Abstract
SAMHD1 limits HIV-1 infection in non-dividing myeloid cells by decreasing intracellular dNTP pools. HIV-1 restriction by SAMHD1 in these cells likely prevents activation of antiviral immune responses and modulates viral pathogenesis, thus highlighting a critical role of SAMHD1 in HIV-1 physiopathology. Here, we explored the function of SAMHD1 in regulating cell proliferation, cell cycle progression and apoptosis in monocytic THP-1 cells. Using the CRISPR/Cas9 technology, we generated THP-1 cells with stable SAMHD1 knockout. We found that silencing of SAMHD1 in cycling cells stimulates cell proliferation, redistributes cell cycle population in the G1/G0 phase and reduces apoptosis. These alterations correlated with increased dNTP levels and more efficient HIV-1 infection in dividing SAMHD1 knockout cells relative to control. Our results suggest that SAMHD1, through its dNTPase activity, affects cell proliferation, cell cycle distribution and apoptosis, and emphasize a key role of SAMHD1 in the interplay between cell cycle regulation and HIV-1 infection.
Keywords: SAMHD1, monocytic cells, gene knockout, HIV-1, dNTP, restriction, cell cycle, apoptosis
Introduction
SAM domain- and HD domain-containing protein 1 (SAMHD1) is the first deoxynucleoside triphosphate triphosphohydrolase (dNTPase) identified in mammalian cells (Goldstone et al., 2011). SAMHD1 induces the hydrolysis of dNTPs and, in concert with cellular ribonucleotide reductase, functions as a key regulator of intracellular dNTP homeostasis. In 2011, two laboratories independently identified SAMHD1 as a host restriction factor inhibiting human immunodeficiency virus type 1 (HIV-1) infection in non-dividing myeloid cells (Hrecka et al., 2011; Laguette et al., 2011). The viral protein X (Vpx) uniquely expressed by lentiviruses such as HIV-2 and many strains of simian immunodeficiency virus, but not by HIV-1, has been shown to restore HIV-1 infection in myeloid cells (Guyader et al., 1989; Yu et al., 1991). The underlying molecular mechanisms remained unknown until the discovery that Vpx targets SAMHD1 for proteasomal degradation, thus counteracting its restriction activity and supporting HIV-1 infection (Hrecka et al., 2011; Laguette et al., 2011). We and others showed that SAMHD1 dNTPase activity is responsible for the extremely low dNTP concentrations associated with the kinetic delay in HIV-1 reverse transcription, and that Vpx accelerates the viral DNA synthesis by elevating cellular dNTP levels in myeloid cells and resting CD4+ T lymphocytes, thus revealing the mechanistic and regulatory links among SAMHD1, Vpx, cellular dNTPs, and viral reverse transcription kinetics (Baldauf et al., 2012; Kim et al., 2012; Lahouassa et al., 2012; St Gelais et al., 2012). Besides its dNTP hydrolase function, SAMHD1 harbors RNase and nuclease activities, which have been postulated to possibly contribute to HIV-1 restriction (Beloglazova et al., 2013; Choi et al., 2015; Ryoo et al., 2014), although these results require further validation.
In addition to the role of SAMHD1 as a host restriction factor, mutations in the SAMHD1 gene have been linked to a genetic immune disorder called Aicardi-Goutières Syndrome (AGS) (Rice et al., 2009), as well as several types of cancer, of both solid and hematological origins [reviewed in (Kohnken et al., 2015)]. These accumulating lines of evidence suggest the involvement of SAMHD1 in the innate immune response and cancer development through the control of dNTP homeostasis. In the last few years, intense efforts have been carried out in order to define the mechanisms by which SAMHD1 interferes with HIV-1 infection in non-dividing cells (Wu, 2013), and unravel its role in immunological diseases and cancer development. However, the physiological functions of SAMHD1 remain to be fully defined, and elucidation of the underlying mechanisms would help the development of potential therapeutic approaches in the context of HIV-1, AGS and cancer.
Here, we developed a monocytic THP-1 cell line model presenting stable knockout (KO) of the SAMHD1 gene by using the CRISPR/Cas9 genome editing technology. We aimed to characterize the phenotype of THP-1 cells lacking SAMHD1 protein in comparison to control cells expressing the endogenous and functional protein. We focused on the effects of SAMHD1 on cell proliferation, cell cycle regulation and cell death, and their potential correlation with HIV-1 restriction. We found that SAMHD1 silencing leads to increased cell growth, perturbation of the cell cycle and reduced susceptibility to apoptosis. Moreover, we observed increased dNTP levels and enhanced HIV-1 infection in dividing SAMHD1 KO THP-1 cells relative to control cells, thus confirming the role of SAMHD1 in the control of HIV-1 life cycle in myeloid cells. Our results shed light on a functional interplay between SAMHD1-mediated regulation of cell cycle, apoptosis and HIV-1 infection through its dNTPase activity.
Results
Stable silencing of the SAMHD1 gene in THP-1 cells
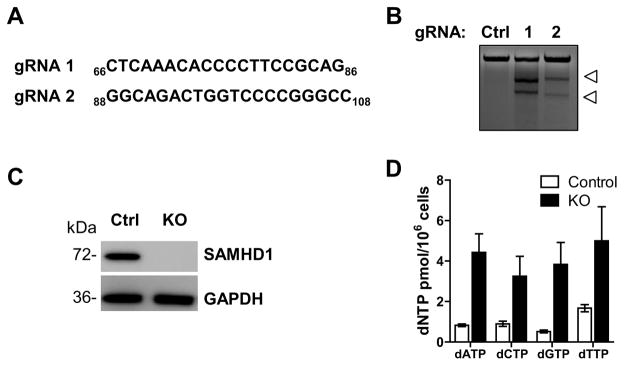
THP-1 cells have been widely used as a cell line model to investigate the functions of primary myeloid-lineage cells such as monocytes, macrophages and dendritic cells (Auwerx, 1991; Berges et al., 2005; Chanput et al., 2014). Here, we employed the CRISPR/Cas9 technology to knockout SAMHD1 in THP-1 cells. We chose the THP-1 cell line for our model system because, opposed to other monocytic cell lines such as U937 cells lacking endogenous SAMHD1 expression, THP-1 cells express similar levels of SAMHD1 compared to primary myeloid cells (Laguette et al., 2011). Two single guide RNAs (gRNA 1 and gRNA 2 indicated in Fig. 1A) targeting unique sequences in exon 1 of the SAMHD1 gene were designed and cloned into a lentiviral vector (Sanjana et al., 2014; Shalem et al., 2014). Polyclonal undifferentiated THP-1 cells were transduced with a lentiviral vector expressing gRNA 1 or gRNA 2, Cas9 and a puromycin resistance marker (Shalem et al., 2014). To assess the ability of the gRNAs to cleave SAMHD1, we used the Surveyor nuclease assay (Guschin et al., 2010; Ran et al., 2013). Compared to untargeted cells, cleavage products were detected only in genomic DNA from polyclonal THP-1 cells transduced with lentiviral vector containing either gRNA 1 or 2 (Fig. 1B, shown by arrows), indicating that the gRNAs successfully targeted the region of interest and, therefore, are valid candidates for knocking out SAMHD1. Immunoblotting analysis confirmed the loss of SAMHD1 expression in three cell clones transduced with two different gRNAs targeting SAMHD1 (KO), while endogenous SAMHD1 expression was detected in the THP-1 clones transduced with the control vector (Fig. 1C and Supplementary Fig. 1A).
