Abstract
Unfolded protein responses (UPR), consisting of three major transducers PERK, IRE1 and ATF6, occur in the midst of a variety of intracellular and extracellular challenges that perturb protein folding in the endoplasmic reticulum (ER). ER stress occurs and is thought to be a contributing factor to a number of human diseases, including cancer, neurodegenerative disorders and various metabolic syndromes. In the context of neoplastic growth, oncogenic stress resulting from dysregulation of oncogenes such as c-Myc, BrafV600E and HRASG12V trigger the UPR as an adaptive strategy for cancer cell survival. PERK is an ER resident type I protein kinase harboring both pro-apoptotic and pro-survival capabilities. PERK, as a coordinator through its downstream substrates, reprograms cancer gene expression to facilitate survival in response to oncogenes and microenvironmental challenges, such as hypoxia, angiogenesis, and metastasis. Herein, we discuss how PERK kinase engages in tumor initiation, transformation, adaption microenvironmental stress, chemoresistance and potential opportunities and potential opportunities for PERK targeted therapy.
Keywords: UPR, PERK, cancer, therapy
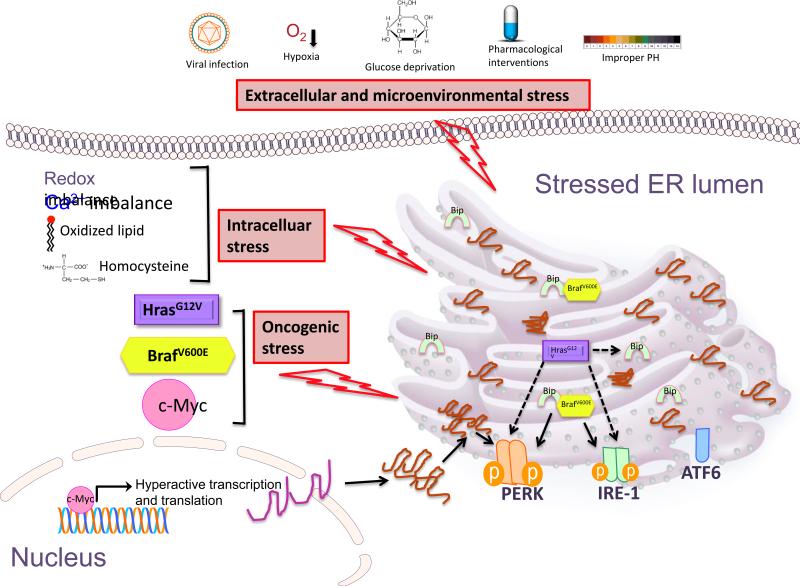
ER stress is intimately involved in a variety of human diseases, including diabetes, neurodegeneration, stroke, viral infection, and heart disease (Kim et al., 2008). Accumulating evidence suggests that chronic ER stress is readily apparent in multiple cancer types including melanoma, multiple myeloma, breast cancer, hepatocellular carcinoma. ER stress is sensed by a coordinated signaling pathway, initiated by signal transducers that span the ER membrane. UPR signaling consists 3 major sensors and their downstream pathways, protein kinase R-like endoplasmic reticulum kinase (PERK), inositol requiring enzyme 1 alpha (IRE1α, ubiquitous), inositol requiring enzyme 1 beta (IRE1β, tissue specific) and activating transcription factor 6 (ATF6) (Figure 1). Molecular chaperone BiP/GRP78 consistently and dynamically binds and arrests them in inactive status in the ER membrane. ER stress triggers the dissociation of these three major sensors from BiP. Released sensors undergo a conformational change necessary for activation and downstream signaling.
Figure 1. Activation of the UPR.
Extracellular stress, intracellular stress as well as oncogene activation trigger ER stress and activate the transducers PERK, IRE-1 and ATF6. Extracullar stress includes viral infection, hypoxia, glucose deprivation, improper PH and pharmalogical interventions. Chemotherapeutic agents such as vemurafinib, paclitaxel and bortezomib induce ER stress in cancer cells. Intracellular stress such as redox imbalance, calcium imbalance, oxidized lipid and homocysteine result in unfolded protein response. Oncogenic stress such as cmyc amplification, BRAF mutation and HRas mutation all trigger ER stress. c-Myc greatly enhances the global transcription and translation, which cause large amount unfolded protein accumulated in the ER and activate PERK signaling. BRAFV600E binds protein chaperon GRP78/Bip and dissociates Bip from the 3 transducers, activating IRE1 and PERK pathways. HRasG12V causes activation of PERK and IRE1, also increases the Bip expression via direct or indirect way, the deep mechanisms are still unknown.
As a central coordinator, the transducers sense stress and initiate a signaling pathway that makes the final life-and-death decisions for the cells undergoing variable stresses. ER stress signaling can restore homeostasis by reducing global protein translation, clearing the misfolded proteins, increase proper protein folding capability, facilitating cell adaption and survival. In contrast, ER stress can also initiate apoptosis by activating cell death genes during unmitigated stress. The competence of cell survival to ER stress is therefore proportional to the intensity and duration of the stress, the extent of the activation of the transducers, the execution of the downstream of effectors, the balance between pro-survival signaling and pro-apoptotic signaling (Chevet et al., 2015).
UPR Signal transducers
IRE1
IRE1 is a type I single pass transmembrane protein that undergoes oligomerization and trans-autophosphorylation under ER stress. IRE1 contains an RNase domain that selectively degrades mRNA, microRNA and ribosomal RNA in an IRE1 dependent decay (RIDD) way. Xbp1 mRNA, a transcriptional regulator, is the major substrate of IRE1. Activation of Xbp1 is the direct result of IRE1-dependent excision of a 26 nucleotide intron from the Xbp1 mRNA thereby generating a spliced Xbp1 mRNA that is more efficiently translated. Spliced Xbp1 modulates gene expression engaged in protein folding, secretion and endoplasmic reticulum-associated degradation (ERAD)(Hetz et al., 2011).
In budding yeast, a single IRE1 is expressed; while yeast IRE1 also harbors protein kinase activity, trans-autophosphorylation is its primary function and this contributes exclusively to its RNase function. As such, enforced dimerization which permits RNase activation is sufficient for IRE1 function (Ron and Hubbard, 2008). In contrast, the function of IRE1 in mammalian cells is more complex and perhaps as a reflection of this, higher eukaryotes harbor two IRE1 genes; IRE1α which is ubiquitously expressed, essential gene and IRE1β which is expressed in a tissue specific manner (Bertolotti et al., 2001; Wang et al., 1998). Unlike yeast, mammalian IRE1 has validated protein kinase substrates that include JNK (Urano et al., 2000) and Ask1 (Nishitoh et al., 2002) via Protein adaptor TRAF2. Phosphorylation of such substrates in turn contributes to cell adaptation to chronic versus acute stress (Lin et al., 2007).
ATF6
ATF6 activation requires export from the ER and translocate to the Golgi apparatus. Under ER stress, ATF6 is translocated to Golgi and processed by protein phosphatase S1P and S2P, where the cytosolic domain of ATF6 goes to nucleus and initiate a specific transcription program involved in protein folding and quality control (Haze et al., 1999; Sitia and Braakman, 2003). ATF6 may contribute to cancer cell dormancy and drug resistance, due to its cell adaptive function (Martino et al., 2013; Schewe and Aguirre-Ghiso, 2008).
