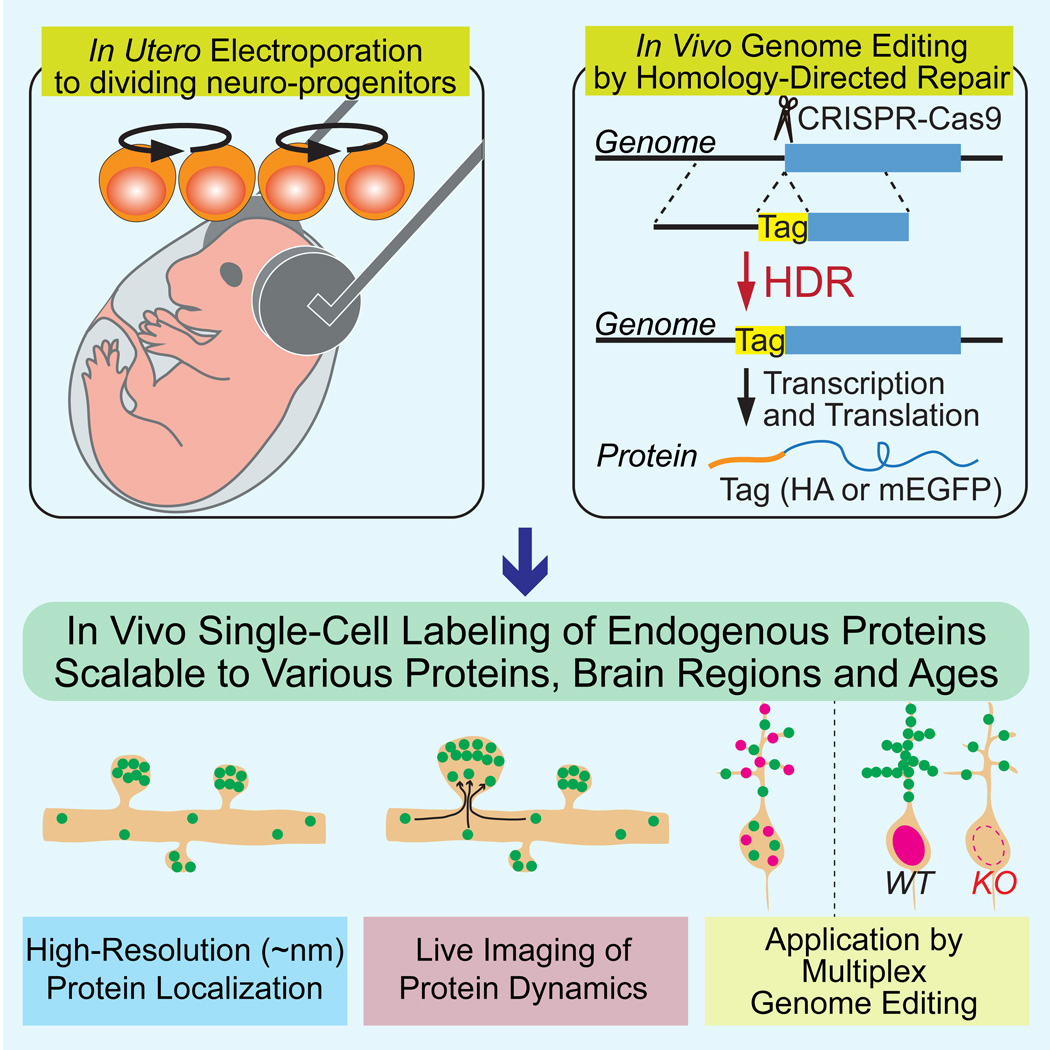
SUMMARY
A scalable and high-throughput method to identify precise subcellular localization of endogenous proteins is essential for integrative understanding of a cell at the molecular level. Here, we developed a simple and generalizable technique to image endogenous proteins with high specificity, resolution and contrast in single cells in mammalian brain tissue. The technique, termed SLENDR, uses in vivo genome editing to insert a sequence encoding an epitope tag or a fluorescent protein to a gene of interest by CRISPR-Cas9-mediated homology-directed repair (HDR). Single-cell, HDR-mediated genome editing was achieved by delivering the editing machinery to dividing neuronal progenitors through in utero electroporation. We demonstrate that SLENDR allows rapid determination of the localization and dynamics of many endogenous proteins in various cell types, regions and ages in the brain. Thus, SLENDR provides a high-throughput platform to map the subcellular localization of endogenous proteins with the resolution of micro- to nanometers in the brain.
Graphical abstract
INTRODUCTION
Precise mapping of a large number of proteins with subcellular resolution is essential to understand cellular processes. Thus, it is critical to develop a rapid and scalable method to determine the localization of proteins with high specificity, resolution and contrast. Conventionally, either immunostaining of endogenous proteins or overexpression of proteins fused with epitope tags or fluorescent proteins have been used to determine protein localization. These methods, however, have significant problems: immunostaining often suffers from the lack of specific antibodies against a protein of interest and the cross-reaction of antibodies with non-targeted proteins; overexpression often causes mistargeting of the expressed protein and potential changes in cell function. To address some of these issues, knock-in mice in which a specific protein is tagged with an epitope tag or fluorescent protein can be used (Yang et al., 2009). However, in dense tissue, such as mammalian brain, it is difficult to obtain images with high contrast in subcellular processes when all cells are labeled. To overcome these problems, several methods have been recently developed for single-cell labeling of endogenous proteins by using recombinant antibody-like proteins or a conditional tag knock-in strategy (Fortin et al., 2014; Gross et al., 2013). However, none of these techniques provides rapid, scalable and high-throughput readouts for the localization of endogenous proteins.
Direct, single-cell manipulation of the genome in vivo to insert a tag sequence to a gene of interest would overcome these limitations, providing rapid, specific and sparse labeling of the gene product. Genome editing based on the clustered regularly interspaced short palindromic repeats (CRISPR)-associated endonuclease Cas9 enables rapid and efficient modification of the genome (Cong et al., 2013; Doudna and Charpentier, 2014; Hsu et al., 2014; Jinek et al., 2012; Sander and Joung, 2014; Wang et al., 2013; Yang et al., 2013). CRISPR-Cas9 induces targeted DNA double-strand breaks in the genome, which are then repaired through either non-homologous end-joining (NHEJ) or homology-directed repair (HDR) pathways (Cox et al., 2015; Doudna and Charpentier, 2014; Hsu et al., 2014; Sander and Joung, 2014; Yang et al., 2013). Although introducing frame-shift knockout mutations through NHEJ at the single-cell level has been established (Straub et al., 2014; Swiech et al., 2015), targeted insertion of a sequence through HDR has not been possible in the mammalian brain in vivo (Heidenreich and Zhang, 2016; Platt et al., 2014; Xue et al., 2014; Yin et al., 2014). This is due to the lack of homologous recombination activity in postmitotic cells and the inefficient delivery of HDR machinery to target cells (Chu et al., 2015; Cox et al., 2015; Heidenreich and Zhang, 2016; Hsu et al., 2014; Maruyama et al., 2015; Saleh-Gohari and Helleday, 2004).
Here, we developed SLENDR (single-cell labeling of endogenous proteins by CRISPR-Cas9-mediated homology-directed repair), a technique that allows HDR-mediated genome editing in the mammalian brain in vivo. Since HDR is known to predominantly occur in the S and G2 phases of the cell cycle (Chu et al., 2015; Heidenreich and Zhang, 2016; Hsu et al., 2014; Maruyama et al., 2015; Saleh-Gohari and Helleday, 2004), we targeted mitotic neuronal progenitors, which presumably have homologous recombination activity. We introduced CRISPR-Cas9-based HDR machinery into progenitor cells in the embryonic mouse brain several days before their final neurogenic divisions using in utero electroporation (IUE) (Nishiyama et al., 2012; Tabata and Nakajima, 2001). We demonstrate that a tag sequence for a short epitope or a longer fluorescent protein can be rapidly and precisely inserted into an endogenous gene of interest in vivo. This method is scalable to many species of proteins in diverse cell types, and permits high resolution imaging with light and electron microscopy both in fixed and live tissue. Thus, SLENDR allows researchers to rapidly and precisely determine the localization and dynamics of endogenous proteins with the resolution of micro- to nanometers in various cell types, regions and ages of the brain, providing a powerful tool suitable for large-scale analysis on a broad spectrum of proteins.
RESULTS
In Vivo Single-Cell Labeling of Endogenous Proteins by Homology-Directed Repair
In order to label a specific protein with a tag in non-dividing brain cells by HDR-mediated gene editing, it is necessary to introduce HDR machinery into progenitor cells before their final cell division. To test whether this strategy, termed SLENDR, can provide the efficiency sufficient for imaging subcellular localization of a protein of interest, we first aimed to insert the HA tag into endogenous CaMKIIα and CaMKIIβ, two major subunits of the dodecameric Ca2+/CaM-dependent kinase II (CaMKII) necessary for neuronal plasticity (Kim et al., 2015; Lee et al., 2009; Lisman et al., 2012).
