence of the T275 degron sequence (CPD) in REV-ERBα. This restriction may help to molecularly corral a single circadian regulator for CDK1-triggered degradation while allowing other repressive clock genes such as REV-ERBβ and CRY to fall under different regulatory cascades that control period (Lamia et al., 2009; Siepka et al., 2007). In contrast, the integration of the CDK1-FBXW7 pathway with the circadian cycle facilitates REV-ERBα degradation coincident with maximum protein levels. Thus, the contrasting roles for FBXW7 and REV-ERBα in regulating clock amplitude, but not clock period, are noteworthy and illustrate how temporal-specific REV-ERBα degradation can mechanistically uncouple these two hallmarks of clock function. Thus, while certain signals control the period of circadian rhythmicity, others modulate amplitude. How these signals integrate to regulate circadian behavior and physiology is an important question for further investigation. Temporal-specific degradation may be key as non-specific turnover of REV-ERBα and REV-ERBβ (and possibly other clock genes) changes period, and not amplitude (DeBruyne et al., 2015).
Given that CDK1 is an evolutionally conserved cell cycle regulator, the pathway delineated in this report sheds light on the coordinated regulatory mechanisms between cell cycle and circadian clock. Increasing lines of evidence suggest that circadian clocks and cell cycle are connected (Hunt and Sassone-Corsi, 2007; Matsuo et al., 2003). It has been proposed that circadian clocks may impose a gating mechanism on the timing of the cell division cycles by controlling many cell cycle regulators such as Wee-1, c-Myc and Cyclins (Hunt and Sassone-Corsi, 2007; Matsuo et al., 2003). DNA damage and CDK inhibitors both serve as zeitgebers to reset the circadian clock (Oklejewicz et al., 2008; Papp et al., 2015) and have been shown to modulate circadian gene expression in mouse liver as well as at the single cell level (Gamsby et al., 2009; Iurisci et al., 2009). Our findings further support this hypothesis, suggesting that these two cycling events share more common regulators than previously appreciated to coordinately maintain diurnal physiological homeostasis. Given that FBXW7α is a bona fide tumor suppressor (Welcker and Clurman, 2008), our discovery that FBXW7α is a critical component of circadian clock regulatory circuitry in peripheral tissue strengthens the link between circadian clock disruption and cancer, and suggests that this could be the basis of aberrant circadian rhythms and metabolism in isolated tumor cells (Sahar and Sassone-Corsi, 2009).
In summary, the discovery of a molecular pathway for controlling REV-ERBα repression defines an unexpected molecular mechanism for re-engaging the transcriptional positive arm of the clock, which, via CDK1 inhibition, suggests the potential to manipulate clock amplitude using small molecule therapy.
Experimental Procedures
See Supplemental Experimental procedures for descriptions of expression constructs, chemicals and antibodies used, and details of cell culture, transfection protocols, retrovirus production and infection, RNA-Seq and animal studies.
Phosopho-specific antibody generation and characterization
A peptide antigen containing phosphorylated T275 of REV-ERBα, generated by the Peptide Core Facility at the Salk Institute, was coupled with KLH carrier protein (Pierce, 77666) and used to immunize rabbits (Pocono Rabbit Farm and Laboratory). Antiserum was affinity purified using antigen-conjugated column (SulfoLink Immobilization column, Pierce).
Recombinant protein preparation and in vitro kinase assays
cDNA encoding full length Rev-erbα and its (198–322) fragment were cloned into pGEX-4T-1 vector and used to transform BL-21 CodonPlus competent cells (Stratagene). GST-tagged proteins were induced by IPTG and purified using Glutathione Sepharose 4B. 2μg of recombinant protein was incubated with 0.2μg of recombinant CDK1/cyclin B (Cell Signaling) in a kinase buffer (Cell Signaling) supplemented with 0.2mM ATP for 30 min in 30°C. The reaction was terminated by LDS sample loading buffer and heated for 10 min before resolution on SDS-PAGE.
