Abstract
Diffuse large B-cell lymphoma is the most common type of non-Hodgkin lymphoma. MicroRNAs (miRNAs) are endogenous small RNA, which can regulate gene expression at the post-transcriptional level. MiRNA profiling has shown a great potential as novel diagnostic and prognostic biomarkers. The present study was performed at the Nemazee Teaching Hospital (Shiraz, Iran) from 2011 to 2013. The aim of this study was to assess the deregulation of miRNAs profiles in DLBL against hyperplasic reactive lymph node as a normal. This could serve as a biomarker for DLBL. The miRCURY LNA™ microarray was used on the total RNA, which was extracted from formalin-fixed paraffin-embedded tissue of 24 de novo diffuse large B-cell lymphoma patients and 14 normal lymph nodes. The greatest changes were detected in miR-4284 and miR-4484 level in patient’s lymphoma samples. These miRNAs can act as a diagnostic biomarker for DLBL.
Keywords: Lymphoma, Large B-Cell, Diffuse, MicroRNAs, Microarray Analysis
What’s Known
Micro RNAs constitute a powerful tool for investigation. They are significant markers for diagnosis and prognosis in specific diseases.
Some RNAs such as miR-180, miR-181, and miR-21 have been introduced in lymphomas.
What’s New
We identified mir-4284 and mir-4484 as novel microRNAs in patients with DLBL samples. They are putative diagnostic biomarkers in DLBL, and they have not been reported in previous studies.
Introduction
MicroRNAs (miRNAs) are small, 19-25 nucleotides, and non-coding RNAs. They perform different biological activities like proliferation, differentiation and apoptosis. MicroRNAs regulate gene expression at post-transcriptional level by translating repression or directing cleavage of miRNAs.1
A change in expression pattern of miRNAs contributes to the pathogenesis of most human malignancies.2 A study demonstrated that some miRNAs are over-expressed while others are markedly reduced in malignant tissues. These findings suggest that miRNAs can act as oncogenes or tumor suppressors.3 Detectable miRNAs in the tissue and other biological samples provide a great source for miRNAs-based biomarkers in human cancers.
Diffuse large B-cell lymphoma (DLBL) is the most common type of non-Hodgkin lymphoma (NHL) with heterogeneous genomic entity. With the use of gene array, DLBL has been subdivided into two distinct subgroups: germinal center B-cell-like (GCB) and activated B-cell-like (ABC).4 Combination therapy that associates cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) with rituximab, (R–CHOP) has become a standard treatment for DLBL leading to complete remission.4
In cancer, miRNAs expression pattern can be highly tissue–specific and clearly have diagnostic and prognostic potential. Oncogenic role of miR-155 in DLBL has denoted an association with NF-KB activity.5 Also, miR-17-92 clusters are over expressed in DLBL and mutated miR-142, which correlates with DLBL.6 miRNAs are a putative biomarkers and therapeutic targets for lymphoma. However, the act of miRNA in DLBL is still not fully understood.
In this study, we investigated the profile of miRNAs in a series of DLBL, using formalin-fixed paraffin-embedded (FFPE) sample, to identify potential miRNAs as a diagnostic biomarker and their target pathway that contributes to the pathogenesis of DLBL.
Materials and Methods
Patients and Specimens
This case control study was performed at the Nemazee Teaching Hospital (Shiraz, Iran) from 2011 to 2013. A formalin-fixed paraffin-embedded (FFPE) series of 24 DLBL were obtained from the archives at the Pathology department of Shiraz University of Medical Sciences. This group consisted of 13 males (with an average age of 40.5 years) and 11 females (with a mean age of 40.1 years). The inclusion criteria were age between 20 to 55 years and designed de novo DLBL. All samples were diagnostic, taken before the patients received therapy. This diagnosis was based on morphology and molecular marker positive for CD20, CD79a, Ki67, Bcl2, and negative or normal patterns for cyclinD1, CD34, CD3, and CD30.