We designed specific single-guide RNAs (sgRNAs) targeting the vicinity of the stop and start codon of CaMKIIα and CaMKIIβ, respectively. We also designed corresponding single-stranded oligodeoxynucleotides (ssODNs) (~200 bases) to integrate the HA tag sequence into the genome just upstream of the stop codon of CaMKIIα and downstream of the start codon of CaMKIIβ (Figures 1A and 1C). To minimize the possibility of gene knockdown in cells where CRISPR-Cas9-mediated DNA double-strand breaks were repaired through the NHEJ pathway, we selected the target sequences throughout this study so that the CRISPR-Cas9 cleavage sites were located either in the non-coding region upstream of the start codon or within 10 bp from the stop codon. We also selected the target sequences and designed ssODNs so that Cas9 could not recognize the loci after HDR was completed (see Supplemental Information). We introduced these constructs (S. pyogenes Cas9 or SpCas9 and sgRNA expressing vectors and ssODNs) together with hyperactive piggyBac transposase and piggyBac transposon vectors expressing monomeric EGFP (mEGFP) as a marker of transfection (Chen and LoTurco, 2012; Yusa et al., 2011) to neuro-progenitor cells using IUE to target pyramidal neurons in the cerebral cortex. The transposon system induces genomic integration of transgenes, preventing the dilution of mEGFP during cell divisions. Following IUE at embryonic day 12 (E12), we performed immunostaining of brain slices at postnatal days 14–48 (P14–48) using anti-HA antibody together with anti-NeuN antibody to label neurons.
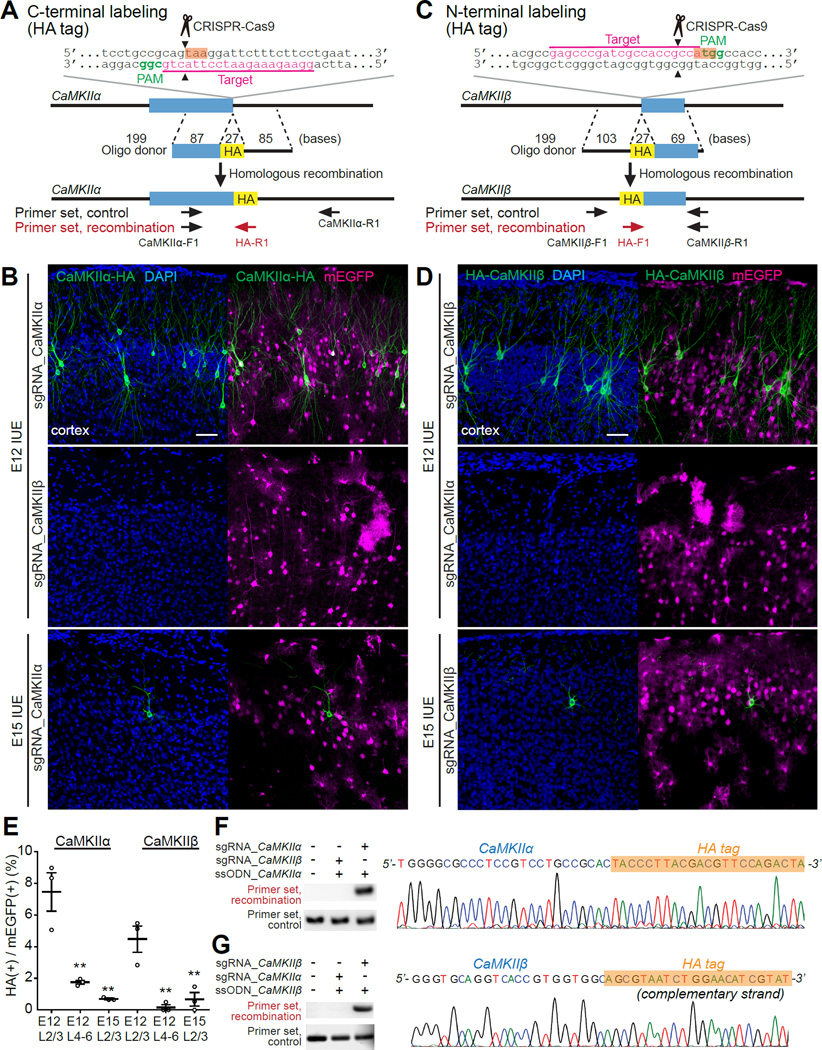
Figure 1. In Vivo Single-Cell Labeling of Endogenous Proteins by SLENDR.
(A and C) Graphical representation of the mouse genomic loci of CaMKIIα (A) and CaMKIIβ (C) showing the target sites for Cas9, sgRNA and ssODNs. The sgRNA targeting sequences are underlined and labeled in magenta. The protospacer-adjacent motif (PAM) sequences are labeled in green. The stop and start codons of CaMKIIα (A) and CaMKIIβ (C), respectively, are marked in orange. The Cas9 cleavage sites are indicated by the black arrowheads. PCR primer sets (control and recombination) for PCR genotyping (E and F) are indicated by the arrows. (B and D) Confocal microscopic images of the cerebral cortex electroporated at E12 (top and middle) and E15 (bottom) showing the DAPI signal (blue) and immunoreactivities for mEGFP (magenta) and the HA tag (green) fused to the C-terminus of endogenous CaMKIIα (B) and N-terminus of endogenous CaMKIIβ (D). Middle panels show negative control experiments in which the sgRNA for CaMKIIα was paired with the ssODNs for CaMKIIβ (B) and vice versa (D).
(E) The efficiency of SLENDR for CaMKIIα and CaMKIIβ (the ratio of the number of HA/mEGFP double-positive neurons to that of mEGFP positive neurons). CaMKIIα, E12 layer (L) 2/3, n = 545 neurons/3 mice; E12 L4–6, n = 275/3; E15 L2/3, n = 713/3. CaMKIIβ, E12 L2/3, n = 716/3; E12 L4-6, n = 379/3; E15 L2/3, n = 367/3. **p < 0.01, Dunnett test, in comparison with E12 L2/3.
(F and G) (left) PCR genotyping using genomic DNA extracted from the electroporated brain. Recombination primer sets (top; F, CaMKIIα-F1 and HA-R1; G, HA-F1 and CaMKIIβ-R1) and control primer sets (bottom; F, CaMKIIα-F1 and CaMKIIα-R1; G, CaMKIIβ-F1 and CaMKIIβ-R1) were used for PCR. (right) DNA sequencing analysis of the PCR products for CaMKIIα-HA (F) and HA-CaMKIIβ (G). The HA tag sequence is marked in orange.
Data are represented as mean ± SEM.
Scale bars, 50 µm.
See also Figure S1 and Tables S1–S3.
In the stained slices, HA signals were observed in a sparse subset of neurons, suggesting that HDR was successfully induced in these cells. Immunofluorescence signal was localized mostly in cytosol and excluded from the nucleus, consistent with previously reported distribution of CaMKII (Lee et al., 2009). Most of HA positive neurons were found in layer 2/3 (Figures 1B, 1D and 1E). Among neurons in transfected area in layer 2/3, about a half of population was mEGFP positive (mEGFP/NeuN: CaMKIIα, 48.2 ± 6.7 %; CaMKIIβ, 40.0 ± 2.9 %). Among these mEGFP positive cells, a small population of the cells were found to be HA positive (HA/mEGFP: CaMKIIα, 7.5 ± 1.2 %; CaMKIIβ, 4.5 ± 0.8 %), providing a few percent of overall knock-in efficiency (HA/NeuN: CaMKIIα, 3.4 ± 0.2 %; CaMKIIβ, 1.8 ± 0.5 %). We also found a smaller population of HA positive neurons in layer 4–6 (HA/mEGFP: CaMKIIα, 1.8 ± 0.1 %; CaMKIIβ, 0.2 ± 0.1 %) (Figures 1B, 1D and 1E). When IUE was performed at E15, a smaller population of layer 2/3 neurons were labeled with the HA tag (HA/mEGFP: CaMKIIα, 0.7 ± 0.1 %; CaMKIIβ, 0.7 ± 0.4 %, HA/NeuN: CaMKIIα, 0.1 ± 0.0 %; CaMKIIβ, 0.2 ± 0.1 %) (Figures 1B, 1D and 1E and Tables S1 and S2). Given that the superficial cortical layers (layer 2/3) are populated with later-born neurons (Chen and LoTurco, 2012), these findings suggest that SLENDR is more efficient when IUE is performed several days before the final neurogenic divisions of targeted cells.
To test the specificity of the construct, we used incorrect ssODNs-sgRNA pairs, like CaMKIIα ssODNs-CaMKIIβ sgRNA or vice versa. Under these conditions, no fluorescence signal was detected in brain slices stained with anti-HA antibody (Figures 1B and 1D). These results indicate that the genome editing is specific to the sequence of the sgRNA. We further confirmed that the expected genome editing occurred at the DNA level by performing PCR amplification of the targeted locus. Electrophoretic analysis revealed the presence of recombined PCR product at a size consistent with the recombined allele in brains transfected with the constructs necessary for HDR (Figures 1F and 1G). In contrast, neither untransfected control brains nor brains transfected with incorrect ssODNs-sgRNA pairs showed the corresponding PCR amplification (Figures 1F and 1G). Furthermore, DNA sequencing of the amplified PCR products indicated that the HA tag sequence was integrated as expected (Figures 1F and 1G). In addition, sgRNAs targeting different sequences in the respective genes produced a similar pattern of HA staining (Figure S1), demonstrating flexibility of construct design. Thus, SLENDR enables specific single-cell labeling of endogenous proteins by either N- or C-terminal epitope tagging.
SLENDR is Scalable to Various Endogenous Proteins
To validate the applicability of SLENDR to visualize the subcellular localization of a broad spectrum of endogenous proteins in brain tissue, we performed SLENDR to fuse the HA tag to either the N- or C-terminus of a variety of proteins including nuclear, cytoskeletal, vesicular, cytosolic and membrane proteins (Figures 2A–2R and S2–S5 and Table S1).
