Abstract
Background
SWEET is a newly identified family of sugar transporters. Although SWEET transporters have been characterized by using Arabidopsis and rice, very little knowledge of sucrose accumulation in the stem region is available, as these model plants accumulate little sucrose in their stems. To elucidate the expression of key SWEET genes involved in sucrose accumulation of sorghum, we performed transcriptome profiling by RNA-seq, categorization using phylogenetic trees, analysis of chromosomal synteny, and comparison of amino acid sequences between SIL-05 (a sweet sorghum) and BTx623 (a grain sorghum).
Results
We identified 23 SWEET genes in the sorghum genome. In the leaf, SbSWEET8-1 was highly expressed and was grouped in the same clade as AtSWEET11 and AtSWEET12 that play a role in the efflux of photosynthesized sucrose. The key genes in sucrose synthesis (SPS3) and that in another step of sugar transport (SbSUT1 and SbSUT2) were also highly expressed, suggesting that sucrose is newly synthesized and actively exported from the leaf. In the stem, SbSWEET4-3 was uniquely highly expressed. SbSWEET4-1, SbSWEET4-2, and SbSWEET4-3 were categorized into the same clade, but their tissue specificities were different, suggesting that SbSWEET4-3 is a sugar transporter with specific roles in the stem. We found a putative SWEET4-3 ortholog in the corresponding region of the maize chromosome, but not the rice chromosome, suggesting that SbSWEET4-3 was copied after the branching of sorghum and maize from rice. In the panicle from the heading through to 36 days afterward, SbSWEET2-1 and SbSWEET7-1 were expressed and grouped in the same clade as rice OsSWEET11/Xa13 that is essential for seed development. SbSWEET9-3 was highly expressed in the panicle only just after heading and was grouped into the same clade as AtSWEET8/RPG1 that is essential for pollen viability. Five of 23 SWEET genes had SNPs that caused nonsynonymous amino acid substitutions between SIL-05 and BTx623.
Conclusions
We determined the key SWEET genes for technological improvement of sorghum in the production of biofuels: SbSWEET8-1 for efflux of sucrose from the leaf; SbSWEET4-3 for unloading sucrose from the phloem in the stem; SbSWEET2-1 and SbSWEET7-1 for seed development; SbSWEET9-3 for pollen nutrition.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-016-0546-6) contains supplementary material, which is available to authorized users.
Keywords: Phloem loading, Phloem unloading, Sugar transporter, Bioethanol, RNA-seq, SNP, Photosynthesis
Background
Sorghum (Sorghum bicolor) accumulates sucrose in the stem. This feature is rare among plants, making sorghum a useful source of bioethanol [1–5]. To enhance bioethanol production, it is important to understand and manipulate sucrose phloem loading, unloading, metabolism, and signaling [6–9] and improve the efficiency of bioethanol production [10–13]. Sucrose content generally depends on its metabolism, transport, and storage [14, 15]. The key genes in sucrose metabolism are sucrose phosphate synthase (SPS) and sucrose synthase (SUS); their products catalyze rate-limiting steps in this metabolic pathway. The key genes in transport and thus sucrose movement between tissues via the phloem are sugars will eventually be exported transporters (SWEET) and sucrose transporters (SUT) [16]. Invertase (INV) is responsible for the degradation of sucrose to glucose and fructose, thus influencing whether sugar molecules are stored as sucrose or starch. These factors synergistically contribute to the stem sucrose content.
SWEET is a newly identified family of sugar transporters [17, 18]. SWEET family genes are duplicated, with a diversity of functions: 21–23 SWEET genes are known in S. bicolor, 17 in Arabidopsis thaliana, 18 in Brachypodium distachyon, 23 in rice (Oryza sativa), 52 in Glycine max, and 24 in Zea mays [19, 20]. Our knowledge of SWEET has been expanded by using model plants such as Arabidopsis [21] and O. sativa [22, 23]. In Arabidopsis thaliana, SWEET proteins are located in the plasma membrane or vacuolar membranes and transport sucrose, glucose, fructose, or 2-deoxyglucose; SWEET genes are expressed in the leaf, root, flower, seed, and/or pollen [21]. The functions of some of the SWEET proteins have been elucidated: for example, AtSWEET11 and 12 are sucrose transporters responsible for the efflux of photosynthesized sucrose from the leaf, and the double mutant accumulates sugar in the leaf [24]. OsSWEET11, which is essential for reproductive development, are used by the pathogenic bacterium to invade its host [25]. However, because these model plants accumulate little sucrose in their stems, no information on the relationship between SWEET and stem sucrose accumulation is available. Expression of the other sorghum sucrose transporter gene family, SUT, differs between Rio (sweet) and BTx623 (grain) sorghum stems [26], but does not differ between Wray (sweet) and Macia (grain) sorghum stems [27]. These findings suggest that SUT expression is not a pivotal rate-limiting factor for sucrose transport. To elucidate the mechanism of sucrose accumulation in the stem, it is therefore important to further characterize sorghum SWEET family genes.
Our aim here was to characterize sorghum SWEET genes using gene expression profiling during the stage of sucrose accumulation. We also used phylogenetic trees to characterize genes and analyzed the synteny of SWEET genes between the sorghum and rice chromosomes. We compared the amino acid sequence of SWEET of sorghum SIL-05 (a sweet sorghum used as a material for bioethanol production; [28] and BTx623 (for which a reference genome sequence is available [29], but which is a grain sorghum with lower sucrose content than SIL-05). We also analyzed the expression of other sugar-related genes: SUT, SPS, SUS, and INV. We then consider all of the results together to discuss the key genes in phloem loading and unloading and thus accumulation of sucrose in sorghum stems.
Results and discussion
Quantification of gene expression during the stage of sucrose accumulation in the stem
In the sweet sorghum SIL-05, total sugar content increased after heading, reaching 18.9 % on day 64 after heading, whereas the glucose and fructose contents decreased slightly from day 17 (Fig. 1). To identify differentially expressed genes, tissue samples were obtained from the leaf, stem, and panicle on days 1, 17, and 36, respectively, after heading. Their RNAs were then sequenced.
Fig. 1.
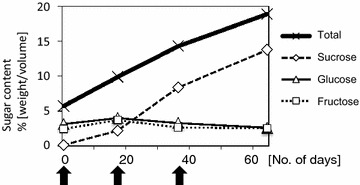
Sugar contents after heading. Y-axis indicates sugar content (weight/volume). X-axis indicates the number of days after heading, and the arrow indicates the point of sampling for RNA-seq
We focused on genes encoding proteins for sugar transport (SWEET, SUT), sugar metabolism (SPS, SUS), and sucrose degradation (INV) (Table 1). We plotted the chromosomal locations of these genes (Fig. 2) on the basis of their chromosomal locations in the BTx623 genome (Table 1). Quantitative trait loci (QTL) for sugar-related traits that were previously analyzed by using a cross of SS79 (a sweet sorghum) × M71 (a grain sorghum) [30] and R9188 (a sweet sorghum) × R9403463-2-1 (a grain sorghum) [31] are also shown in Fig. 2.
