Abstract
Babesia species are obligate intraerythrocytic tick-borne protozoan parasites that are the etiologic agents of babesiosis, a potentially life-threatening, malaria-like illness in humans and animals. Babesia-infected people have been known to suffer from complications including liver problems, severe hemolytic anemia, and kidney failure. As reported by the Food and Drug Administration, 38% of mortality cases observed in transfusion recipients were associated with transfusion transmitted diseases of which babesiosis is the chief culprit. As of now, no tests have been licensed yet for screening blood donors for babesiosis. Current diagnostic tools for babesiosis including enzyme-linked immunosorbent assay, fluorescence in situ hybridization, and polymerase chain reaction are expensive and burdened with multifarious shortcomings. In this research, a low-cost, high-specificity, quick, and easy-to-use insulator-based dielectrophoretic diagnostic tool is developed for characterizing and concentrating Babesia-infected cells in their homogenous mixture with healthy cell population. In this work, a mixture of Babesia-infected (varying parasitemia) and healthy red blood cells (RBCs or erythrocytes) was exposed to non-uniform electric fields in a fabricated microfluidic platform to manipulate and sort the Babesia-infected cells within a minute. At DC voltage configurations of 10 V and 0/6 V in the inlet and the two outlet channels, respectively, the diseased cells were seen to flow in a direction different from the healthy RBCs. Bright field and fluorescence microscopy were utilized to present qualitative differentiation of the healthy erythrocytes from the infected cells. The proposed micro device platform was able to enrich RBCs from 0.1% to ∼70% parasitemia. This device, when finally developed into a point-of-care diagnostic chip, would enhance the detection of Babesia-infected erythrocytes and as well serve as a precursor to babesiosis vaccine development.
I. INTRODUCTION
Babesia species are tick-borne, tick-transmitted apicomplexan haemoprotozoan parasites that are the etiologic agents of babesiosis, in animals and humans. Babesia species have recently emerged as a growing public health concern for humans, primarily in the United States. The initial U.S. case of human babesiosis was first reported from California in 1966.1 Since then, there has been a substantial growth in reported cases made to the Centre for Disease Control and Prevention (CDC). In 2013, the national surveillance conducted in 27 states reported 1762 cases2 with the Northeast and upper Midwest regions as the most endemic.3,4 Babesia species are naturally transmitted to humans and other mammals through the bite of infected ixodid ticks.5 Infection with Babesia produces a spectrum of diseases that can range from asymptomatic to severe, life-threatening illnesses.1 Some patients who apparently resolve infections based on symptoms, via self-cure or chemotherapy, can permanently maintain low level parasitemia,6 which is often very difficult to detect, even by the state-of-art sensitive real-time polymerase chain reaction (PCR) assays.7,8 These asymptomatic, chronically infected persons, therefore, are probably the main source of secondary transmission of Babesia, i.e., by blood transfusion.3
As of today, a variety of interventions have been made to prevent the transmission of pathogenic agents by blood donation or transfusion. One such screening technique is based on indirect immunofluorescent antibody (IFA) testing;9 a method that detects both IgM (immunoglobulin M) and IgG antibodies in Babesia-infected red blood cells (RBCs).10,11 The presence of IgG antibodies is an indication of present or past infections, including those in which the infection may have cleared.10 However, IFA is resistant to automation and is not readily amenable to the high throughput need for testing of blood although it displays high sensitivity, specificity, and reproducibility.10
Peripheral blood smear also has been widely used.12 This method is only useful at the acute stage of infection when the parasitemia levels are at their highest.2,5 At the chronic phase of infection, detection is rarely observed on blood smears due to the low percentage of parasitized erythrocytes (PPE). In contrast to peripheral blood smears, PCR assay is considered more sensitive for detecting the presence of the parasites in both acute infections13,14 and, to a lesser extent, chronic Babesia infections. However, parasitemia diminishes with time and detection by PCR is difficult after about 2 months.6 EIA/enzyme-linked immunosorbent assay (ELISA), which uses recombinant antigens, has also been developed,3,15 but the time frame needed to obtain the results of the test is considerably substantial. The afore-mentioned shortcomings depict a dire need for better blood screening methodologies especially at donation centers.
A notable observation during the asexual growth cycle of Babesia parasites in a natural host is the attachment, penetration, and internalization of the host RBCs by Babesia's extracellular merozoites.1,16 After internalizing the host RBC, they asexually multiply and come out of the RBC by rupturing it. The invasion causes ridge formation on the surface of the RBCs as well as modification to the adhesive, mechanical, structural, and functional properties of the RBC.17 This research hypothesizes that these invasions affect the electrophysiological properties of the infected RBCs. This is based on the report that the invasion of RBCs by Plasmodium falciparum (a closely related Apicomplexan-Aconoidasidaic protozoan pathogen) affected the electro-physiological properties of the Plasmodium-infected RBCs.18 For this reason, we theorize that the difference in the dielectric properties between the infected and healthy RBCs could be utilized to ensure their dielectrophoretic separation. Among many microfluidic techniques available to manipulate cells, dielectrophoresis (DEP) has been proved to sort cells based on the subtle differences observed in their electrical properties.19–21
DEP, a non-destructive electrokinetic transport technique, manipulates cells by creating non-uniformity in the electric field in the microchannel. Aside manipulation, the tool has also been used for the separation and detection of bioparticles.22–24 Traditional (classical) DEP uses embedded microelectrodes positioned in a spatially non-uniform manner to achieve particle separation, trapping, and focusing by applying AC electric fields to induce motion.22–24 However, challenges with bubble formation due to electrolysis, electrode fouling and delamination, sample contamination, and decaying electric field as the distance from the electrode surface progresses, were major factors that led to the introduction of insulator-based DEP (iDEP).22,25 In iDEP, electrodes are usually placed far outside the channel (in inlet and outlet ports) in order to mitigate fouling and other disadvantages of classical DEP. DC fields are then applied using these electrodes: a phenomenon termed as DC-iDEP. Recent advancement in iDEP has witnessed the application of AC fields at diverse frequencies.26,27 In DC-iDEP or low frequency AC-iDEP, electrical characteristics of the cell membrane are usually leveraged for cellular clarification but high-frequency AC-iDEP seeks to explore the contributions of the cytoplasmic and/or nucleoplasmic peculiarities26 depending on the range of frequencies in consideration. Several researchers have applied iDEP to successfully achieve their set targets.23 However, as of current state-of-art, this is the first research that is being reported to use iDEP in separating Babesia-infected RBCs within a homogenous sample containing healthy RBCs too.
