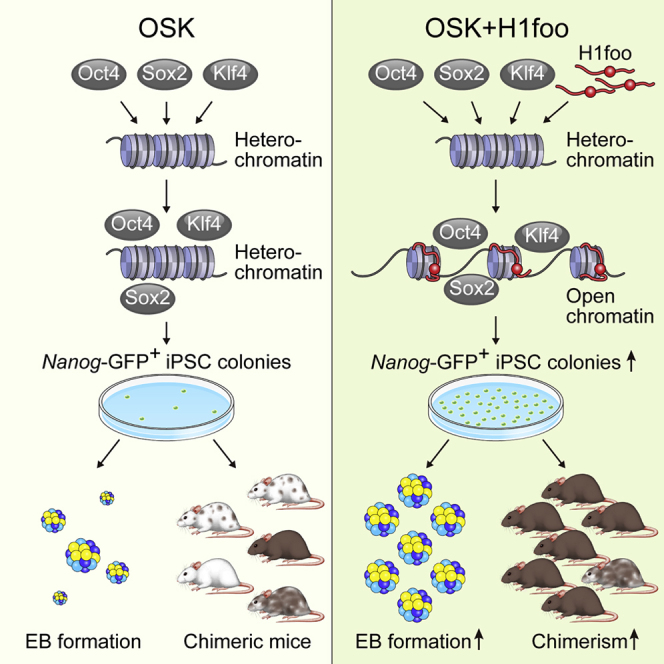
Summary
Embryonic stem cells (ESCs) are a hallmark of ideal pluripotent stem cells. Epigenetic reprogramming of induced pluripotent stem cells (iPSCs) has not been fully accomplished. iPSC generation is similar to somatic cell nuclear transfer (SCNT) in oocytes, and this procedure can be used to generate ESCs (SCNT-ESCs), which suggests the contribution of oocyte-specific constituents. Here, we show that the mammalian oocyte-specific linker histone H1foo has beneficial effects on iPSC generation. Induction of H1foo with Oct4, Sox2, and Klf4 significantly enhanced the efficiency of iPSC generation. H1foo promoted in vitro differentiation characteristics with low heterogeneity in iPSCs. H1foo enhanced the generation of germline-competent chimeric mice from iPSCs in a manner similar to that for ESCs. These findings indicate that H1foo contributes to the generation of higher-quality iPSCs.
Graphical Abstract
Highlights
-
•
H1foo enhanced the efficiency of iPSC generation
-
•
H1foo promoted in vitro differentiation characteristics with low heterogeneity
-
•
H1foo enhanced the generation of germline-competent chimeric mice
In this article, Yuasa and colleagues show that the mammalian oocyte-specific linker histone H1foo has beneficial effects on iPSC generation. Induction of H1foo with Oct4, Sox2, and Klf4 enhanced the efficiency of iPSC generation and improved the quality of iPSCs with low heterogeneity in vitro and in vivo. These findings indicate that H1foo contributes to the generation of higher quality iPSCs.
Introduction
Induced pluripotent stem cells (iPSCs) can be generated from somatic cells by introducing Oct4, Sox2, Klf4, and c-Myc (Takahashi and Yamanaka, 2006). This can be achieved by reprogramming the transcription network and epigenetic signature of the parental somatic cells. iPSCs have several benefits for basic research, drug innovation, and regeneration therapy. However, recent studies have reported genetic and epigenetic variations with iPSCs (Hussein et al., 2011, Kim et al., 2010, Polo et al., 2010, Ruiz et al., 2012, Stadtfeld et al., 2010, Taapken et al., 2011), which influences gene expression and could lead to functional diversity within iPSC replicates (Liang and Zhang, 2013). In fact, several studies have reported the heterogeneous differentiation potential among generated iPSC clones compared with those of embryonic stem cells (ESCs) (Feng et al., 2010, Hu et al., 2010, Narsinh et al., 2011). It is important that every iPSC clone shows high quality without variation for basic research and clinical purposes. Many attempts have been made to solve these problems by various methods (Gafni et al., 2013), but not all iPSCs exhibit quality as high as that of ESCs.