PERK
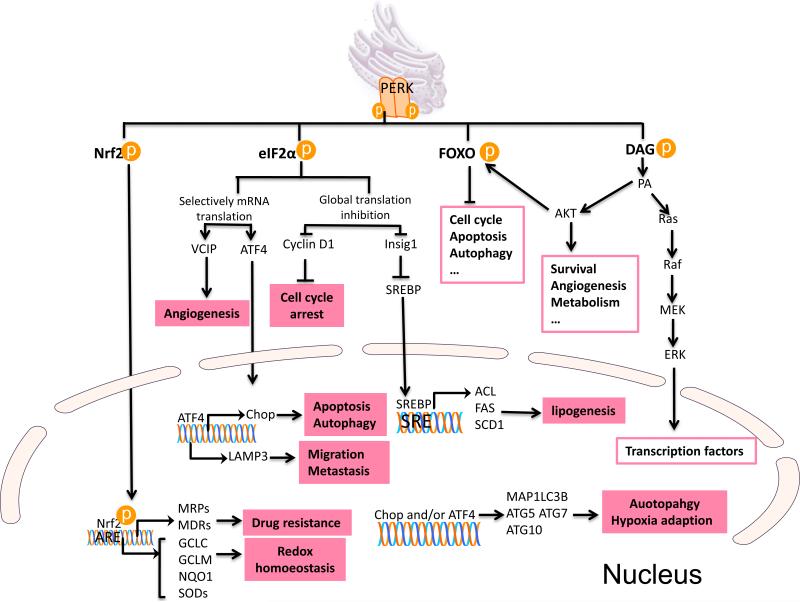
While PERK like IRE1 is a single pass type I protein kinase, it contrasts with IRE1 in that it is a dedicated protein kinase. Analogous with IRE1, titration of BiP by misfolded proteins permits PERK oligomerization, autophosphorylation and consequent phosphorylation of downstream substrates. PERK can phosphorylate a growing number of substrates including eIF2α (Harding et al., 1999), Nrf2 (Cullinan et al., 2003) and FOXO (Zhang et al., 2013), and the lipid diacylglycerol (DAG) (Bobrovnikova-Marjon et al., 2012; Pytel et al., 2015) (Figure 2). One of the key regulatory functions of PERK is its role as a regulator of protein translation; a reflection of direct phosphorylation of translation initiation factor 2α (eIF2α). eIF2α regulates the binding of the methionyl tRNA to the ribosome. Phosphorylation of eIF2α at Ser51 disrupts activation of the 43S translation initiation complex formation, thereby reducing the rate of general protein translation (Harding et al., 1999). The PERK-dependent reduction of protein translation is thought to limit nascent protein transport to ER lumen reducing potential chaperone load thereby permitting chaperones to clear misfolded aggregates. Intriguingly, translation of select proteins is increased; examples ATF4 and cIAP1/2 and, due to the presence of an upstream short open reading frame (uORF) (Hamanaka et al., 2009; Yaman et al., 2003). C/EBP homologous protein (CHOP), activated by ATF4 accumulation, plays multiple function as a transcriptional factor in ER stress signaling pathway, such as apoptosis and autophagy. For instance, CHOP activates proapoptotic Bax, leading to mitochondria mediated apoptosis (Szegezdi et al., 2006); CHOP and ATF4 cooperatively modulate expression of genes encoding regulators of autophagy including p62, Atg5, Atg7, and Atg10 (B'Chir et al., 2013; Szegezdi et al., 2006). In addition, ATF4 triggers increased expression of GADD34, a component of the PP1 phosphatase that dephosphorylates eIF2α (Ma and Hendershot, 2003), thereby mediating a negative feedback loop that limits PERK signaling.
Figure 2. PERK signaling promotes cancer cell survival and aggressiveness.
PERK phosphorylates its protein substrates Nrf2, eIF2α, FOXO and lipid substrate DAG. PERK phosphorylated Nrf2 dissociates from Keap1 and imported to nucleus, activating the transcription of drug resistance gene and redox enzyme genes. MRPs, multidrug resistance-associated proteins; MDRs, multidrug resistance proteins; GCLC, glutamate cysteine ligase catalytic subunit; GCLM, glutamate cysteine ligase modifier subunit; NQO1, NAD(P)H:quinone oxidoreductase 1; SODs, superoxide dismutases. PERK phosphorylated eIF2α exerts global protein inhibition, including cell cycle regulator cyclin D1 and lipid biosynthesis modulator insig1 (insulin inducible gene 1), which sequesters SREBP 1c (Sterol Regulatory Element Binding Protein) on the membrane to prevent its maturation. ACL, ATP citrate lyase; FAS, fatty acid synthase; SCD1, stearyl-CoA desaturase-1. eIF2α also selectively promotes some gene expression via alternative upstream open reading frame, such as ATF4 and VCIP. ATF4 activates CHOP transcription. ATF4, Chop alone or together regulate a broad range of genes, including those that regulate apoptosis, autophagy and hypoxia adaption. ATF4 regulated LAMP3 (Lysosomal-associated membrane protein 3) mediates migration and metastasis under ER stress. PERK activation of FOXO and phosphorylation DAG regulates the AKT function.
PERK-dependent phosphorylation of nuclear factor erythroid 2-related factor 2 (Nrf2) on Threonine 80 promotes Nrf2 dissociation from Keap1, a scaffolding protein that contributes to Nrf2 degradation, thereby allowing Nrf2 translocation to the nucleus and activation downstream. Nrf2 as a master transcriptional factor of redox homeostasis limits ROS damage and renders cells chemoresistance (Cullinan et al., 2003) (Figure 2). The target genes of Nrf2 include antioxidant enzymes NQO1, GCLC et al., and drug transporters such as MRP2, MRP3 et al., conferring Nrf2 crucial function to maintain the redox homeostasis and resistance to chemotherapy (Ma, 2013).
A third PERK substrate FOXO1 is phosphorylated on S243 in Drosophila (Zhang et al., 2013) (Figure 2); this residue is conserved in human FOXO1 within cluster of phosphorylatable serines, Ser298, Ser301, Ser308 and Ser311 cluster, which regulates the nuclear translocation of FOXO1 (Zhang et al., 2013). Interestingly, PERK also regulates FOXO1 activity indirectly by participating in AKT activation, which is an upstream negative regulator of FOXO1 activity (Bobrovnikova-Marjon et al., 2012). The direct and indirect regulation of FOXO activity by PERK generates a negative feedback mediating insulin resistance and other functions.
Finally, PERK is also known to possess lipid kinase activity, phosphorylating the lipid diacylglycerol (DAG) (Figure 2) (Bobrovnikova-Marjon et al., 2012). DAG is the precursor to generate a central node lipid second massager phosphatidic acid (PA), which engages multiple mitogenic pathways and promotes tumorigenesis (Bruntz et al., 2014).
Oncogenic drivers, the UPR, and PERK
Tumors must proliferate and survive in a microenvironment that should be incompatible with rapid growth and expansion. The tumor environment is typically poorly vascularized resulting in limitations in key nutrients including oxygen, glucose, and growth factors (Harris, 2002). Such limitations trigger a number of stress responses, including the UPR (Figure 1). While engagement of these pathways can trigger cell death and prevent accumulation of damaged cells or simply aberrant growth, pathways such as the UPR are also adaptive and can be co-opted to enhance tumor cell survival and ultimately tumor progression. Such a process can be considered “non-oncogene addiction” and led many to consider whether such adaptive pathways are potential therapeutic targets. In this section, we focus on the contributions of PERK to tumor progression.
c-Myc is a potent oncogene that drives tumorigenesis in part through its capacity to increase ribozyme expression, biogenesis and ultimately protein synthesis (Grandori et al., 2005). As a result, c-Myc expression is associated with a dramatic increase in PERK activity both in mouse models and human lymphoma that harbor a c-Myc translocation (Hart et al., 2012). Myc expressing cells appear exquisitely dependent upon PERK-dependent autophagy and in the absence of PERK the reduction in autophagy favors apoptosis. PERK activity regulates ULK1 and ATG5 expression, both of which are essential for sustaining the high level of autophagy. This evidence couples the c-Myc increased protein synthesis and the activation of PERK-eIF2α-ATF4 UPR branch, suggesting UPR plays essential roles for c-Myc mediated cell survival and tumor progression via induction of cytoprotective autophagy.