Table 1.
Sorghum SWEET genes, other sugar-related genes, and constitutively expressed genes
| Function | Gene name | Gene ID (ver. 2.1) | Gene ID (ver. 1.4) | Chromosomal location |
|---|---|---|---|---|
| Sugar transporter | SbSWEET1-1 | Sobic.001G373600 | Sb01g035490 | Chr 1: 58,985,432–58,988,278 |
| SbSWEET1-2 | Sobic.001G377600 | Sb01g035840 | Chr 1: 59,380,534–59,384,540 | |
| SbSWEET2-1 | Sobic.002G259300 | Sb02g029430 | Chr 2: 64,413,792–64,416,541 | |
| SbSWEET3-1 | Sobic.003G015200 | Sb03g001520 | Chr 3: 1,356,535–1,358,800 | |
| SbSWEET3-2 | Sobic.003G038700 | Sb03g003470 | Chr 3: 3,617,943–3,620,183 | |
| SbSWEET3-3 | Sobic.003G038800 | Sb03g003480 | Chr 3: 3,622,464–3,625,366 | |
| SbSWEET3-4 | Sobic.003G149000 | Sb03g012930 | Chr 3: 15,675,845–15,681,441 | |
| SbSWEET3-5 | Sobic.003G182800 | Sb03g024250 | Chr 3: 48,309,667–48,324,107 | |
| SbSWEET3-6 | Sobic.003G213000 | Sb03g027260 | Chr 3: 54,756,647–54,760,169 | |
| SbSWEET3-7 | Sobic.003G269300 | Sb03g032190 | Chr 3: 60,633,184–60,636,494 | |
| SbSWEET3-8 | Sobic.003G377700 | Sb03g041740 | Chr 3: 69,215,104–69,218,784 | |
| SbSWEET4-1 | Sobic.004G133500 | Sb04g012910 | Chr 4: 20,553,590–20,558,352 | |
| SbSWEET4-2 | Sobic.004G133600 | Sb04g012920 | Chr 4: 20,691,080–20,696,805 | |
| SbSWEET4-3 | Sobic.004G136600 | Sb04g015420 | Chr 4: 35,162,670–35,166,305 | |
| SbSWEET4-4 | Sobic.004G157100 | Sb04g021000 | Chr 4: 49,118,793–49,122,120 | |
| SbSWEET5-1 | Sobic.005G123500 | Sb05g018110 | Chr 5: 44,351,922–44,354,521 | |
| SbSWEET7-1 | Sobic.007G191200 | Sb07g026040 | Chr 7: 61,176,996–61,180,220 | |
| SbSWEET8-1 | Sobic.008G094000 | Sb08g013620 | Chr 8: 36,493,752–36,496,643 | |
| SbSWEET8-2 | Sobic.008G094300 | Sb08g013840 | Chr 8: 36,993,118–36,995,615 | |
| SbSWEET8-3 | Sobic.008G094400 | Sb08g014040 | Chr 8: 37,249,178–37,251,607 | |
| SbSWEET9-1 | Sobic.009G080900 | Sb09g006950 | Chr 9: 11,309,919–11,312,702 | |
| SbSWEET9-2 | Sobic.009G143500 | Sb09g020860 | Chr 9: 50,116,198–50,119,686 | |
| SbSWEET9-3 | Sobic.009G252000 | Sb09g030270 | Chr 9: 58,680,303–58,682,170 | |
| Sugar transporter | SbSUT3 | Sobic.001G254000 | Sb01g022430 | Chr 1: 28,168,652–28,172,476 |
| SbSUT1 | Sobic.001G488700 | Sb01g045720 | Chr 1: 68,703,383–68,709,450 | |
| SbSUT5 | Sobic.004G190500 | Sb04g023860 | Chr 4: 53,509,428–53,512,882 | |
| SbSUT4 | Sobic.004G353600 | Sb04g038030 | Chr 4: 67,476,512–67,481,811 | |
| SbSUT6 | Sobic.007G214500 | Sb07g028120 | Chr 7: 63,062,892–63,066,154 | |
| SbSUT2 | Sobic.008G193300 | Sb08g023310 | Chr 8: 55,332,646–55,338,922 | |
| Sugar degradation | INV1 | Sobic.001G099700 | Sb01g008910 | Chr 1: 7,615,347–7,617,621 |
| INV2 | Sobic.003G440900 | Sb03g047060 | Chr 3: 73,993,613–73,997,226 | |
| INV3 | Sobic.004G004800 | Sb04g000620 | Chr 4: 439,003–443,225 | |
| INV4 | Sobic.006G255600 | Sb06g031930 | Chr 6: 60,211,622–60,214,854 | |
| Sugar synthesis | SPS1 | Sobic.003G403300 | Sb03g043900 | Chr 3: 71,135,755–71,141,978 |
| SPS2 | Sobic.004G068400 | Sb04g005720 | Chr 4: 5,592,102–5,599,224 | |
| SPS3 | Sobic.005G089600 | Sb05g007310 | Chr 5: 12,955,276–12,961,424 | |
| SPS4 | Sobic.009G233200 | Sb09g028570 | Chr 9: 57,284,130–57,297,240 | |
| SPS5 | Sobic.010G205100 | Sb10g025240 | Chr 10: 54,483,016–54,493,428 | |
| Sugar synthesis | SUS1 | Sobic.001G378300 | Sb01g035890 | Chr 1: 59,452,295–59,460,141 |
| SUS2 | Sobic.004G357600 | Sb04g038410 | Chr 4: 67,754,722–67,766,746 | |
| SUS3 | Sobic.010G276700 | Sb10g031040 | Chr 10: 60,830,697–60,835,335 | |
| Control | actin | Sobic.008G047000 | Sb08g003970 | Chr 8: 4,615,047–4,617,619 |
| EF1alpha | Sobic.010G182100 | Sb10g023330 | Chr 10: 51,879,475–51,882,620 | |
| GAPDH | Sobic.010G262500 | Sb10g029870 | Chr 10: 59,688,771–59,701,308 |
Chromosomal locations are based on the reference genome Sbicolor_v2.1_255
Fig. 2.