Fig. 1. SAMHD1 knockout in THP-1 cells by CRISPR/Cas9.
(A) CRISPR/Cas9 gRNA sequences used to produce the THP-1 SAMHD1 KO cells. Indicated sequences target the exon 1 of the SAMHD1 gene (nucleotide number of exon 1 indicated as subscript). (B) Surveyor nuclease assay confirming SAMHD1 gRNA constructs. Genomic DNA was isolated from a stably transduced and selected polyclonal population of THP-1 cells, followed by PCR amplification of the SAMHD1 targeted region. Amplicons were slowly reannealed. Surveyor nuclease-mediated cleavage of the heteroduplexes generated was detected in DNA from cells transfected with either gRNA 1 or 2 (white arrows), confirming that the gRNAs efficiently target SAMHD1. (C) SAMHD1 protein expression in non-differentiated THP-1 SAMHD1 control (clone 1) and KO cells (KO clone 1, derived from transduction with gRNA 2) confirms efficient knockout of the SAMHD1 gene. GAPDH was used as loading control. (D) SAMHD1 KO cells show increased dNTP levels compared to control cells. Intracellular dNTP concentration in non-differentiated THP-1 cells expressing or not SAMHD1 was determined by single nucleotide incorporation assay. Error bars represent standard deviation of duplicate biological samples.
As SAMHD1 is a key regulator of intracellular dNTP homeostasis (Ballana and Este, 2015), we analyzed dNTP levels in non-differentiated SAMHD1 KO and control cells by using our previously described dNTP assay (Diamond et al., 2004). Intracellular dNTP pools increased 3- to 6-fold in SAMHD1 KO compared to control cells, as shown in Fig. 1D (KO clone 1, derived from gRNA 2, and control clone 1, referred respectively as KO and control from here on) and Supplementary Fig. 1B. Our data are consistent with previously published results showing a significant increase in the dNTP levels in primary human macrophages or dendritic cells in which efficient SAMHD1 degradation was induced by Vpx (Hollenbaugh et al., 2014b; Kim et al., 2012; St Gelais et al., 2012), thus further confirming that the complete silencing of SAMHD1 expression in KO cells leads to up-regulation of the intracellular dNTP pool.
Silencing of SAMHD1 affects cell proliferation and alters cell cycle status
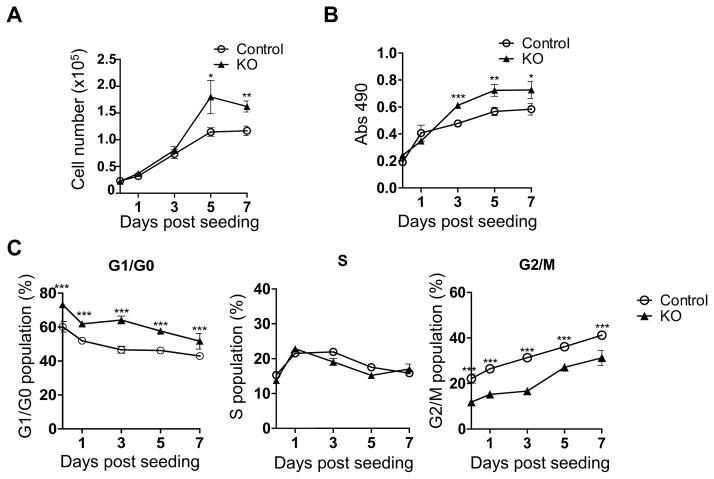
Overexpression of SAMHD1 has been described to reduce the proliferation of HeLa cells and the lung cancer cell line A549 (Clifford et al., 2014; Wang et al., 2014). To investigate the effect of SAMHD1 on THP-1 cell growth, we analyzed the proliferation of non-differentiated control and KO cells by trypan blue exclusion and 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assays. By performing a time course experiment, we observed enhanced growth rate and viability of KO cells compared to control cells, with a significant increase of the number of live cells at days 5 and 7 after seeding (1.6 and 1.4-fold, respectively, Fig. 2A), while a significant increase of cell proliferation and metabolism (1.3-fold) was observed from day 3 to day 7 (Fig. 2B). These results prompted us to investigate whether SAMHD1 may induce perturbation of the cell cycle. Flow cytometry analysis performed in asynchronous cells over a period of 7 days showed that knockout of SAMHD1 was associated with significantly increased G1/G0 cell population in comparison to control cells, while a decrease was observed in the fraction of cells in G2/M phase (Fig. 2C). A similar cell cycle profile was obtained in cells synchronized in G1 phase by serum starvation (data not shown), further suggesting that SAMHD1 delays cell cycle progression most likely through accumulation of the cells in G2/M. Notably, additional two SAMHD1 KO cell clones (derived from gRNA 1 or 2) showed similar phenotypes compared to control cells (Supplementary Fig. 1C–D), thus confirming that the effects on cell proliferation and cell cycle status can be specifically ascribed to SAMHD1 silencing. Although the investigation of the molecular mechanisms associated with these effects is beyond the scope of this study, these data suggest that SAMHD1 negatively impacts cell cycle progression, leading to reduced cell growth and proliferation.
Fig. 2. THP-1 SAMHD1 knockout cells have increased cell proliferation and altered cell cycle status.
(A) Live cell counting. SAMHD1 control and KO cells were cultured for 7 days and, at the indicated time points, the number of viable cells was determined by trypan blue exclusion. Error bars represent standard deviation of quadruple samples. Statistical analysis was performed with the unpaired T-test with Welch’s correction (*P=0.0268, **P=0.0011). (B) Proliferation of SAMHD1 control and KO cells was measured in time course by MTS assay. Error bars represent standard deviation of four replicates. Statistical analysis was performed using the unpaired T-test with Welch’s correction (*P=0.0130, **P=0.0053, ***P=0.0002). (C) Alteration of the cell cycle by SAMHD1. Flow cytometry analysis of cell cycle progression in SAMHD1 KO cells in comparison to control cells was performed by propidium iodide staining. Error bars represent standard deviation of triplicate samples. Statistical analysis was performed using the Two-way ANOVA (**P<0.01, ***P<0.001).