BRAF/NRas mutations are the most common driver mutations in melanoma (Pollock et al., 2003). Oncogenic BRAFV600E triggers chronic ER stress, resulting in higher basal level of autophagy and resistance of apoptosis in melanoma. In melanoma, IRE1/ASK1/JNK accounts for autophagy induced by BRAFV600E (Corazzari et al., 2014). BRAFV600E also activates PERK as demonstrated by eIF2α phosphorylation and ATF4 induction in melanoma cell lines. Whether PERK contributes to the pro-survival autophagy in the BRAF mutant melanoma cells is not clear (Corazzari et al., 2014). One attractive hypothesis would be that PERK mediated autophagy and facilitates BRAFV600E-dependent tumorigenesis in melanoma. Indeed, a second group addressed this question in the context of BRAFV600E inhibitor resistant melanoma cells (Ma et al., 2014).
BRAFV600E inhibition by vemurafenib represents a major advance in the melanoma treatment; unfortunately, almost all the patients got relapsed eventually and developed the BRAFV600E inhibition resistance (Flaherty et al., 2010). BRAFV600E inhibitor treatment triggers translocation of mutant BRAFV600E to the ER resulting in sequestration of BiP/Grp78 thereby activating PERK. PERK in turn was suggested to mediate through cytoprotective autophagy (Figure 1). Ultimately, PERK inhibitors appear to sensitize BRAFV600E mutant melanoma cells to BRAFV600E inhibitor treatment (Ma et al., 2014). One might speculate that combining vemurafenib with a PERK inhibitor or an autophagy inhibitor might produce robust clinical efficacy.
The HER2/Neu oncogene can also induce PERK in the context of mammary carcinoma. In mouse models of HER2/Neu-dependent mammary adenocarcinoma, deletion of PERK reduced growth of both the primary tumor and reduced metastasis by 50% (Bobrovnikova-Marjon et al., 2010). In this model, PERK loss had no impact on tumor incidence suggesting it is required for progression rather than transformation. The impact on progression was attributed to maintenance of redox homeostasis, via Nrf2, in this system rather than autophagy (Bobrovnikova-Marjon et al., 2010).
PERK impacts accumulation of select oncoproteins and tumor suppressors
Initial work on ER stress revealed a close association with cell cycle arrest. Subsequent work demonstrated PERK-dependent signals mediate arrest in G1 phase; a feature of reduced cyclin D1 translation (Brewer and Diehl, 2000; Hamanaka et al., 2005). PERK activation and eIF2α phosphorylation suppresses general protein translation, including cyclin D1 translation. Due to its short half-life, cyclin D1 expression is greatly attenuated during ER stress. Attenuated expression of cyclin D1 causes the impaired activity of cyclinD1-CDK4 complex, followed by p21Cip1 redistribution to and inhibition of CDK2 thereby ensuring cell cycle arrest at G1 phase. Enforced expression of cyclin D1 mutants that are refractory to ubiquitin-dependent degradation, overcomes cell cycle arrest triggered by ER stress (Brewer et al., 1999) but in so doing sensitize cells to apoptosis.
Independent of cyclin D1, PERK mediated UPR triggers p53 accumulation and cell cycle arrest via ribosomal subunits: Hdm2 interaction (Zhang et al., 2006). PERK mediated eIF2α phosphorylation causes the reduced polysome formation and it is likely also increased free ribosomes. Therefore, these free ribosomal proteins, specifically rpL5/rpL11/rpL23 bind Hdm2 and sequester Hdm2 in the nucleus, which causes the impaired E3 ligase activity of Hdm2 and consequently p53 turnover slowdown. Accumulation of p53 also contributes to p21Cip1 induction and cell cycle arrest in G1/S phase. Biogenesis of ribosome is energy consuming in physiological condition. It is reasonable to speculate that cells under harsh condition such as nutrition deprivation develop the strategy shutting off general protein synthesis, ribosome biogenesis and stopping cell cycle progressing to obtain an opportunity restoring the homeostasis. p53 accumulation beyond key threshold will trigger apoptotic gene expression, such as PUMA and NOXA, and inducing apoptosis (Li et al., 2006). ER stress has also been suggested to accelerate p53 degradation via GSK3β (Pluquet et al., 2005; Qu et al., 2004). Still other work suggests that ER stress leads to an initial downregulation of p53 followed by its recovery at later stages (Thomas et al., 2013). Emerging evidence suggests ER stress and PERK induces p47, a N-terminal truncated p53 translated from the 2nd translational initiation site in p53 mRNA. PERK activation initiates a cap-independent p53 translation and consequently accumulating p53/47. p47 in turn triggers arrest in G2 phase (Bourougaa et al., 2010). Ultimately, p53 contributes to regulation of the G1/S transition cell cycle, while p47 regulates the G2 cell cycle transition via upregulation 14-3-3σ.
Does UPR activation and p53 regulation impact cell cycle checkpoints? Indeed, ER stress mediated protein translation attenuation has been implicated in Chk1 phosphorylation (Thomas et al., 2013). Since p53 is phosphorylated and activated by Chk1 (Polager and Ginsberg, 2009), and p53 can impact recovery from protein translation inhibition by modulating GADD34, the phosphatase of eIF2α, p53 mediated G1/S arrest and Chk1 mediated G2 phase transition interactively affect each other. The G2/M and G1 arrest could occur sequentially during ER stress (Thomas et al., 2013). Taken together, PERK mediated cyclin D1 translation attenuation, p53 and its downstream cell cycle effectors (such as p21), p53 isoforms p53/p47 and Chk1 modulate cell cycle exit in response to ER stress and ultimately contribute to tumor cell fate.
PERK and tumor angiogenesis
Angiogenesis a process necessary for expanding the vascular network forming and provide the area of adequate oxygen and nutrients, is a crucial step for tumor development. The level of neoplastic angiogenesis reflects the degree of aggressiveness of tumors (Nishida et al., 2006). Anti-angiogenic inhibitors have been developed as potential therapeutic agents, targeting angiogenesis alone does not have long term beneficial for survival (Nishida et al., 2006). Work from a number of groups has linked PERK with tumor angiogenesis. PERK+/+ tumors exhibit microvessel formation from endothelia cells, while PERK−/− tumors have reduced vasculature (Blais et al., 2006). One key factor that may contribute is VCIP (VEGF and type I collagen inducible protein). VCIP plays pivotal roles in VEGF and bFGF induced capillary morphogenesis (Wary and Humtsoe, 2005). Cells under hypoxia express VCIP via translational regulation through 5’UTR internal initiation, which is exclusively dependent upon PERK, highlighting the importance of PERK pathway in tumor angiogenesis under hypoxia condition (Blais et al., 2006).