The reprogramming of somatic cells was originally demonstrated by producing cloned frogs using somatic cell nuclear transfer (SCNT) into Xenopus oocytes (Gurdon et al., 1958). Reprogramming of mammalian somatic cells has also been achieved using SCNT into oocytes, including those of mice and humans (Noggle et al., 2011, Wakayama et al., 1998). The procedure of SCNT into oocytes is similar to iPSC generation with respect to the time course of extinction of the parental gene-expression profile and activation of pluripotency (Egli et al., 2011). Both somatic cell reprogramming processes involve dynamic rearrangement of the epigenetic profile (Apostolou and Hochedlinger, 2013, Hussein et al., 2014). These findings suggest that the constituents of oocytes include a reprogramming-promoting factor. The linker histone H1 family binds to linker DNA and generates higher-order chromatin structures to control gene expression. Most members of the linker histone family consist of somatic linker histones that condense chromatin; therefore, these structures generally repress global gene-transcription activity (Hebbar and Archer, 2008, Steinbach et al., 1997). Mammalian oocytes contain the maternal-specific linker histone H1foo, a homolog of the Xenopus linker histone B4. H1foo is specifically expressed during the germinal vesicle stage and until the late two-cell or early four-cell stage, coincident with the early wave of zygotic genome reactivation (Gao et al., 2004, Tanaka et al., 2003, Tanaka et al., 2005). In Xenopus SCNT, somatic linker histones in transplanted nuclei are rapidly exchanged for linker histone B4, and the transplanted nuclei begin to swell and initiate decondensation (Byrne et al., 2003, Jullien et al., 2010); furthermore, in mouse SCNT the same phenomenon is observed with H1foo (Becker et al., 2005). Unlike other somatic linker histones, B4 and H1foo do not restrict the accessibility of the linker DNA, but decondense the chromatin and permit transcriptional activation (Hayakawa et al., 2012, Saeki et al., 2005).
Based on these studies, we hypothesized that H1foo has a beneficial effect on iPSC generation. Here, we show that H1foo enhanced the generation of mouse iPSCs when co-expressed with Oct4, Sox2, and Klf4. Furthermore, H1foo promoted several in vitro differentiation characteristics with low heterogeneity in iPSCs that were similar to those of ESCs. Specifically, H1foo enhanced germline-competent chimeric mouse generation. These findings indicate that H1foo contributes to the generation of higher-quality iPSCs.
Results
Exogenous Expression of H1foo Promotes Qualified iPSC Generation
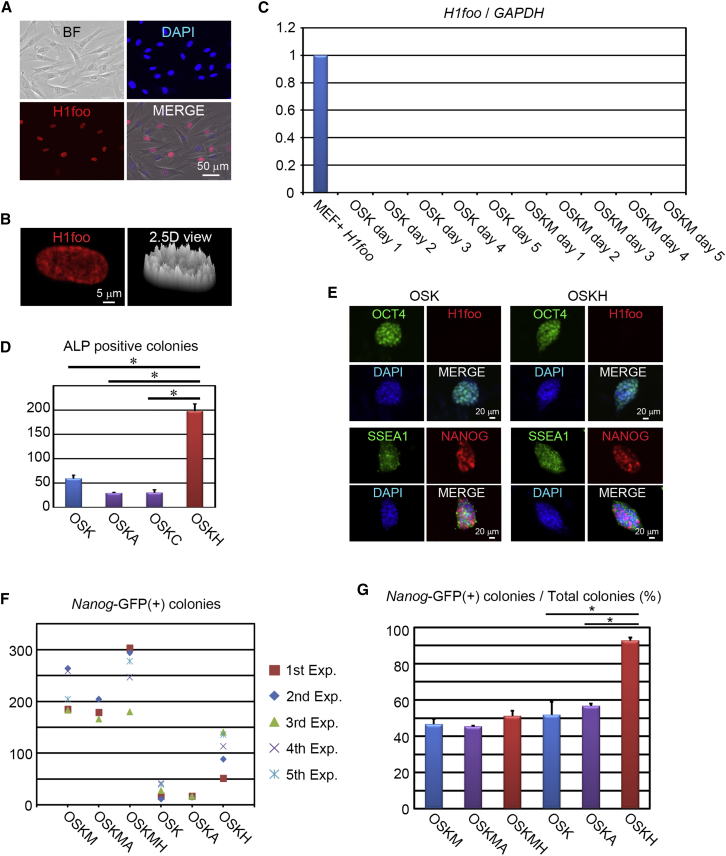
We examined the effect of exogenous H1foo on somatic cell reprogramming by introducing H1foo during iPSC generation with Oct4, Sox2, and Klf4 (OSK) or Oct4, Sox2, Klf4, and c-Myc (OSKM). Retrovirus vector-mediated exogenous H1foo was exclusively expressed in the nucleus of adult mouse tail-tip fibroblasts (Figures 1Aand S1A) and highly expressed peripherally in the nucleus (Figure 1B). SCNT into oocytes induced swelling of nuclei and chromatin decondensation (Gao et al., 2004, Tamada et al., 2006, Teranishi et al., 2004), so we investigated the effect of H1foo on nuclei (Figure S1B). We found that H1a, H1c, and H1foo had no significant effect on nuclear swelling (Figures S1C and S1D). However, interestingly, only H1foo reduced the intensively stained area, namely the heterochromatin area (Figure S1E). Next, we addressed whether intrinsic H1foo might be expressed during iPSC generation from mouse embryonic fibroblasts (MEFs) by introducing OSK or OSKM. However, we did not observe detectable intrinsic H1foo expression (Figure 1C). Co-expression of H1foo with OSK (OSKH) significantly enhanced the number of alkaline phosphatase-positive ESC-like colonies compared with OSK, OSK and H1a (OSKA), or OSK and H1c (OSKC) (Figure 1D). The OSKH-iPSCs expressed pluripotency markers similarly to control iPSCs (OSK), and H1foo was silenced (Figure 1E). We then examined the effect of H1foo on qualified iPSC generation using tail-tip fibroblasts from Nanog-GFP transgenic adult mice (Okita et al., 2007) (Figure S1G). Interestingly, H1foo also maximally promoted Nanog-GFP-positive colony generation (8-fold) during iPSC generation by OSK (Figure 1F). Notably, H1foo specifically enhanced GFP-positive colonies as opposed to GFP-negative colonies (Figure 1G).