Silencing of SAMHD1 results in reduced spontaneous activation of apoptosis
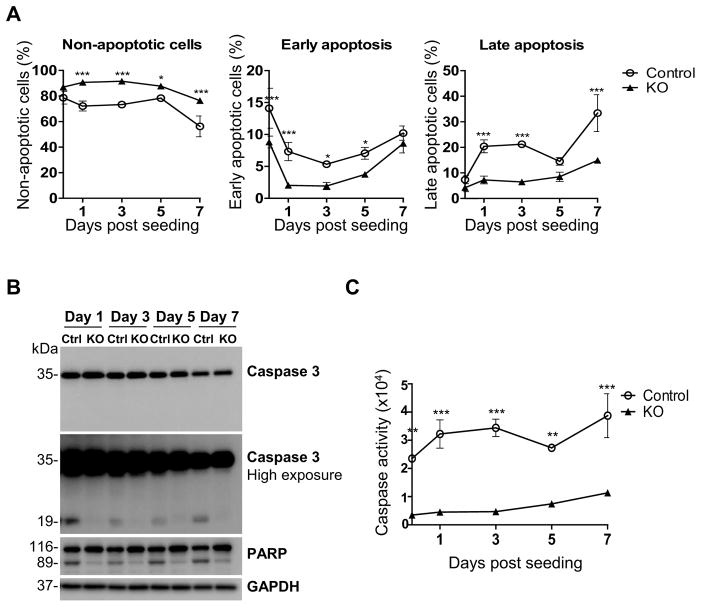
We next evaluated whether reduced proliferation of control cells is correlated with increased cell death. We analyzed apoptosis induction by Annexin-V staining, which allowed us to distinguish cells in early and late apoptosis due to the exposure of phosphatidylserine residues on the cell membrane, an event occurring at early stages during apoptotic cell death (Verhoven et al., 1995). As shown in Fig. 3A and Supplementary Fig. 2A, a higher percentage of early or late apoptotic cells was observed, respectively, at 1 or 3 days post-seeding in SAMHD1 expressing control cells compared to KO cells, consistently with more pronounced cell proliferation (Fig. 2B and Supplementary Fig. 1C) and G1/G0 accumulation (Fig. 2C and Supplementary Fig. 1D) detected in KO clones. In line with the data in Fig. 2A showing more robust cell viability, flow cytometry analysis of apoptosis demonstrated a higher percentage of the live KO cell population compared to control cells (Fig. 3A, left panel). Moreover, cells expressing SAMHD1 showed increased activation of caspase 3, as demonstrated by detection of cleaved active forms of caspase 3 and cleavage of poly (ADP-ribose) polymerase (PARP), a known target of active caspase 3 and marker of apoptosis (Boulares et al., 1999) (Fig. 3B and Supplementary Fig. 2B), and by direct measurement of caspase 3/7 activities using a luminescence-based assay (Fig. 3C and Supplementary Fig. 2C). Compared to control cells, SAMHD1 KO cells showed very low or absent caspase activation (e.g., 7-fold decrease at day 3 in Fig. 3C). These data suggest that SAMHD1 silencing may render the cells less prone to apoptosis compared to normal cells.
Fig. 3. Reduced apoptosis in SAMHD1 knockout THP-1 cells compared to control cells.
(A) SAMHD1 control and KO cells were seeded in 12-well plate and, at day 0, 1, 3, 5 and 7, apoptosis was measured by flow cytometry via cell staining with PE-Annexin V (to detect phosphatidylserine exposure) and 7-AAD (to distinguish viable from non-viable cells). The left panel shows non-apoptotic, live cells (negative for Annexin V and 7-AAD staining); middle panel represents cells in early apoptosis, which are positive to Annexin V but negative to 7-AAD staining; the “Late apoptosis” panel on the right shows the percentage of cells exposing phosphatidylserine on the surface (positive for Annexin V) and with damaged cell membrane (positive for 7-AAD). Error bars indicate the standard deviation of triplicate samples. Statistical analysis was performed using the Two-way ANOVA (*P<0.05, **P<0.01, ***P<0.001). (B) Immunoblotting analysis of caspase 3 and PARP cleavage as markers of apoptosis induction. GAPDH was used as loading control. (C) Caspase 3/7 activities is significantly more pronounced in control cells compared to KO cells, as measured by a luminescent-based assay. Error bars represent standard deviation of two independent experiments, each performed in quadruplicates. Two-way ANOVA was performed for statistical analysis. (**P<0.01, ***P<0.001).
Effect of SAMHD1 knockout and overexpression on HIV-1 infection in THP-1 cells
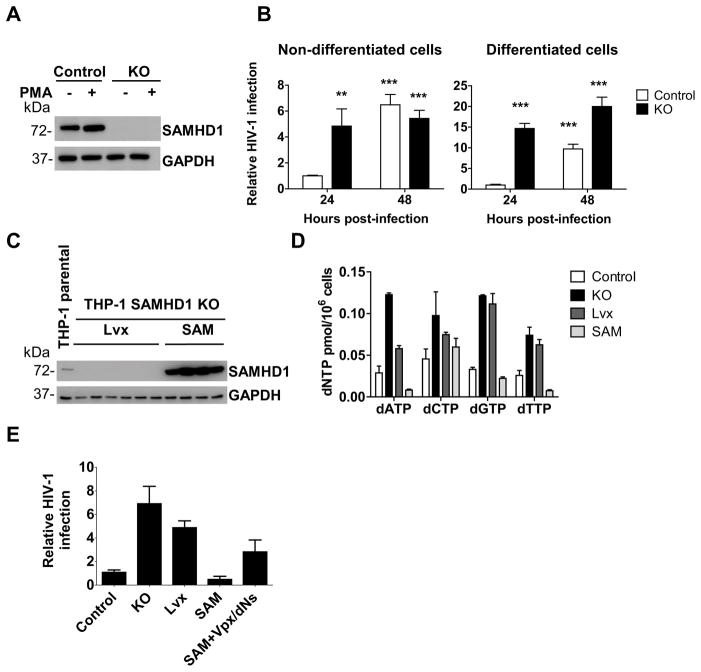
SAMHD1 is a known restriction factor of HIV-1 infection in non-dividing cells such as primary monocytes, dendritic cells, macrophages and resting CD4+ T-cells, where SAMHD1 is highly expressed (Baldauf et al., 2012; Berger et al., 2012; Hrecka et al., 2011; Laguette et al., 2011). SAMHD1-mediated HIV-1 restriction in phorbol 12-myristate 13-acetate (PMA)-differentiated, non-dividing macrophage-like cells (such as THP-1 or U937 cell lines) has been previously reported (Laguette et al., 2011; Lahouassa et al., 2012), but its effect on virus infection efficiency in dividing monocytic cell lines has not been carefully characterized. To explore this aspect and identify a potential link between SAMHD1 restriction activity and its effects on cell proliferation, cell cycle and apoptosis, we first analyzed SAMHD1 expression in control and KO cells differentiated or not with PMA treatment. Immunoblotting analysis showed a modest increase of SAMHD1 protein level in PMA-differentiated non-cycling control cells, in line with our published results (St Gelais et al., 2014) (Fig. 4A). Next, non-differentiated and differentiated THP-1 control and SAMHD1 KO cells were infected with a single-cycle luciferase reporter HIV-1 pseudotyped with the vesicular stomatitis virus protein G (HIV-1-Luc/VSV-G) (Wang et al., 2016). Luciferase activity was measured at 24 and 48 hours post-infection (hpi) as an indication of infection efficiency. At 24 hpi, cycling KO cells were more susceptible to HIV-1 infection compared to control cells (5-fold increase, Fig. 4B left panel), indicating that SAMHD1 is able to counteract HIV-1 infection in dividing THP-1 monocytic cells. At 48 hpi, HIV-1 infection efficiency was similar in the two cell lines, irrespective of SAMHD1 expression. Further investigation is required to unravel the mechanism responsible for the loss of SAMHD1 restriction activity in these cells at this time point. Conversely, in non-cycling SAMHD1 KO cells, HIV-1 infection was more efficient at both time points (14- and 20-fold increase at 24 and 48 hpi, respectively) (Fig. 4B, right panel), confirming that silencing of SAMHD1 in THP-1 cells results in the expected HIV-1 restriction phenotype as described in published studies (Laguette et al., 2011; Lahouassa et al., 2012; St Gelais et al., 2014).