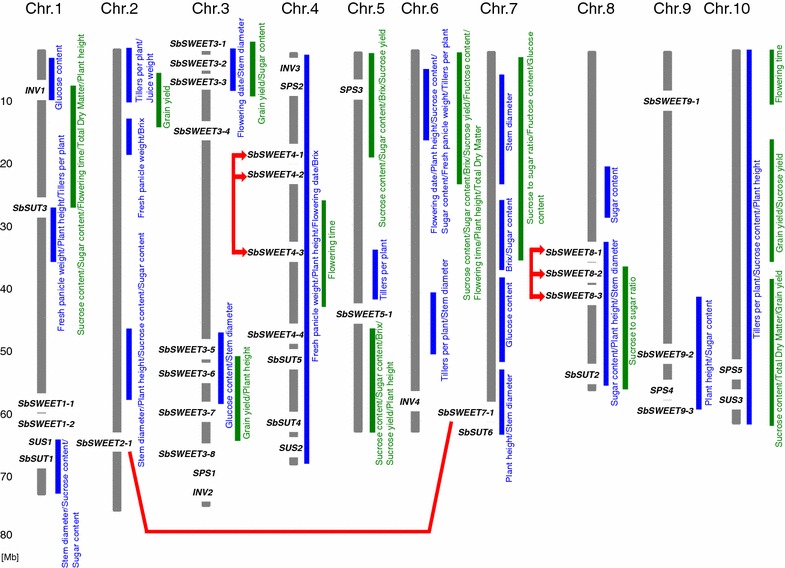
Chromosomal locations of sugar-related genes in sorghum. Chromosomal locations of SWEET, SUT, INV, SPS, and SUS genes are shown. The positions are based on the reference genome BTx623. Duplicated genes focused in this study are linked by red lines. QTLs for sugar-related traits in sweet × grain sorghums are also shown on the basis of two independent QTL analyses: SS79 (sweet sorghum) × M71 (grain sorghum) (green bars [30]; and R9188 (sweet sorghum) × R9403463-2-1 (grain sorghum) (blue bars [31]). Numbers in the column at the left indicate the physical lengths of the chromosome [Mb megabase]
FPKM values were calculated to compare the expression level of each gene. The FPKM of 23 SWEET genes, 6 SUT genes, 5 SPS genes, 3 SUS genes, and 4 INV genes are shown in Fig. 3. We then focused on the highly and/or differentially expressed genes.
Fig. 3.
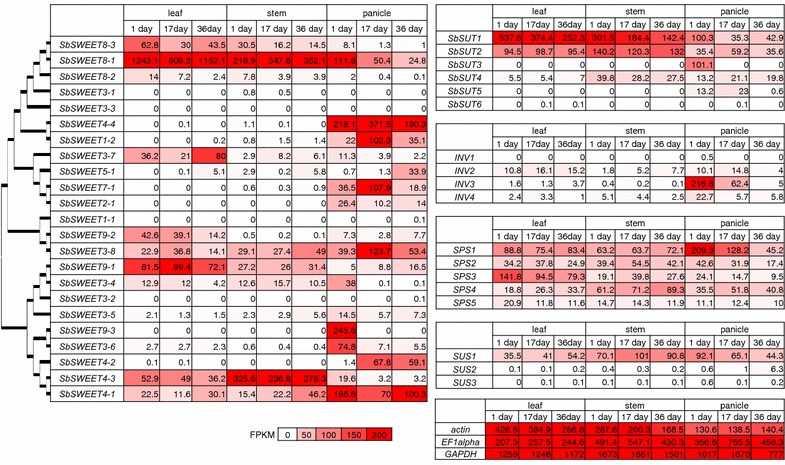
FPKM values of sugar-related genes at the sucrose accumulation stage. FPKM (fragments per kilobase of exon per million mapped sequence reads) values reflect the quantities of existing RNA of each paralog in the cells or tissues. FPKM values for SWEET, SUT, INV, SPS, SUS, actin, elongation factor 1-alpha (EF1alpha), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) are shown as heatmaps. Actin, EF1alpha, and GAPDH are constitutively expressed controls. The number in each box indicates the FPKM value of each gene. Boxes at the bottom indicate the reference color intensities of FPKM values. Samples were extracted from the leaf, stem, or panicle on days 1, 17, and 36 after heading (the stage of sucrose accumulation in the stem). Phylogenetic trees of 23 putative sorghum SWEET genes are also shown on the left side
Identification of SNPs in SIL-05 and BTx623
Single nucleotide polymorphisms (SNPs) were found between SIL-05 and BTx623. Five SWEET genes had SNPs that caused nonsynonymous amino acid substitutions between SIL-05 and BTx623 (Table 2). These amino acid substitutions might affect the transport activity of SWEET proteins.
Table 2.
SNPs and amino acid substitutions in SWEET proteins between the cultivars SIL-05 and BTx623
| Gene name | Chromosomal location | Nucleotide | Amino acid | ||
|---|---|---|---|---|---|
| BTx623 | SIL-05 | BTx623 | SIL-05 | ||
| SbSWEET1-2 | Chr 1: 59,381,183 | A | T | F | Y |
| SbSWEET3-7 | Chr 3: 60,636,090 | G | A | V | I |
| SbSWEET4-1 | Chr 4: 20,557,328 | G | C | L | V |
| SbSWEET4-3 | Chr 4: 35,163,171 | C | G | E | D |
| SbSWEET9-3 | Chr 9: 58,680,805 | C | G | G | A |
Genes potentially responsible for sucrose accumulation in the stem of sorghum
SbSWEET8-1 (Sobic.008G094000)
We examined the genes potentially responsible for sucrose synthesis and efflux from the leaf. SbSWEET8-1 was extremely highly expressed in the leaf from the start of heading through to 36 days after heading (Fig. 3). It was grouped in the same clade as AtSWEET11 and AtSWEET12 (Fig. 4). AtSWEET11 and AtSWEET12 play a role in the efflux of photosynthesized sucrose to the apoplast in the leaves of Arabidopsis [21]. Although there were three tandemly duplicated SWEET genes (SbSWEET8-1, SbSWEET8-2, and SbSWEET8-3; Fig. 2) in the same clade (Fig. 4), SbSWEET8-1 was the only one expressed at extremely high levels (FPKM > 800 in the leaf; Fig. 3). We therefore consider that it plays a major role in sucrose efflux from the leaf (Fig. 5a, b). Moreover, the gene encoding the enzymes SPS3 (Sobic.005G089600) was highly expressed in the leaf. SbSUT1 (Sobic.001G488700), a gene encoding an SUT transporter, was highly expressed in the leaf from the start of heading through to 36 days after heading, suggesting that sucrose is taken up and concentrated in the sieve element–companion cell complex (Fig. 5b). These data suggested that sucrose is newly synthesized and actively exported from the leaf at this stage (Fig. 5a).
Fig. 4.