Remarkably, evidence emerged recently demonstrated that the pro-angiogenesis factor VEGF could induce UPR response via PERK and ATF6 to promote endothelia cell survival and angiogenesis (Karali et al., 2014). Interestingly, in this case, regulation reflects ER stress-independent UPR signaling through the PLCγ and mTORC1. VEGF rapidly activates all three UPR mediators without the displacement BiP from UPR transducers. PLCγ inhibitors, PLCγ deletion and mTORC1 inhibition effectively inhibited PERK, IRE1 and ATF6 activation. It has been proposed that PLCγ may bind to mTOR triggering its translocation to the ER membrane (Karali et al., 2014; Wang and Kaufman, 2014). The UPR signaling induced by VEGF is essential for the endothelia cells maintain the mTORC2 activity and ensues the AKT phosphorylation on Ser473. As a result, the VEGF phosphorylates AKT on Thr308 via PI3KPDK1 pathway, phosphorylates AKT on Ser473 via PLCγ/mTORC1/ATF6 PERK/mTORC2, achieving the maximal activation of AKT. Overall, VEGF induced ATF6 and PERK, but not IRE1 activation, promote endothelia cell survival and ensure its other functions. Of note, the CHOP activation, which represents the proapoptotic aspect of PERK signaling, is undetectable (Karali et al., 2014). Based on this, it is reasonable to speculate other growth factors in the tumor microenvironment could induce cancer cell angiogenesis via UPR. The VEGF triggered UPR signaling extends the knowledge of physiological function of UPR components as well as the stress alleviation function under stress condition for tumor cells.
PERK adapts cancer cells microenvironmental stresses (low glucose-hypoxia)
Nutrients such as glucose, the key 6 carbon sugar, is relatively low in the tumor microenvironment a consequence of poor and disordered vascular structure (Gullino et al., 1967; Hirayama et al., 2009). Cancer cells are highly addicted to glucose utilization, a consequence of the Warburg effect. This effect can be attributed to a shift from oxidative phosphorylation to glycolysis; this shift to glycolysis is now known to provide key substrates for anabolic metabolism while maintaining high energy for cell growth and proliferation (Vander Heiden et al., 2009). In this context, activated PERK can facilitate cancer cell survival under low glucose condition partially by AKT activation and hexokinase II mitochondria translocation (Hou et al., 2015).
In the solid tumor microenvironment tolerance to hypoxia is crucial for tumor cell survival. The oxygen concentration around solid tumor microenvironment and within the tumor proper is variable again a reflection of poor vasculature or dynamic blood flow. Hypoxia triggers reprogramming of metabolic gene expression which in turn reduces oxygen consumption and improves energy utilization. The hypoxic response is largely dependent on HIF-1 and HIF-2 (Ratcliffe, 2007). In addition to HIF mediated hypoxia adaption, the global protein synthesis reduction modulated by UPR is crucial to coordinate the hypoxia tolerance (Koumenis et al., 2007).
Hypoxia induces the PERK-eIF2α axis and loss of PERK compromises tumor cell survival under hypoxia condition in vitro (Koumenis et al., 2002). Additionally, tumor xenograft studies revealed that hypoxia triggers PERK signaling. Dominant negative PERK or eIF2α Ser51A impairs tumor growth in xenograft models and this correlates with increased apoptosis in hypoxic areas of the tumor (Bi et al., 2005). Additional evidence linked UPR responses to protection of tumor cells from hypoxia via induction of MAP1LC3B and ATG5 linking PERK-dependent regulation of autophagy with the hypoxic response (Kouroku et al., 2007; Rouschop et al., 2010). Increased expression of MAP1LC3B and ATG5 is dependent upon ATF4 and CHOP. Direct analysis revealed that both CHOP and ATF4 bind directly the respective promoters, leading to their induction in mRNA level. Collectively, these studies suggest that PERK signaling mediated hypoxia tolerance in tumor development. Targeting PERK pathway may sensitize hypoxia tumor cells to apoptosis.
PERK also stimulates tumor migration and metastasis under hypoxia conditions (Nagelkerke et al., 2013). Tumors under hypoxia condition tend to be more metastatic and malignant, correlated with poor prognosis (Chan and Giaccia, 2007). Whether PERK facilitates hypoxia activated metastasis becomes intriguing. Lysosomal associated membrane protein3 (LAMP3) has been identified greatly induced under hypoxia condition, mediating metastasis in breast cancer, cervix cancer and multiple cancer types (Mujcic et al., 2013; Nagelkerke et al., 2013; Nagelkerke et al., 2014). PERK mediated UPR mediates LAMP3 expression, where knockdown of PERK, ATF4 or overexpression GADD34 all compromised LAMP3 expression under hypoxia condition (Mujcic et al., 2009) suggesting a direct link between PERK and hypoxia stimulated metastatic spread.
Of note, there is increasing evidence suggests that autophagy is pro-survival for cancer cells within the tumor microenvironment (Amaravadi et al., 2011). PERK-dependent reprogramming of autophagic gene expression to promote cancer cell survival, adapts cells to harsh conditions such as hypoxia and oxidative tumor microenvironment, induces vascularization and migration, confers cancer cells aggressiveness and chemoresistance. Taken together, the autophagy induced by PERK is crucial for PERK mediated tumorigenesis.
PERK and redox homoeostasis
In addition to regulation of cytoprotective autophagy, PERK also regulates cell redox homeostasis. In the context of ROS accumulation within tumors, PERK action is clearly adaptive (Bobrovnikova-Marjon et al., 2010). PERK deficiency in the cancer cells results in increased ROS accumulation, a consequence of reduced Nrf2 activation. Nrf2, a direct PERK substrate, maintains the redox homeostasis by regulating expression of the enzymes (eg. NQO1 and GCLC) which produce antioxidant glutathione (Cullinan and Diehl, 2004; Cullinan et al., 2004; Cullinan et al., 2003). Nrf2 activation provides a cytoprotective effects on cells to counteract the ER stress perturbed cellular redox balance at the very beginning. Interestingly, CHOP depletes the glutathione and negatively correlated with Nrf2 expression, suggesting the Nrf2 may not mitigate the prolonged stress and ultimately cells commit to apoptosis (Cullinan and Diehl, 2004).
Regulation of Lipid biogenesis by PERK
Alterations in lipid metabolism is an under appreciated characteristic of tumorigenesis (Santos and Schulze, 2012). Work investigating the impact of PERK on mammary development demonstrated that PERK modulates expression of key lipogenic enzymes such as FAS, ACL and SCD1(Bobrovnikova-Marjon et al., 2008). Analysis of PERK function in the mammary gland revealed PERK-dependent control of lipid and free fatty acid production. With regard to mechanism, sterol regulatory element binding protein (SREBP1c), a major regulator of lipid metabolic genes, is induced and activated in a PERK-eIF2α dependent pathway. SREBP1c activation reflects PERK-dependent translational silencing of Insig1, a membrane anchor that prevents SREBP1 Golgi-translocation and processing (Bobrovnikova-Marjon et al., 2008). As discussed above, PERK possesses lipid kinase activity wherein it generates Phosphatidic acid (PA) through phosphorylation of diacylglycerol (DAG) as substrate (Bobrovnikova-Marjon et al., 2012). PERK lipid kinase activity is dependent on p85α subunit a regulatory subunit of phosphoinosotide kinase 3. PERK-dependent activation of AKT, mTOR and Erk1/2 in response to ER stress is dependent upon PA generation, highlighting the extensive roles of PERK in mitogenic signaling during tumorigenesis.
PERK-dependent regulation of metastasis
Tumor metastasis, a key aspect of the malignant phenotype, remains poorly understood at the molecular level and as such remains beyond current therapeutic intervention modalities. Most of the life threatening cancers experiences the invasive-metastatic cascade. Not surprisingly given its capacity to regulate cell survival in response to microenvironmental stress, PERK, has been implicated in metastatic spread. PERK contributes to metastasis of HER2/Neu adenocarcinoma (Bobrovnikova-Marjon et al., 2010). While the underlying mechanism was not elucidated in this work, work from additional groups have proposed regulation of metastasis through the PERK-eIF2α-ATF4-LAMP3 axis in cervix cancer, breast cancer and head and neck squamous cell carcinoma (Mujcic et al., 2013; Nagelkerke et al., 2013; Nagelkerke et al., 2014).