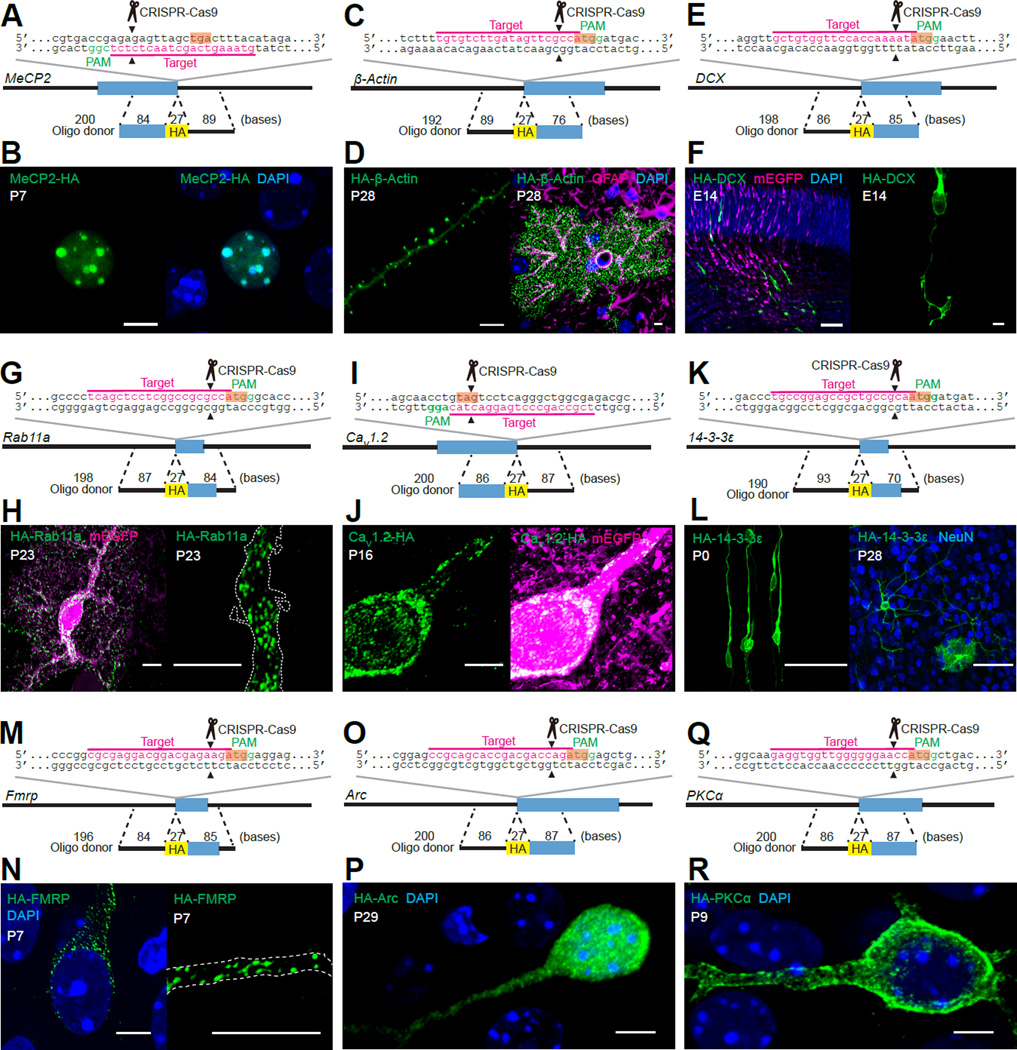
Figure 2. SLENDR is Scalable to Various Endogenous Proteins.
(A, C, E, G, I, K, M, O and Q) Graphical representation of the mouse genomic loci of MeCP2 (A), β-Actin (C), DCX (E), Rab11a (G), CaV1.2 (I), 14-3-3ε (K), Fmrp (M), Arc (O) and PKCα (Q) showing the target sites for Cas9, sgRNA and ssODNs. The sgRNA targeting sequences are underlined and labeled in magenta. The PAM sequences are labeled in green. The stop (A and I) and start (C, E, G, K, M, O and Q) codons are marked in orange. The Cas9 cleavage sites are indicated by the black arrowheads.
(B, F, H, J, N, P and R) Confocal microscopic images of the cerebral cortex showing the DAPI signal (blue) and immunoreactivities for mEGFP (magenta) and the HA tag (green) fused to the C-terminus of MeCP2 (B) and CaV1.2 (J) and the N-terminus of DCX (F), Rab11a (H), FMRP (N), Arc (P) and PKCα (R). The dashed line represents the shape of the dendrite (H and N). (D) Images of the cerebral cortex showing the DAPI signal (blue) and immunoreactivities for GFAP (magenta, an astrocyte marker) and the HA tag (green) fused to the N-terminus of β-Actin. (L) Images of the cerebral cortex at P0 and P28 showing immunoreactivities for NeuN (blue, a neuron marker) and the HA tag (green) fused to the N-terminus of 14-3-3 ε.
Scale bars, 5 µm (B, D, F, right, H, J, N, P and R); 50 µm (F, left; L).
See also Figures S2–S5 and Tables S1 and S3.
We first fused the HA tag to MeCP2, a chromatin-associated protein that regulates gene transcription (Chen et al., 2001). MeCP2 was selected for the following reasons. First, the endogenous subcellular localization can be imaged by traditional immunostaining approaches using a specific, characterized antibody. Second, MeCP2 is known to accumulate in heterochromatin in the nucleus, allowing for the contrast needed for staining of the endogenous protein in tissue. Therefore, double-immunostaining for the SLENDR inserted HA tag and endogenous MeCP2 enabled further validation of the SLENDR approach. As expected, we found that MeCP2-HA and HA-MeCP2 were distributed exclusively in the nucleus at P7 (Figures 2A, 2B, S4B and S4C). In addition, MeCP2-HA was accumulated in the heterochromatin regions (labeled with DAPI) (Figure 2B) and well co-localized with endogenous MeCP2 (Figure S4A), confirming the specificity of SLENDR. We also evaluated potential effects of SLENDR in transfected cells that did not undergo HDR. Since many mEGFP-positive cells were expected to undergo the error-prone, NHEJ-mediated repair of Cas9-induced DNA double-strand breaks, we designed the targeting sgRNA to place the cleavage site in the 5’-untranslated region of MeCP2 gene (Figure S4B). This strategy was designed to minimize potential effects on expression of the targeted gene in these cells. Indeed, the intensities of endogenous MeCP2 detected by an antibody against the C-terminus of MeCP2 was similar between mEGFP-positive and negative cells (Figure S4D; p = 0.19, Student’s t test). This indicated that the endogenous expression of MeCP2 was not significantly disturbed by the NHEJ-mediated repair.
Next, we applied SLENDR to insert the HA tag to endogenous β-Actin, a major cytoskeletal protein in dendritic spines. As expected, immunostaining using the anti-HA antibody showed that HA-β-Actin was highly accumulated in dendritic spines in layer 2/3 pyramidal neurons and microfilaments in astrocytes in the cortex at P28 (Yuste and Bonhoeffer, 2004) (Figures 2C and 2D). In addition, the high contrast images demonstrate that SLENDR enables the precise evaluation of the number and morphology of dendritic spines without the potential morphological phenotypes caused by overexpression of fluorophore tagged-actin or actin binding proteins (Riedl et al., 2008).
To evaluate the timescale of HDR after introducing SLENDR constructs, we targeted Doublecortin (DCX), a protein that is expressed early in brain development. DCX is a microtubule-associated protein expressed in postmitotic migrating and differentiating neurons in the developing brain (Gleeson et al., 1999). Intriguingly, HA-DCX was detected in the cortical migrating neurons and accumulated in the growth cone as early as 60 h after IUE at E12 (Figures 2E and 2F), suggesting that HDR occurred rapidly, possibly within one or two days after IUE. HA-DCX was detected at both E14 and E18 (Figures 2F and Figure S5B), suggesting SLENDR is suitable for studying protein localization in the embryonic brain. In addition, we evaluated the effect of NHEJ on the expression of DCX by immunostaining using an antibody against the C-terminus of the protein. The images showed that 98.7 % of the mEGFP positive neurons exhibited DCX expression (n = 151 cells) (Figure S5A). Together with the negligible NHEJ effects on MeCP2 expression (Figure S4D), these data suggest that our strategy of targeting the Cas9-mediated cleavage at 5’-untranslated regions minimizes the effect of NHEJ on the expression of the target gene for N-terminal tagging.
We further applied SLENDR to a variety of endogenous proteins with distinct subcellular localizations. Rab11a, a small GTPase involved in the endosomal recycling of proteins, was localized to numerous, small dispersed vesicles throughout the soma and dendrites in cortical neurons and astrocytes at P23, consistent with the pattern expected from the localization of recycling endosomes (Hutagalung and Novick, 2011) (Figures 2G, 2H and S5C). CaV1.2, the α1C subunit of the L-type voltage-gated calcium channel, was distributed in clusters on cell bodies and proximal dendrites in layer 2/3 pyramidal neurons at P16 (Figures 2I and 2J) (Hell et al., 1993). Immunofluorescence signal of CaV1.2-HA was also detected in the nucleus. This signal may represent the C-terminal fragment of CaV1.2, which functions as a calcium channel associated transcription regulator (Gomez-Ospina et al., 2006). 14-3-3ε, a signaling protein involved in neuronal migration and synaptic plasticity, was diffusely distributed in the cytoplasm both at P0 and P28 in neurons and astrocytes (Toyo-oka et al., 2014) (Figures 2K and 2L). FMRP, a polyribosome-associated RNA-binding protein that regulates translation of a large number of mRNAs (Contractor et al., 2015), was found in puncta, which likely reflect FMRP-associated mRNA granules, in the soma as well as dendrites in pyramidal neurons at P7 and P28 (Figures 2M, 2N and S5D). Arc, an immediate early gene product involved in synaptic plasticity, was localized both in the nucleus and cytoplasm at P29 (Figure 2O and 2P) (Korb et al., 2013; Shepherd and Bear, 2011). Finally, we found that α isoform of protein kinase C (PKC), a member of a family of serine/threonine kinases implicated in a wide range of cellular functions (Steinberg, 2008), was distributed mostly on the plasma membrane of the soma and dendrites at P9 (Figures 2Q and 2R). Interestingly, PKCα was less accumulated on the plasma membrane at P27, suggesting that endogenous PKCα may be more active at the developing stage (Figure S5E–S5G). Thus, SLENDR enables us to determine the distribution pattern of proteins that has been undefined or controversial by conventional methods.