In this novel work, it is demonstrated for the first time that when a mixture of Babesia-infected RBCs and healthy RBCs are subjected to non-uniform electric fields in a microchannel embedded with insulated saw-tooth shaped obstacles, they can be substantially concentrated and separated. The results obtained from this work demonstrate the potency of iDEP microfluidic platform as an electrokinetic portable point-of-care tool for screening donors' blood for possible protozoan infections (i.e., babesiosis in this research) at donation centers where there is a significant need. Utilizing iDEP technology in concentrating Babesia-infected RBCs could also generate the high parasitemia desired in preliminary research works on microbial attenuation; one of the steps in the development of vaccine.
II. THEORY
According to the Newton's second law of motion, electrokinetic and dielectrophoretic forces are some essential external forces that dictate the direction of movement of cells, flowing under non-uniform electric field, within a microchannel. Electrokinetic (EK) forces account for electrophoretic (EP) and electro-osmotic (EO) forces due to the material of the microchannel itself and the suspending buffer medium. When suspended cells are made to flow in a microchannel, the flow can be electro-osmotically driven owing to the formation of electric double layer of counter ions at the walls.28 When an electric field is applied across the channel, the ions in the double layer move towards the electrode of opposite polarity. This creates motion of the fluid near the walls and transfers via viscous forces into convective motion of the bulk fluid: a phenomenon called electro-osmotic pumping. Under this flow condition, the cells experience hydrodynamic drag force (Eq. (1)), which represents a balance between stokes frictional force (acting on the interface between the fluid and the particle) and electro-osmotic force29
| (1) |
where dynamic viscosity of the medium in which the cells will be suspended, a = radius of the cell, and , are cell and electro-osmotic flow velocities, respectively. In iDEP-based microchannel, hurdles are created within the microchannel to generate non-uniform electric field using DC or low frequency AC supply. The strength of the electric field is known to be highest at the peak of the hurdle (Figure 1). Cells suspended in the medium (buffer) are momentarily polarized at the peak of the hurdle owing to the strength of the electric field at that point. Depending on the polarizability of the cells relative to the medium, the dielectrophoretic force (DEP) (Eq. (2)) acting at this hurdle tip (Figure 1) can cause cells to be separated
| (2) |
where V = volume of the particle, = permittivity of the medium, = conductivity of the cell. = conductivity of the medium, and = gradient of electric field magnitude. The term represents the Clausius-Mossotti factor, which determines the movement of the cells away or toward high field region. The extent of separation is usually a function of the amount of the induced dielectrophoretic force experienced by the cells. Therefore, to design an effective electrokinetic-based microfluidic platform, it is important to analyze the effects of the above parameters on cells translocation using mathematical models and numerical simulations. Also, the mobility equations (Eqs. (3)–(5)), which seek to relate the strength of the electric field to velocity have also been utilized in tracking bioparticle along a microfluidic channel
| (3) |
| (4) |
| (5) |
where is the medium viscosity (Pa s), , , and are, respectively, electroosmotic, electrophoretic, and dielectrophoretic mobilities. is the electric field, and are the permittivity of the vacuum, permittivity of the medium, zeta potentials of the particle, and zeta potential of the channel material respectively. Velocity of the cell is usually a function of the drag force acting within the microchannel (Eq. (1)).
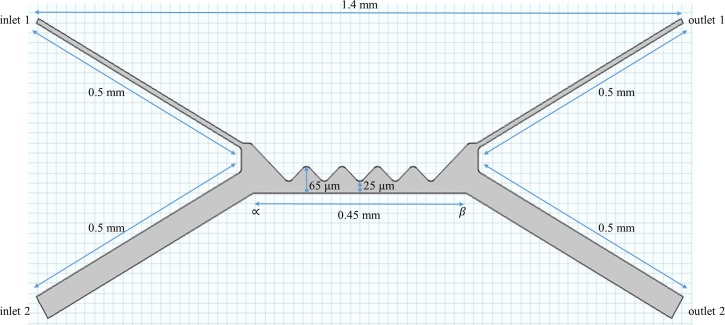
FIG. 1.
The schematic representation of the microdevice with two inlet and two-outlet ports. The entire device is about 1.4 mm long with embedded saw-tooth geometry to create non-uniformity in the electric field.
III. MATERIALS AND METHODS
A. Computational simulation
Simulation of the trajectories or motion of both the healthy and infected RBCs was done using COMSOL Multiphysics 5.0 (COMSOL, Inc., Burlington, MA, USA) commercial software package. The essence of simulation was to optimize the geometry of the iDEP-based microdevice as well as the operating voltage that would be sufficient to generate the appropriate non-uniform electric field gradient such that the cells experience varying dielectrophoretic forces based on their electrophysiological properties. The data used for the infected RBCs in the simulation were obtained for P. falciparum, the etiologic agent for malaria in human. Due to the fact that both Babesia and Plasmodium have been studied to show very similar characteristics with respect to pathogenesis and clinical course30 and are of similar life cycle,31 they have been reported to show very similar characteristics, hence, would yield comparable electrical manipulation response.31 The boundary conditions were based on the non-uniformity caused by the electric field gradient within the channel and the channel wall was assumed as insulated.