Fig 4. Knockout of SAMHD1 increases HIV-1 infection of non-differentiated and differentiated THP-1 cells.
(A) THP-1 SAMHD1 control and KO cells were grown in the absence or presence of 100 ng/ml of PMA for 24 hours to induce differentiation into macrophage-like cells. Immunoblotting analysis using a specific SAMHD1 antibody confirmed efficient knockout of SAMHD1 in KO cells. (B) HIV-1 infection is increased in KO cells compared to control cells. Control and SAMHD1 KO cells, differentiated or not with PMA as described in (A), were infected with a single-cycle HIV-1-Luc/VSV-G at a multiplicity of infection (MOI) of 1. At 24 and 48 hours post-infection, HIV-1 infection efficiency was measured by luciferase assay. Luciferase values were normalized for protein concentration. The infection level in the control cells at 24 hours post-infection was set as 1 and relative values are shown. A representative experiment performed with four replicates per sample is presented. Statistical analysis was performed with the unpaired T-test with Welch’s correction (**P=0.0019, ***P<0.0001). (C) SAMHD1 overexpression in KO cells. KO cells were transduced with LVX-IRES-mCherry control lentiviral vector (Lvx) or LVX-SAMHD1-IRES-mCherry vector (SAM). Cells were sorted by flow cytometry and SAMHD1 expression was analyzed in six Lvx and four SAM clones after differentiation with PMA by immunoblotting using a SAMHD1 specific antibody. GAPDH was used as loading control. (D) Rescue of SAMHD1 in KO cells restores SAMHD1-mediated depletion of dNTP levels. Intracellular dNTP concentration was measured in differentiated cells as described in Fig. 1D. (E) Effect of SAMHD1 KO and overexpression on HIV-1 restriction. Cells were infected with a GFP-expressing HIV-1 virus and, after 72 hours, the percentage of GFP-positive cells was measured by flow cytometry. While silencing of SAMHD1 results in more efficient HIV-1 infection, overexpression of SAMHD1 in KO cells restores SAMHD1 restriction effects. Counteraction of SAMHD1-mediated viral restriction was detected in SAM cells co-treated with Vpx and dNs.
To confirm that the effects on dNTP intracellular concentration (Fig. 1D) and HIV-1 infection (Fig. 4B) observed in KO cells were due to the absence of SAMHD1 expression, the mutated SAMHD1 gene resistant to the CRISPR/Cas9 targeting was re-introduced into the KO cells by transducing the KO cell line derived from gRNA 2 with either control vector (Lvx) or SAMHD1-expressing (SAM) lentiviruses. The clones (six for Lvx and four for the SAMHD1 expression vector) were analyzed for SAMHD1 expression after differentiation with PMA (Fig. 4C). As expected, SAMHD1 protein was not detected in the six Lvx clones, whereas four SAM clones displayed robust SAMHD1 expression. Notably, SAMHD1 expression levels in the SAM clones were significantly higher than those in THP-1 parental and control cells (Fig. 4C and data not shown). Overexpression of SAMHD1 reversed the increased dNTP concentration phenotype observed in the KO and Lvx cells (Fig. 4D). Interestingly, dATP, dGTP and dTTP levels in the SAMHD1 cells were lower than those detected in control cells (Fig. 4D). This could be due to the increased expression of SAMHD1 in the SAM clones compared to the control cells.
Furthermore, we investigated the effect of SAMHD1 overexpression on HIV-1 infection of differentiated cells. PMA-treated THP-1 cells were infected with VSV-G-pseudotyped HIV-1-GFP (Diamond et al., 2004) and analyzed for GFP expression by flow cytometry at 72 hpi. As shown in Fig. 4E, KO and Lvx THP-1 cells lacking SAMHD1 displayed elevated HIV-1 infection compared to control cells (7- and 5-fold increase, respectively), correlating the loss of SAMHD1 with enhanced HIV-1 replication. HIV-1 infection was limited in SAM cells, which were also more effective at HIV-1 restriction than the control cells (Fig. 4E), consistently with the higher SAMHD1 expression and lower dNTP concentration observed in the SAM cells (Fig. 4C and 4D). These data indicate the tight interplay among SAMHD1 expression levels, dNTP concentrations, and HIV-1 infection efficiency, which we and others have previously observed in primary human macrophages or dendritic cells (Baldauf et al., 2012; Kim et al., 2012; Laguette et al., 2011; Lahouassa et al., 2012; St Gelais et al., 2012). Lastly, we sought to test whether the SAM clones were sensitive to Vpx-mediated SAMHD1 degradation and the treatment with deoxynucleosides (dNs), which independently elevate intracellular dNTP levels through the salvage pathway of dNTP synthesis (Lahouassa et al., 2012). We used a combined Vpx/dNs treatment as neither treatment alone resulted in a significant increase of HIV-1 infection efficiency (data not shown), most likely due to the very high expression levels of SAMHD1 in the SAM clones. As expected, SAMHD1-mediated suppression of HIV-1 infection of SAM cells was rescued by the combined Vpx/dNs treatment (Fig. 4E).