Notably, we never observed HA-positive cells when we used incorrect ssODNs-sgRNA pairs (Figure S2 and Figure S4). In addition, precise genome editing was confirmed by PCR amplification of the targeted loci followed by DNA sequencing of the PCR products (Figure S3). Furthermore, for each target protein, all HA-positive cells in the same region showed similar HA-staining pattern. These results collectively demonstrate that SLENDR enables us to label endogenous proteins specifically. Taken together, SLENDR is a highly generalizable technique that can be applied to various species of proteins in the brain from embryonic to adult stages for high-quality mapping of subcellular localization.
Nanometer-Resolution Analysis by SLENDR
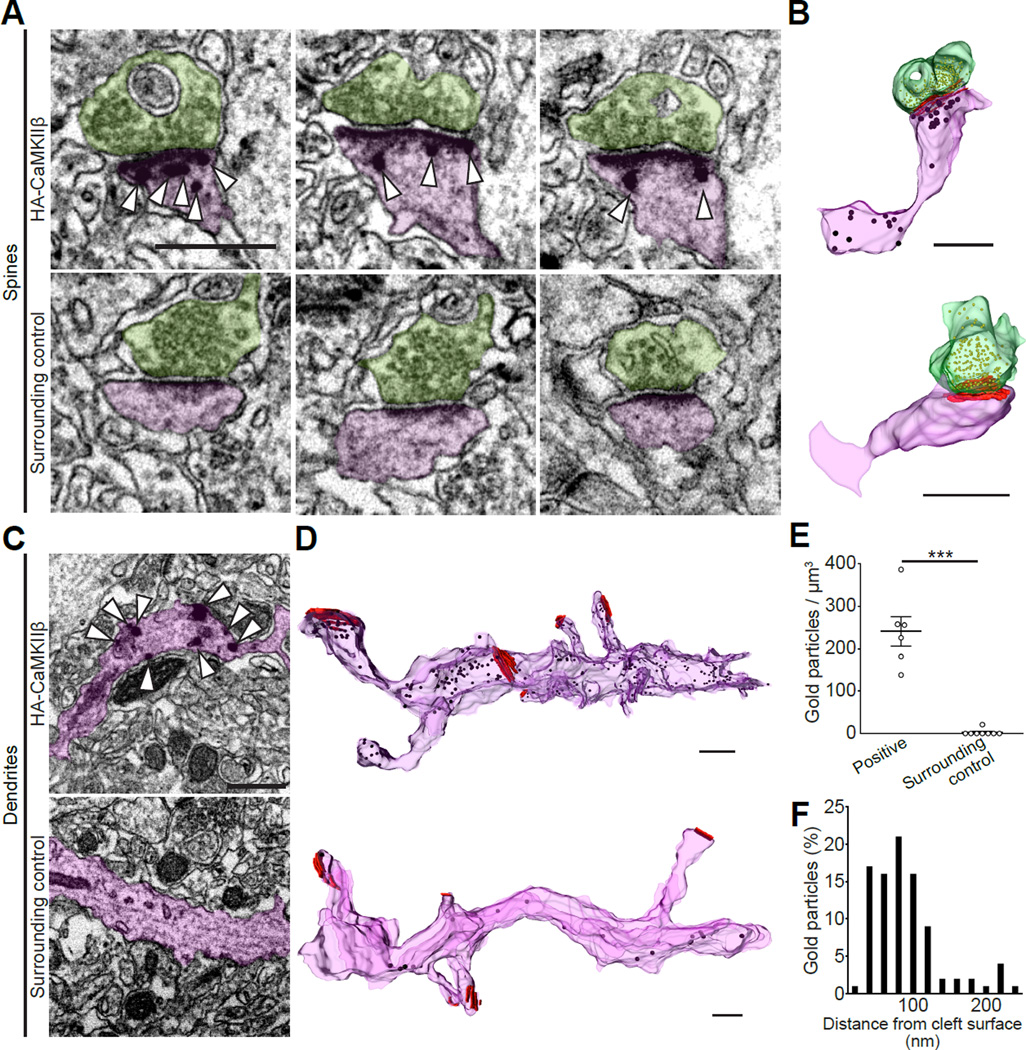
Immunoelectron microscopy allows nanoscale visualization of endogenous proteins with defined ultrastructures in cells. However, the lack of reliable antibodies compatible with electron microscopy imaging limits its application to a variety of proteins. Thus, we tested whether SLENDR could be applied to ultrastructural imaging of endogenous proteins using immunoelectron microscopy. To do so, we used cortical tissue in which endogenous CaMKIIβ is fused with HA using SLENDR (Figures 1C and 1D). We applied pre-embedding staining technique: we incubated the tissue with HA-antibody and secondary antibody conjugated with gold, followed by silver enhancement and tissue embedding. We then prepared serial thin-section (50 nm) of the tissue using the automatic tape-collecting ultramicrotome (ATUMtome). ATUMtome permits rapid and automated cutting and collection of serial thin sections onto a continuous reel of tape (Kasthuri et al., 2015). We imaged a number of serial sections on the tape by scanning electron microscopy and reconstructed three-dimensional images of SLENDR generated knock-in cells (Figure 3A-3D). We found that CaMKIIβ was localized near the postsynaptic density (PSD) (mode ~80 nm) in dendritic spines (Figure 3F). Finally, the specificity of the immunogold labeling was examined by comparing the labeling density in HA-positive and surrounding neurons in the same specimen (Figure 3E; HA-positive, 241.3 ± 34.5 particles / µm3; surrounding control, 2.6 ± 2.6 particles / µm3; p < 0.001, Student’s t test). These experiments demonstrate that SLENDR is useful for nanometer scale localization of endogenous proteins.
Figure 3. Nanometer-Scale Analysis of Endogenous Proteins by SLENDR.
(A–D) Electron microscopic images of dendritic spines (A) and shafts (C) in the cerebral cortex showing immunogold labeling for the HA tag (arrowheads) fused to the N-terminus of CaMKIIβ in HA positive (top) and surrounding control (bottom) cells. Three-dimensional reconstructions of corresponding spines (B) and dendrites (D). Analyzed spines and dendrites are marked in purple and presynaptic terminals are marked in green. Presynaptic vesicles and postsynaptic densities are marked in yellow and red, respectively (B and D).
(E) Density of immunogold particles in HA-positive spines (n = 6) and surrounding control spines (n = 8). ***p < 0.001, Student’s t test.
(F) The frequency distribution of distances between individual gold particles and cleft surfaces (n = 92 particles/ 6 spines).
Data are represented as mean ± SEM.
Scale bars, 500 nm.
See also Table S3.
SLENDR is Scalable to Various Cell Types in Various Brain Regions
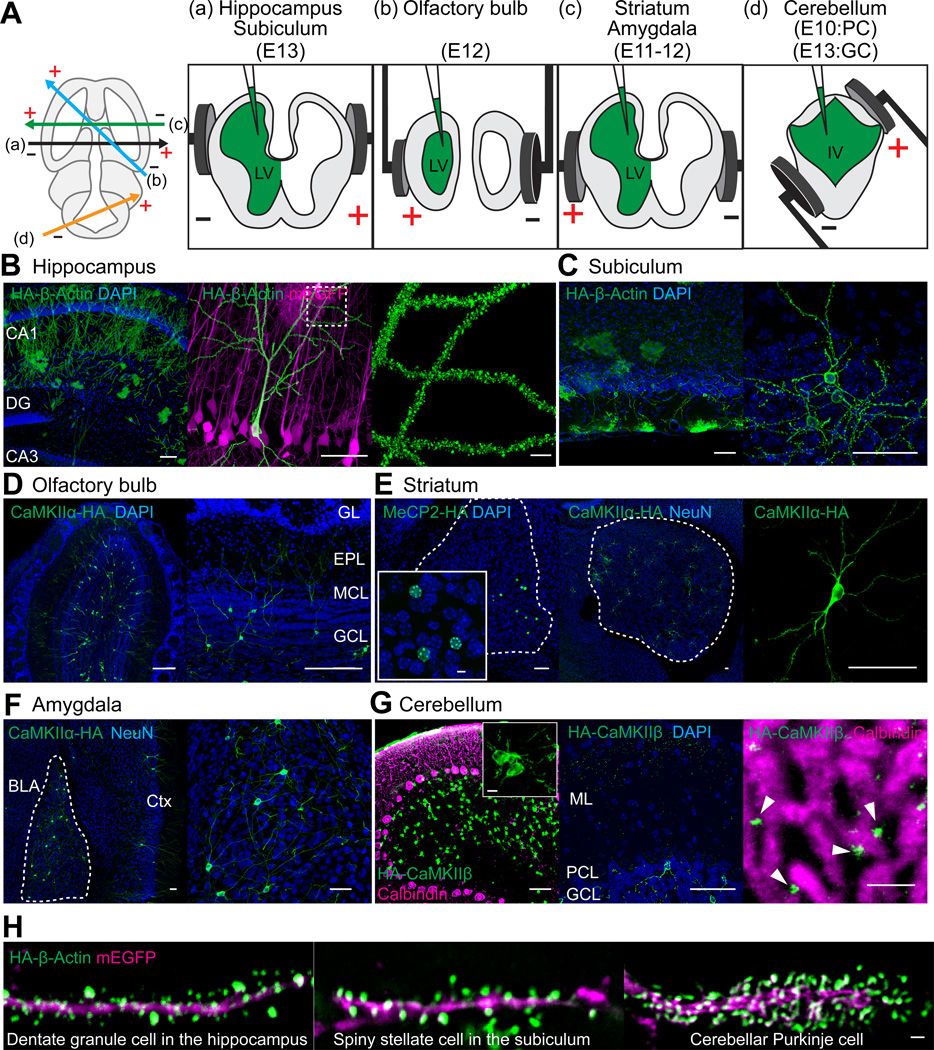
We then tested if SLENDR can be applied to different cell types in different brain regions by targeting distinct progenitor cells present at different locations and timings in the developing brain (Borrell et al., 2005; Chen and LoTurco, 2012; Kitazawa et al., 2014; Nishiyama et al., 2012; Soma et al., 2009). By adjusting the timing and electroporation angle (Figure 4A), various HA-tagged endogenous proteins (β-Actin, CaMKIIα, CaMKIIβ and MeCP2) were observed in various cell types in widespread brain regions (Table S2), including CA1 pyramidal neurons and dentate granule cells in the hippocampus (Figures 4B and 4H), spiny stellate cells in the subiculum (Figures 4C and 4H), granule cells in the olfactory bulb (Figure 4D), medium spiny neurons in the striatum (Figure 4E), basolateral amygdala neurons (Figure 4F), and granule and Purkinje cells in the cerebellum (Figures 4G and 4H). HA-CaMKIIβ in parallel fibers originating from cerebellar granule cells was detected along the dendrites of Purkinje cells in the molecular layer (Figure 4G).