Flow dynamics of the suspended cells was modeled using the synergy of Navier-Stokes and continuity equations while assuming incompressible creeping flow and no slip boundary conditions. The density of water (1050 kg/m3) and viscosity (0.001 Pa s) were also used because the buffer was assumed to have conditions similar to water (at ambient temperature) but for its pH and conductivity. Electric field regime combined the generalized ohms law with the Gauss' law and continuity equation to handle the stationary electric current in conductive media. The transport of diluted species (healthy and infected RBCs) was also accounted for using the mass conversation equation. Fluid flow, mass transport, and electric flow fields were solved separately, and the interface condition was matched up iteratively to get the solutions.
Based on the observation from the simulation, a new geometry for the microfluidic device was sequentially designed until continuous separation was observed. Table I shows the properties used in the computational simulation.
TABLE I.
The electrophysiological properties used in simulating the trajectories of the RBCs. The values of relative permittivity and conductivity of infected and uninfected cells were as reported by Gascoyne et al.32 Conductivity of medium , viscosity of medium , dielectric constant of medium (at 25 °C) , permittivity of vacuum , electrophoretic (EO) mobility (calculated).
| Properties | Healthy RBC | Infected RBC |
|---|---|---|
| Zeta potential (mV) | ||
| Radius ( | ||
| Concentration (mol/m3) | 1.0 | 0.01 |
| DEP mobility (a | ||
| EP mobility (a | ||
| Clausius-Mossotti factora |
Calculated.
B. Device layout
The geometry of the 1.4 mm microdevice is as shown in Figure 1. It consists of two inlet and outlet arms each of which measures 0.5 mm long. The insulating region is made up of an array of hurdles, which are slightly filleted at the tip to provide the appropriate electric field strength for generating the negative dielectrophoretic force required for sorting the cells. The length |∝β| represents the length of the hurdle region. The distance 25 μm between the peak of the hurdle and surface of the length |∝β| was set to make the cells experience adequate dielectrophoretic force. Inlet 2 (containing only the buffer) helps in adding impetuous to the electroosmotic contribution within the channel, thus assists in focusing the RBCs towards the separation zone.
C. Fabrication process
A 3 M silicon wafer (fabricated by Trianja Technologies, Inc., TX, USA) with a pattern made through wet etching was used to cast the Polydimethylsiloxane (PDMS) device platform. The etched silicon wafer referred here as the master had 35 designs per chip. Rapid prototyping using the commercially fabricated master was initiated by mixing PDMS monomer (sylgard 184 silicon elastomer base) and a curing agent (sylgard 184 silicon elastomer curing agent) in 10:1 w/w ratio followed by degassing the mixture using Dekker Vacuum Pump/Degassing set-up for about 20 min. The air-free PDMS slurry mixture was poured onto the master, which was contained in a 10 cm polystyrene Petri dish. The polymer was cured in (Blue M) automated oven at 80 °C for 1 h, and then peeled off from the master. The master was ensured leveled during the curing process so as to disregard any geometrical variations. Peeled PDMS device with indented channels were punched with a 3 mm Miltex biopsy puncher to obtain the ports/reservoirs for the inlet and outlet channels. The device was then cut and fitted onto a 0.17 mm thick corning borosilicate micro cover glass (size 24 mm × 40 mm). The PDMS was finally exposed to plasma treatment by the use of Harrick PDC-32G plasma cleaner/sterilizer at 300 mTorr, for 1-min to irreversibly bond onto a clean cover slide and to close the microchannels. The plasma treatment also ensured microdevice hydrophilicity generation, due to which external devices were not required to pump in the samples into the microchannel. The sealed device was placed in the petri dish with channels filled with deionized water to maintain its hydrophilicity.
D. Cell culturing and preparation
1. Cell culturing
The in vitro culturing technique for Mo7 strain of Babesia bovis involved cultivating B. bovis infected erythrocytes in a microaerophilous phase (MASP) using 24 well suspension plates, at 10% (v/v) packed cell volume (PCV), and incubated at 37 °C in a 5% CO2 in air humidified atmosphere. Cultures were maintained in M-199 culture medium (Gibco, 22340020) supplemented with 50 μg/ml gentamicin (Gibco, 15710-049), 1% (v/v) fungizone (Gibco, 15290-026), 20 mM N-Tris(Hydroxymethyl) Methyl-2-aminoethane sulfonic acid (Sigma-Aldrich, T5691) and 40% (v/v) bovine serum. Subcultivation was performed by splitting/dilution with fresh normal bovine erythrocytes and M-199 medium when the achieved parasitemia levels were about 2%–3%. Parasitemia was monitored by microscopic examination of Giemsa stained thin smears under a 100× microscope oil objective. The method described above was as specified by Michael G. Levy and Miodrag Ristic group.33 DNA from infected erythrocytes was extracted and tested for gene specific to Babesia pathogens of cattle including B. bovis and B. bigemina. However, B. bovis was the only species detected in the tissue culture used in the experiments.
2. Experimental cell-sample preparation
50 g/ml dextrose buffer solution was prepared by dissolving 25 g of dextrose crystalline solids (weighed with 204 Mettler Electronic Weighing Balance) in 50 ml de-ionized water. The conductivity and pH of the medium (buffer) were measured to be 0.052 S/m and 7.04, respectively, using Accumet XL 200 Ph/mV/conductivity meter. 10% parasitemia erythrocytes were centrifuged at 1500 rpm for 5-min. 1 μl (packed cell volume) of this cell sample was measured and serially diluted with non-infected (normal) erythrocytes until 0.0001% parasitemia was achieved. This was done to determine the lowest percentage parasitemia that the microdevice can detect. Within the confinement of a 1300 Series A2 Bio-Safety Cabinet, 1 μl of each of the cell samples was measured and transferred into a 1 ml micro test-tube containing 600 μl low-conductivity freshly prepared buffer. The buffer was of low-conductivity to forestall any possible heat generation that could lyse the RBCs during their exposure to DC voltages. Cells were used for experiment approximately 30 min after culturing. This was done to ensure that the cell membrane, upon which DC-iDEP relies, retained its characteristics.