Taken together, these results show that SAMHD1 knockout and overexpression in THP-1 cells modulate HIV-1 infection by regulating the intracellular dNTP concentration, thus biochemically and virologically mimicking what is observed with primary monocyte-derived macrophages or dendritic cells, and can be a convenient tool to further study the precise mechanisms of SAMHD1-mediated retroviral restriction.
Discussion
SAMHD1 is a cellular restriction factor that limits HIV-1 infection in non-dividing myeloid cells and resting CD4+ T cells, by inducing the hydrolysis of dNTPs, which inhibits the viral reverse transcription process (Baldauf et al., 2012; Lahouassa et al., 2012). Non-productive infection of myeloid cells by SAMHD1 likely prevents activation of anti-viral immune responses, which may allow HIV-1 to escape immune recognition and establish persistent infection (Wu, 2012). Therefore, complete elucidation of the physiological functions of SAMHD1 in myeloid cells is important for the development of potential anti-viral approaches.
In this study, we generated a stable SAMHD1 KO THP-1 cell line to explore the effect of silencing SAMHD1 on cell proliferation, cell-cycle progression, and apoptosis. Beside its role in HIV-1 restriction, SAMHD1 is known to influence the proliferation of several cell types by regulating dNTP homeostasis (Kohnken et al., 2015). We characterized the phenotype of dividing SAMHD1 KO THP-1 cells in comparison to control cells, and found that SAMHD1 reduces cell proliferation and metabolism, in line with previous published work showing that overexpression of SAMHD1 negatively affects the proliferation of cancer cells (Clifford et al., 2014; Wang et al., 2014). Moreover, we observed that silencing of SAMHD1 redistributes cells mainly in the G1/G0 phase of the cell cycle. Our results are consistent with published data showing that SAMHD1 knock-down in proliferating human fibroblast cell lines (Franzolin et al., 2013) and that SAMHD1 mutations in fibroblasts from AGS patients (Kretschmer et al., 2015) result in accumulation of the cells in G1, decreased percentage of cells in G2 and increase of the intracellular dNTP pools. In addition to its effect on cell cycle progression, we found that SAMHD1 regulates apoptosis. Indeed, KO cells appear to be more resistant to spontaneous apoptotic cell death relative to control cells, as demonstrated by the reduced activation of caspase 3/7 and absent cleavage of the apoptotic marker PARP. Importantly, the effects of SAMHD1 silencing on cell growth, proliferation, cell cycle progression and apoptosis induction were observed in an additional control and two KO clones, thus suggesting that the phenotype described is unlikely due to off-target effects of the CRISPR/Cas9 system, and further confirming a key role of SAMHD1 in these processes.
Notably, changes in cell cycle progression were consistently associated with altered expression of cyclin D3. Our unpublished results revealed that the cyclin D3 mRNA level is significantly down-regulated in KO cells compared to control cells (data not shown). This data is consistent with accumulation of SAMHD1 KO cells in G1/G0, as cyclin D3 controls cell cycle progression at G1/S phase (Bartkova et al., 1998; Herzinger and Reed, 1998). Further investigation will help elucidate a potential interplay of cyclin D3 with SAMHD1-mediated control of cell proliferation.
We found that disturbances in cell cycle progression induced by SAMHD1 silencing are associated with increased intracellular dNTP concentrations and more productive HIV-1 infection of KO versus control cells. Previous studies from our and other groups showed that overexpressed SAMHD1 does not restrict HIV-1 infection in dividing HeLa and HEK293T cells (St Gelais et al., 2012; St Gelais et al., 2014; Welbourn and Strebel, 2016). Here we provide first evidence that SAMHD1 limits HIV-1 infection in actively proliferating THP-1 cells, and that the absence of SAMHD1 renders the cells more susceptible to HIV-1 infection. Increased HIV-1 infection efficiency could also be detected in differentiated cells, thus confirming the established HIV-1 restriction ability of SAMHD1 in non-dividing cells (Lahouassa et al., 2012), and further validating our THP-1 SAMHD1 KO model as a useful tool to study SAMHD1 functions in monocytic cells. Of note, while the enhancement of HIV-1 infection in differentiated KO cells is detected at both time points tested (24 and 48 hpi), we observed similar viral infection efficiency in non-differentiated control and SAMHD1 KO cells at 48 hpi. We exclude the possibility that this effect could be due to increased SAMHD1 T592 phosphorylation status, which is known to negatively regulate its HIV-1 restriction activity (Cribier et al., 2013; White et al., 2013), at 48 hpi (data not shown). Further analysis is needed to identify the responsible mechanisms. Importantly, by performing SAMHD1 knock-in experiments, we could reverse the phenotype observed in KO cells, confirming that the increase in dNTP levels and HIV-1 infection efficiency was mainly due to the absence of SAMHD1. Moreover, in cells overexpressing SAMHD1, its expression or activity could be impaired by co-treatment with Vpx and dNs, which are known to counteract SAMHD1-mediated HIV-1 restriction (Laguette et al., 2011; Lahouassa et al., 2012). Of note, SAMHD1 overexpressing cells showed even lower dNTP concentrations and more efficient HIV-1 restriction compared to control cells, which could be due to different expression levels of SAMHD1 protein in the two cell lines.
Correlation between cell cycle progression and SAMHD1 expression levels and its HIV restriction function remains to be confirmed (Franzolin et al., 2013; Kretschmer et al., 2015; Pauls et al., 2014; Yan et al., 2015). A complete picture of the mechanisms by which SAMHD1 influences cell cycle regulation is still missing, and it would be interesting to further investigate how cell cycle regulation affects its HIV-1 restriction activity in dividing cells, as well as to identify the role of cell cycle-related proteins during retroviral infection in myeloid cells.
In summary, by using a novel THP-1-derived cell model, we suggest that SAMHD1 can act as a regulator of the cell cycle and apoptosis-inducing factor in cycling THP-1 monocytic cells. These effects correlated with HIV-1 restriction, thus proposing a functional link between cell cycle control and HIV-1 restriction by SAMHD1. The new THP-1 cell model that we generated can expedite preliminary in vitro studies and future in vivo translational investigations, thus helping elucidate the cellular functions of SAMHD1 and mechanisms of retroviral restriction.