Figure 4. SLENDR is Scalable to Various Cell Types in Various Brain Regions.
(A) Schematic illustration of IUE for targeting distinct brain regions. The relative position of the electrodes (+, positive pole; −, negative pole) are shown in the transverse section of the brain to target different brain areas (left). The position of electrode paddles and the injected DNA (green) are shown in the coronal section of the brain to target hippocampus and subiculum (a), olfactory bulb (b), striatum and amygdala (c) and cerebellum (d). LV, lateral ventricle; IV, fourth ventricle; PC, Purkinje cell; GC, granule cell.
(B) Confocal microscopic images of the hippocampus showing the DAPI signal (blue) and immunoreactivities for mEGFP (magenta) and the HA tag (green) fused to β-Actin. DG, dentate gyrus.
(C–F) Images of the subiculum (C), olfactory bulb (D), striatum (E) and amygdala (F) showing the DAPI signal (C–E, blue) and immunoreactivities for NeuN (E, F, blue) and the HA tag (green) fused to β-Actin (C), CaMKIIα (D to F) and MeCP2 (E). GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell layer; GCL, granule cell layer; BLA, basolateral amygdala; Ctx, cerebral cortex.
(G) Images of the cerebellum showing the DAPI signal (blue) and immunoreactivities for calbindin D-28k (magenta, a Purkinje cell marker) and the HA tag (green) fused to CaMKIIβ. ML, molecular layer; PCL, Purkinje cell layer; GCL, granule cell layer.
(H) Images of the dendrites of a dentate granule cell in the hippocampus, a spiny stellate cell in the subiculum and a Purkinje cell in the cerebellum showing immunoreactivities for mEGFP (magenta) and the HA tag (green) fused to β-Actin.
Scale bars, 50 µm (B–G); 5 µm (B, right; E, inset; G, inset and right); 1 µm (H).
See also Tables S2 and S3.
As we showed that staining of endogenous HA-β-Actin enables clear visualization of dendritic spine morphology in neurons (Figure 2D), we imaged HA-β-Actin in various cell types in the brain to compare the structure of dendritic spines. Morphological diversity of dendritic spines existed among different cell types including dentate granule cells (Figure 4H) and CA1 pyramidal neurons (Figure 4B) in the hippocampus, layer 2/3 pyramidal neurons in the cortex (Figure 2D), spiny stellate cells in the subiculum (Figure 4H) and cerebellar Purkinje cells (Figure 4H). Thus, SLENDR enables comparison of subcellular localization of endogenous proteins in various neuron subtypes and brain regions across developmental stages, providing regional and developmental specific information about protein localization.
Labeling Multiple Endogenous Proteins by SLENDR
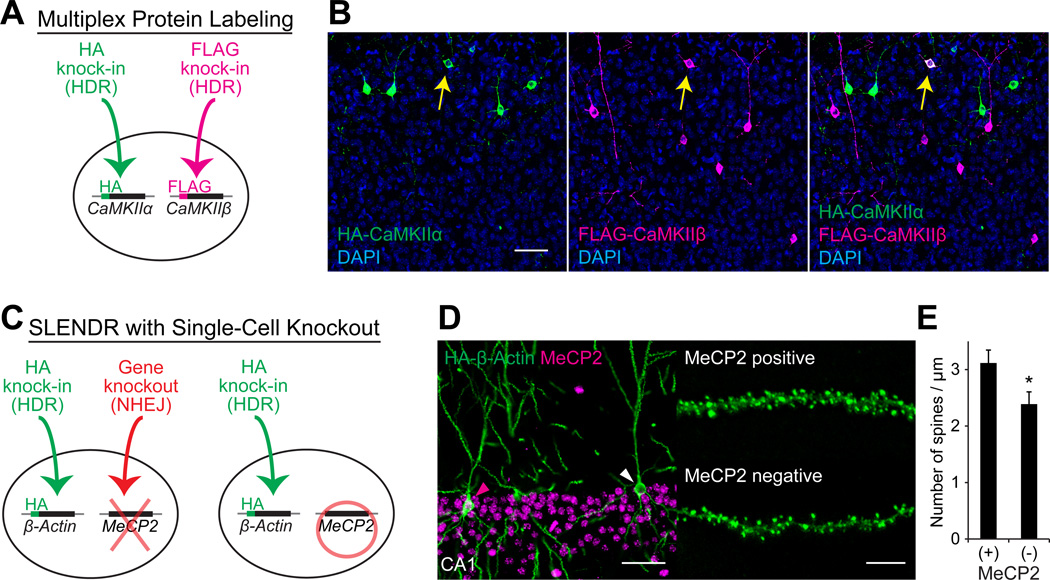
Multiplex labeling of different endogenous proteins with different tags would enable high resolution co-localization assays. Since CRISPR-Cas9 is able to target multiple genes simultaneously (Cong et al., 2013; Heidenreich and Zhang, 2016; Swiech et al., 2015), we tested the ability of SLENDR to target two different proteins in the same cell (Figure 5A). We performed simultaneous labeling of CaMKIIα and CaMKIIβ with the HA and FLAG tag, respectively, by co-introducing SLENDR constructs for HA-CaMKIIα and FLAG-CaMKIIβ into the developing cortex at E13 by IUE (Figure 5A). We performed immunostaining of brain slices at P14 using anti-HA and anti-FLAG antibodies. HA-CaMKIIα and FLAG-CaMKIIβ were detected in a number of layer 2/3 neurons (HA-positive/mEGFP-positive, 4.5 ± 0.4 %; FLAG-positive/mEGFP-positive, 4.9 ± 0.6 %; n = 577 cells), and a significant fraction of neurons exhibited both HA and FLAG signals (HA and FLAG double-positive/mEGFP-positive, 0.7 ± 0.1 %; n = 577 cells) (Figure 5B). This double labeling efficiency was higher than the simple multiplication of each labeling efficiency (4.5% × 4.9% = 0.2%), demonstrating the practicality of the method for co-localization assay. Thus, SLENDR allows labeling of two different species of proteins with different tags in single cells, providing a valuable tool for co-localization assays of a pair of endogenous proteins.
Figure 5. Application of SLENDR by Multiplex Genome Editing.
(A) Schematics of multiplex labeling of different endogenous proteins with different tags. The HA and FLAG sequences are inserted to CaMKIIα and CaMKIIβ, respectively, in the same cell through HDR-mediated genome editing.
(B) Multiplex labeling of endogenous CaMKIIα and CaMKIIβ. Confocal microscopic images of the cerebral cortex at P14 showing the DAPI signal (blue) and immunoreactivities for the HA tag (green) and the FLAG tag (magenta) fused to the N-terminus of endogenous CaMKIIα and CaMKIIβ, respectively. The yellow arrows indicate HA and FLAG double-positive layer 2/3 neurons.
(C) Schematics of combining SLENDR with NHEJ-mediated gene knockout. The HA sequence is inserted to β-Actin through HDR-mediated genome editing and a frame-shift mutation is induced in MeCP2 through NHEJ-mediated genome editing in the same cell (left). In our strategy, some cells in the same tissue undergo only HDR-mediated genome editing (right), allowing comparison of the expression and localization of endogenous proteins within the same brain slice.
(D) Immunofluorescence images of MeCP2 (magenta) and HA-β-Actin (green) in the hippocampus CA1 region. Magenta arrowhead, MeCP2-positive; white arrowhead, MeCP2-negative. Representative images of secondary dendrites of MeCP2-positive and negative neurons (right).
(E) The averaged density of spines on secondary or tertiary apical dendrites in MeCP2-positive (n = 527 spines/ 6 neurons) and negative (n = 437/5) neurons. *p < 0.05, Student’s t test.
Data are represented as mean ± SEM.
Scale bars, 50 µm (B and D, left); 5 µm (D, right).
See also Table S3.