Rev-erbα protein complex purification and mass spectrometry
Protein complex purification was carried out as described (Ikura et al., 2000; Nakatani and Ogryzko, 2003). Briefly, 293T cells transfected with pcDNA3-Rev-erbα expression plasmid were lysed in NETN buffer containing 170mM NaCl supplemented with protease and phosphatase inhibitors. The clarified lysates sequentially purified with anti-Flag antibody (Sigma, A2220) and anti-HA antibody conjugated beads. The final eluent was precipitated using trichloroacetic acid (TCA) for mass spectrometry analysis (additional details in Supplemental Experimental procedures).
Cell-based circadian assay
Cell-based circadian assays were performed as described (Zhang et al., 2009). Briefly, U2OS cells harboring Bmal1 promoter controlled luciferase reporter were reverse transfected either with Cdk1 or Fbxw7 siRNAs in 96-well plates. Three days after transfection, media was replaced with 180μl HEPES-buffered explant medium supplemented with luciferin (1 mM) and B-27 supplements, and bioluminescence recorded at 36°C. REV-ERBα null fibroblasts harboring Bmal1-promoter controlled reporter (Liu et al., 2008) were transduced with retrovirus encoding either WT Rev-erbα or T275A mutant. After changing to fresh explant medium at ambient temperature, luciferase activities were monitored in the LumiCycle (Actimetrics). The normalized amplitude of circadian Bmal1-dLuc oscillation from WT and T275A mutant expressing cells were calculated and plotted. Bmal1 expression was determined by quantitative PCR (Cho et al., 2012).
Metabolite analysis
Hepatic and serum triglycerides were measured using commercial kits (Wako Chemicals). Serum insulin, glucagon, PAI-1, GLP-1 levels were measured by Bio-plex mouse diabetes assay kit (Bio-Rad).
Supplementary Material
Acknowledgments
We thank Drs S Reed, G Wahl and B Vogelstein for valuable reagents. We thank Y Dai, L Ong and C Brondos for technical and administrative assistance. RME is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and March of Dimes Chair in Molecular and Developmental Biology and is supported by NIH Grants DK057978, DK090962, HL088093, and HL105278; Glenn Foundation; Leona M and Harry B Helmsley Charitable Trust 2012-PG-MED-002; Ipsen/Biomeasure; Ellison Medical Foundation; and Samuel Waxman Cancer Research Foundation. XZ is supported by grants from Susan G Komen Breast Cancer Research Foundation (PDF0601100) and the Glenn Foundation for Medical Research. KL is supported by NIH Grant DK097164. S.A.K is supported by NIH Grant R01GM074868. J.R.Y is supported by NIH P41 GM103533 and R01 MH067880.
Footnotes
Author Contributions
X.Z., A.R.A., R.T.Y., M.D., and R.M.E. designed and supervised the research. X.Z., T.H., X.H., H.C., L.-W.C., K.L., S.L., and E.B. performed research. X.Z., T.H., X.H., S.L., A.R.A., C.L., R.T.Y., J.R.Y., S.A.K., M.D., and R.M.E. analyzed data. X.Z., A.R.A., R.T.Y., M.D., and R.M.E. wrote the manuscript.