Figure 1.
Exogenous Expression of H1foo Promotes iPSC Generation
(A) Immunostaining for H1foo (red), including nuclei stained with DAPI (blue) and bright-field (BF) images. Scale bar, 50 μm.
(B) Immunostaining for H1foo (red) with a 2D image (left) and a 2.5D image (right). The density of H1foo is depicted by the height in the 2.5D view. Scale bar, 5 μm.
(C) qRT-PCR analysis of endogenous H1foo expression during reprogramming, normalized by H1foo expression in H1foo-overexpressed MEFs (n = 3 independent experiments).
(D) Alkaline phosphatase-positive iPSC colony formations for OSK, OSK with H1a (OSK + H1a; OSKA), OSK with H1c (OSK + H1c; OSKC), and OSK with H1foo (OSK + H1foo; OSKH). Error bars represent the SEM (n = 3 independent experiments). ∗p < 0.05.
(E) Immunostaining for pluripotency markers (OCT4 [green], SSEA1 [green], and Nanog [red]), including H1foo (red) and nuclei stained with DAPI (blue) in OSK-iPSCs and OSKH-iPSCs. Scale bars, 20 μm.
(F) Number of Nanog-GFP-positive iPSC colonies in each experiment (n = 5 independent experiments).
(G) Proportion of Nanog-GFP-positive colonies to the total number of ESC-like colonies. Error bars represent the SEM (n = 5 independent experiments). ∗p < 0.05.
Characteristics of OSKH-iPSC Generation
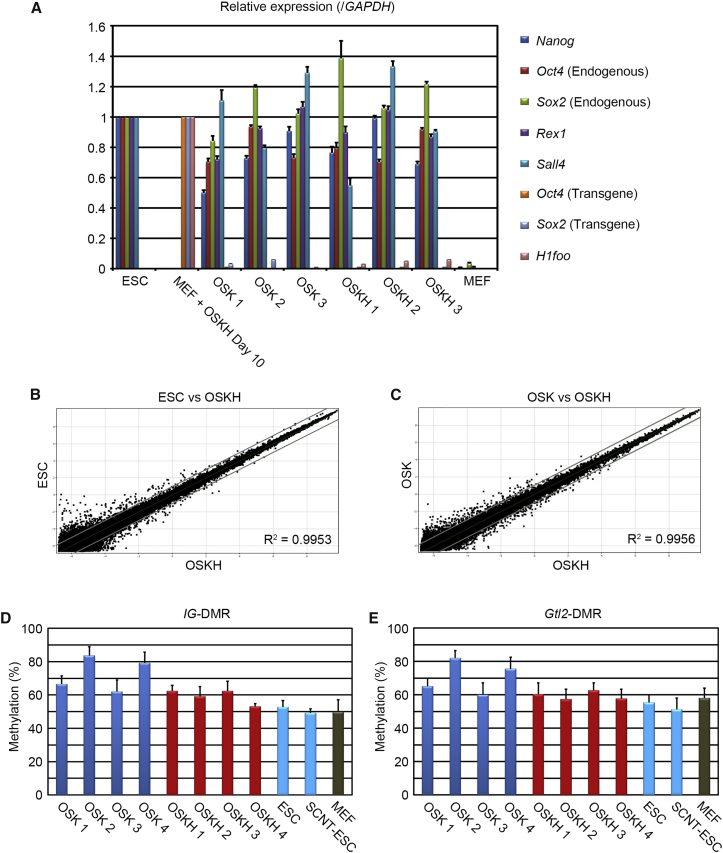
Next, we examined the iPSC characteristics produced by OSK and OSKH. The OSKH-iPSCs expressed pluripotency markers similar to those of OSK-iPSCs. Meanwhile the transgenes, including H1foo, were silenced (Figure 2A). Regarding the growth rate of iPSCs, there was no significant difference between OSK- and OSKH-treated cells (Figure S1F). We investigated the differences in global gene-transcriptome profiles among ESCs, three replicates of OSK-iPSCs, and four replicates of OSKH-iPSCs. All cell types were remarkably similar and showed a correlation coefficient (R2) of 0.99 (Figures 2B and 2C). We then examined DNA demethylation in the promoter regions of pluripotency marker genes (Figure S1H) and the differentiation potencies by teratoma formation (Figure S1I). Furthermore, we focused on genes differentially expressed between OSK-iPSCs and OSKH-iPSCs. We chose eight differentially expressed genes, which were statistically significant with more than a 2-fold difference between OSK-iPSCs and OSKH-iPSCs (p < 0.05). Many of the genes showed more similar expression patterns between OSKH-iPSCs and ESCs than between OSK-iPSCs and ESCs (Figures S2A–S2H), but the expression levels of these transcripts were heterogeneous among the replicates in the same cell type, indicating that those genes would not have a definitive role in stem cell properties. Although a