SLENDR in Combination with Single-Cell Knockout
The ability to examine endogenous subcellular protein localization in the context of a knockout of a different protein would provide functional insight into the interaction between the visualized and deleted proteins. In this regard, combining SLENDR with NHEJ-based single-cell knockout would be of particular interest, since it would allow for the study of cell-autonomous gene function and to compare normal and knockout cells in the same tissue (Zong et al., 2005). Taking advantage of the multiplexity of CRISPR-Cas9, we simultaneously introduced SLENDR constructs to insert the HA tag to β-Actin and CRISPR constructs to induce NHEJ-mediated gene knockout of MeCP2 in progenitors of hippocampal neurons at E13 (Figure 5C) (Incontro et al., 2014; Straub et al., 2014; Swiech et al., 2015). Visualization of HA-tagged β-Actin by SLENDR enables visualization of dendritic spine morphology, providing a useful tool to study effects of MeCP2 gene deletion on dendritic spines in single cells.
While knockout efficiency of NHEJ-mediated genome editing has been reported to be very high in neurons (~70–100 %) (Heidenreich and Zhang, 2016; Incontro et al., 2014; Straub et al., 2014; Swiech et al., 2015), immunohistochemistry with anti-MeCP2 antibody showed that MeCP2 was absent in 48.8 % of mEGFP-positive CA1 pyramidal neurons at P28. This reduced efficiency was probably due to the dilution of the knockout constructs during cell divisions (Chen and LoTurco, 2012; Yusa et al., 2011). As expected, both MeCP2 negative and positive neurons were observed in HA-β-Actin positive CA1 neurons in the same slices (Figures 5D). Notably, the knockout efficiency in HA-β-Actin positive neurons (24.1%) was substantially lower than that in whole population (48.8%). This is presumably because the knockout constructs are diluted in a population of cells that divides extensively, and this population of cells likely provides more efficient HDR. A loss of function mutation in MeCP2, which leads to Rett syndrome in humans, has been reported to impair maturation of dendritic spines (Chen et al., 2001; Swiech et al., 2015). Therefore, we compared the density of dendritic spines between MeCP2 negative and positive neurons (Figure 5D and 5E). Spine density in the apical dendrite of CA1 neurons was comparable in MeCP2 positive neurons to that reported in these neurons in a previous electron microscopic study (Harris et al., 1992). Importantly, spine density was significantly lower in MeCP2 negative neurons, consistent with previous studies (Figure 5E;p = 0.046, Student’s t test) (Swiech et al., 2015).
Although high-quality antibodies against endogenous proteins are required to distinguish knockout and control cells, our strategy combining SLENDR with NHEJ-mediated gene knockout provides a useful means to investigate cell-autonomous effects of a gene of interest on endogenous proteins.
Live Imaging of Endogenous Proteins by SLENDR
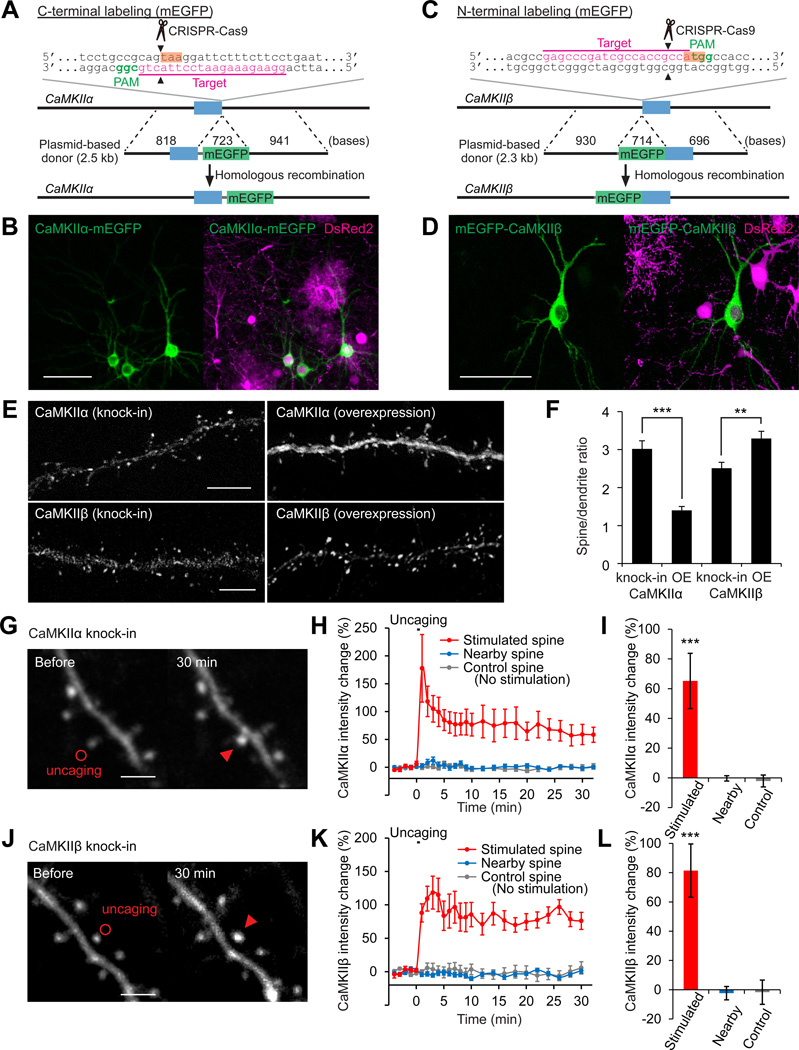
Labeling endogenous proteins with fluorescent proteins would allow us to image protein dynamics in live cells without overexpression artifacts, further expanding the applicability of SLENDR. Thus, we tested if SLENDR can be used to insert a long sequence encoding a fluorescent protein into a gene of interest. For this, we targeted mEGFP to the sequence just upstream of the stop codon of CaMKIIα or downstream of the start codon of CaMKIIβ in the genome (Figures 6A and 6C). We used a plasmid-based template (~2.3–2.5 kb) containing the ~0.7 kb sequence of mEGFP (see Supplemental Information). The SLENDR constructs together with the transposon vectors expressing DsRed2 were introduced to progenitors of layer 2/3 neurons through IUE at E12. At P14–28, we found that a sparse subset of layer 2/3 neurons exhibited mEGFP fluorescence fused to endogenous CaMKIIα and CaMKIIβ (Figures 6B and 6D). The efficiency of mEGFP knock-in was lower than that of HA knock-in (mEGFP-positive/DsRed2-positive, < 1%), consistent with previous studies showing that a long sequence insertion through HDR is less efficient (Cox et al., 2015). The specificity of mEGFP knock-in was confirmed by control experiments using correct and incorrect template-sgRNA pairs and DNA sequencing following PCR amplification of the targeted locus (Figure S6).
Figure 6. Localization and Dynamics of Endogenous Proteins Labeled with mEGFP by SLENDR.
(A and C) Graphical representation of the mouse genomic loci of CaMKIIα (A) and CaMKIIβ (C) showing the targeting sites for Cas9, sgRNA and HDR donor plasmid. The sgRNA targeting sequences are underlined and labeled in magenta. The PAM sequences are labeled in green. The stop and start codons of CaMKIIα (A) and CaMKIIβ (C) are marked in orange.
(B and D) Confocal microscopic images of the somatosensory cortex showing the fluorescence of DsRed2 (magenta) and mEGFP (green) fused to CaMKIIα/β.
(E) Images of apical secondary dendrites in layer 2/3 fixed at P14 showing mEGFP-tagged endogenous (knock-in) or overexpressed (OE) CaMKIIα/β.
(F) The spine/dendrite ratio of the peak intensities of mEGFP-tagged CaMKIIα/β. CaMKIIα, knock-in, n = 51/6 (spines/neurons); OE, n = 40/5. CaMKIIβ, knock-in, n = 58/4; OE, n = 83/5. (G-L) Two-photon microscopic images before and 30 min after glutamate uncaging showing mEGFP-tagged endogenous CaMKIIα/β in layer 2/3 neurons (G and J). Red circles, uncaging spots; red arrowheads, stimulated spines. Averaged time courses (H and K) and sustained values (I and L; averaged over 20–30 min) of CaMKIIα/β intensity change in the stimulated (red; H and I, n = 11/8; K and L, n =9/7), nearby (2–5 µm from the stimulated spines; blue; H and I, n = 31/8; K and L, n =31/7) and control spines with no stimulation (grey; H and I, n = 23/5; K and L, n =13/2). ***p < 0.001, Student’s t test (F) and Dunnett test (I and L).
Data are represented as mean ± SEM.
Scale bars, 50 µm (B and D); 5 µm (E); 2 µm (G and J).
See also Figure S6 and Table S3.
To further verify the benefit of imaging endogenous proteins, we compared the distribution pattern of endogenous and overexpressed mEGFP-tagged CaMKII in layer 2/3 cortical neurons at P14. Since CaMKIIβ has an actin-binding domain (Lisman et al., 2012), overexpressed CaMKIIβ tends to accumulate in spines to a greater extent than overexpressed CaMKIIα. Notably, the degree of enrichment of endogenous CaMKIIα and CaMKIIβ in spines, as measured by the spine/dendrite ratio of the peak intensity, was significantly higher and lower than that of overexpressed CaMKIIα and CaMKIIβ, respectively (Figures 6E and 6F; CaMKIIα, p < 0.001; CaMKIIβ, p = 0.004, Student’s t test). This is perhaps because overexpressed subunits tend to form homomeric enzymes, while endogenous CaMKIIα and CaMKIIβ tend to form heteromers (Lisman et al., 2012). Thus, the localization of endogenous (heteromeric) subunits would be enriched in spines to an extent that is between homomeric CaMKIIα and CaMKIIβ.