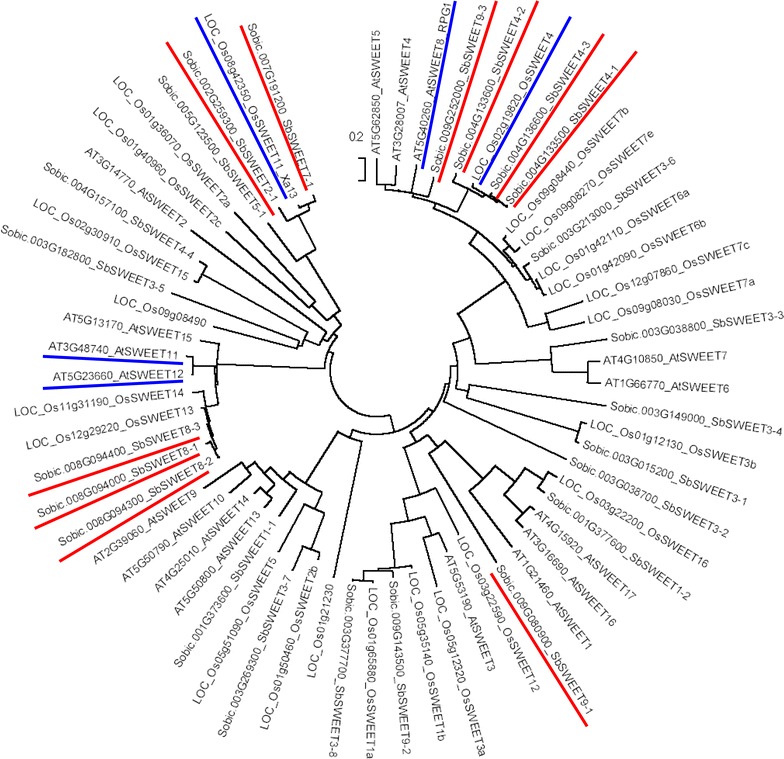
Evolutionary tree of SWEET. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. This analysis involved 63 amino acid sequences (23 of Sorghum bicolor, 17 of Arabidopsis thaliana, 23 of Oryza sativa). Sorghum SWEET genes focused in this study are underlined by red and functionally validated Arabidopsis or rice SWEET genes are underlined by blue
Fig. 5.
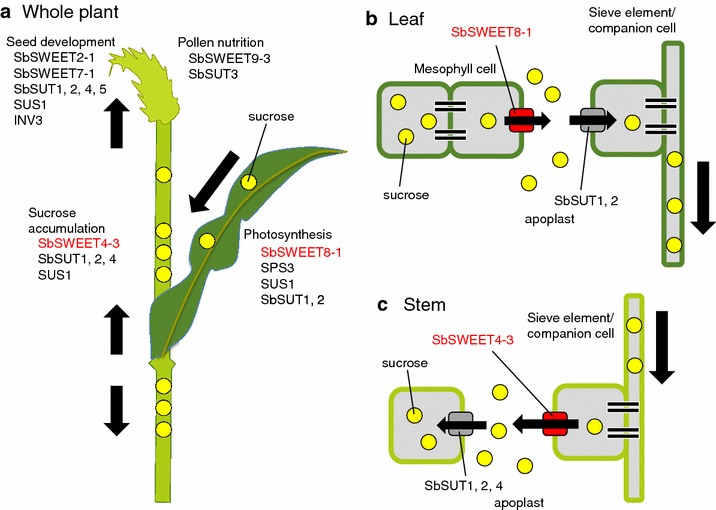
Schematic models of gene expression and roles of SWEET proteins in phloem loading and unloading. a Representative genes highly expressed in each tissue during the sucrose accumulation stage; those likely involved in phloem loading of sucrose in the leaf and unloading and accumulation in the stem are shown. b Sucrose efflux associated with SWEET proteins in the leaf. Sucrose is synthesized in leaf mesophyll cells and diffuses through the plasmodesmata. SWEET proteins facilitate sucrose efflux into the apoplast. Subsequently, sucrose is taken up and concentrated in the sieve element–companion cell complex by SUT sucrose symporters. Sucrose is transported through the sieve elements out of the leaves to the stem, roots, and seeds. SbSWEET8-1 (Sobic.008G094000) may play a role in the efflux of photosynthesized sucrose to the leaf apoplast. This model was constructed on the basis of an analogy to that in Arabidopsis. c SWEET-dependent sucrose accumulation in the stem. Synthesized sucrose is transported from the leaf through the sieve element, and SWEET proteins might facilitate sucrose efflux into the stem apoplast. SbSWEET4-3 (Sobic.004G136600) is a sugar transporter that might contribute to phloem unloading
SWEET9-1 (Sobic.009G080900) was expressed at higher levels in the leaf than in the stem and panicles from the start of heading through to 36 days after heading (Fig. 3). SWEET9-1 had no potential orthologs in Arabidopsis or rice (Fig. 4). Because the expression level of SWEET8-1 was massively higher in the leaf through to 36 days after heading than that of SWEET9-1, SWEET8-1 may function mainly in phloem loading in the leaf.
SbSWEET4-3 (Sobic.004G136600)
We next examined the genes potentially responsible for sucrose accumulation in the stem. SbSWEET4-3 was expressed more highly in the stem than in the other tissues during the stage of sucrose accumulation (Fig. 3). Although SWEET expression is diverse in the various tissues of sorghum, this potent expression in the stem is unique to SbSWEET4-3 (Fig. 3). SbSWEET4-1, SbSWEET4-2 and SbSWEET4-3 were located on chromosome 4 (Fig. 2) and categorized into the same clade (Fig. 4). The products of these three SbSWEET genes had high levels of amino acid identity with each other (Additional file 1: Fig. S1A), but the tissue specificity was different: SbSWEET4-3 was expressed mainly in the stem, whereas SbSWEET4-1 and SbSWEET4-2 were expressed mainly in the panicles (Fig. 3). We therefore hypothesized that SbSWEET4-3 is a sugar transporter with specific roles in the stem.
We compared the coded sequence and expression of SbSWEET4-3 between SIL-05 and BTx623. One amino acid substitution (D229E) was found between SIL-05 and BTx623 (Table 2). The aspartic acid (D) residue at 229 is conserved as D in the paralogs of SIL-05 (in the case of SbSWEET4-1, SbSWEET4-2, and SbSWEET4-3) (Additional file 1: Fig. S1A), and in other SWEET homologs in Brachypodium distachyon, Oryza sativa, Setaria italica, and Zea mays (Additional file 1: Fig. S1B). D229 has also been found in SbSWEET4-3 of other sweet sorghums (Cowley and Top76-6 [32]). We therefore consider that the D residue is necessary for efficient sucrose transport, although some SWEETs might function in tissues other than the stem. Because of the D229E substitution, SbSWEET4-3 in BTx623 might have relatively low sucrose transport activity. Moreover, the relatively high level of expression in the stem of SIL-05 (more than ten times that in the panicle; Fig. 3) differs from that in BTx623: the expression levels of SbSWEET4-3 in the stem of BTx623 at the time of anthesis (150 days after sowing) are as low as those in the panicle [33]. We therefore consider that the amino acid substitution at 229 and the higher level of expression of SbSWEET4-3 in the stem than in the panicle might explain the higher sucrose accumulation in the stem of SIL-05 than in that of BTx623.