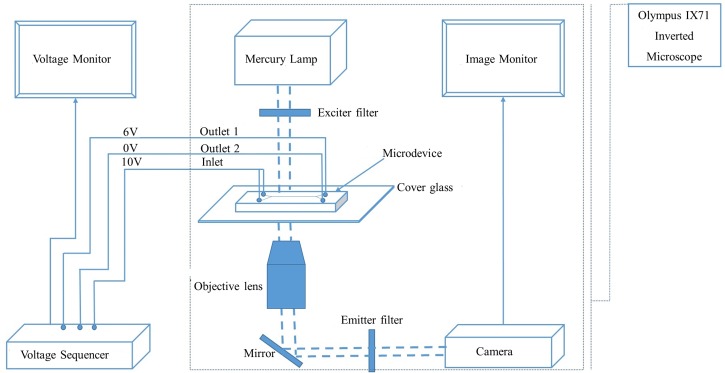
E. Experimental set up
The experimental set-up consists of an integration of the LabSmith HVS448 high voltage sequencer with an IX71 Olympus inverted microscope as shown in Figure 2. The microdevice, mounted on the IX71 Olympus inverted microscope was entirely filled with the low conductivity dextrose medium and 0.008 in. diameter pure platinum electrodes were inserted into the inlet and outlet reservoirs as means of electrical connections. At the inlet channel 1 (Figure 1), the buffer solution was sucked off and replaced with a mixture of healthy and infected-RBCs (5.5% PPE) suspended in the dextrose medium. Having ensured no pressure head, DC voltages were applied using HVS448 high voltage sequencer. Inlets 1 and 2 (Figure 2) were connected to the same voltage source (10 V) while the outlet 2 was connected to the ground (0 V). The voltage at outlet 1 (Figure 2) was swept from 5.5 to 6.5 V to visualize the trajectories of the cells within the microchannel. Sequential sorting of the cells was observed and appropriate images captured. The experiment was conducted in two phases: pre-separation staining (before introducing the sample into microchannel) and post-separation staining stage (after the experimental run- at the outlet ports). In the first phase, cell samples were labeled with green fluorescent protein (GFP) (only infected RBCs got stained owing to the presence of nucleated Babesia cells within the healthy RBCs) and experimented for optimal detection of the traditional fluorescein isothiocyanate (FITC). This pre-separation staining method enabled the visualization of the green-labeled Babesia nucleus as the infected RBCs migrated within the iDEP-based microchannel. In the second phase (post-separation staining phase), microscopic examinations were carried out under bright field by staining the separated cells at the outlet ports. Separated RBCs in each of the outlet ports were stained with 3-stage Siemens diff quik stain kit set to visualize the corresponding proportions of healthy and Babesia-infected RBCs. Quantitative analyses were carried out, and the percentage parasitized erythrocytes (PPE) were calculated at both outlet ports. The whole process was repeated six (6) times for each of the samples (10.0%–0.0001% parasitemia).
FIG. 2.
The schematic diagram of the experimental set-up showing the integration of the voltage sequencer with the Olympus IX71 Inverted Microscope.
IV. RESULTS AND DISCUSSION
The application of iDEP to concentrate and sort biological cells is not only about seeing the cells move to different destinations after experiencing DEP force. It goes further to the examination of the cell contents at such destinations. This section shows the results obtained from both simulation and experiment and further provides insights as to how the two result streams compare.
A. Computational results
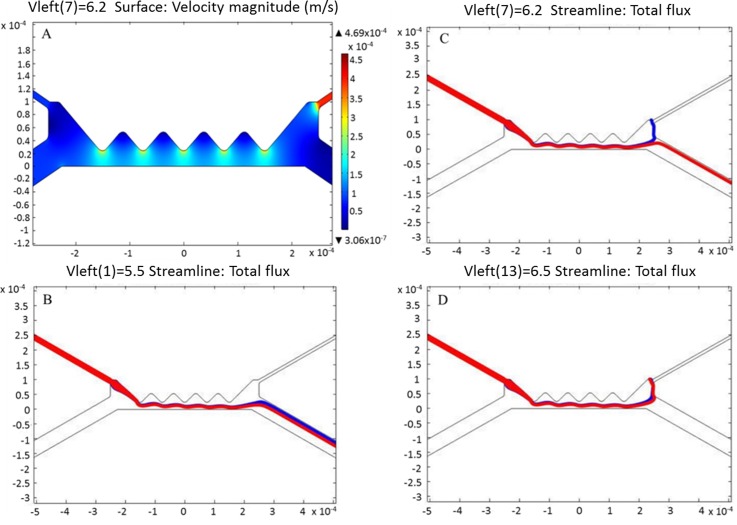
The governing equations were solved for fluid flow, mass transport, and electric field using COMSOL Multiphysics v5.1. In order to have an effective dielectrophoretic force, the simulation was made with conditions that consider both electrophoretic and electro-osmotic flow. This generated a substantial balance between the drag force and the dielectrophoretic force, which utilized the appropriate electric field strength at the peak of the saw-tooth shaped insulating hurdles embedded within the microchannel. Figure 3(a) shows the surface velocity magnitude of the flow within the microchannel. Evidently, the fluid velocity at the channel wall at a given period of time is sufficiently low. The fluid flow is, thus, at sufficient pace to enable the suspended erythrocytes experience appropriate dielectrophoretic force that separates them according to their inherent dielectric properties. The applied potential difference between the inlet and the two separate outlet ports was swept to determine the optimum DC voltage range necessary for the dielectrophoretic separation. At low voltage (below 6.2 V), the generated electric field strength was not sufficient to effect the separation of the cells. As shown in Figure 3(b), both healthy (red) and infected erythrocytes (blue) moved into the same port after passing through the insulating hurdles. In Figure 3(c), the applied voltage, 6.2 V, was adequate to make the erythrocytes experience the desired DEP force. At voltage beyond this sorting voltage, the potential to move into the upper right-hand outlet channel was so high that all cells, healthy and infected, were drawn into it (Figure 3(d)).