We next performed live imaging to monitor the synapse-specific translocation of CaMKII after the stimulation of a single dendritic spine (Bosch et al., 2014; Lee et al., 2009; Lisman et al., 2012; Nishiyama and Yasuda, 2015). We prepared organotypic cortical slice cultures from mice in which endogenous CaMKIIα or CaMKIIβ was labeled with mEGFP by SLENDR. When we applied a train of two-photon glutamate uncaging pulses (1 Hz, 30 s) at single spines, mEGFP-tagged CaMKIIα and CaMKIIβ were rapidly accumulated in the stimulated spines and remained for more than 30 min. The translocation was highly restricted to the stimulated spine, with no significant increase in fluorescence intensity in the surrounding spines (Figures 6G–6L; CaMKIIα, p < 0.001; CaMKIIβ, p < 0.001; Dunnett test). These results demonstrate that SLENDR enables monitoring of the dynamics of endogenous proteins in living tissue.
DISCUSSION
In this study, we have developed SLENDR, which allows in vivo genome editing in the mammalian brain for single-cell labeling of endogenous proteins. We demonstrated that SLENDR is a simple and efficient technique to rapidly determine the subcellular localization of endogenous proteins with the resolution of micro- to nanometers in brain tissue. Importantly, the technique is generalizable to a broad spectrum of proteins and various cell types in widespread brain regions. SLENDR also can be used for multiplex labeling of different proteins or for mosaic analysis by combining labeling with single-cell knockout. Furthermore, SLENDR is capable of inserting a long sequence such as that encoding mEGFP in vivo and thus enables live imaging of endogenous proteins during biological processes in the brain.
HDR-mediated genome editing has been a challenge in the brain due to the lack of HDR activity in postmitotic neurons and the inefficient delivery of HDR machinery into the brain. This has limited its application in the field of neuroscience (Heidenreich and Zhang, 2016). We circumvented these problems by targeting mitotic progenitors in the embryonic brain using IUE. In our experiments, the knock-in efficiency for the HA tag insertion was sufficient to image the subcellular localization of a protein of interest in single cells in brain tissue. We also showed that SLENDR can be used to knock-in a long mEGFP sequence, albeit at lower efficiency. To extend the SLENDR technique to broader applications for multiplexed labeling and fluorescent protein fusion, it may be necessary to increase the efficiency of HDR.
It is known that the efficiency of HDR highly depends on the cell cycle and is limited by the competition with NHEJ (Chu et al., 2015; Lin et al., 2014; Maruyama et al., 2015). Consistent with this, we found that SLENDR was efficient in a limited time window when neuronal progenitors were still dividing. CRISPR-Cas9-mediated HDR occurred within a few days after IUE (Figure 2F) and the HDR efficiency was significantly reduced when IUE was performed near the stage of the final cell divisions of target cells (Figures 1B and 1D). In this regard, the direct delivery of pre-assembled Cas9 protein-guide RNA ribonucleoprotein complexes (RNPs), rather than expressing these components from plasmids, might increase the time window for SLENDR, as RNPs have recently been reported to provide rapid nuclease action with high efficiency and low off-target effects for genome editing (Kim et al., 2014; Lin et al., 2014). Alternatively, genetic or pharmacological inhibition of the NHEJ pathway may increase the HDR efficiency (Chu et al., 2015; Maruyama et al., 2015), although potential side effects must be addressed.
We validated the specificity of the SLENDR-mediated sequence insertion in the genome in several ways. First, we designed sgRNAs by unbiased genome-wide analysis to minimize the potential off-target cleavages by Cas9 (Ran et al., 2013). Second, we performed control experiments by using incorrect sgRNAs and found no cells expressing the HA tag. Third, we used distinct sgRNAs targeting the identical gene and observed similar HA staining patterns. Fourth, the localization of HA staining was consistent with that previously reported based on immunohistochemical, biochemical or electron microscopic studies. Fifth, the localization of HA staining was similar from cell to cell. In addition, although off-target cleavage by Cas9 may be induced in the genome, HDR is unlikely to occur at the off-target sites, because homologous recombination is known to be a highly sequence-specific event. The specificity of SLENDR could be further enhanced by using recently reported Cas9 variants with minimum or no off-target effects while retaining comparable on-target cleavage activity (Kleinstiver et al., 2016; Slaymaker et al., 2016).
One of the merits of SLENDR is that high-quality antibodies can be used for the detection of tags. In addition, once staining conditions have been optimized for a tag, SLENDR can be applied to image various tagged proteins without extensive optimization. Furthermore, since SLENDR allows protein labeling in a sparse subset of cells in the tissue, the specificity of immunostaining can be easily validated by examining surrounding negative control cells in the same specimen. These features are particularly advantageous for immunoelectron microscopy imaging, which often requires extensive optimization of staining conditions and good control samples (e.g. knockout mice).
We have demonstrated that introducing a single epitope tag by SLENDR is sufficient to detect relatively low abundant proteins such as CaV1.2 and Arc (Figures 2J and 2P). The sensitivity could be further increased by inserting multiple copies of epitopes or more antigenic probes such as Spaghetti-monster (Viswanathan et al., 2015). Overall, because many studies are constrained by the lack of high-quality antibodies against proteins of interest, SLENDR will provide a generalizable and reliable platform for exploring localization of uncharacterized endogenous proteins using light and electron microscopy.
Multiplexing CRISPR-Cas9 mediated processes provided some insight into how HDR and NHEJ occur following double strand breaks. We found that the cells labeled with a tag with SLENDR, or in other words the cells that underwent HDR, showed less NHEJ-mediated knockout. This result suggests that HDR occurs more frequently in a population of highly dividing cells. Consistent with this, the efficiency of double-labeling via multiple HDRs was higher than that obtained by the simple multiplication of each labeling efficiency. The high efficiency of double-labeling by SLENDR opens the possibility of co-localization assay for a pair of endogenous proteins with high resolution and contrast. Given relatively consistent knock-in efficiency (Table S1), the SLENDR-based double labeling technique would be applicable to many pairs of proteins.
There are a few limitations in SLENDR. First, the technique might lead to unintended sequence changes through insertion/deletion mutations mediated by NHEJ in the transfected cells. This problem may cause a large impact particularly in the case of N-terminal tagging, since altered sequence near the N-terminal region could change the transcription and translation of the target gene. Therefore, throughout this study, we attempted to minimize the effect of on-target NHEJ by choosing target sequences at 5’-untranslated region or near the stop codon for N- or C-terminal tagging, respectively. As expected from this design, the expression of DCX was detected in 98.7 % of the mEGFP positive neurons following SLENDR for the N-terminal tagging of DCX (Figure S5A). In addition, the expression level of MeCP2 was not significantly affected for the N-terminal tagging of MeCP2 (Figure S4D). However, potential effects of NHEJ have to be carefully evaluated for each gene depending on the purpose of experiments. For example, when the target is a secreted or a type I membrane protein with a signal sequence, one should select a target sequence for CRISPR-Cas9 mediated cleavage to minimize the possibility of deletion or mislocalization of the gene products. Second, immunodetection of a fused epitope tag may be difficult for some proteins, because the accessibility of antibodies may be sterically limited. For example, we failed to detect the HA signal with immunostaining in tissue in which the HA tag was inserted to the C-terminus of PSD-95 with SLENDR, whereas PCR detected the HA knock-in allele at the DNA level (data not shown). This is likely due to high protein density of PSD, which is known to often prevent antibodies to access to targeted proteins in the structure (Fukaya and Watanabe, 2000). This limitation of immunostaining can be overcome by fusing a fluorescent protein tag with SLENDR and directly observing the fluorescence (Fortin et al., 2014). These limitations would not detract the impact of SLENDR, as the technique should be applicable to most proteins with little optimization as shown in this study.
Since the 5’-NGG protospacer-adjacent motif (PAM) of SpCas9 abundantly exists in the genome and other CRISPR endonucleases with different PAMs are also available to target different sites in the genome (Cong et al., 2013; Hsu et al., 2014; Zetsche et al., 2015), most proteins should be suitable for SLENDR. Furthermore, its throughput and cost-effectiveness are much higher than previous single-cell protein labeling methods (Fortin et al., 2014; Gross et al., 2013). Thus, SLENDR should allow large-scale, potentially genome-wide, determination of precise subcellular localization of endogenous proteins in various cell types and ages, providing a new level of understanding of protein and cellular function in the brain.
EXPERIMENTAL PROCEDURES
Full experimental details can be found in the Supplemental Experimental Procedures.
Animals
All experimental procedures were approved by the Max Planck Florida Institute for Neuroscience Institutional Animal Care and Use Committee and were performed in accordance with guidelines from the US National Institutes of Health. Swiss Webster mice were obtained from Charles River. The day on which the vaginal plug was detected was designated as embryonic day 0 (E0). The first 24 h after birth was referred to as postnatal day 0 (P0). Both of male and female mice were used.
DNA Constructs
The human codon-optimized SpCas9 and sgRNA expression plasmid was a gift from F. Zhang (pX330, Addgene plasmid # 42230) (Cong et al., 2013). To generate sgRNA-expressing plasmids, a pair of annealed oligos (~20 bp) was ligated into the sgRNA scaffold of pX330 (Ran et al., 2013). We purchased single-stranded oligodeoxynucleotides (ssODNs) for HDR from Integrated DNA Technologies. To fuse mEGFP (monomeric EGFP, A206K) to the C- or N-terminus of CaMKIIα or CaMKIIβ, respectively, plasmid-based donor templates for HDR were prepared. Other details are described in the Supplemental Experimental Procedures.