definitive marker that reflects the quality of iPSCs has not been found, aberrant epigenetic silencing of the Dlk1-Dio3 gene cluster could indicate developmental potency, particularly because it contributes to the ratio of chimerism in mouse iPSCs (Stadtfeld et al., 2010). We analyzed the methylation status of an intergenic differentially methylated region (IG-DMR) that is located between the Dlk1 and Gtl2 genes and a Gtl2 differentially methylated region (Gtl2-DMR) (Figures 2D and 2E). Both loci in OSKH-iPSCs were highly demethylated, similarly to ESCs and MEFs. We next investigated the expression of several transcripts that increase or diminish in the early reprogramming phase (Lujan et al., 2015). Interestingly, OSKH-induced MEFs showed significant upregulation of several early reprogramming markers (Figures S2I, S2J, and S2K) and downregulation of fibroblast markers (Figures S2L and S2M) in comparison with OSK- or OSKA-induced MEFs. We confirmed that OSKH correctly traces the same pathway already reported.
Figure 2.
Characteristics of OSKH-Induced iPSCs
(A) qRT-PCR analysis of pluripotency genes and H1foo in iPSCs. Error bars represent the SEM (n = 3 independent experiments).
(B and C) Pairwise scatterplots of global gene-expression cDNA microarray patterns of OSKH-iPSCs (n = 4) compared with ESC (n = 1) (B) or OSK-iPSCs (n = 3) (C). The gray lines indicate log2 2-fold changes in gene-expression levels between the paired cell types.
(D and E) Degree of DNA methylation at IG-DMR and Gtl2-DMR in four OSK iPSC clones, four OSKH-iPSC clones, and ESCs, as well as MEFs analyzed by pyrosequencing. Error bars represent the SEM (n = 3 independent experiments).
H1foo Enhances the In Vitro Differentiation Potential
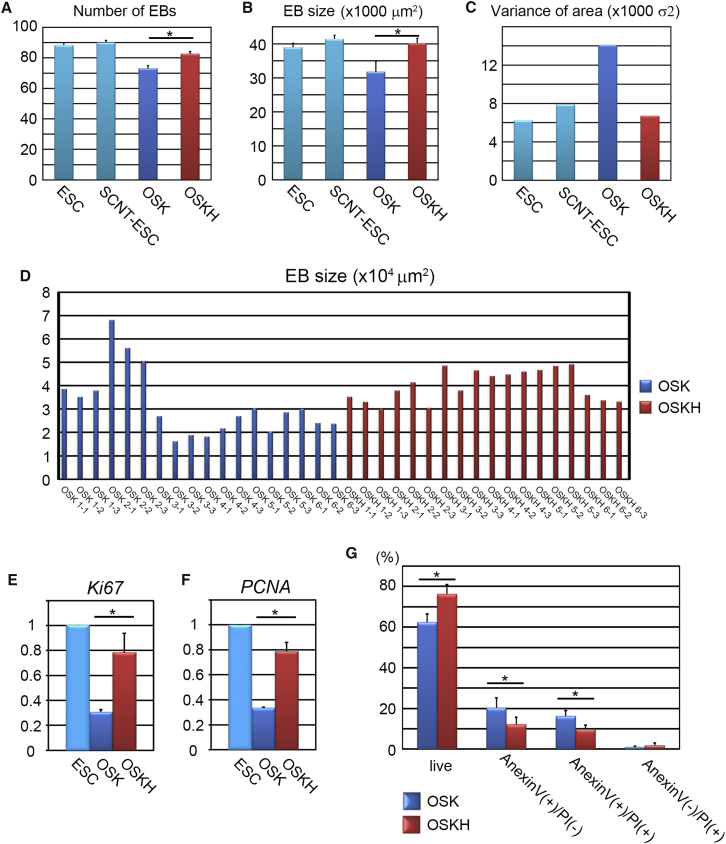
Next, we asked whether H1foo could improve the quality of iPSCs. Often iPSCs highly express pluripotency markers but show poor differentiation efficiency in vitro, including characteristics such as a low number of embryoid bodies (EBs) and frazzled EBs with poor differentiation potential. Moreover, this undesired trend is highly apparent in low-passage iPSCs and is correlated to the remnant source cell epigenetic memories (Kim et al., 2010, Polo et al., 2010). We examined the differentiation potency by EB formation in low-passage (P5) OSK-iPSCs, OSKH-iPSCs, and two types of ESCs as controls. First, we examined the number and the size of EBs at 5 days after differentiation (Figure S3A), which were more similar between OSKH-iPSCs and ESCs than between OSK-iPSCs and ESCs (Figures 3A and 3B). Furthermore, the variance of EB size was smaller in OSKH-iPSCs than in OSK-iPSCs (Figures 3C and 