The 14 control FFPEs were diagnosed as hyperplastic lymph nodes matched with the case. They included 10 males (with an average age of 37.5 years) and 4 females (with an average age of 38.1 years). They had non-tumoral samples and normal pattern of immunohistochemistry. All samples were chosen for re-assessment of the original pathological diagnosis by an expert pathologist. All patients had to sign the consent form before participating in the study. The project was approved by the Ethics Committee of Shiraz University of Medical Sciences. The exclusion criterion was any history of malignancy for the control group.
RNA Extraction
Total RNA was isolated from all samples using TRIzol reagent based on manufacturer’s instruction (Invitrogen, Carlsbad, CA, USA). The quality was assessed using a bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA) and after a deparaffinization step, RNA quality was assessed taking into account 260/280 ratio and 260/230 ratio by NanoDrop 2000 (Thermo Scientific, USA). All samples had RIN values of 7.
Labeling and Hybridizations
500 ng total RNA from both the samples and reference were labeled with Hy3™ and Hy5™ fluorescent label, respectively, using miRCURY LNA™ microRNA Hi-Power labeling kit, and Hy3™/Hy5™ (Exiqon, Denmark) in accordance with the manufacturer’s instructions. The Hy3™-labeled samples and a Hy5™-labeled reference RNA sample were mixed pairwise and hybridized to the mercury LNA™ microRNA array 7th Gen (Exiqon, Denmark), which contains capture probes targeting all microRNAs for human, mouse, or rat registered in the miRBASE 18.0. The hybridization was performed according to the miRCURY LNA™ microRNA array instruction manual using Tecan’s HS 4800™ hybridization station (Tecan, Austria). The microRNA array slides were scanned by the Agilent G2565BA microarray scanner system (Agilent Technologies, Inc., USA) and the image analysis was carried out using the ImaGene® 9 (miRCURY LNA™ microRNA array analysis software, Exiqon, Denmark). In order to correct for the background signals, Normexp with an offset value of 10 was used and the global LOWESS (locally weighted scatterplot smoothing) regression algorithm was applied (figure 1).
Figure 1.
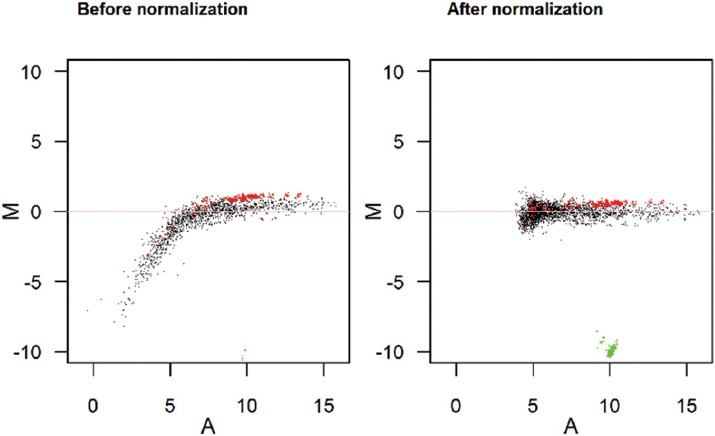
M/A plot show normalization by the use of spike-ins and LOWESS. Green: Hy3 controls; Red: Spike-ins; Black: All other probes.
The microRNA profiling has been identified as a subset of 50 top microRNAs. For studies where at least three replicates have been supplied for each sample, the array includes P values. For the adjusted P<0.05 were considered as significant.
Statistical analysis was performed using t-test, the ±1 fold changes of patients miRNAs versus normal, and P-value less than 0.05 was considered statistically significant.
Results
We determined miRNAs profiling in 24 DLBL patients and 14 lymph node reactive micro arrays using an unsupervised hierarchical clustering model. Principal component analysis (PCA) was performed on the 50 microRNAs that mostly varied across all samples irrespective of their biological group (figure 2).
Figure 2.