Sustained PERK-eIF2α-ATF4 activation has been implicated in the Epithelia to mesenchymal (EMT) transition, a process that contributes to tumor progression and metastasis (Feng et al., 2014). Tumor cells acquiring the EMT, characterized as suppression of epithelia markers and upregulation of mesenchymal markers (such as snail and twist), become more invasive and drug resistant. Carcinoma cells with mesenchymal traits generate the properties of cancer stem cells, seeding more efficiently in both the primary and metastatic tumors (Mani et al., 2008). UPR signaling is activated during EMT process (Feng et al., 2014). Specifically, EMT triggers PERK-eIF2α signaling. The correlation between EMT and PERK activation also confirmed in primary breast cancer, colon cancer, gastric cancer, lung cancer as well as metastatic cancers spanning hundreds of clinical samples (Feng et al., 2014).
PERK contributes to chemoresistance
Multi-drug resistance remains an obstacle for chemotherapy. Amplification or mutation of ATP-binding cassette (ABC) transporters is implicated in increasing the efflux of drugs. Additional mechanisms for drug resistance include the upregulation of antioxidant enzymes, which could relieve the cancer cells from ROS produced from the chemotherapeutic chemicals. Nrf2 is a master regulator of antioxidant enzymes and glutathione biosynthetic enzymes (McMahon et al., 2001; Wang et al., 2008). Given that PERK regulates Nrf2, one might anticipate an impact on drug resistance. Indeed, PERK-Nrf2 axis associates with multi-drug resistance in de-differentiation cancer cells (Del Vecchio et al., 2014).
De-differentiation is associated poor prognosis and multi-drug resistance (Del Vecchio et al., 2014). De-differentiated cancer cells have constitutively pre-activated PERK-Nrf2 signaling, which is different from differentiated cells. In differentiated tumor cells, Nrf2 is actviated by oxidative stress through PERK mediated UPR pathway. However, in de-differentiated tumor cells, the PERK-Nrf2 is constitutive and activates a series antioxidant genes expression prior to the onset of oxidative stress, conferring cells with drug resistance. Evidence suggests that inhibition of PERK-Nrf2 could re-sensitize such cells to chemotherapy offering new avenues for therapeutic intervention.
PERK signaling through eIF2α may also contribute to drug and chemoresistance. PERK-eIF2α signaling is enhanced in chronic myeloid leukemia (CML) progression and associated with imatinib resistance. Blocking the PERK pathway impaires the proliferation and cologenic capacities of CML cells and sensitizes them to imatinib-induced apoptosis (Kusio-Kobialka et al., 2012).
Non-coding RNAs contribute to cell fate regulation by PERK
The investigation of small non-coding RNAs, also called microRNAs (miRNAs) has greatly advanced our knowledge of cancer biology. MicroRNAs play fundamental roles in many aspects of cancer development (Price and Chen, 2014) and the contributions of microRNAs to cell fate following ER stress have only recently gained attention. High throughput profiles on ER stress revealed that more than 80 microRNAs are significantly changed under ER stress, some of which contribute to the cell fate decision (Read et al., 2014). For example, miR-7a is upregulated in response to ER inducer thapsigargin and tunicamycin. The in vitro stimulated ischemia also induces both UPR response (confirmed by CHOP activation) and miR-7a expression (Read et al., 2014).
PERK regulates expression of two intronic micro-RNAs, miR-204 and miR-211(Chitnis et al., 2012). MiR-204 and miR-211 share the same seed sequence, although miR-204 is embedded within intronic sequence of the calcium channel trpm3, while miR-211 is embedded within intron 2 of trpm1. Unlike canonic microRNA that function by targeting 3’UTRs in the cytoplasm, miR-211/miR-204 transcriptionally regulate stress-dependent CHOP expression via the 5’UTR and promoter region. Here, miR-211/miR-204 nucleates a repressive complex referred to as RITS (RNA Induced Transcription Silencing) on the 5’UTR of rare CHOP transcripts. MiR-204 and miR-211 complexes contain an Argonaut protein and EZH2. Following miR-mediated binding to nascent transcript (Eulalio et al., 2008), this complex deposits repressive histone marks on promoter chromatin reducing RNA polymerase occupancy and reducing transcription. Intriguingly, the accumulation and turnover of miR-204 and miR-211 are rapid; offering cells a short window to antagonize CHOP mediated prematured apoptosis and restore the cellular homeostasis (Chitnis et al., 2012).
CHOP directly regulates microRNAs, including miR-708, which resides within the host gene Odz4 (Behrman et al., 2011). miR-708 serves as a potential therapeutic agent for metastatic breast cancer as it targets endoplasmic reticulum protein neuronatin to reduce calcium mediated cell migration (Ryu et al., 2013). However, whether PERK-CHOP regulates metastasis via miR-708 is elusive. miR-30c-2* regulates Xbp1 expression, induced by ER stress specifically through PERK pathway. This is another example for microRNA bridging the UPR pathways PERK and IRE1 crosstalk (Byrd et al., 2012). PERK also suppresses micro-RNA expression. miR-106b-25 cluster, as well as its host gene Mcm7, are negatively regulated by PERK-ATF4 and Nrf2 (Poliseno et al., 2010). miR-106b-25 cluster suppression contributes to apoptosis under severe ER stress, required for pro-apoptotic Bim activation (Gupta et al., 2012).
There is increasing attention in Long non-coding RNA (lncRNA) in cancer research, albeit with little knowledge of their function and working mechanisms. LncRNAs, RNA >200 bp nucleotides with no protein coding capacity, have the potential to play crucial roles in chromosome modification, transcription regulation and post-transcriptional modulation (Mercer et al., 2009). Recent work linked ER stress with expression of the lncRNA Malat1 (Bhattacharyya and Vrati, 2015). Flavivirus infection, which triggers UPR signaling in host cells, induces PERK-dependent Malat1 expression. Malat1, originally identified from metastatic cancer cells, has been reported to promote tumor growth and metastasis in multiple cancers through regulating alternative splicing or binding to other transcription factors, such as EZH2 (Yoshimoto et al., 2015). Although the ER stress associated transcription factors for Malat1 and its target genes in the context of ER stress remain to be ascertained, given the neoplastic features of Malat1, we would anticipate that PERK induced lncRNA Malat1 might participate in PERK mediated tumorigenesis. More lncRNAs are waiting to be explored in the context of UPR and carcinogenesis.
Targeting PERK for cancer treatment
Genetic work revealing a prominent role for PERK in tumor progression, angiogenesis and metastasis, stimulated a search for small molecule PERK inhibitors with the hope of therapeutic efficacy. GSK2606414, a first generation of PERK inhibitor, targets PERK in its inactive DFG conformation at the ATP binding site. GSK2606414 inhibits PERK autophosphorylation in vitro and reduces growth of tumor xenograft (Axten et al., 2012). Second generation GSK2656157 showed modest improvement in pharmacological effects. Oral administration of GSK2656157 inhibited multiple tumor xenograft growth in a dose-dependent manner (Atkins et al., 2013; Axten et al., 2013).