FIG. 3.
Computation velocity and particle flux due to the channel geometry and applied DC voltage. (a) Surface velocity magnitude within the microchannel. (b) Total flux of erythrocytes at the pre-separation stage (at voltage below 6.2 V) (c) Total flux of erythrocytes at the separation stage (at 6.2 V) (d) Total flux of erythrocytes at the post-separation stage (at voltage beyond 6.2 V). Red-color flux represents the healthy (normal) erythrocytes while the blue-color flux depicts the Babesia-infected erythrocytes.
B. Experimental validation of the sorting voltage
The dependence of iDEP force on cell size, field gradient, cell, and medium conductivities was leveraged by varying only the field gradient (since it is a function of the applied voltage). Sweeping the applied voltage, therefore, from 5.5 to 6.5 V enabled the identification of the exact range of non-uniform electric field gradient necessary for the separation between healthy and Babesia-infected RBCs population (Table II). Before 5.9 V and after 6.1 V, it can be said that the associated field strength was not sufficient for the cells to experience the required iDEP force since other factors (cell size, cell, and medium conductivities) on which the dielectrophoretic force depends, have been fixed. The application of 5.9–6.1 V DC occasioned the desired separation even though, at this time, the extent of separation could not be ascertained. However, backflow, which might have resulted from dipole re-orientation, was observed at different periods. Therefore, experiment run time was fixed at 50 s with the inlet-outlet 10–6/0 V (Figure 2) configurations to ensure maximum separation of cells.
TABLE II.
Effects of voltage sweep (in volts) in the direction of flux using dextrose buffer of conductivity 0.052 s/m. (No separation indicates that both healthy and infected RBCs were moving to the same outlet port.) PPE of the inlet sample = 10%, inlet voltage = 10 V and outlet voltage 1 = 0 V. Note: Similar trend was observed for all inlet samples (10%–0.1%).
| Outlet 2 (V) | Sorting | Initial backflow period (s) | Outlet 2 (V) | Sorting | Initial backflow period (s) |
|---|---|---|---|---|---|
| 5.5 | No separation | 0 | 6.1 | Cells separated | 44 |
| 5.6 | No separation | 0 | 6.2 | No separation | 0 |
| 5.7 | No separation | 0 | 6.3 | No separation | 0 |
| 5.8 | No separation | 0 | 6.4 | No separation | 0 |
| 5.9 | Cells separated | 41 | 6.5 | No separation | 0 |
| 6 | Cells separated | 53 |
C. Optical validation
The optical examination of the post separation outlet-ports contents at the inlet-outlet voltage configuration of 10–6/0 V is presented here. The images were captured for both fluorescence and bright-field regimes.
1. Fluorescence
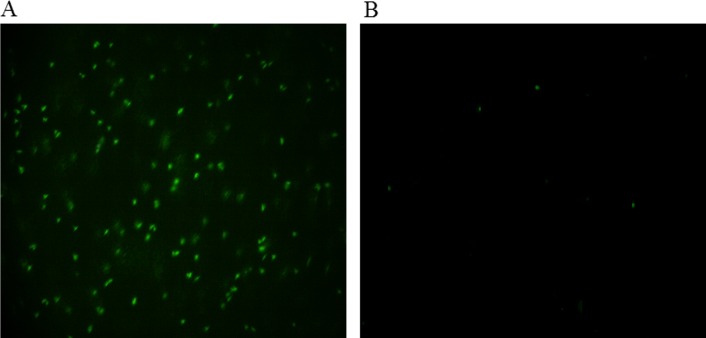
To ascertain the directional influx of the healthy and infected cells into their different ports (from the simulation results in Figure 3(c)), fluorescence microscopy with green-fluorescent-protein (GFP) pre-separation labeling was employed (Sec. III E). It was easy to trace the direction of movement of the Babesia-infected erythrocytes from the color they emitted, since the GFP binds to the nucleus of the Babesia pathogen (located within the infected-RBCs). The post-separation image examination from the pictures obtained during experimental run revealed that the labeled cell populations were indeed Babesia-infected erythrocytes due to the green emissions, which were highly concentrated in the outlet port 1 (Figure 4(a)). Some green-colored cells were seen in outlet-port 2, and this indicated that the separation was not total (Figure 4(b)).
FIG. 4.
Images of the post-separation GFP-stained Babesia-infected and healthy erythrocytes when the inlet % parasitemia was 10%. (a) Microdevice outlet port 1 (rich in Babesia-infected erythrocytes). (b) Microdevice outlet port 2 (lean in Babesia-infected erythrocytes). Magnification: 100×. The green spots indicate the Babesia parasites inside RBCs.
2. Bright field
Bright field imaging and quantification of the extent of separation was carried out using a post-separation diff-quik cell staining technique (Sec. III E for method details). This method enables the visualization of the Babesia parasite (B. Bovis) within the RBCs. Babesia-infected erythrocytes were enriched in one of the outlet ports in a similar manner as observed in the fluorescence microscopy. Figure 5 shows the images of each of the outlet ports obtained by the inverted microscope at 100×. Visualizing the Babesia parasites inside the RBCs (Figure 5) is an indication that the cells were not lysed and that the operating conditions were suitable for the sorting process. In essence, it can be said that there was no appreciable heat generation during the application of voltages across the whole length of the microdevice.
FIG. 5.