We analyzed the synteny of SWEET genes between the sorghum and rice chromosomes using the Plant Genome Duplication Database [34]. The region of chromosome 4 of sorghum had synteny with chromosome 2 of rice, but large insertions/deletions were present, and SbSWEET4-3 had no corresponding SWEET genes on chromosome 2 of rice (Fig. 6). In the region corresponding to SbSWEET4-3, there were three genes instead of putative SWEET homologs; the functions of LOC_Os02g26294 and LOC_Os02g26300 are unknown, and LOC_Os02g26310 functions as a leucine-rich repeat receptor-like protein kinase (Fig. 6). We considered that LOC_Os02g19820 (OsSWEET4) of rice was the ortholog of SbSWEET4-1. LOC_Os02g19820 was expressed in leaf, stem, and tissues in the panicle [35]. As the N-terminal region of SbSWEET4-3 was similar to that of SbSWEET4-1 (Additional file 1: Fig.S1A), we thus considered that SbSWEET4-3 was duplicated from SbSWEET4-1 after the branching of sorghum and rice (Fig. 6).
Fig. 6.
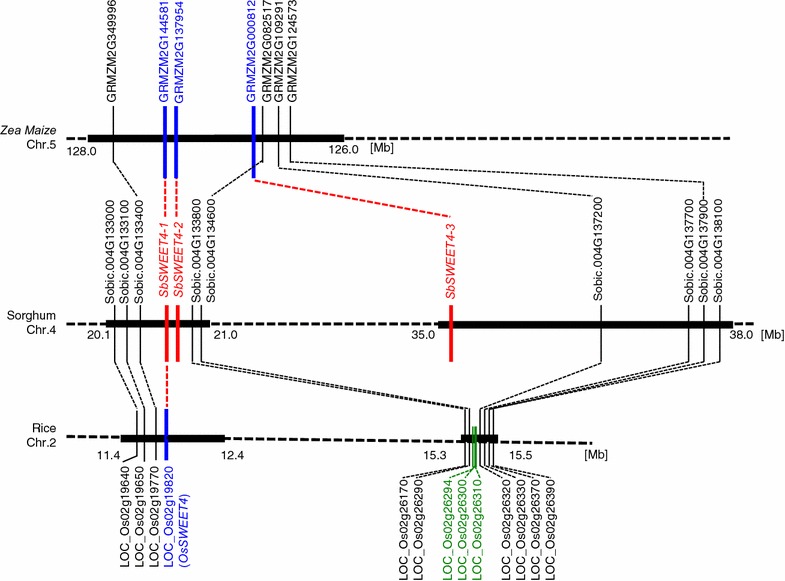
Synteny of SWEET genes between chromosome 4 of sorghum, chromosome 5 of maize, and chromosome 2 of rice. Three highly similar sorghum SWEET genes were annotated on 20.1–38.0 Mb region of chromosome 4 (SbSWEET4-1, SbSWEET4-2 and SbSWEET4-3: in red). Zea maize has three genes corresponding to SbSWEET4-1, SbSWEET4-2, and SbSWEET4-3 on chromosome 5, (GRMZM2G144581, GRMZM2G137954 and GRMZM2G000812, respectively: in blue). SbSWEET4-1 corresponds to the rice gene LOC_Os02g19820 (in blue). SbSWEET4-2 and SbSWEET4-3 were likely duplicated from SbSWEET4-1 after the divergence of sorghum and maize from rice. SbSWEET4-3 has no corresponding gene on chromosome 2 of rice. Three rice genes (in green) are annotated instead of the SWEET homologs. Numbers on map indicate locations on the chromosome (Mb megabase)
The region also had synteny with chromosome 5 of Zea mays. The three sorghum SWEET paralogs (SbSWEET4-1, SbSWEET4-2 and SbSWEET4-3) corresponded to the three respective maize SWEET genes (GRZM2G144581, GRZM2G137954 and GRMZM2G000812, respectively; Fig. 6). A comprehensive phylogenetic tree including 777 putative SWEET genes of 131 species supports this correspondence [19]. The tissue specificity of the genes was similar to that in sorghum: GRZM2G144581 is expressed mainly in the embryo; GRZM2G137954 in the seed and endosperm; GRMZM2G000812 in the stem 0–30 days after pollination (DAP) [36]. The stem-specific expression after DAP strongly supports the hypothesis that GRMZM2G000812/SbSWEET4-3 has specific roles in the stem. Therefore, gene duplication occurred before the branching of sorghum and maize. Why, then, does maize not accumulate as much sucrose in the stem as sorghum? One hypothesis is that there is a difference in the stem’s capacity to accumulate juice. Some cultivars (such as SIL-05) have a juicy parenchyma in the stem that can effectively accumulate sugar juice, but sorghum cultivars with a dry pith do not [37]. We therefore consider that because maize has a dry pith in the stem, it does not effectively accumulate sugar juice there. We consider that the trait of juicy stem in sorghum is necessary for the accumulation of large amounts of sucrose in this tissue.
We therefore consider that SbSWEET4-3 plays a pivotal role in sweet sorghums because of its potent expression in the stem, the amino acid substitution between SIL-05 and BTx623, and the absence of an orthologous gene in the syntenic region of O. sativa. SbSWEET4-3 is a strong candidate for a sucrose transporter that unloads sucrose from the phloem to the stem apoplast during the sucrose accumulation stage (Fig. 5a, c).
Other SWEET genes
On the basis of their analogy to Arabidopsis or rice genes, we examined SWEET genes with functions other than sucrose accumulation.
SbSWEET2-1 (Sobic.002G259300) and SbSWEET7-1 (Sobic.007G191200) were expressed only in the panicle from the start of heading through to 36 days afterward (Fig. 3); these genes are in the same clade as rice OsSWEET11/Xa13 (Fig. 4). OsSWEET11/Xa13 is expressed in the panicle and is essential for reproductive development [23, 25], suggesting that these SbSWEET2-1 and SbSWEET7-1 have roles in seed development (Fig. 5a).