The combination use of PERK inhibitor and proteasome inhibitor may also be a promising treatment for multiple cancers. Proteasome inhibitors such as carfilzomib and oprozomib exhibit significant clinical success in patients suffering from multiple myeloma and mantle cell lymphoma (McBride et al., 2015; O'Connor et al., 2005). Mechanistically, the proapoptotic impact of proteasome inhibitors is at least partially UPR-dependent (Dong et al., 2009; Nawrocki et al., 2005; Obeng et al., 2006). However, as PERK has pro-survival activities through ATF4 and Nrf2, PERK inhibition may foster improved clinical response (Obeng et al., 2006; Zang et al., 2012). It is attractive to speculate the combination use of PERK inhibitor could sensitize tumors to the proteasome inhibitors, reducing the dose of the drug and limiting the potential side effects.
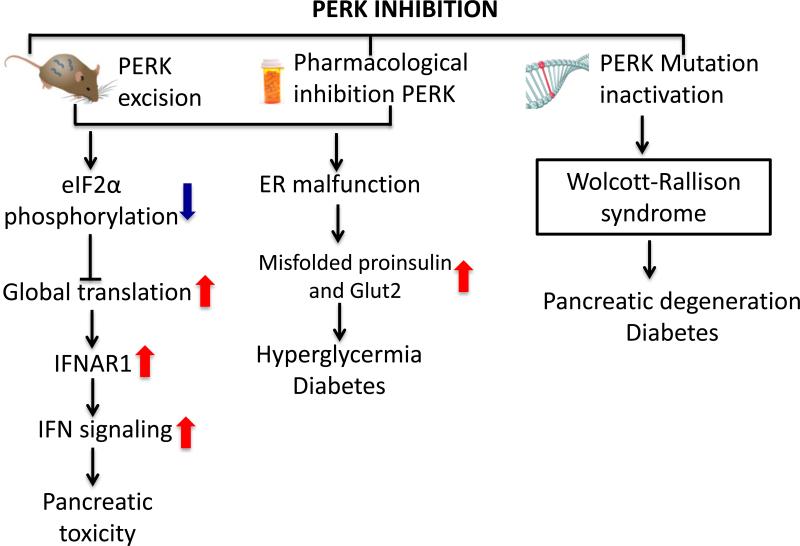
While PERK inhibitors remain an exciting anti-tumor strategy, there are likely dose limiting toxicities associated with pancreatic damage (Figure 3). Genetic ablation of PERK triggers damage to both endocrine and exocrine pancreas resulting in a severe diabetic phenotype (Gao et al., 2012). Importantly, PERK function is required regardless of age. Support for this conclusion stems from eIF2α Ser51/Ala mutant knock-in mice which also exhibit severe glucose intolerance and diabetic phenotypes (Cavener et al., 2010).
Figure 3. Pancreatic toxicity of PERK inhibition.
PERK inhibition by either genetic deletion or pharmacological inhibition causes IFN signaling increase in pancreas and accumulation of misfolded insulin, proinsulin and glucose transporters Glut2, leading to hyperglycermia and diabetes. In human, PERK inactivation mutation results in Wlocott-Rallison syndrome, which is characterized with pancreatic degeneration and diabetes.
While initial work suggested pancreatic damage observed in PERK deficient mice was cell autonomous, recent work has implicated non-cell autonomous IFN signaling in the deleterious effects of the PERK inhibition (Yu et al., 2015). IFNAR1, which encodes the interferon receptor1, is downregulated by PERK signaling. Global PERK deletion or pancreatic specific PERK deletion causes pancreas failure in mice in virtue of the high expression of IFNAR1 and hyperactivation of IFN signaling. Elevated IFNs are associated with pancreatitis and type I diabetes mellitus in human (Vassileva et al., 2003). Importantly, antagonizing the increased IFN receptor activity was suggested as a way to mitigate the PERK inhibitor deleterious effects. Indeed, the anti-IFNAR1 antibody relieved the pancreatic toxicity, partially rescued the pancreas mass and stabilized glucose homeostasis in mice treated with PERK inhibitor GSK2606414 (Yu et al., 2015). A better understanding of the mechanisms of PERK inhibitor side effects is needed to develop the therapeutic strategy.
Concluding remarks
Great progress has been made in the past two decades to enhance our understanding of PERK contributions to cell fate and tumorigenesis. The contribution of PERK in cancer development is complicated, providing both the pro-survival and also pro-apoptotic signaling. It is not clear what dictates PERK pro-apoptotic signaling from pro-survival signaling. PERK function is dependent on tissue types and the microenvironment; for example, PERK excision did not affect the proliferation of mammary epithelia cells, whereas it potentiates tumor cell expansion (Bobrovnikova-Marjon et al., 2010). Moreover, PERK function differs greatly in distinct tumor stages. For example, PERK serves as a barrier for HRasG12V driven melanoma initiation (Denoyelle et al., 2006), but promotes cytoprotective autophagy in the melanoma progression and chemoresistance (Ma et al., 2014). The amount of active PERK may also determine inclusion of specific the downstream effectors and output. Furthermore, the crosstalk among PERK, IRE1 and ATF6 also comprehensively integrates the stress signaling and the output is dependent on the coordination among these three downstream pathways and their effectors.
Current evidence supports a model wherein PERK contributes to genomic instability, adaption to tumor microenvironment, aggressiveness and chemoresistance. For these reasons, PERK mediated cancer development integrates signaling networks and involved in multiple physiological functions. PERK inhibitors exhibit the potential in the anti-tumor therapies, but the minimization of side effects has to be further delineated. A deeper understand of how to combine PERK inhibitors with other drugs to compensate the pancreatic cytotoxicity is fundamental important for a viable cancer therapy.
Acknowledgements
This work was supported by National Institutes of Health grants P01 CA104838 (JAD).