3D). OSKH-iPSCs expressed a higher content of proliferation markers than OSK-iPSCs (Figures 3E and 3F). However, a significant difference was not observed in the differentiation marker profiles among ESCs, OSK-iPSCs, and OSKH-iPSCs (Figure S3B). We also examined the population of apoptotic cells during differentiation. Apoptosis in OSKH-iPSCs was suppressed compared with OSK-iPSCs (Figures 3G and S3C). Taken together, the data suggest that H1foo causes iPSCs to be more adaptable to in vitro differentiation conditions and promotes the homogeneity of EBs.
Figure 3.
In Vitro Differentiation Potential in OSKH-iPSCs
(A) Number of EBs on day 5 after differentiation. Error bars represent the SEM (n = 3 independent experiments). ∗p < 0.05.
(B) Size of EBs on day 5 after differentiation. Error bars represent the SEM (n = 3 independent experiments). ∗p < 0.05.
(C) Variance of EB sizes from each ESCs and iPSCs (n = 3 independent experiments).
(D) Variation of EB sizes. Each bar represents one experiment.
(E and F) Cell-proliferation markers Ki67 (E) and proliferating cell nuclear antigen (PCNA; F) on day 2 after differentiation. Error bars represent the SEM (n = 3 independent experiments). ∗p < 0.05.
(G) Apoptotic cell distribution determined by fluorescence-activated cell sorting analysis of cells labeled with annexin V-fluorescein isothiocyanate and propidium iodide (PI) on day 1 after differentiation induction. Error bars represent the SEM (n = 3 independent experiments). ∗p < 0.05.
H1foo Enhances the In Vivo Differentiation Potential
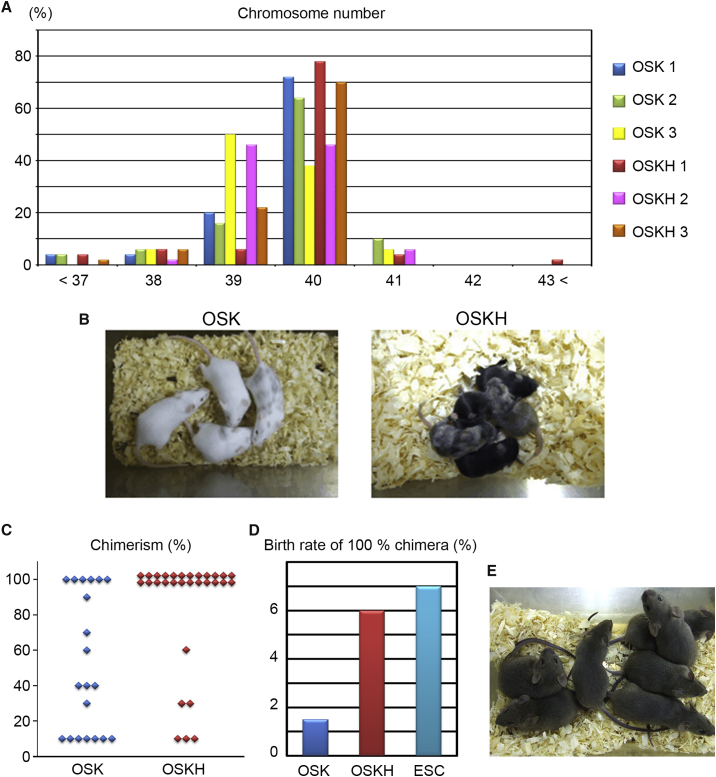
We finally investigated the in vivo differentiation potential of OSK-iPSCs and OSKH-iPSCs using a co-culture aggregation method (Eakin and Hadjantonakis, 2006). First we examined the chromosome number, which showed no significant difference in chromosomal abnormality between OSK-iPSCs and OSKH-iPSCs (Figure 4A). Each iPSC clone was aggregated with 100 ICR mouse embryos at the eight-cell stage, and chimeric embryos were transferred into the uteri of pseudopregnant females. Notably, OSKH-iPSCs generated more live chimeras, with higher chimerism, than OSK-iPSCs (Figures 4B, 4C, S4B, and S4C). Moreover, some OSKH-iPSC clones generated more 100% chimeras than ESCs (Figure S4B). Germline transmission potential is one of the most stringent hallmarks of pluripotent stem cells. We examined the germline transmission from 100% chimeric mice by in vitro fertilization. OSKH-iPSCs generated many pups with colored coats, confirming the favorable germline transmission potential of OSKH-iPSCs (Figures 4E, S4D, and S4E). We did not find any phenotypic abnormalities in these pups.