SbSWEET9-3 (Sobic.009G252000) was highly expressed in the panicle only just after heading, after which their expression decreased (Fig. 3). These SWEET genes are grouped into the same clade as AtSWEET8/RPG1 (Fig. 4). AtSWEET8/RPG1 is essential for pollen viability through the transport of glucose across the plasma membranes of tapetum cells and pollen cells [21, 38]. We thus consider that SbSWEET9-3 are involved in the transport of glucose and contribute to pollen nutrition in sorghums (Fig. 5a).
SUT
SUT genes are in another sugar transporter family. SUT paralogs had tissue-specific expression in sorghum cultivars. In SIL-05, SbSUT1 and SbSUT2 were expressed highly in the leaf; SbSUT1, SbSUT2, and SbSUT4 in the stem; and SbSUT1, SbSUT2,SbSUT3,SbSUT4, and SbSUT5 in the panicle. SbSUT3 was expressed in the panicle just after heading (Fig. 3). In Rio (a sweet sorghum), SUT2 and SUT5 are expressed relatively highly in the stem, whereas in BTx623, SUT5, and SUT6 are expressed relatively highly in the inflorescence sink [26]. This tissue-specific expression suggests that SUT paralogs function in different sinks—i.e., in either the stem (for sucrose accumulation) or the grain or inflorescence (for starch synthesis or pollen nutrition). However, SUT genes are not differentially expressed between Wray (a sweet sorghum) and Macia (a grain sorghum) [27]. Therefore, the high level of expression of SUT genes in the stem [26] might be specific to Rio, and not a general feature of sweet sorghums.
INV
Invertase converts sucrose to glucose and fructose. All four sorghum INV genes were hardly expressed, or not expressed in the stem (Fig. 3). Given this absence of INV activity, loaded sucrose would not be hydrolyzed to glucose and fructose, and sucrose would therefore accumulate in the apoplast (Fig. 5). This is consistent with a previous analysis of INV enzymatic activity in the sorghum stem: INV activity in sorghum differs from that in sugarcane, as sugarcane also transfers sucrose to storage parenchyma, with hydrolysis to hexoses by cell-wall INV in the stem [39, 40]. In the panicle just after heading, one sorghum INV3 (Sobic.004G004800) was highly expressed (Fig. 3); this is consistent with a previous report of the occurrence of cell-wall INV activity in developing seeds [41], suggesting that sorghum INV contributes to starch synthesis in developing seeds.
SUS
SUS is a sucrose-cleaving enzyme that provides UDP-glucose and fructose [42]. SUS1 was expressed in all tissues at the sugar accumulation stage (Fig. 3). What is the effect of the sucrose-cleaving enzyme SUS1 in the stem of SIL-05? One hypothesis is that SUS1 provides energy and materials (e.g., cellulose) for construction of the sink structure of the internodes, which in turn increases the sucrose accumulation capacity in the stem of SIL-05. The SUS gene in sugarcane (called SS) is expressed at high levels in immature (developing) internodes, but at low levels in mature internodes [42]. A second hypothesis is that SUS1 production increases the hexose content of the stem of SIL-05. The hexose (glucose and fructose) content of the stem of SIL-05 (2–3 % each; Fig. 1) is uniquely high among high-Brix sorghums [43]. SUS1 expression was higher in the stem of SIL-05 (Fig. 3) than in that of BTx623 [33]. The relatively high level of expression of SUS1 in the stem might therefore relate to the high hexose content of SIL-05.
Other genes that might contribute to sucrose accumulation in stems
Expression diversity in sorghum cultivars might be responsible for the characteristic differences in sucrose accumulation between sweet and grain sorghums. Between BTx623 and Keller (a sweet sorghum), 3436 genes are differentially expressed, although 80 % of these differentially expressed genes have orthologs in rice [44]. Sugar-related traits have also been analyzed by quantitative trait locus (QTL) analysis using a cross of SS79 (a sweet sorghum) × M71 (a grain sorghum) [30] or R9188 (a sweet sorghum) × R9403463-2-1 (a grain sorghum) [31]. These traits have been assigned to the ten sorghum chromosomes (Fig. 2), but the genes responsible for the traits have not yet been identified. Here, we showed the chromosomal locations of SWEET, SUT, SPS, SUS, and INV (Fig. 2). Some of these genes might be identical to those that were the targets of these previous QTL analyses. However, these genes are not located on the short arm of chromosome 6, even though a QTL analysis [30] indicated that this region was associated with sugar content (Fig. 2). Genes for heading date, plant height, stem diameter, tiller number per plant, panicle weight, and juice weight might also contribute to the final sugar content in sorghum stems. Therefore, genes for these sugar-related agronomic traits will need to be analyzed in the future.
Conclusions
We determined the expression of key SWEET genes for phloem loading and unloading (and thus sucrose accumulation) in sorghum stems. We consider that SbSWEET8-1 plays a key role in the efflux of photosynthesized sucrose from the leaf and that SbSWEET4-3 is a sugar transporter that unloads sucrose from the phloem to the stem apoplast during the sucrose accumulation stage. We also consider that SbSWEET2-1 and SbSWEET7-1 play a key role in seed development and SbSWEET9-3 in pollen nutrition. These SWEET genes will be the targets for technological improvement in the production of biofuels.
Methods
Plant materials and quantification of stem sugar content
The sorghum cultivar SIL-05 (line number 89) was obtained from Shinshu University in Nagano, Japan. Stem sugar content was measured during the stage at which sucrose is considered to accumulate in the stem (1, 17, 36 and 64 days after heading). The volume/weight of total sugar content, sucrose, fructose, and glucose was measured by capillary electrophoresis and calculated using protocols previously described [43].
RNA sequencing
RNA was extracted from the second leaf from the flag leaf, the stem (internode only), and the panicle during the stage of sucrose accumulation in the stem (1, 17, and 36 days after heading). Each tissue was immediately frozen in liquid nitrogen and mixed to minimize the effect of transcriptome unevenness among plants. RNA quality was calculated with a Bioanalyzer 2100 algorithm (Agilent Technologies, Palo Alto, CA, USA); high-quality (RNA Integrity Number >8) RNA was used. Sequencing of each 100 bp using an Illumina Hiseq 2000 sequencer (Illumina, San Diego, CA, USA) has been described previously [45, 46].
Data analysis
Low-quality nucleotides (<Q15) from both the 5′- and the 3′-ends, and adaptors, were trimmed using Cutadapt version 1.0 (https://cutadapt.readthedocs.org/en/stable/). Reads were aligned against sorghum rRNA gene sequences [47] using Bowtie 2 version 2.0.0 beta6 [48]; aligned reads were removed. The remaining reads were aligned to the sorghum reference genome of BTx623 (Sbicolor_v2.1_255) [29] using TopHat version 2.0.4 [49] and Cufflinks version 2.2.0 [50]. FPKM (fragments per kilobase of exon per million mapped sequence reads) values were calculated for each gene model annotated in Phytozome ver.10.3 [51].