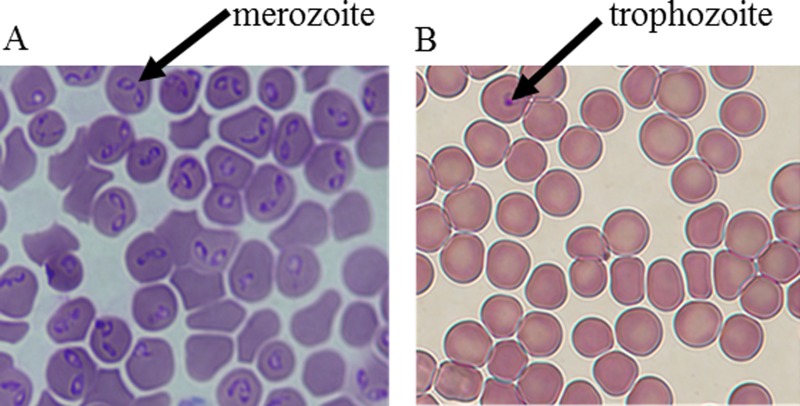
Images obtained at 100× magnification using post-separation diff-quik-stain kit showing Babesia-infected and healthy erythrocytes. Only the parasites were stained. Here, the cells were stained after they had been separated. (a) Microdevice outlet port 1 (rich in Babesia-infected erythrocytes). (b) Microdevice outlet port 2 (lean in Babesia-infected erythrocytes). The arrows show the parasite (nucleus-like structure) residing in the RBCs. The pair-shaped parasites are the merozoites while the round-shaped parasites are the trophozoites. Any RBC that contains the parasite is considered infected. The images were obtained when the inlet parasitemia was ∼5%.
D. Quantitative analysis
Quantification of the proportion of parasitized RBCs in each of the ports is essential in determining the efficiency of the microdevice. This was done to account for the specificity (the proportion of sorting and concentration of the RBCs) of the microdevice as well as it is sensitivity (the limit of inlet concentration that the microdevice can clarify). Infected cell population of 10% parasitemia was sequentially diluted with healthy RBCs, and experiments were carried out on each of the resultant concentrations.
As presented in Table III, the microdevice was sensitive to inlet parasitemia from 10% to as low as 0.1%. Between these values, 67%–70% of the infected RBCs were recovered in outlet 1 (Figure 1). The average percentage parasitemia in this outlet 1 depicts that outlet 1 is specific to the infected RBCs. Outlet 2, which should have no infected RBCs according to simulation, still has between 0.1 and 2% parasitemia even though it is largely specific to healthy RBCs. Dilution of 0.1% inlet parasitemia to 0.05% inlet parasitemia yielded no separation as the inlet cells behaved as if no infected cells were presented, hence, moved in totality to outlet 2. Further attempt to dilute and experiment with cells up to 0.00001% inlet parasitemia also yielded no separation. This implies that the microdevice can only sort cell populations whose parasitemia is ≥0.1%. On the assumption that 1 of RBCs contains RBCs, it means that the microdevice is only sensitive to blood sample, which has 5000 parasites per . As of today, PCR have been established to detect as low as 1–5 intraerythrocytic parasites per .34 but the processing time to obtain this result is 24 h and about 5 of sample is required. The result presented with our proposed microdevice however uses very less sample (∼1 ) and processing time of <1 min.
TABLE III.
Percentage recovery of infected RBCs for various concentrations of the inlet samples.
| Sample | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPE (inlet) | 10 | 5 | 4 | 3 | 2 | 1 | 0.5 | 0.1 | 0.05 | 0.01 | 0.005 | 0.0001 | 0.00001 |
| PPE (outlet1) | 70 | 68 | 68 | 68 | 68 | 67 | 67 | 67 | 0 | 0 | 0 | 0 | 0 |
| PPE (outlet 2) | 2 | 1.2 | 0.9 | 0.5 | 0.2 | 0.1 | 0.1 | 0.1 | 0.05 | 0.01 | 0.005 | 0.0001 | 0.00001 |
Kuzman et al.35 reported that a change in the elastic properties of the cell membrane would occur when the pH of the cell's suspending medium is changed. Changing the properties of the cell membrane could affect the DEP force that the cell would experience when passed through a non-uniform electric field. Therefore, throughout the experiment, the suspending medium pH of 7.04 was maintained not only to preserve the associated properties of the cell but also to provide a natural thriving environment for the RBCs. When a solution which has intracellular action mechanism is added to a suspension of viable cells, the change in the cells' dielectric properties is small.36 In this work, we introduced pre-separation green fluorescent protein (GFP) staining of cells to aid the visualization of the green-labeled Babesia cells. The post-separation quantitative analyses revealed that the percentage-parasitized erythrocytes obtained from both pre-separation and post-separation staining sets were substantially similar. This indicates that the degrees of sorting observed were pure functions of DEP effects and were, by no means, dependent on the employed stains. One point to note here is the variation in the percentage parasitemia. As reported,6 one of the defects of the current diagnostic tools for babesiosis is the inability to detect Babesia in low-parasitemia sample. Our experiment revealed that at varied parasitemia content, cell differentiation would occur in as much as the right voltage configuration is used. Nevertheless, more research works need to be done regarding the range at which low or high parasitemia content should domicile.
V. CONCLUSION
This is the first study to have reported the application of insulator-based dielectrophoresis (iDEP) to separate or enrich Babesia-infected erythrocytes. The results presented here show that at voltage configuration of 10, 0, 6 V in the inlet and the two outlet reservoirs, respectively, Babesia-infected RBCs could be isolated from their healthy counterparts. Studying the dielectrophoretic separation of Babesia-infected RBCs is the first of many steps required to develop a viable point-of-care diagnostic devices needed at blood donation centers to screen donors' blood for the notorious babesiosis disease. As demonstrated, the required sorting voltage is 6 V. This gives an indication that the point-of-care diagnostic device could be battery powered when completed. The entire separation process was completed within one minute; an indication that iDEP is a fast electrokinetic tool for identifying the dreaded parasite. DEP has been a useful tool in sorting particles, especially for bioparticles like cells. The properties associated with sorting are largely dependent on morphology, conductivity, size, and surface characteristics of the cells. Dielectric properties of the cells play a major role in determining the destination of the cells within any microfluidic channel experiencing non-uniform electric field. Simulating this sorting process before the real-time experimental endeavors is a worthwhile practice.