Materials and methods
Generation of SAMHD1 knockout THP-1 cells
Two single guide RNAs (gRNAs) targeting unique sequences of the SAMHD1 gene were designed and cloned into the lentiCRISPR v1 plasmid, which contains a puromycin resistance cassette and Cas9 nuclease (Sanjana et al., 2014; Shalem et al., 2014). Lentiviral vectors were produced by co-transfection of polyclonal THP-1 cells with the lentiCRISPR v1/gRNA, d8.74 and pMD2 plasmids using the cationic polymer polyethylenimine (PEI) method (Boussif et al., 1995). Viral supernatant was collected and used to transduce polyclonal undifferentiated THP-1 cells. Both gRNA constructs were functionally tested for their ability to mediate genomic insertion, deletion or inversions by isolating the genomic DNA from stably transduced and selected polyclonal population of THP-1 cells and performing a Surveyor nuclease assay as per manufacturer’s instructions (Guschin et al., 2010; Ran et al., 2013). Puromycin resistant cells were selected from each gRNA vector transduction, and then sorted by fluorescence-activated cell sorting (FACS) into 96-well plates for clonal expansion.
Generation of SAMHD1 overexpressing cell line
Silent mutations in the SAMHD1 gene were introduced by site directed mutagenesis (QuikChange Lightning kit, Agilent Genomics) to make it resistant to the gRNA 2-mediated CRISPR/Cas9 cleavage. The following primers were used: 5′-GAAGCTGATTGGTCACCTGGACTAGAACTCCATCCCGACTAC-3′ and 5′-GTAGTCGGGATGGAGTTCTAGTCCAGGTGACCAATCAGCTTC-3′. This Cas9 resistant SAMHD1 gene was cloned into LVX-IRES-mCherry vector (Clontech). Lentiviral vectors containing either LVX-IRES-mCherry control (Lvx, expressing only mCherry) or LVX-SAMHD1-IRES-mCherry (SAM, expressing SAMHD1 and mCherry) were produced by co-transfection of HEK293FT cells using the PEI method (Boussif et al., 1995). Viral supernatant was used to transduce the gRNA 2 KO cells. The mCherry expressing THP-1 cells were single-cell sorted by FACS into 96-well plates and clonally expanded.
Cell culture
THP-1-derived control, SAMHD1 KO, Lvx and SAM cell clones were maintained in RPMI-1640 (ATCC), supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 1 μg/ml puromycin at 37°C, 5% CO2. To induce differentiation into non-dividing, macrophage-like cells, cells were treated with 100 ng/ml PMA for 24 or 72 hours. HEK293FT cells were grown in DMEM containing 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin. GHOST/X4/R5 cells used for virus titration purposes were cultured as described (Wang et al., 2016).
Live cell counting
Control or SAMHD1 KO THP-1 cells (2.5×104 cells per well) were seeded in four replicates in 96-well plate in 100 μl of culture media. At the indicated time points, trypan blue was added to each well and live cell numbers were counted.
MTS assay for cell proliferation
Cell proliferation was measured according to the colorimetric CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega). Control or SAMHD1 KO THP-1 cells (2.5×104 cells per well) were seeded in four replicates in 96-well plate in 100 μl of culture media. Wells containing only media were used for background measurement. At the indicated time points, plates were incubated for 1 hour at 37ºC with 20 μl of MTS reagent and absorbance at 490 and 690 nm (specific and non-specific readings, respectively) was read with a plate reader. Cell proliferation was calculated as the average of 490 nm absorbance values, corrected for non-specific and background readings, of four replicates per sample.
Cell cycle analysis by propidium iodide staining
Control and SAMHD1 KO THP-1 cells were seeded, in triplicate, in 12-well plates at a density of 2×105 cells per well. At day 0, 1, 3, 5 and 7 after seeding, cells were counted, harvested, fixed in 70% ice-cold ethanol, and stained with Guava Cell Cycle Reagent (EMD Millipore), according to the manufacturer’s instructions. Cell cycle data was then acquired on the Guava flow cytometer instrument and analyzed using Cytosoft 4.2.
Detection of apoptosis by Annexin-V staining
Control and SAMHD1 KO THP-1 cells were seeded as described for the cell cycle analysis. At day 0, 1, 3, 5 and 7 after seeding, cells were counted, washed in ice-cold PBS and resuspended in 1X Binding Buffer at a concentration of 1×106 cells/ml. Cells were then stained with PE-Annexin V and 7-Amino-Actinomycin (7-AAD) using the PE Annexin V Apoptosis Detection Kit I (BD Pharmingen), according to the manufacturer’s instructions. Apoptosis data was acquired on the Guava flow cytometer, data analysis and gating was performed using FlowJo software.
Caspase 3/7 activation assay
Control and SAMHD1 KO THP-1 cells (5×105 per well) were seeded in 6-well plates. At day 0, 1, 3, 5 and 7 after seeding, cells were counted and 1×104 cells per well were transferred into a 96-well plate. Caspase 3/7 activity was determined using the luminescent Caspase-Glo 3/7 assay kit (Promega), according to manufacturer’s instructions. Briefly, plates were incubated for 1 hour at room temperature with 100 μl/well of Caspase-Glo 3/7 reagent, and luciferase activity was measured with a plate reader (Perkin Elmer). Wells containing only cell culture media were used to measure background luminescence, which was subtracted from experimental values.
Immunoblotting analysis
Cells were harvested, washed with PBS and lysed in cell lysis buffer (Cell Signaling) containing protease inhibitor (Sigma-Aldrich) as described (Wang et al., 2016). Cell extracts were resolved by SDS-PAGE and subjected to immunoblotting analysis using the following antibodies: rabbit polyclonal anti-SAMHD1 antibody (ProSci, #1224), rabbit polyclonal anti-caspase 3 antibody (Cell Signaling, #9662S), rabbit polyclonal anti-PARP antibody (Cell Signaling, #9542). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) detection (AbD serotec) was used as loading control. Immunoblotting images were captured and analyzed by the Luminescent Image analyzer (LAS 4000) as previously described (Wang et al., 2016).
HIV-1 production and infection assays
Single cycle, luciferase reporter HIV-1-Luc/VSV-G was produced and titrated as previously described (Wang et al., 2016). Control or SAMHD1 KO THP-1 cell lines were infected with HIV-1-Luc/VSV-G at a multiplicity of infection (MOI) of 1 infectious unit per cell and infection efficiency was determined by measuring luciferase activity at 24 and 48 hpi using a luciferase reporter kit (Promega), according to manufacturer’s instructions. Luciferase values were normalized to total protein concentration determined by bicinchoninic acid assay (BCA, Pierce). VSV-G pseudotyped D3HIV-GFP virus was produced as previously described with slight modification (Diamond et al., 2004). Briefly, HEK293FT cells were co-transfected with VSV-G protein and D3HIV-GFP, in which env and nef genes are deleted and replaced with GFP. Viral supernatant was collected at 48 hpi and concentrated (~200-fold) by ultracentrifugation at 22,000 rpm for 2 hours at 4°C. All PMA-differentiated THP-1 cells were infected with equal amounts of D3 vector by spinfection at 400 g for 30 min. Cells were analyzed for GFP expression 72 hpi by flow cytometry (Miltenyi MACSQuant).