Categorization of the sorghum SWEET gene family
We chose 23 putative sorghum SWEET genes in the BTx623 reference genome using the EggNOG database [19]. Even though the number of SWEET family genes differs depending on the database (e.g., 21 homologs, [20]; 22 in phytozome 10.3, [51]), the members are nearly consistent. Evolutionary analyses were conducted in MEGA7 [52]. The evolutionary history was inferred using the neighbor-joining method [53]. The evolutionary distances were computed using the Poisson correction method [54] and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated. The data on chromosomal synteny were based on the Plant Genome Duplication Database [34].
Authors’ contributions
HM carried out the RNA extraction, sequencing, and data analysis and drafted the manuscript. SK participated in the breeding of the sorghum SIL-05 (line number 89) and provided the seeds. HK grew the sorghum plant, carried out the quantification of their sucrose content, and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Mr. Takayuki Yazawa of NIAS for advice on bioinformatics and Ms. Ikumi Tsujii of NIAS for sampling and RNA extraction.
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
The datasets supporting the conclusions of this article are available in the DDBJ (DNA Data Bank of Japan) Sequence Read Archive (Accession No. DRA004664).
Consent for publication
Not applicable.
Ethical approval and consent to participate
Not applicable.
Funding
This work was supported by Kieikai Research Foundation (2014).
Abbreviations
- SPS
sucrose phosphate synthase
- SUS
sucrose synthase
- SWEET
sugars will eventually be exported transporters
- SUT
sucrose transporter
- INV
invertase
- SNP
single nucleotide polymorphism
Additional file
10.1186/s13068-016-0546-6 Amino acid alignment of SWEET proteins. (A) Alignment of SWEET4-1, SWEET4-2, and SWEET4-3 of SIL-05 and BTx623. Red arrows indicate amino acids that differ between SIL-05 and BTx623: V172L of SWEET4-1 and D229E of SWEET4-3. (B) Alignment of putative orthologs of SWEET4-1 among plants. The aspartic acid at 229 of SWEET4-1 (corresponding to position 229 of sorghum SWEET4-3; red arrow) is conserved among putative SWEET orthologs in Brachypodium distachyon, Oryza sativa, Setaria italica, and Zea mays.
Contributor Information
Hiroshi Mizuno, Phone: +81-29-838-6002, Email: hmizuno@affrc.go.jp.
Shigemitsu Kasuga, Email: skasuga@shinshu-u.ac.jp.
Hiroyuki Kawahigashi, Email: shiwak@affrc.go.jp.
References
- 1.Calvino M, Messing J. Sweet sorghum as a model system for bioenergy crops. Curr Opin Biotechnol. 2012;23(3):323–329. doi: 10.1016/j.copbio.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 2.de Siqueira Ferreira S, Nishiyama MY, Jr, Paterson AH, Souza GM. Biofuel and energy crops: high-yield Saccharinae take center stage in the post-genomics era. Genome Biol. 2013;14(6):210. doi: 10.1186/gb-2013-14-6-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JH, Li SZ, Han B, Yu MH, Li GM, Jiang Y. A novel cost-effective technology to convert sucrose and homocelluloses in sweet sorghum stalks into ethanol. Biotechnol Biofuels. 2013;6:174. doi: 10.1186/1754-6834-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai H, Dunn JB, Wang ZC, Han J, Wang MQ. Life-cycle energy use and greenhouse gas emissions of production of bioethanol from sorghum in the United States. Biotechnol Biofuels. 2013;6:141. doi: 10.1186/1754-6834-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feltus FA, Vandenbrink JP. Bioenergy grass feedstock: current options and prospects for trait improvement using emerging genetic, genomic, and systems biology toolkits. Biotechnol Biofuels. 2012;5:80. doi: 10.1186/1754-6834-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun DM. SWEET! The pathway is complete. Science. 2012;335(6065):173–174. doi: 10.1126/science.1216828. [DOI] [PubMed] [Google Scholar]
- 7.Braun DM, Wang L, Ruan YL. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot. 2014;65(7):1713–1735. doi: 10.1093/jxb/ert416. [DOI] [PubMed] [Google Scholar]
- 8.Bolouri Moghaddam MR, Van den Ende W. Sugars, the clock and transition to flowering. Front Plant Sci. 2013;4:22. doi: 10.3389/fpls.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 10.Couger MB, Youssef NH, Struchtemeyer CG, Liggenstoffer AS, Elshahed MS. Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp. strain C1A. Biotechnol Biofuels. 2015;8:208. doi: 10.1186/s13068-015-0390-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin AP, Palmer WM, Byrt CS, Furbank RT, Grof CPL. A holistic high-throughput screening framework for biofuel feedstock assessment that characterises variations in soluble sugars and cell wall composition in Sorghum bicolor. Biotechnol Biofuels. 2013;6:186. doi: 10.1186/1754-6834-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sykes RW, Gjersing EL, Foutz K, Rottmann WH, Kuhn SA, Foster CE, Ziebell A, Turner GB, Decker SR, Hinchee MAW, et al. Down-regulation of p-coumaroyl quinate/shikimate 3ʹ-hydroxylase (C3ʹ H) and cinnamate 4-hydroxylase (C4H) genes in the lignin biosynthetic pathway of Eucalyptus urophylla × E. grandis leads to improved sugar release. Biotechnol Biofuels. 2015;8:1. doi: 10.1186/s13068-015-0316-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petti C, Harman-Ware AE, Tateno M, Kushwaha R, Shearer A, Downie AB, Crocker M, DeBolt S. Sorghum mutant RG displays antithetic leaf shoot lignin accumulation resulting in improved stem saccharification properties. Biotechnol Biofuels. 2013;6:146. doi: 10.1186/1754-6834-6-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludewig F, Flugge UI. Role of metabolite transporters in source-sink carbon allocation. Front Plant Sci. 2013;4:231. doi: 10.3389/fpls.2013.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qazi HA, Paranjpe S, Bhargava S. Stem sugar accumulation in sweet sorghum—activity and expression of sucrose metabolizing enzymes and sucrose transporters. J Plant Physiol. 2012;169(6):605–613. doi: 10.1016/j.jplph.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 16.Hedrich R, Sauer N, Neuhaus HE. Sugar transport across the plant vacuolar membrane: nature and regulation of carrier proteins. Curr Opin Plant Biol. 2015;25:63–70. doi: 10.1016/j.pbi.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Chen LQ. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201(4):1150–1155. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- 18.Eom JS, Chen LQ, Sosso D, Julius BT, Lin IW, Qu XQ, Braun DM, Frommer WB. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol. 2015;25:53–62. doi: 10.1016/j.pbi.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44(D1):D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao MZ, Sonah H, Song L, Lin L, Chaudhary J, et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 2015;16:520. doi: 10.1186/s12864-015-1730-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84(84):865–894. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 22.Yuan M, Zhao JW, Huang RY, Li XH, Xiao JH, Wang SP. Rice MtN3/saliva/SWEET gene family: evolution, expression profiling, and sugar transport. J Integr Plant Biol. 2014;56(6):559–570. doi: 10.1111/jipb.12173. [DOI] [PubMed] [Google Scholar]