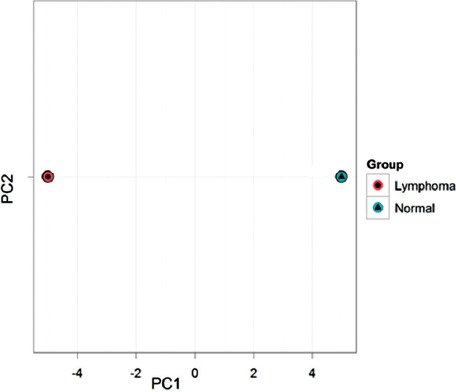
The principal component analysis performed on all samples and on the top 50 microRNAs of DLBL and normal.
The absolute value of the log fold changes was larger than 1 out of the total number of microRNAs. As shown in figure 3, it is represented as a heat map diagram. All calculations were done in the R/Bioconductor software.
Figure 3.
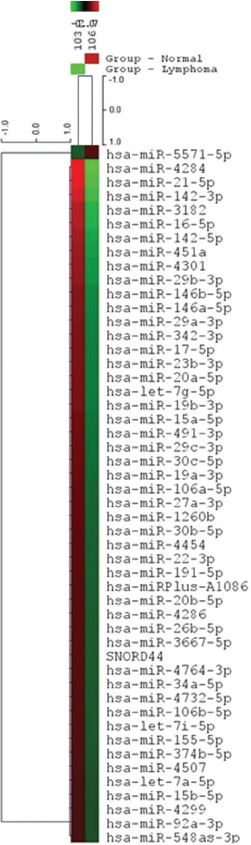
Heat map and unsupervised hierarchical clustering show miRNAs micro array of DLBL against normal samples. Red color display an expression below the reference channel and green color display the expression higher than the reference channel.
Discussion
The role and identity of lymphoma-associated microRNA are not fully understood. Consequently, this study aimed at investigating microRNAs in DLBL, which is the most frequently occurring non-Hodgkin lymphoma using miRCURYLNA™ microRNA array. It is obvious that the tissue samples are fixed in formalin after surgery. This fixation cannot have a significant effect on the miRNAs expressions, which are visible using archived FFPE specimens, and it may be used as a great biomarker and novel target for cancer research.7 Different methods have been used to profile miRNA expression, including northern blotting with radio nucleotide probes, oligonucleotide macro arrays, quantitative PCR-based amplification of precursor or mature RNAs, bead-base profiling and micro array based locked nucleic acids (LNA). We performed miRNAs array by LNA, which is an efficient method to screen the expression of the number of miRNAs, reliable in obtaining expression profiles, and more sensitive in comparison with similar methods.8
We showed that miRNA expression patterns could clearly help to distinguish between DLBL and lymph node reactive. We also identified some increased and decreased micro RNAs from 24 DLBL patients against 14 Lymph nodes reactive. Especially, up-regulated miR-4284, miR-21, miR142-3P, miR-155, miR-3182, miR-16, 142-5P, miR-451, miR-4301, and miR-29b and down-regulated miR-4484, miR-143-3p, and miR-125 were detected (figure 4).
Figure 4.
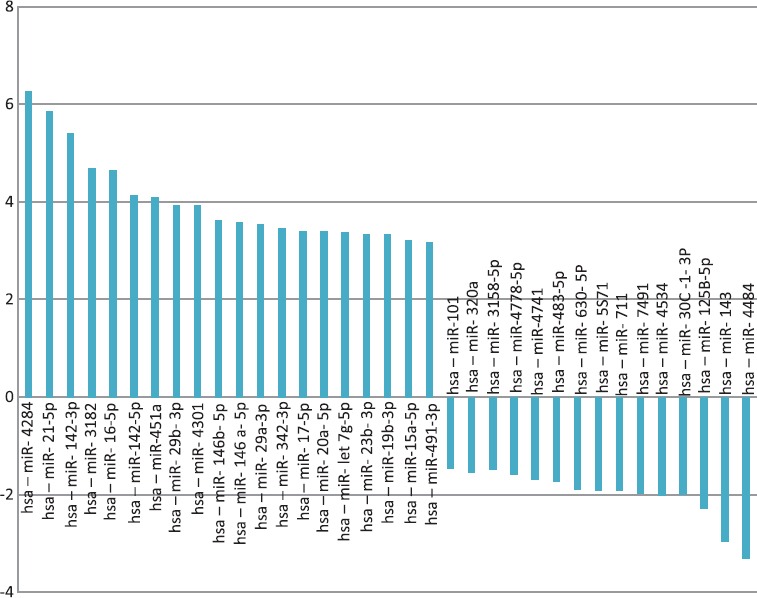
The most alteration shows microRNAs expressions of DLBL patients against normal.