Figure 4.
In Vivo Differentiation Potential in OSKH-iPSCs
(A) Chromosome number of each iPSC.
(B) Chimeric mice generated from OSK-iPSCs or OSKH-iPSCs.
(C) Number of agouti coat colored chimeric mice derived from each of four replicates of iPSCs.
(D) Birth rate of 100% chimeric mice (100% chimeric mice/embryos transferred).
(E) The coat color of pups from a 100% chimeric OSKH mouse shows germline transmission.
Discussion
H1foo promoted the generation of Nanog-GFP-positive colonies when it was co-expressed with OSK. Notably, the proportion of GFP-positive colonies was improved to 90%. OSKH-iPSCs demonstrated differentiation potency more similar to that of ESCs than did OSK-iPSCs, especially with respect to in vitro EB formation, chimerism, and germline transmission in vivo. We also examined the effect of H1foo with Oct4, Sox2, Klf4, and c-Myc, but the number of alkaline phosphatase-positive colonies and Nanog-GFP colonies, and the proportion of GFP-positive colonies were not significantly different to those of OSKM-iPSCs. c-Myc promotes iPSC generation but is not essential for reprogramming. On the other hand, c-Myc lowers the proportion of Nanog-GFP-positive colonies and increases the tumorigenicity of cells (Nakagawa et al., 2008). Therefore, it is preferable to omit c-Myc in iPSC generation, although it is still commonly used to promote reprogramming efficiency. We showed here that H1foo could be substituted for c-Myc in terms of reprogramming efficiency and showed superiority with respect to qualifying iPSCs.
Recent studies have demonstrated that oocyte constituents play a key role in somatic cell reprogramming in SCNT. Co-expression of maternal-specific factors in oocytes, such as Glis1 (Maekawa et al., 2011) and TH2A/TH2B (Shinagawa et al., 2014), enhances the reprogramming efficiency of iPSC generation. Investigation of maternal-specific factors in oocytes has great potential for innovating somatic cell reprogramming and deciphering the reprogramming mechanisms (Gurdon and Wilmut, 2011). H1foo is specifically expressed during the germinal vesicle stage and is essential for oocyte maturation (Furuya et al., 2007, Gao et al., 2004). Interestingly, exogenous expression of H1foo in ESCs leads to the prevention of differentiation in vitro due to continuous pluripotency gene activation (Hayakawa et al., 2012). In our study, H1foo was properly silenced in generated iPSCs, which induced successful reprogramming but did not hinder the differentiation potency of OSKH-iPSCs.
The detailed molecular mechanisms regarding how H1foo enhances the reprogramming efficiency in iPSC generation and why OSKH-iPSCs exhibit improved quality remain elusive. The higher-order chromatin structure is crucially dependent on architectural chromatin proteins, including the family of linker histone proteins. Although somatic cells contain numerous linker histone variants, only one, H1foo, is present in mouse oocytes (Tanaka et al., 2001). In the mouse egg, somatic linker histones in sperm-derived chromatin are rapidly replaced by H1foo after fertilization (Tanaka et al., 2001). In SCNT oocytes, the somatic linker histone H1c in the donor chromatin is also rapidly replaced by H1foo in mice (Gao et al., 2004, Teranishi et al., 2004). In Xenopus SCNT, oocyte-specific linker histone B4 loading to genome-wide somatic chromatin is required for successful reprogramming (Jullien et al., 2010, Jullien et al., 2014, Miyamoto et al., 2007). In the early phase of the reprogramming process, global loss of histone H3 lysine 27 trimethylation (H3K27me3) occurs and epigenetic modification affects the status of heterochromatin (Hussein et al., 2014). In our study, H1foo reduced the heterochromatin area, which is consistent with previous reports that H1foo keeps chromatin looser than somatic H1 and other linker histones, and may support the generation of a more suitable chromatin state for reprogramming. These data suggest that dominant occupancy of oocyte-specific linker histone in donor chromatin may be required for successful reprogramming and might erase the parental epigenetic status. To determine whether innate H1foo would cooperatively induce reprogramming during iPSC generation by OSK, we examined H1foo expression during iPSC generation by OSK. We did not detect H1foo expression, which suggests that H1foo is not essential for OSK-dependent reprogramming. Therefore, we did not perform loss-of-function experiments such as H1foo knockdown by small interfering RNA.