- 23.Yuan M, Wang SP. Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013;6(3):665–674. doi: 10.1093/mp/sst035. [DOI] [PubMed] [Google Scholar]
- 24.Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335(6065):207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- 25.Chen LQ, Hou BH, Lalonde S, Takanaga H, Hartung ML, Qu XQ, Guo WJ, Kim JG, Underwood W, Chaudhuri B, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milne RJ, Byrt CS, Patrick JW, Grof CPL. Are sucrose transporter expression profiles linked with patterns of biomass partitioning in Sorghum phenotypes? Front Plant Sci. 2013;4:223. doi: 10.3389/fpls.2013.00223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bihmidine S, Baker RF, Hoffner C, Braun DM. Sucrose accumulation in sweet sorghum stems occurs by apoplasmic phloem unloading and does not involve differential sucrose transporter expression. BMC Plant Biol. 2015;15:186. doi: 10.1186/s12870-015-0572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno R, Ichinose H, Honda M, Takabatake K, Sotome I, Takai T, Maehara T, Okadome H, Isobe S, Gau M, et al. Use of whole crop sorghums as a raw material in consolidated bioprocessing bioethanol production using Flammulina velutipes. Biosci Biotechnol Biochem. 2009;73(7):1671–1673. doi: 10.1271/bbb.90099. [DOI] [PubMed] [Google Scholar]
- 29.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457(7229):551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 30.Shiringani AL, Frisch M, Friedt W. Genetic mapping of QTLs for sugar-related traits in a RIL population of Sorghum bicolor L. Moench. Theor Appl Genet. 2010;121(2):323–336. doi: 10.1007/s00122-010-1312-y. [DOI] [PubMed] [Google Scholar]
- 31.Ritter KB, Jordan DR, Chapman SC, Godwin ID, Mace ES, McIntyre CL. Identification of QTL for sugar-related traits in a sweet × grain sorghum (Sorghum bicolor L. Moench) recombinant inbred population. Mol Breed. 2008;22(3):367–384. doi: 10.1007/s11032-008-9182-6. [DOI] [Google Scholar]
- 32.Luo H, Zhao WM, Wang YQ, Xia Y, Wu XY, Zhang LM, Tang BX, Zhu JW, Fang L, Du ZL, et al. SorGSD: a sorghum genome SNP database. Biotechnol Biofuels. 2016;9:37. doi: 10.1186/s13068-016-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makita Y, Shimada S, Kawashima M, Kondou-Kuriyama T, Toyoda T, Matsui M. MOROKOSHI: transcriptome database in Sorghum bicolor. Plant Cell Physiol. 2015;56(1):e6. doi: 10.1093/pcp/pcu187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee TH, Tang HB, Wang XY, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 2013;41(D1):D1152–D1158. doi: 10.1093/nar/gks1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato Y, Antonio BA, Namiki N, Takehisa H, Minami H, Kamatsuki K, Sugimoto K, Shimizu Y, Hirochika H, Nagamura Y. RiceXPro: a platform for monitoring gene expression in japonica rice grown under natural field conditions. Nucleic Acids Res. 2011;39:D1141–D1148. doi: 10.1093/nar/gkq1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekhon RS, Lin HN, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. Genome-wide atlas of transcription during maize development. Plant J. 2011;66(4):553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 37.Mace ES, Jordan DR. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench) Theor Appl Genet. 2010;121(7):1339–1356. doi: 10.1007/s00122-010-1392-8. [DOI] [PubMed] [Google Scholar]
- 38.Sun MX, Huang XY, Yang J, Guan YF, Yang ZN. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013;26(2):83–91. doi: 10.1007/s00497-012-0208-1. [DOI] [PubMed] [Google Scholar]
- 39.HoffmannThoma G, Hinkel K, Nicolay P, Willenbrink J. Sucrose accumulation in sweet sorghum stem internodes in relation to growth. Physiol Plant. 1996;97(2):277–284. doi: 10.1034/j.1399-3054.1996.970210.x. [DOI] [Google Scholar]
- 40.Wang JP, Nayak S, Koch K, Ming R. Carbon partitioning in sugarcane (Saccharum species) Front Plant Sci. 2013;4:201. doi: 10.3389/fpls.2013.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain M, Chourey PS, Li QB, Pring DR. Expression of cell wall invertase and several other genes of sugar metabolism in relation to seed development in sorghum (Sorghum bicolor) J Plant Physiol. 2008;165(3):331–344. doi: 10.1016/j.jplph.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Verma AK, Upadhyay SK, Verma PC, Solomon S, Singh SB. Functional analysis of sucrose phosphate synthase (SPS) and sucrose synthase (SS) in sugarcane (Saccharum) cultivars. Plant Biol. 2011;13(2):325–332. doi: 10.1111/j.1438-8677.2010.00379.x. [DOI] [PubMed] [Google Scholar]
- 43.Kawahigashi H, Kasuga S, Okuizumi H, Hiradate S, Yonemaru J. Evaluation of Brix and sugar content in stem juice from sorghum varieties. Grassl Sci. 2013;59(1):11–19. doi: 10.1111/grs.12006. [DOI] [Google Scholar]
- 44.Jiang SY, Ma ZG, Vanitha J, Ramachandran S. Genetic variation and expression diversity between grain and sweet sorghum lines. BMC Genomics. 2013;14:18. doi: 10.1186/1471-2164-14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizuno H, Kawahigashi H, Kawahara Y, Kanamori H, Ogata J, Minami H, Itoh T, Matsumoto T. Global transcriptome analysis reveals distinct expression among duplicated genes during sorghum-interaction. BMC Plant Biol. 2012;12:121. doi: 10.1186/1471-2229-12-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mizuno H, Yazawa T, Kasuga S, Sawada Y, Ogata J, Ando T, Kanamori H, Yonemaru J, Wu J, Hirai MY, et al. Expression level of a flavonoid 3′-hydroxylase gene determines pathogen-induced color variation in sorghum. BMC Res Notes. 2014;7:761. doi: 10.1186/1756-0500-7-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouyang S, Buell CR. The TIGR Plant Repeat Databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32(Database issue):D360–363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goodstein DM, Shu SQ, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40(D1):D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saitou N, Nei M. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 54.Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press; 1965. pp. 97–166. [Google Scholar]