Intracellular dNTP measurement
Cellular dNTP levels were determined by a single nucleotide RT incorporation assay as previously described (Diamond et al., 2004). Four distinct 19-mer DNA templates, each with a distinct nucleotide (N) at the 5′ end (5′-NTGGCGCCCGAACAGGGAC-3′), were separately annealed to an 18-mer primer (5′-GTCCCTGTTCGGGCGCCA-3), 32P-labelled at its 5′ end. Reactions contained 200 fmol template/primer, 4 μL of purified RT (HIV-1 HXB2), 25 mM Tris–HCl, pH 8.0, 2 mM dithiothreitol, 100 mM KCl, 5 mM MgCl2, and 10 μM oligo(dT), and cellular dNTP extracts (diluted to be within linear range of the assay, 2–50%) in a final volume of 20 μL/reaction. Reactions were incubated at 37°C for 5 min and then quenched with 10 μL of 40 mM EDTA and 99% (vol/vol) formamide at 95°C for 2 min. The reactions were resolved on a 14% urea-PAGE gel (AmericanBio, Inc.) and analyzed using Pharos FX molecular imager (Biorad). The images were analyzed using ImageLab software.
Vpx and dNs treatment
Virus-like particles containing Vpx were generated as previously described (Hollenbaugh et al., 2014a). PMA differentiated THP-1 were transduced with Vpx containing VLPs 18 hours prior to infection with HIV-1 GFP. At 4 hours prior to infection, cells were also treated with 2.5 mM dNs. Cells were subsequently washed with PBS twice and infected as described above.
Statistical analysis
Data were analyzed using the Student T test or Two-way ANOVA followed with Bonferroni test in Graphpad 5.0. Statistical significance was defined at P <0.05.
Supplementary Material
Supplementary Fig. 1. Effects of SAMHD1 silencing on dNTP levels, cell proliferation and cell cycle progression of THP-1 cells. (A) Immunoblotting analysis confirmed efficient SAMHD1 knockout in non-differentiated THP-1 SAMHD1 control and KO cell clones. GAPDH was used as loading control. (B) Intracellular dNTP levels in non-differentiated THP-1 control and SAMHD1 KO clones by single nucleotide incorporation assay. (C) MTS and (D) cell cycle analyses were performed as described in Fig. 2B–C in the indicated SAMHD1 control and KO clones. Control clones 1 and 2 were produced by transducing THP-1 parental cells with control CRISPR lentiviral vector. KO clone 1, derived from transduction with gRNA 2; KO clones 2 and 3 were derived from transduction with gRNA 1.
Supplementary Fig. 2. SAMHD1 induces spontaneous apoptosis in THP-1 cells. (A) Apoptosis detection by Annexin V/7-AAD double staining as described in Fig. 3A was performed in additional THP-1 SAMHD1 control and KO cell clones. (B) Caspase 3 activation and PARP cleavage detection by immunoblotting. GAPDH was used as loading control. (C) Activation of caspase 3/7 in SAMHD1 control and KO clones over a period of 7 days.
Acknowledgments
This work was supported by NIH grants AI104483 (L.W.), AI049781 (B.K.), GM104198 (B.K.), and MH100999 (R.F.S.). This project was supported in part by the Emory+Children’s Pediatric Research Center Flow Cytometry Core and the Emory University Integrated Cellular Imaging Microscopy Core of the Emory+Children’s Pediatric Research Center. L.W. is also supported in part by the Public Health Preparedness for Infectious Diseases Program of The Ohio State University and by NIH grants (CA181997 and AI120209).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Baek Kim, Email: baek.kim@emory.edu.
Li Wu, Email: wu.840@osu.edu.
References
- Auwerx J. The human leukemia cell line, THP-1: a multifacetted model for the study of monocyte-macrophage differentiation. Experientia. 1991;47:22–31. doi: 10.1007/BF02041244. [DOI] [PubMed] [Google Scholar]
- Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nature medicine. 2012;18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballana E, Este JA. SAMHD1: at the crossroads of cell proliferation, immune responses, and virus restriction. Trends in microbiology. 2015;23:680–692. doi: 10.1016/j.tim.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Lukas J, Strauss M, Bartek J. Cyclin D3: requirement for G1/S transition and high abundance in quiescent tissues suggest a dual role in proliferation and differentiation. Oncogene. 1998;17:1027–1037. doi: 10.1038/sj.onc.1202016. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Flick R, Tchigvintsev A, Brown G, Popovic A, Nocek B, Yakunin AF. Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. The Journal of biological chemistry. 2013;288:8101–8110. doi: 10.1074/jbc.M112.431148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Turpin J, Cordeil S, Tartour K, Nguyen XN, Mahieux R, Cimarelli A. Functional analysis of the relationship between Vpx and the restriction factor SAMHD1. The Journal of biological chemistry. 2012;287:41210–41217. doi: 10.1074/jbc.M112.403816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges C, Naujokat C, Tinapp S, Wieczorek H, Höh A, Sadeghi M, Opelz G, Daniel V. A cell line model for the differentiation of human dendritic cells. Biochem Biophys Res Commun. 2005;333:896–907. doi: 10.1016/j.bbrc.2005.05.171. [DOI] [PubMed] [Google Scholar]
- Boulares AH, Yakovlev AG, Ivanova V, Stoica BA, Wang G, Iyer S, Smulson M. Role of poly(ADP-ribose) polymerase (PARP) cleavage in apoptosis. Caspase 3-resistant PARP mutant increases rates of apoptosis in transfected cells. The Journal of biological chemistry. 1999;274:22932–22940. doi: 10.1074/jbc.274.33.22932. [DOI] [PubMed] [Google Scholar]
- Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanput W, Mes JJ, Wichers HJ. THP-1 cell line: an in vitro cell model for immune modulation approach. Int Immunopharmacol. 2014;23:37–45. doi: 10.1016/j.intimp.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Choi J, Ryoo J, Oh C, Hwang S, Ahn K. SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology. 2015;12:46. doi: 10.1186/s12977-015-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford R, Louis T, Robbe P, Ackroyd S, Burns A, Timbs AT, Wright Colopy G, Dreau H, Sigaux F, Judde JG, Rotger M, Telenti A, Lin YL, Pasero P, Maelfait J, Titsias M, Cohen DR, Henderson SJ, Ross MT, Bentley D, Hillmen P, Pettitt A, Rehwinkel J, Knight SJ, Taylor JC, Crow YJ, Benkirane M, Schuh A. SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood. 2014;123:1021–1031. doi: 10.1182/blood-2013-04-490847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by Cyclin A2/CDK1 Regulates Its Restriction Activity toward HIV-1. Cell reports. 2013;3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. The Journal of biological chemistry. 2004;279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzolin E, Pontarin G, Rampazzo C, Miazzi C, Ferraro P, Palumbo E, Reichard P, Bianchi V. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstone DC, Ennis-Adeniran V, Hedden JJ, Groom HC, Rice GI, Christodoulou E, Walker PA, Kelly G, Haire LF, Yap MW, de Carvalho LP, Stoye JP, Crow YJ, Taylor IA, Webb M. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480:379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Guyader M, Emerman M, Montagnier L, Peden K. VPX mutants of HIV-2 are infectious in established cell lines but display a severe defect in peripheral blood lymphocytes. The EMBO journal. 1989;8:1169–1175. doi: 10.1002/j.1460-2075.1989.tb03488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzinger T, Reed SI. Cyclin D3 is rate-limiting for the G1/S phase transition in fibroblasts. The Journal of biological chemistry. 1998;273:14958–14961. doi: 10.1074/jbc.273.24.14958. [DOI] [PubMed] [Google Scholar]