miR-4284 and miR-4484 were identified as novel microRNAs in patients of DLBL samples. Our results show, miR-4284 and miR-4484 putative as diagnostic biomarkers in diffuse large B-cell lymphoma, although they have not been reported in previous studies.
miRNA target prediction was performed using the software analysis tools (included in the websites: http:/microrna.sanger.ac.uk/Software/Rfam/mirna, mirTarbase and proteins), which were coded by target genes followed by KEGG pathway to realize the cell fate. We inspected main targets of these miRNAs that may have a role in DLBL.
Gene of miR-4284 is located on chromosome 7. Based on bioinformatic studies, a main target gene for miR-4284 is tumor necrosis factor receptor-associated factor 4 (TRAF4), which is recruited to the active TGF-β receptor complex 4 (TRAF4) as a key component mediating pro-oncogenic TGF-β-induced SMAD and non-SMAD signaling. TGF-β receptor-TRAF4 interaction triggers Lys 63-linked TRAF4 polyubiquitylation and subsequent activation of the TGF-β-activated kinase (TAK). TRAF4 is needed for TGF-β-induced migration, mesenchymal to epithelial transition, and breast cancer metastasis. Increased TRAF4, correlate with elevated levels of phosphorylated SMAD2 and phosphorylated TAK1 as well as poor prognosis among breast cancer patients.9 Moreover, SMADs signaling has a dual role in cancer cells, i.e. regulation of cell death and proliferation (table 1). Up-regulation of miR-4284 in lymphoid tissue, such as lymph node, induces TRAF4, SMAD2 and TAK1 activity and can transform normal lymphoid tissue to DLBL.10
Table 1.
The target gene predicted for miR-4284 and miR-4484 as well as their role in cells
| Target genes | Roles in cells | |
|---|---|---|
| miR-4284 | TRAF4, BCL 10, HDAC, HOXA1, PTEN | Apoptosis, differentiation, proliferation |
| miR-4484 | AATF, PTPN14, PI3K, PTEN | Apoptosis |
The expression pattern of SMADs proteins in DLBL showed that TGF-β signaling pathway is mostly operative in this type of lymphoma through these proteins. Disturbance of TGF-β signaling pathway in lymphoma may lead to the development of lymphoma.10
In addition, a significant target for this microRNA is already predicted MLL gene. MLL plays a role in myeloid, lymphoid leukemia and lymphoma. We found that miR-4284 regulate BCL 10, HDAC and Hox A1. BCL 10 induces apoptosis to active NF-kappa B. HDAC plays a key role in the regulation of eukaryotic gene expression. HDAC associate with retinoblastoma tumor-suppressor protein; this complex is an important modulator for cell proliferation and differentiation. Hox proteins are temporally regulated during embryonic development. This protein has a role in DNA-binding transcription factor, which regulate cell morphogenesis and differentiation.11 miR-4284 can elevate MLL, BCL 10, and HDAC that play a role in lymphogenesis by blocking apoptosis and increasing proliferation (table 1).