References
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins C, Liu Q, Minthorn E, Zhang SY, Figueroa DJ, Moss K, Stanley TB, Sanders B, Goetz A, Gaul N, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer research. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, et al. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). Journal of medicinal chemistry. 2012;55:7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- Axten JM, Romeril SP, Shu A, Ralph J, Medina JR, Feng Y, Li WH, Grant SW, Heerding DA, Minthorn E, et al. Discovery of GSK2656157: An Optimized PERK Inhibitor Selected for Preclinical Development. ACS medicinal chemistry letters. 2013;4:964–968. doi: 10.1021/ml400228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B'Chir W, Maurin AC, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic acids research. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman S, Acosta-Alvear D, Walter P. A CHOP-regulated microRNA controls rhodopsin expression. The Journal of cell biology. 2011;192:919–927. doi: 10.1083/jcb.201010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Wang X, Novoa I, Jungreis R, Schlessinger K, Cho JH, West AB, Ron D. Increased sensitivity to dextran sodium sulfate colitis in IRE1beta-deficient mice. The Journal of clinical investigation. 2001;107:585–593. doi: 10.1172/JCI11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Vrati S. The Malat1 long non-coding RNA is upregulated by signalling through the PERK axis of unfolded protein response during flavivirus infection. Scientific reports. 2015;5:17794. doi: 10.1038/srep17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi M, Naczki C, Koritzinsky M, Fels D, Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. The EMBO journal. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K, Koumenis C, Harding HP, Ron D, Holcik M, et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Molecular and cellular biology. 2006;26:9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, Diehl JA. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29:3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, Romero M, Cavener DR, Thompson CB, Diehl JA. PERK-dependent regulation of lipogenesis during mouse mammary gland development and adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Pytel D, Riese MJ, Vaites LP, Singh N, Koretzky GA, Witze ES, Diehl JA. PERK utilizes intrinsic lipid kinase activity to generate phosphatidic acid, mediate Akt activation, and promote adipocyte differentiation. Molecular and cellular biology. 2012;32:2268–2278. doi: 10.1128/MCB.00063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias MM, Marullo S, Fahraeus R. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Molecular cell. 2010;38:78–88. doi: 10.1016/j.molcel.2010.01.041. [DOI] [PubMed] [Google Scholar]
- Brewer JW, Diehl JA. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12625–12630. doi: 10.1073/pnas.220247197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:8505–8510. doi: 10.1073/pnas.96.15.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruntz RC, Lindsley CW, Brown HA. Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacological reviews. 2014;66:1033–1079. doi: 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd AE, Aragon IV, Brewer JW. MicroRNA-30c-2* limits expression of proadaptive factor XBP1 in the unfolded protein response. The Journal of cell biology. 2012;196:689–698. doi: 10.1083/jcb.201201077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener DR, Gupta S, McGrath BC. PERK in beta cell biology and insulin biogenesis. Trends in endocrinology and metabolism: TEM. 2010;21:714–721. doi: 10.1016/j.tem.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DA, Giaccia AJ. Hypoxia, gene expression, and metastasis. Cancer metastasis reviews. 2007;26:333–339. doi: 10.1007/s10555-007-9063-1. [DOI] [PubMed] [Google Scholar]
- Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer discovery. 2015;5:586–597. doi: 10.1158/2159-8290.CD-14-1490. [DOI] [PubMed] [Google Scholar]
- Chitnis NS, Pytel D, Bobrovnikova-Marjon E, Pant D, Zheng H, Maas NL, Frederick B, Kushner JA, Chodosh LA, Koumenis C, et al. miR-211 is a prosurvival microRNA that regulates chop expression in a PERK-dependent manner. Molecular cell. 2012;48:353–364. doi: 10.1016/j.molcel.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corazzari M, Rapino F, Ciccosanti F, Giglio P, Antonioli M, Conti B, Fimia GM, Lovat PE, Piacentini M. Oncogenic BRAF induces chronic ER stress condition resulting in increased basal autophagy and apoptotic resistance of cutaneous melanoma. Cell death and differentiation. 2014 doi: 10.1038/cdd.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Diehl JA. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following endoplasmic reticulum stress. The Journal of biological chemistry. 2004;279:20108–20117. doi: 10.1074/jbc.M314219200. [DOI] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Molecular and cellular biology. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Molecular and cellular biology. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Vecchio CA, Feng Y, Sokol ES, Tillman EJ, Sanduja S, Reinhardt F, Gupta PB. De-Differentiation Confers Multidrug Resistance Via Noncanonical PERK-Nrf2 Signaling. PLoS Biol. 2014;12:e1001945. doi: 10.1371/journal.pbio.1001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, Pointer JN, Gruber SB, Su LD, Nikiforov MA, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nature cell biology. 2006;8:1053–1063. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- Dong H, Chen L, Chen X, Gu H, Gao G, Gao Y, Dong B. Dysregulation of unfolded protein response partially underlies proapoptotic activity of bortezomib in multiple myeloma cells. Leukemia & lymphoma. 2009;50:974–984. doi: 10.1080/10428190902895780. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Feng YX, Sokol ES, Del Vecchio CA, Sanduja S, Claessen JH, Proia TA, Jin DX, Reinhardt F, Ploegh HL, Wang Q, et al. Epithelial-to-mesenchymal transition activates PERK-eIF2alpha and sensitizes cells to endoplasmic reticulum stress. Cancer discovery. 2014;4:702–715. doi: 10.1158/2159-8290.CD-13-0945. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Sartori DJ, Li C, Yu QC, Kushner JA, Simon MC, Diehl JA. PERK is required in the adult pancreas and is essential for maintenance of glucose homeostasis. Molecular and cellular biology. 2012;32:5129–5139. doi: 10.1128/MCB.01009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nature cell biology. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- Gullino PM, Grantham FH, Courtney AH. Glucose consumption by transplanted tumors in vivo. Cancer research. 1967;27:1031–1040. [PubMed] [Google Scholar]
- Gupta S, Read DE, Deepti A, Cawley K, Gupta A, Oommen D, Verfaillie T, Matus S, Smith MA, Mott JL, et al. Perk-dependent repression of miR-106b-25 cluster is required for ER stress-induced apoptosis. Cell death & disease. 2012;3:e333. doi: 10.1038/cddis.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Bennett BS, Cullinan SB, Diehl JA. PERK and GCN2 contribute to eIF2alpha phosphorylation and cell cycle arrest after activation of the unfolded protein response pathway. Molecular biology of the cell. 2005;16:5493–5501. doi: 10.1091/mbc.E05-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Bobrovnikova-Marjon E, Ji X, Liebhaber SA, Diehl JA. PERK-dependent regulation of IAP translation during ER stress. Oncogene. 2009;28:910–920. doi: 10.1038/onc.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nature reviews Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- Hart LS, Cunningham JT, Datta T, Dey S, Tameire F, Lehman SL, Qiu B, Zhang H, Cerniglia G, Bi M, et al. ER stress-mediated autophagy promotes Myc-dependent transformation and tumor growth. The Journal of clinical investigation. 2012;122:4621–4634. doi: 10.1172/JCI62973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Molecular biology of the cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiological reviews. 2011;91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- Hirayama A, Kami K, Sugimoto M, Sugawara M, Toki N, Onozuka H, Kinoshita T, Saito N, Ochiai A, Tomita M, et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer research. 2009;69:4918–4925. doi: 10.1158/0008-5472.CAN-08-4806. [DOI] [PubMed] [Google Scholar]
- Hou X, Liu Y, Liu H, Chen X, Liu M, Che H, Guo F, Wang C, Zhang D, Wu J, et al. PERK silence inhibits glioma cell growth under low glucose stress by blockage of p-AKT and subsequent HK2's mitochondria translocation. Scientific reports. 2015;5:9065. doi: 10.1038/srep09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali E, Bellou S, Stellas D, Klinakis A, Murphy C, Fotsis T. VEGF Signals through ATF6 and PERK to promote endothelial cell survival and angiogenesis in the absence of ER stress. Molecular cell. 2014;54:559–572. doi: 10.1016/j.molcel.2014.03.022. [DOI] [PubMed] [Google Scholar]
- Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nature reviews Drug discovery. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Bi M, Ye J, Feldman D, Koong AC. Hypoxia and the unfolded protein response. Methods in enzymology. 2007;435:275–293. doi: 10.1016/S0076-6879(07)35014-3. [DOI] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Molecular and cellular biology. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell death and differentiation. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- Kusio-Kobialka M, Podszywalow-Bartnicka P, Peidis P, Glodkowska-Mrowka E, Wolanin K, Leszak G, Seferynska I, Stoklosa T, Koromilas AE, Piwocka K. The PERK-eIF2alpha phosphorylation arm is a pro-survival pathway of BCR-ABL signaling and confers resistance to imatinib treatment in chronic myeloid leukemia cells. Cell cycle (Georgetown, Tex) 2012;11:4069–4078. doi: 10.4161/cc.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee B, Lee AS. Endoplasmic reticulum stress-induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. The Journal of biological chemistry. 2006;281:7260–7270. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annual review of pharmacology and toxicology. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G, et al. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. The Journal of clinical investigation. 2014;124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hendershot LM. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. The Journal of biological chemistry. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino MB, Jones L, Brighton B, Ehre C, Abdulah L, Davis CW, Ron D, O'Neal WK, Ribeiro CM. The ER stress transducer IRE1beta is required for airway epithelial mucin production. Mucosal immunology. 2013;6:639–654. doi: 10.1038/mi.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride A, Klaus JO, Stockerl-Goldstein K. Carfilzomib: a second-generation proteasome inhibitor for the treatment of multiple myeloma. American journal of health-system pharmacy : AJHP : official journal of the American Society of Health-System Pharmacists. 2015;72:353–360. doi: 10.2146/ajhp130281. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer research. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mujcic H, Nagelkerke A, Rouschop KM, Chung S, Chaudary N, Span PN, Clarke B, Milosevic M, Sykes J, Hill RP, et al. Hypoxic activation of the PERK/eIF2alpha arm of the unfolded protein response promotes metastasis through induction of LAMP3. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6126–6137. doi: 10.1158/1078-0432.CCR-13-0526. [DOI] [PubMed] [Google Scholar]
- Mujcic H, Rzymski T, Rouschop KM, Koritzinsky M, Milani M, Harris AL, Wouters BG. Hypoxic activation of the unfolded protein response (UPR) induces expression of the metastasis-associated gene LAMP3. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2009;92:450–459. doi: 10.1016/j.radonc.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Nagelkerke A, Bussink J, Mujcic H, Wouters BG, Lehmann S, Sweep FC, Span PN. Hypoxia stimulates migration of breast cancer cells via the PERK/ATF4/LAMP3-arm of the unfolded protein response. Breast cancer research : BCR. 2013;15:R2. doi: 10.1186/bcr3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagelkerke A, Sweep FC, Stegeman H, Grenman R, Kaanders JH, Bussink J, Span PN. Hypoxic regulation of the PERK/ATF4/LAMP3-arm of the unfolded protein response in head and neck squamous cell carcinoma. Head & neck. 2014 doi: 10.1002/hed.23693. [DOI] [PubMed] [Google Scholar]
- Nawrocki ST, Carew JS, Dunner K, Jr., Boise LH, Chiao PJ, Huang P, Abbruzzese JL, McConkey DJ. Bortezomib inhibits PKR-like endoplasmic reticulum (ER) kinase and induces apoptosis via ER stress in human pancreatic cancer cells. Cancer research. 2005;65:11510–11519. doi: 10.1158/0008-5472.CAN-05-2394. [DOI] [PubMed] [Google Scholar]
- Nishida N, Yano H, Nishida T, Kamura T, Kojiro M. Angiogenesis in cancer. Vascular health and risk management. 2006;2:213–219. doi: 10.2147/vhrm.2006.2.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, Inoue K, Hori S, Kakizuka A, Ichijo H. ASK1 is essential for endoplasmic reticulum stress-induced neuronal cell death triggered by expanded polyglutamine repeats. Genes & development. 2002;16:1345–1355. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor OA, Wright J, Moskowitz C, Muzzy J, MacGregor-Cortelli B, Stubblefield M, Straus D, Portlock C, Hamlin P, Choi E, et al. Phase II clinical experience with the novel proteasome inhibitor bortezomib in patients with indolent non-Hodgkin's lymphoma and mantle cell lymphoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:676–684. doi: 10.1200/JCO.2005.02.050. [DOI] [PubMed] [Google Scholar]
- Obeng EA, Carlson LM, Gutman DM, Harrington WJ, Jr., Lee KP, Boise LH. Proteasome inhibitors induce a terminal unfolded protein response in multiple myeloma cells. Blood. 2006;107:4907–4916. doi: 10.1182/blood-2005-08-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluquet O, Qu LK, Baltzis D, Koromilas AE. Endoplasmic reticulum stress accelerates p53 degradation by the cooperative actions of Hdm2 and glycogen synthase kinase 3beta. Molecular and cellular biology. 2005;25:9392–9405. doi: 10.1128/MCB.25.21.9392-9405.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nature reviews Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Science signaling. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, Moses TY, Hostetter G, Wagner U, Kakareka J, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- Price C, Chen J. MicroRNAs in Cancer Biology and Therapy: Current Status and Perspectives. Genes & Diseases. 2014;1:53–63. doi: 10.1016/j.gendis.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pytel D, Majsterek I, Diehl JA. Tumor progression and the different faces of the PERK kinase. Oncogene. 2015 doi: 10.1038/onc.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, Koumenis C, Taya Y, Yoshimura A, Koromilas AE. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes & development. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe PJ. HIF-1 and HIF-2: working alone or together in hypoxia? The Journal of clinical investigation. 2007;117:862–865. doi: 10.1172/JCI31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read DE, Gupta A, Ladilov Y, Samali A, Gupta S. miRNA signature of unfolded protein response in H9c2 rat cardiomyoblasts. Cell & bioscience. 2014;4:56. doi: 10.1186/2045-3701-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Hubbard SR. How IRE1 reacts to ER stress. Cell. 2008;132:24–26. doi: 10.1016/j.cell.2007.12.017. [DOI] [PubMed] [Google Scholar]
- Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. The Journal of clinical investigation. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, McDonnell K, Choi H, Gao D, Hahn M, Joshi N, Park SM, Catena R, Do Y, Brazin J, et al. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer cell. 2013;23:63–76. doi: 10.1016/j.ccr.2012.11.019. [DOI] [PubMed] [Google Scholar]
- Santos CR, Schulze A. Lipid metabolism in cancer. The FEBS journal. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- Schewe DM, Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10519–10524. doi: 10.1073/pnas.0800939105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO reports. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SE, Malzer E, Ordonez A, Dalton LE, van 't Wout EF, Liniker E, Crowther DC, Lomas DA, Marciniak SJ. p53 and translation attenuation regulate distinct cell cycle checkpoints during endoplasmic reticulum (ER) stress. The Journal of biological chemistry. 2013;288:7606–7617. doi: 10.1074/jbc.M112.424655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileva G, Chen SC, Zeng M, Abbondanzo S, Jensen K, Gorman D, Baroudy BM, Jiang Y, Murgolo N, Lira SA. Expression of a novel murine type I IFN in the pancreatic islets induces diabetes in mice. Journal of immunology (Baltimore, Md : 1950) 2003;170:5748–5755. doi: 10.4049/jimmunol.170.11.5748. [DOI] [PubMed] [Google Scholar]
- Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nature reviews Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- Wang XJ, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, Chen W, Yi X, Zheng W, Wondrak GT, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. The EMBO journal. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wary KK, Humtsoe JO. Anti-lipid phosphate phosphohydrolase-3 (LPP3) antibody inhibits bFGF- and VEGF-induced capillary morphogenesis of endothelial cells. Cell communication and signaling : CCS. 2005;3:9. doi: 10.1186/1478-811X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaman I, Fernandez J, Liu H, Caprara M, Komar AA, Koromilas AE, Zhou L, Snider MD, Scheuner D, Kaufman RJ, et al. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Yoshimoto R, Mayeda A, Yoshida M, Nakagawa S. MALAT1 long non-coding RNA in cancer. Biochimica et biophysica acta. 2015 doi: 10.1016/j.bbagrm.2015.09.012. [DOI] [PubMed] [Google Scholar]
- Yu Q, Zhao B, Gui J, Katlinski KV, Brice A, Gao Y, Li C, Kushner JA, Koumenis C, Diehl JA, et al. Type I interferons mediate pancreatic toxicities of PERK inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:15420–15425. doi: 10.1073/pnas.1516362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Thomas SM, Chan ET, Kirk CJ, Freilino ML, DeLancey HM, Grandis JR, Li C, Johnson DE. The next generation proteasome inhibitors carfilzomib and oprozomib activate prosurvival autophagy via induction of the unfolded protein response and ATF4. Autophagy. 2012;8:1873–1874. doi: 10.4161/auto.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. The Journal of biological chemistry. 2006;281:30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- Zhang W, Hietakangas V, Wee S, Lim SC, Gunaratne J, Cohen SM. ER stress potentiates insulin resistance through PERK-mediated FOXO phosphorylation. Genes & development. 2013;27:441–449. doi: 10.1101/gad.201731.112. [DOI] [PMC free article] [PubMed] [Google Scholar]