This work used B. bovis samples, which is assumed would be considerably similar to the B. microti, the pathogen that affects human. As observed from the sorting voltage values required to characterize the infected cells, we have a close agreement between the simulation (6.2 V) and experiments (6.0 V), which proves that simulation is a useful guide in determining the electric field strength that will be sufficient enough to occasion the desired sorting. Relative error analyses performed on the experimental data revealed an admissible confidence of >95%, and this gives credence to the entire dielectrophoretic process. It, therefore, becomes a hydra-headed endeavor to really determine where the course-effect representation of the simulation-experiment discrepancy should domicile. Worthy of note is the fact that the values of the electrophysiological properties used in the simulation were for P. falciparum—the pathogen that causes malaria. In malaria-endemic regions, however, the predisposition of misdiagnosing babesiosis for malaria is very high. It might be worthwhile to determine the crossover frequencies of the hundreds of Babesia strains and characterize them accordingly. This will be the focus of our research group in the future. The results of our proof-of-concept experiment have shown that the internalization of the Babesia cells within the RBCs actually affected the electrical properties of the RBCs. At the outset, it was only hypothesized that the electrical properties of the cells would have changed as a result of the Babesia attack. Dielectrophoresis has substantiated this claim, and researchers could count on this electrical-property-change confirmation.
Many opportunities for advancing this project abound. First, the device was able to concentrate infected RBCs (10%–0.1% PPE) to ∼70%. Both B. bovis stages: merozoites and trophozoites that occur during the infection of bovine erythrocytes were separated from health RBCs. However, the observation that trophozoites were the main parasite in outlet port 2 might be a precursor to some differential DEP response between trophozoites and merozoites. In order words, the parasites in outlet port 1 were predominantly merozoites while the 0.1%–2% parasites in outlet port 2 were substantially trophozoites.
There is a need to improve this capture rate (∼70%). Not only would this improved capture rate enhance post separation sensing, it will also increase the number of Babesia pathogen that can be extracted from the infected RBCs after separation. The extracted pathogen can be used for experiments on babesiosis vaccine formulation. In Figure 1, the gap between the tip of the hurdle and the opposite site of the microchannel is 25 μm. RBCs are known to have a diameter of 7 μm. This indicates that at a time, about three (3) RBCs might find their ways into this 25 μm-space. The result of this is some form of shielding in which one RBC experiences maximum available DEP force at the hurdle while other RBCs are presumably shielded from maximum dielectrophoretic impact. Therefore, future work will look into the possibility of reducing this 25 μm space through device optimization strategy in COMSOL Multiphysics. However, there will be a need to strike a balance between the numerically optimized microdevice and the feasibility of fabricating the simulated microdevice.
Finally, the current result has shown the capability of the proposed device to convert low parasitemia of ∼0.1% to higher parasitemia (∼70%). This makes it easier for either biosensor or PCR to detect the parasite. Hence, the device, in its current form, can be used to concentrate samples before they are fed into PCR for analyses. The act will circumvent the challenge of persistently low parasitemia, which tend to make PCR utilize large sample volumes.
ACKNOWLEDGMENTS
This work was funded by the University of Idaho, Office of Research and Economic Development (ORED) Grant. Cell culturing was fully supported by Audrey Lau and Paul Lace, United States Department of Agriculture (USDA), Department of Veterinary Medicine, Washington State University, Pullman, WA. Imaging was assisted by Dr. James Moberly and Dr. Eric Aston, Department of Chemical and Materials Engineering, University of Idaho, Moscow, ID and Dr. Ann Norton at the National Institutes of Health (NIH) Imaging Center, Department of Biology, University of Idaho, Moscow, ID.
References
- 1. Gorenflot A., Moubri K., Precigout E., Carcy B., and Schetters T. P., Ann. Trop. Med. Parasitol. 92, 489–501 (1998). 10.1080/00034989859465 [DOI] [PubMed] [Google Scholar]
- 2.United States Center for Disease Control and Prevention, “ Babesiosis surveillance,” Weekly Rep. 61, 505–509 (2012). [Google Scholar]
- 3. Kogut S. J., Thill C. D., Prusinski M. A., Lee J.-H., Backneson P. B., Coleman J. L., Anand M., and White D. J., Emerging Infect. Dis. 11, 476–478 (2005). 10.3201/eid1103.040599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meldrum S. C., Birkhead G. S., White D. J., Benach J. L., and Morse D. L., Clin. Infect. Dis. 15, 1019–1023 (1992). 10.1093/clind/15.6.1019 [DOI] [PubMed] [Google Scholar]