Data deposition: RNA-Seq data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database, Accession # SRP059440.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelmant G, Begue A, Stehelin D, Laudet V. A functional Rev-erb alpha responsive element located in the human Rev-erb alpha promoter mediates a repressing activity. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3553–3558. doi: 10.1073/pnas.93.8.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–667. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai A, Lehman MN, Lloyd JM, Wise PM. Transplantation of fetal suprachiasmatic nuclei into middle-aged rats restores diurnal Fos expression in host. Am J Physiol. 1997;272:R422–428. doi: 10.1152/ajpregu.1997.272.1.R422. [DOI] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne JP, Baggs JE, Sato TK, Hogenesch JB. Ubiquitin ligase Siah2 regulates RevErbalpha degradation and the mammalian circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:12420–12425. doi: 10.1073/pnas.1501204112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Gamsby JJ, Loros JJ, Dunlap JC. A phylogenetically conserved DNA damage response resets the circadian clock. J Biol Rhythms. 2009;24:193–202. doi: 10.1177/0748730409334748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet M, Gomez-Abellan P. Timing of food intake and obesity: a novel association. Physiol Behav. 2014;134:44–50. doi: 10.1016/j.physbeh.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle. 2010;9 doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumidi L, Grechez A, Dumont J, Cottel D, Kafatos A, Moreno LA, Molnar D, Moschonis G, Gottrand F, Huybrechts I, et al. Impact of REV-ERB alpha gene polymorphisms on obesity phenotypes in adult and adolescent samples. Int J Obes (Lond) 2013;37:666–672. doi: 10.1038/ijo.2012.117. [DOI] [PubMed] [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell. 2013;152:1106–1118. doi: 10.1016/j.cell.2013.01.054. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol. 2010;8:e1000559. doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20746–20751. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Hurd MW, Zimmer KA, Lehman MN, Ralph MR. Circadian locomotor rhythms in aged hamsters following suprachiasmatic transplant. Am J Physiol. 1995;269:R958–968. doi: 10.1152/ajpregu.1995.269.5.R958. [DOI] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Iurisci I, Filipski E, Sallam H, Harper F, Guettier C, Maire I, Hassan M, Iacobelli S, Levi F. Liver circadian clock, a pharmacologic target of cyclin-dependent kinase inhibitor seliciclib. Chronobiol Int. 2009;26:1169–1188. doi: 10.3109/07420520903209942. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumadaki S, Karasawa T, Matsuzaka T, Ema M, Nakagawa Y, Nakakuki M, Saito R, Yahagi N, Iwasaki H, Sone H, et al. Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7alpha (Fbw7alpha) causes hepatosteatosis through Kruppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor gamma2 (PPARgamma2) pathway but not SREBP-1c protein in mice. The Journal of biological chemistry. 2011;286:40835–40846. doi: 10.1074/jbc.M111.235283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Chen WF, Yue Z, Chen D, Sowcik M, Sehgal A, Zheng X. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell. 2012;11:428–438. doi: 10.1111/j.1474-9726.2012.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oklejewicz M, Destici E, Tamanini F, Hut RA, Janssens R, van der Horst GT. Phase resetting of the mammalian circadian clock by DNA damage. Curr Biol. 2008;18:286–291. doi: 10.1016/j.cub.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Onoyama I, Suzuki A, Matsumoto A, Tomita K, Katagiri H, Oike Y, Nakayama K, Nakayama KI. Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. The Journal of clinical investigation. 2011;121:342–354. doi: 10.1172/JCI40725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp SJ, Huber AL, Jordan SD, Kriebs A, Nguyen M, Moresco JJ, Yates JR, Lamia KA. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. Elife. 2015;4 doi: 10.7554/eLife.04883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013;342:1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Ramkisoensing A, Meijer JH. Synchronization of Biological Clock Neurons by Light and Peripheral Feedback Systems Promotes Circadian Rhythms and Health. Front Neurol. 2015;6:128. doi: 10.3389/fneur.2015.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Sancar G, Brunner M. Circadian clocks and energy metabolism. Cellular and molecular life sciences : CMLS. 2014;71:2667–2680. doi: 10.1007/s00018-014-1574-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–922. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature. 2012;485:62–68. doi: 10.1038/nature11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Jothi R, Birault V, Jetten AM. RORgamma directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40:8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Yin L. Circadian rhythms in liver physiology and liver diseases. Comprehensive Physiology. 2013;3:917–940. doi: 10.1002/cphy.c120017. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Yagita K, Tamanini F, Yasuda M, Hoeijmakers JH, van der Horst GT, Okamura H. Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J. 2002;21:1301–1314. doi: 10.1093/emboj/21.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW, 3rd, Janes J, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell. 2009;139:199–210. doi: 10.1016/j.cell.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.