- Hollenbaugh JA, Tao S, Lenzi GM, Ryu S, Kim DH, Diaz-Griffero F, Schinazi RF, Kim B. dNTP pool modulation dynamics by SAMHD1 protein in monocyte-derived macrophages. Retrovirology. 2014a;11:63. doi: 10.1186/s12977-014-0063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. The Journal of biological chemistry. 2012;287:21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohnken R, Kodigepalli KM, Wu L. Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Molecular cancer. 2015;14:176. doi: 10.1186/s12943-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer S, Wolf C, Konig N, Staroske W, Guck J, Hausler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, Dahl A, Rapp A, Cardoso MC, Shevchenko A, Lee-Kirsch MA. SAMHD1 prevents autoimmunity by maintaining genome stability. Annals of the rheumatic diseases. 2015;74:e17. doi: 10.1136/annrheumdis-2013-204845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature immunology. 2012;13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Munoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menendez-Arias L, Clotet B, Keppler OT, Marti R, Posas F, Ballana E, Este JA. Cell Cycle Control and HIV-1 Susceptibility Are Linked by CDK6-Dependent CDK2 Phosphorylation of SAMHD1 in Myeloid and Lymphoid Cells. Journal of immunology. 2014;193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GI, Bond J, Asipu A, Brunette RL, Manfield IW, Carr IM, Fuller JC, Jackson RM, Lamb T, Briggs TA, Ali M, Gornall H, Couthard LR, Aeby A, Attard-Montalto SP, Bertini E, Bodemer C, Brockmann K, Brueton LA, Corry PC, Desguerre I, Fazzi E, Cazorla AG, Gener B, Hamel BC, Heiberg A, Hunter M, van der Knaap MS, Kumar R, Lagae L, Landrieu PG, Lourenco CM, Marom D, McDermott MF, van der Merwe W, Orcesi S, Prendiville JS, Rasmussen M, Shalev SA, Soler DM, Shinawi M, Spiegel R, Tan TY, Vanderver A, Wakeling EL, Wassmer E, Whittaker E, Lebon P, Stetson DB, Bonthron DT, Crow YJ. Mutations involved in Aicardi-Goutieres syndrome implicate SAMHD1 as regulator of the innate immune response. Nature genetics. 2009;41:829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo J, Choi J, Oh C, Kim S, Seo M, Kim SY, Seo D, Kim J, White TE, Brandariz-Nunez A, Diaz-Griffero F, Yun CH, Hollenbaugh JA, Kim B, Baek D, Ahn K. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nature medicine. 2014;20:936–941. doi: 10.1038/nm.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology. 2012;9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Hach JC, White TE, Diaz-Griffero F, Yount JS, Wu L. Identification of Cellular Proteins Interacting with the Retroviral Restriction Factor SAMHD1. Journal of virology. 2014;88:5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoven B, Schlegel RA, Williamson P. Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T lymphocytes. The Journal of experimental medicine. 1995;182:1597–1601. doi: 10.1084/jem.182.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, St Gelais C, de Silva S, Zhang H, Geng Y, Shepard C, Kim B, Yount JS, Wu L. Phosphorylation of mouse SAMHD1 regulates its restriction of human immunodeficiency virus type 1 infection, but not murine leukemia virus infection. Virology. 2016;487:273–284. doi: 10.1016/j.virol.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Lu FZ, Shen XY, Wu Y, Zhao LT. SAMHD1 is down regulated in lung cancer by methylation and inhibits tumor cell proliferation. Biochemical and biophysical research communications. 2014;455:229–233. doi: 10.1016/j.bbrc.2014.10.153. [DOI] [PubMed] [Google Scholar]
- Welbourn S, Strebel K. Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology. 2016;488:271–277. doi: 10.1016/j.virol.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell host & microbe. 2013;13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. SAMHD1: a new contributor to HIV-1 restriction in resting CD4+ T-cells. Retrovirology. 2012;9:88. doi: 10.1186/1742-4690-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. Cellular and Biochemical Mechanisms of the Retroviral Restriction Factor SAMHD1. ISRN Biochem. 2013;2013:11. doi: 10.1155/2013/728392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Hao C, DeLucia M, Swanson S, Florens L, Washburn MP, Ahn J, Skowronski J. CyclinA2-Cyclin-dependent Kinase Regulates SAMHD1 Protein Phosphohydrolase Domain. The Journal of biological chemistry. 2015;290:13279–13292. doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XF, Yu QC, Essex M, Lee TH. The vpx gene of simian immunodeficiency virus facilitates efficient viral replication in fresh lymphocytes and macrophage. Journal of virology. 1991;65:5088–5091. doi: 10.1128/jvi.65.9.5088-5091.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Effects of SAMHD1 silencing on dNTP levels, cell proliferation and cell cycle progression of THP-1 cells. (A) Immunoblotting analysis confirmed efficient SAMHD1 knockout in non-differentiated THP-1 SAMHD1 control and KO cell clones. GAPDH was used as loading control. (B) Intracellular dNTP levels in non-differentiated THP-1 control and SAMHD1 KO clones by single nucleotide incorporation assay. (C) MTS and (D) cell cycle analyses were performed as described in Fig. 2B–C in the indicated SAMHD1 control and KO clones. Control clones 1 and 2 were produced by transducing THP-1 parental cells with control CRISPR lentiviral vector. KO clone 1, derived from transduction with gRNA 2; KO clones 2 and 3 were derived from transduction with gRNA 1.
Supplementary Fig. 2. SAMHD1 induces spontaneous apoptosis in THP-1 cells. (A) Apoptosis detection by Annexin V/7-AAD double staining as described in Fig. 3A was performed in additional THP-1 SAMHD1 control and KO cell clones. (B) Caspase 3 activation and PARP cleavage detection by immunoblotting. GAPDH was used as loading control. (C) Activation of caspase 3/7 in SAMHD1 control and KO clones over a period of 7 days.