- 5. Homer M. J., Aguilar-Delfin I., Telford S. R., Krause P. J., and Persing D. H., Clin. Microbiol. Rev. 13, 451–469 (2000). 10.1128/CMR.13.3.451-469.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krause P. J., Spielman A., Telford S. R., Sikand V. K., McKay K., Christianson D., Pollack R. J., Brassard P., Magera J., Ryan R., and Persing D. H., New Engl. J. Med. 339, 160–164 (1998). 10.1056/NEJM199807163390304 [DOI] [PubMed] [Google Scholar]
- 7. Leiby D. A., Chung A. P., Gill J. E., Houghton R. L., Persing D. H., Badon S., and Cable R. G., Transfusion 45, 1804–1810 (2005). 10.1111/j.1537-2995.2005.00609.x [DOI] [PubMed] [Google Scholar]
- 8. Vannier E., Borggraefe I., Telford S. R., Menon S., Brauns T., Spielman A., Gelfand J. A., and Wortis H. H., J. Infect. Dis. 189, 1721–1728 (2004). 10.1086/382965 [DOI] [PubMed] [Google Scholar]
- 9. Chisholm E. S., Ruebush T. K., Sulzer A. J., and Healy G. R., Am. J. Trop. Med. Hyg. 27, 14–19 (1978). [DOI] [PubMed] [Google Scholar]
- 10. Krause P. J., Telford S. R., Ryna R., Conrad P. A., and Wilson M., J. Infect. Dis. 169, 923–926 (1994). 10.1093/infdis/169.4.923 [DOI] [PubMed] [Google Scholar]
- 11. Krause P. J., Ryan R., Telford S., Persing D., and Spielman A., J. Clin. Microbiol. 34(8), 2014–2016 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Healy G. R. and Ruebush T. K., Am. Soc. Clin. Pathol. 73, 107–109 (1980). 10.1093/ajcp/73.1.107 [DOI] [PubMed] [Google Scholar]
- 13. Brandt F., Healy G. R., and Welch M., J. Parasitol. 63, 934–937 (1977). 10.2307/3279918 [DOI] [PubMed] [Google Scholar]
- 14. Gleason N. N., Healy G. R., Western K. A., Benson G. D., and Schultz M. G., J. Parasitol. 56, 1256–1257 (1970). 10.2307/3277582 [DOI] [PubMed] [Google Scholar]
- 15. Houghton R. L., Homer M. J., Reynolds L. D., Sleath P. R., Lodes M. J., Berardi V., Leiby D. A., and Persing D. H., Transfusion 42, 1488–1496 (2002). 10.1046/j.1537-2995.2002.00215.x [DOI] [PubMed] [Google Scholar]
- 16. Hunfeld K. P., Hilderbrandt A., and Gray J. S., Int. J. Parasitol. 38, 1219–1237 (2008). 10.1016/j.ijpara.2008.03.001 [DOI] [PubMed] [Google Scholar]
- 17. Hutchings C. L., Li A., Fernandez K. M., Fletchet T., Jackson L. A., Molloy J. B., Jorgensen W. K., Lim C. T., and Cooke B. M., Mol. Microbiol. 65, 1092–1105 (2007). 10.1111/j.1365-2958.2007.05850.x [DOI] [PubMed] [Google Scholar]
- 18. Gascoyne P., Pethig R., Satayavivad J., Becker F. F., and Ruchirawat M., Biochim. Biophys. Acta 1323, 240–252 (1997). 10.1016/S0005-2736(96)00191-5 [DOI] [PubMed] [Google Scholar]
- 19. Srivastava S., Gencoglu A., and Minerick A., Anal. Bioanal. Chem. 399, 301–321 (2011). 10.1007/s00216-010-4222-6 [DOI] [PubMed] [Google Scholar]
- 20. Srivastava S. K., Baylon-Cardiel J. L., Lapizco-Encinas B. H., and Minerick A. R., J. Chromatogr. A 1218, 1780–1789 (2011). 10.1016/j.chroma.2011.01.082 [DOI] [PubMed] [Google Scholar]
- 21. Srivastava S. K., Artemiou A., and Minerick A. R., Electrophoresis 32(18), 2530–2540 (2011). 10.1002/elps.201100089 [DOI] [PubMed] [Google Scholar]
- 22. Srivastava S. K., Daggolu P. R., Burgess S. C., and Minerick A. R., Electrophoresis 29, 5033–5046 (2008). 10.1002/elps.200800166 [DOI] [PubMed] [Google Scholar]
- 23. Jubery T. Z., Srivastava S. K., and Dutta P., Electrophoresis 35, 691–713 (2014). 10.1002/elps.201300424 [DOI] [PubMed] [Google Scholar]
- 24. Ivory C. F. and Srivastava S. K., Electrophoresis 32, 2323–2330 (2011). 10.1002/elps.201100115 [DOI] [PubMed] [Google Scholar]
- 25. Adekanmbi E. O. and Srivastava S., Lab Chip 16, 2148–2167 (2016). 10.1039/C6LC00355A [DOI] [PubMed] [Google Scholar]
- 26. Farmehini V., Rohani A., Su Y.-H., and Swami N. S., Lab Chip 14, 4183–4187 (2014). 10.1039/C4LC00801D [DOI] [PubMed] [Google Scholar]
- 27. Su Y.-H., Tsegaye M., Varhue W., Liao K.-T., Abebe L. S., Smith J. A., Guerrant R. L., and Swami N. S., Analyst 139, 66–73 (2014). 10.1039/C3AN01810E [DOI] [PubMed] [Google Scholar]
- 28. Hawkins B. G. and Kirby B. J., Electrophoresis 31, 3622–3633 (2010). 10.1002/elps.201000429 [DOI] [PubMed] [Google Scholar]
- 29. Mohammad S. C., Hamid L., Hesamodin K., and Mohsen J. S., Electrophoresis 35, 3523–3532 (2014). 10.1002/elps.201400349 [DOI] [PubMed] [Google Scholar]
- 30. Krause P. J., Daily J., Telford S. R., Vannier E., Lantos P., and Spielman A., Trends Parasitol. 23, 605–609 (2007). 10.1016/j.pt.2007.09.005 [DOI] [PubMed] [Google Scholar]
- 31. Kuttel C., Nascimento E., Demierre N., Silva T., Braschler T., Renaud P., and Oliva A. G., Acta Trop. 102, 63–68 (2007). 10.1016/j.actatropica.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 32. Gascoyne P., Mahidol C., Ruchirawat M., Satayavivad J., Watcharasit P., and Becker F. F., Lab Chip 2, 70–75 (2002). 10.1039/b110990c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Levy M. G. and Ristic M., Science 207, 1218–1220 (1980). 10.1126/science.7355284 [DOI] [PubMed] [Google Scholar]
- 34. Moody A., Clin. Microbiol. Rev. 15, 66–78 (2002). 10.1128/CMR.15.1.66-78.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuzman D., Znidarcic T., Gros M., Vrhovec S., Svetina S., and Zeks B., Pfligers Arch. 440, R193–R194 (2000). 10.1007/s004240000061 [DOI] [PubMed] [Google Scholar]
- 36. Patel P. M., Bhat A., and Markx G. H., Enzyme Microb. Technol. 43, 523–530 (2008). 10.1016/j.enzmictec.2008.09.006 [DOI] [Google Scholar]