H1foo induced successful reprogramming for iPSC generation in a stringent assay, thus contributing to chimerism and germline transmission. Although in vivo experiments cannot be performed in humans, it is important to generate high-quality iPSCs without variation among different iPSC lines.
Experimental Procedures
Details of all procedures are available in Supplemental Experimental Procedures.
All experiments were performed in accordance with the Keio University Animal Care Guidelines and were approved by the Ethics Committee of Keio University (20-041-4), which conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Author Contributions
S.Y. designed the research. A.K., S.Y., F.S., Y.S., T.S., D.K., S.K., M.T., S.T., H.H., T.E., Y.T., S.M., S.T., H.O., K.Y., H.W., M.O., and R.K. performed the research. T.N. conducted the electron microscopic observation. A.K., S.Y., F.S., Y.M., H.O., K.Y., M.T., and K.F. analyzed the data. A.K. and S.Y. wrote the paper.
Acknowledgments
Nanog-GFP-IRES-puro transgenic mice were kindly provided from Dr. Shinya Yamanaka. Several ESCs were kindly donated: R1 ESC from Dr. John C. Roder, B6J-23ˆ(UTR) ESC from Dr. Fumihiro Sugiyama, and SCNT-ESC (B6mt-1) from Riken BioResource Center. This study was supported in part by research grants from Grants-in-Aid for Scientific Research (JSPS KAKENHI grant numbers 24117716, 26670408, 15H01521, 15K14431), the Highway Program for Realization of Regenerative Medicine from Japan Science and Technology Agency, the Program for Intractable Diseases Research utilizing Disease-specific iPSCs from the Japan Agency For Medical Research and Development (AMED), Translational Research Network Program from AMED, and Keio University Medical Science Fund. H.O. is a Founding Scientist and a paid SAB of San Bio Co. Ltd.
Published: May 26, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.04.015.
Accession Numbers
The accession number for the microarray data reported in this paper is GEO: GSE79515.
Supplemental Information
References
- Apostolou E., Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M., Becker A., Miyara F., Han Z., Kihara M., Brown D.T., Hager G.L., Latham K., Adashi E.Y., Misteli T. Differential in vivo binding dynamics of somatic and oocyte-specific linker histones in oocytes and during ES cell nuclear transfer. Mol. Biol. Cell. 2005;16:3887–3895. doi: 10.1091/mbc.E05-04-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne J.A., Simonsson S., Western P.S., Gurdon J.B. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 2003;13:1206–1213. doi: 10.1016/s0960-9822(03)00462-7. [DOI] [PubMed] [Google Scholar]
- Eakin G.S., Hadjantonakis A.-K. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Nat. Protoc. 2006;1:1145–1153. doi: 10.1038/nprot.2006.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D., Chen A.E., Saphier G., Ichida J., Fitzgerald C., Go K.J., Acevedo N., Patel J., Baetscher M., Kearns W.G. Reprogramming within hours following nuclear transfer into mouse but not human zygotes. Nat. Commun. 2011;2:488. doi: 10.1038/ncomms1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Lu S.J., Klimanskaya I., Gomes I., Kim D., Chung Y., Honig G.R., Kim K.S., Lanza R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- Furuya M., Tanaka M., Teranishi T., Matsumoto K., Hosoi Y., Saeki K., Ishimoto H., Minegishi K., Iritani A., Yoshimura Y. H1foo is indispensable for meiotic maturation of the mouse oocyte. J. Reprod. Dev. 2007;53:895–902. doi: 10.1262/jrd.19008. [DOI] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A.A., Manor Y.S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gao S., Chung Y.G., Parseghian M.H., King G.J., Adashi E.Y., Latham K.E. Rapid H1 linker histone transitions following fertilization or somatic cell nuclear transfer: evidence for a uniform developmental program in mice. Dev. Biol. 2004;266:62–75. doi: 10.1016/j.ydbio.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B., Wilmut I. Nuclear transfer to eggs and oocytes. Cold Spring Harbor Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon J.B., Elsdale T.R., Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Ohgane J., Tanaka S., Yagi S., Shiota K. Oocyte-specific linker histone H1foo is an epigenomic modulator that decondenses chromatin and impairs pluripotency. Epigenetics. 2012;7:1029–1036. doi: 10.4161/epi.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbar P.B., Archer T.K. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J. Biol. Chem. 2008;283:4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein S.M., Batada N.N., Vuoristo S., Ching R.W., Autio R., Narva E., Ng S., Sourour M., Hamalainen R., Olsson C. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- Hussein S.M., Puri M.C., Tonge P.D., Benevento M., Corso A.J., Clancy J.L., Mosbergen R., Li M., Lee D.S., Cloonan N. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- Jullien J., Astrand C., Halley-Stott R.P., Garrett N., Gurdon J.B. Characterization of somatic cell nuclear reprogramming by oocytes in which a linker histone is required for pluripotency gene reactivation. Proc. Natl. Acad. Sci. USA. 2010;107:5483–5488. doi: 10.1073/pnas.1000599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jullien J., Miyamoto K., Pasque V., Allen G.E., Bradshaw C.R., Garrett N.J., Halley-Stott R.P., Kimura H., Ohsumi K., Gurdon J.B. Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming. Mol. Cell. 2014;55:524–536. doi: 10.1016/j.molcel.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M.J., Ji H., Ehrlich L.I. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E., Zunder E.R., Ng Y.H., Goronzy I.N., Nolan G.P., Wernig M. Early reprogramming regulators identified by prospective isolation and mass cytometry. Nature. 2015;521:352–356. doi: 10.1038/nature14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M., Yamaguchi K., Nakamura T., Shibukawa R., Kodanaka I., Ichisaka T., Kawamura Y., Mochizuki H., Goshima N., Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474:225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Furusawa T., Ohnuki M., Goel S., Tokunaga T., Minami N., Yamada M., Ohsumi K., Imai H. Reprogramming events of mammalian somatic cells induced by Xenopus laevis egg extracts. Mol. Reprod. Dev. 2007;74:1268–1277. doi: 10.1002/mrd.20691. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Narsinh K.H., Sun N., Sanchez-Freire V., Lee A.S., Almeida P., Hu S., Jan T., Wilson K.D., Leong D., Rosenberg J. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J. Clin. Invest. 2011;121:1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noggle S., Fung H.L., Gore A., Martinez H., Satriani K.C., Prosser R., Oum K., Paull D., Druckenmiller S., Freeby M. Human oocytes reprogram somatic cells to a pluripotent state. Nature. 2011;478:70–75. doi: 10.1038/nature10397. [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T., Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Liu S., Figueroa M.E., Kulalert W., Eminli S., Tan K.Y., Apostolou E., Stadtfeld M., Li Y., Shioda T. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz S., Diep D., Gore A., Panopoulos A.D., Montserrat N., Plongthongkum N., Kumar S., Fung H.L., Giorgetti A., Bilic J. Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:16196–16201. doi: 10.1073/pnas.1202352109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H., Ohsumi K., Aihara H., Ito T., Hirose S., Ura K., Kaneda Y. Linker histone variants control chromatin dynamics during early embryogenesis. Proc. Natl. Acad. Sci. USA. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa T., Takagi T., Tsukamoto D., Tomaru C., Huynh L.M., Sivaraman P., Kumarevel T., Inoue K., Nakato R., Katou Y. Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell. 2014;14:217–227. doi: 10.1016/j.stem.2013.12.015. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach O.C., Wolffe A.P., Rupp R.A. Somatic linker histones cause loss of mesodermal competence in Xenopus. Nature. 1997;389:395–399. doi: 10.1038/38755. [DOI] [PubMed] [Google Scholar]
- Taapken S.M., Nisler B.S., Newton M.A., Sampsell-Barron T.L., Leonhard K.A., McIntire E.M., Montgomery K.D. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat. Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tamada H., Van Thuan N., Reed P., Nelson D., Katoku-Kikyo N., Wudel J., Wakayama T., Kikyo N. Chromatin decondensation and nuclear reprogramming by nucleoplasmin. Mol. Cell Biol. 2006;26:1259–1271. doi: 10.1128/MCB.26.4.1259-1271.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Hennebold J.D., Macfarlane J., Adashi E.Y. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kihara M., Meczekalski B., King G.J., Adashi E.Y. H1oo: a pre-embryonic H1 linker histone in search of a function. Mol. Cell Endocrinol. 2003;202:5–9. doi: 10.1016/s0303-7207(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kihara M., Hennebold J.D., Eppig J.J., Viveiros M.M., Emery B.R., Carrell D.T., Kirkman N.J., Meczekalski B., Zhou J. H1FOO is coupled to the initiation of oocytic growth. Biol. Reprod. 2005;72:135–142. doi: 10.1095/biolreprod.104.032474. [DOI] [PubMed] [Google Scholar]
- Teranishi T., Tanaka M., Kimoto S., Ono Y., Miyakoshi K., Kono T., Yoshimura Y. Rapid replacement of somatic linker histones with the oocyte-specific linker histone H1foo in nuclear transfer. Dev. Biol. 2004;266:76–86. doi: 10.1016/j.ydbio.2003.10.004. [DOI] [PubMed] [Google Scholar]
- Wakayama T., Perry A.C.F., Zuccotti M., Johnson K.R., Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.