In Utero Electroporation
In utero electroporation was performed as previously described (Borrell et al., 2005; Chen and LoTurco, 2012; Imamura and Greer, 2015; Kitazawa et al., 2014; Nishiyama et al., 2012; Soma et al., 2009; Tabata and Nakajima, 2001). The final concentration of each plasmid (pX330-derivatives, pPB-CAG-mEGFP, pPB-CAG-DsRed2, pPB-CAG-tdTomato and pCAG-hyPBase), the ssODNs for HDR and the double-stranded DNA template for mEGFP insertion were 1 µg/µl, 20 µM and 1 µg/µl, respectively (Table S3). The position and angle of the electrode was set as previously described with some modifications (see Figure 4A). Other details are described in the Supplemental Experimental Procedures.
Histology
Mice were fixed by transcardial perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer and coronal sections (50 or 100 µm in thickness) were prepared using a vibratome (VT1200, Leica). Immunohistochemistry was performed as previously described (Fukaya and Watanabe, 2000; Nishiyama et al., 2012). Other details are described in the Supplemental Experimental Procedures.
Preembedding Immunoelectron Microscopy
An HA-CaMKIIβ knock-in mouse was transcardially perfused with 4% paraformaldehyde and 0.1% glutaraldehyde in 0.1 M phosphate buffer. The fixed brain was sectioned into 50 µm thick slices with a vibratome (VT 1200, Leica), and sections with positive DsRed2 labeling were chosen. Tissue processing for preembedding immunoelectron microscopy was performed as previously described (Parajuli et al., 2012). After processing, a region containing HA positive neurons was trimmed out, and 50 nm thick sections were cut and collected onto a kapton tape by ATUMtome (RMC/Boeckeler) (Kasthuri et al., 2015; Schalek et al., 2011). The kapton tape was placed on a silicon wafer, and a layer of 5 nm thick carbon was coated on the wafer surface by the high vacuum sputter coater (ACE600, Leica). The sections on the wafer were imaged under a scanning electron microscope (SEM, Merlin VP Compact, Zeiss) assisted with Atlas 5 AT software (Zeiss). Other details are described in the Supplemental Experimental Procedures.
Two-Photon Glutamate Uncaging
Organotypic cortical slice cultures were prepared from mEGFP-CaMKIIα/β knock-in mice as described previously (Stoppini et al., 1991; Yamamoto et al., 1989). Secondary apical dendritic branches of layer 2/3 neurons were imaged at 15–17 days in vitro under a custom-built two-photon microscope with two Ti:Sapphire lasers (Chameleon, Coherent) (Lee et al., 2009). MNI-caged L-glutamate (4-methoxy-7-nitroindolinyl-caged L-glutamate, Tocris) was uncaged with a train of 6 ms laser pulses (3.5–4 mW under the objective, 30 times at 1 Hz) near a spine of interest. Experiments were performed at 32 °C. Images were analyzed with MATLAB (MathWorks) and ImageJ. Other details are described in the Supplemental Experimental Procedures.
Statistical Analysis
All statistical values were presented as mean ± SEM. The Student’s t test was used when two independent samples were compared. Dunnett test was used for multiple comparisons. Statistical analysis was performed with GraphPad Prism 6. Differences between data sets were judged to be significant at p < 0.05.
Supplementary Material
Acknowledgments
The authors thank Sanger Institute for the gift of the piggyBac transposon vector plasmids (pPB-CAG.EBNXN and pCMV-hyPBase) and Y.Hayashi for CaMKIIα and CaMKIIβ cDNA; L. Colgan, M. Yuzaki, S. Soderling and M. Kano for critical reading of the manuscript; the Yasuda lab members for discussion; J. Richards, K. Liu and J. Chen for technical assistance; and D. Kloetzer for laboratory management. This work was supported by the National Institute of Health (R01MH080047 and DP1NS096787 to R.Y.) and the Human Frontier Science Program (long-term fellowship to T.M.). The authors declare no competing financial interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
T.M. and J.N. conceived the SLENDR idea. T.M., J.N. and R.Y. designed the experiments. T.M. and J.N. performed most experiments and data analysis. Y.S. and N.K. performed electron microscopy imaging and data analysis. T.M., J.N. and R.Y. wrote the paper. All the authors discussed the results and commented on the manuscript.
REFERENCES
- Borrell V, Yoshimura Y, Callaway EM. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 2005;143:151–158. doi: 10.1016/j.jneumeth.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Bosch M, Castro J, Saneyoshi T, Matsuno H, Sur M, Hayashi Y. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82:444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, LoTurco J. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J Neurosci Methods. 2012;207:172–180. doi: 10.1016/j.jneumeth.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contractor A, Klyachko VA, Portera-Cailliau C. Altered neuronal and circuit excitability in Fragile X syndrome. Neuron. 2015;87:699–715. doi: 10.1016/j.neuron.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- Fortin DA, Tillo SE, Yang G, Rah JC, Melander JB, Bai S, Soler-Cedeno O, Qin M, Zemelman BV, Guo C, et al. Live imaging of endogenous PSD-95 using ENABLED: a conditional strategy to fluorescently label endogenous proteins. J Neurosci. 2014;34:16698–16712. doi: 10.1523/JNEUROSCI.3888-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Watanabe M. Improved immunohistochemical detection of postsynaptically located PSD-95/SAP90 protein family by protease section pretreatment: a study in the adult mouse brain. J Comp Neurol. 2000;426:572–586. [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel CaV1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross GG, Junge JA, Mora RJ, Kwon HB, Olson CA, Takahashi TT, Liman ER, Ellis-Davies GC, McGee AW, Sabatini BL, et al. Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron. 2013;78:971–985. doi: 10.1016/j.neuron.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidenreich M, Zhang F. Applications of CRISPR-Cas systems in neuroscience. Nat Rev Neurosci. 2016;17:36–44. doi: 10.1038/nrn.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel a1 subunits. J Cell Biol. 1993;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura F, Greer CA. Segregated labeling of olfactory bulb projection neurons based on their birthdates. Eur J Neurosci. 2015;41:147–156. doi: 10.1111/ejn.12784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vazquez-Reina A, Kaynig V, Jones TR, et al. Saturated reconstruction of a volume of neocortex. Cell. 2015;162:648–661. doi: 10.1016/j.cell.2015.06.054. [DOI] [PubMed] [Google Scholar]
- Kim K, Lakhanpal G, Lu HE, Khan M, Suzuki A, Kato-Hayashi M, Narayanan R, Luyben TT, Matsuda T, Nagai T, et al. A temporary gating of actin remodeling during synaptic plasticity consists of the interplay between the kinase and structural functions of CaMKII. Neuron. 2015;87:813–826. doi: 10.1016/j.neuron.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa A, Kubo K, Hayashi K, Matsunaga Y, Ishii K, Nakajima K. Hippocampal pyramidal neurons switch from a multipolar migration mode to a novel “climbing” migration mode during development. J Neurosci. 2014;34:1115–1126. doi: 10.1523/JNEUROSCI.2254-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016 doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korb E, Wilkinson CL, Delgado RN, Lovero KL, Finkbeiner S. Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nat Neurosci. 2013;16:874–883. doi: 10.1038/nn.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama J, Hayashi Y, Nomura T, Miura E, Kakegawa W, Yuzaki M. Selective and regulated gene expression in murine Purkinje cells by in utero electroporation. Eur J Neurosci. 2012;36:2867–2876. doi: 10.1111/j.1460-9568.2012.08203.x. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Yasuda R. Biochemical computation for spine structural plasticity. Neuron. 2015;87:63–75. doi: 10.1016/j.neuron.2015.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli LK, Nakajima C, Kulik A, Matsui K, Schneider T, Shigemoto R, Fukazawa Y. Quantitative regional and ultrastructural localization of the CaV2.3 subunit of R-type calcium channel in mouse brain. J Neurosci. 2012;32:13555–13567. doi: 10.1523/JNEUROSCI.1142-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh-Gohari N, Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalek R, Kasthuri N, Hayworth K, Berger D, Tapia J, Morgan J, Turaga S, Fagerholm E, Seung H, Lichtman J. Development of high-throughput, high-resolution 3D reconstruction of large-volume biological tissue using automated tape collection ultramicrotomy and scanning electron microscopy. Microscopy and Microanalysis. 2011;17:966–967. [Google Scholar]
- Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma M, Aizawa H, Ito Y, Maekawa M, Osumi N, Nakahira E, Okamoto H, Tanaka K, Yuasa S. Development of the mouse amygdala as revealed by enhanced green fluorescent protein gene transfer by means of in utero electroporation. J Comp Neurol. 2009;513:113–128. doi: 10.1002/cne.21945. [DOI] [PubMed] [Google Scholar]
- Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88:1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS One. 2014;9:e105584. doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33:102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Toyo-oka K, Wachi T, Hunt RF, Baraban SC, Taya S, Ramshaw H, Kaibuchi K, Schwarz QP, Lopez AF, Wynshaw-Boris A. 14-3-3ε and ζ regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J Neurosci. 2014;34:12168–12181. doi: 10.1523/JNEUROSCI.2513-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S, Williams ME, Bloss EB, Stasevich TJ, Speer CM, Nern A, Pfeiffer BD, Hooks BM, Li WP, English BP, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015;12:568–576. doi: 10.1038/nmeth.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Chen S, Yin H, Tammela T, Papagiannakopoulos T, Joshi NS, Cai W, Yang G, Bronson R, Crowley DG, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514:380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Kurotani T, Toyama K. Neural connections between the lateral geniculate nucleus and visual cortex in vitro . Science. 1989;245:192–194. doi: 10.1126/science.2749258. [DOI] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Siao CJ, Nagappan G, Marinic T, Jing D, McGrath K, Chen ZY, Mark W, Tessarollo L, Lee FS, et al. Neuronal release of proBDNF. Nat Neurosci. 2009;12:113–115. doi: 10.1038/nn.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A. 2011;108:1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T. Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci. 2004;5:24–34. doi: 10.1038/nrn1300. [DOI] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.