Moreover miR-4484 regulates AATF by interacting with MRP3 K12/DLK, which is involved in the induction of cell apoptosis. PTPN 14 is also from a protein tyrosine phosphatase (PTP) family. PTP is a signaling molecule with roles in many cellular activities and lymphatic development. Additionally, PI3K is a specific target of miR-4484; it plays a role in cell proliferation and oncogene. Researchers showed that down-regulated miR-4484 in DLBL may cause suppression of AATF and PTPN14. These down-regulations may induce and mediate transformation of normal cells to cancerous form.12
Gene expression array and immunohistochemical studies have shown two molecular subgroups of DLBL, GC, and ABC. miRNAs profile is a strong tool that can differentiate between lymphoma and DLBL subtype. A miR17-92 cluster consists of miR-17, miR-19B, miR-20A, and miR-92A expressed at higher levels in ABC subtype, which is more aggressive than GC. We cannot discriminate DLBL subgroups by miRNAs in this study because we did not perform gene array analysis of patients.13 Many clinical factors, included in the International Prognostic Index have failed to recognize the R-CHOP resistant DLBL patients. miR-4284 and miR-4484 may serve as potential biomarkers for the prognosis of DLBL. miRNAs could influence changes during chemotherapy and/or radiotherapy.14
Additionally, in this study, we found a decrease in miR-143 and miR-125B-5p and an increase in miR-21 and miR-142, which were reported previously in DLBL and some cancers.15 If we had genetic data of DLBL, we would have related miRNAs expression to subtypes of DLBL.
Conclusion
We identified some miRNAs up- or down-regulated in DLBL. The greatest changes in miR-4284 and miR-4484 level may indeed act as a tissue and prognostic biomarker for DLBL. In addition, we will follow these aberrantly miRNAs changes in serum of DLBL patients to find serum biomarker. Although we carried out the array on these samples, but it should be done in large scale and the results must be confirmed by q-PCR.
Acknowledgment
This work was supported by the Department of Hematology, School of Medical Sciences, Tarbiat Modares University (Tehran, Iran), and a grant (number 91-187) from the Organ Transplant Research Center, Shiraz University of Medical Sciences (Shiraz, Iran). We appreciate their assistance in retrieving samples and clinical data.
Conflict of Interest: None declared.
References
- 1.Macfarlane LA, Murphy PR. MicroRNA:Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537–61. doi: 10.2174/138920210793175895. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pillai RS. MicroRNA function:multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–61. doi: 10.1261/rna.2248605. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 4.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 5.Rai D, Karanti S, Jung I, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–44. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 7.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–74. doi: 10.1261/rna.642907. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenvang J, Silahtaroglu AN, Lindow M, Elmen J, Kauppinen S. The utility of LNA in microRNA-based cancer diagnostics and therapeutics. Semin Cancer Biol. 2008;18:89–102. doi: 10.1016/j.semcancer.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Zhou F, Garcia de Vinuesa A, de Kruijf EM, Mesker WE, Hui L, et al. TRAF4 promotes TGF-beta receptor signaling and drives breast cancer metastasis. Mol Cell. 2013;51:559–72. doi: 10.1016/j.molcel.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Ikushima H, Miyazono K. TGFbeta signalling:a complex web in cancer progression. Nat Rev Cancer. 2010;10:415–24. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 11.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812:592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Sandhu SK, Croce CM, Garzon R. Micro-RNA Expression and Function in Lymphomas. Adv Hematol. 2011;2011:347137. doi: 10.1155/2011/347137. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culpin RE, Proctor SJ, Angus B, Crosier S, Anderson JJ, Mainou-Fowler T. A 9 series microRNA signature differentiates between germinal centre and activated B-cell-like diffuse large B-cell lymphoma cell lines. Int J Oncol. 2010;37:367–76. doi: 10.3892/ijo_00000685. [DOI] [PubMed] [Google Scholar]
- 14.Zheng RL, Jiang YJ, Wang X. Role of microRNAs on therapy resistance in Non-Hodgkin’s lymphoma. Int J Clin Exp Med. 2014;7:3818–32. [ PMC Free Article] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwanhian W, Lenze D, Alles J, Motsch N, Barth S, Doll C, et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 2012;1:141–55. doi: 10.1002/cam4.29. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]


