Abstract
The human high-affinity copper transporter 1 (hCtr1) transports both Cu(I) and cisplatin (cDDP). Because Cu deficiency is lethal yet Cu overload is poisonous, hCtr1 expression is transcriptionally upregulated in response to Cu deficiency but is downregulated under Cu replete conditions in controlling Cu homeostasis. The up- and down-regulation of hCtr1 is regulated by Specific protein 1 (Sp1), which itself is also correspondingly regulated under these Cu conditions. hCtr1 expression is also upregulated by cDDP via upregulation of Sp1. The underlying mechanisms of these regulations are unknown. Using gel-electrophoretic mobility shift assays, we demonstrated here that Sp1-DNA binding affinity is reduced under Cu replete conditions but increased under reduced Cu conditions. Similarly, Sp1-DNA binding affinity is increased by cDDP treatment. This in vitro system demonstrated, for the first time, that regulation of Sp1/hCtr1 expression by these agents is modulated by the stability of Sp1-DNA binding, the first step in the Sp1-mediated transcriptional regulation process.
Keywords: Cu(II), cisplatin, Specific protein (Sp1), high-affinity copper transporter (hCtr1), copper homeostasis
Graphical abstract
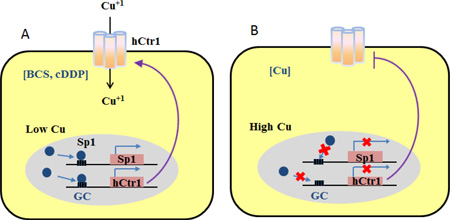
Proposed models depicting Specific protein 1 (Sp1)-mediated transcriptional regulation of Sp1 itself and the human high-affinity copper transporter 1 (hCtr1) expression by bathocuproine sulfonate (BCS, a copper chelator) and cisplatin (cDDP) [A] and by copper [B].
Specificity protein 1 (Sp1) is a ubiquitously expressed transcription regulator that plays important roles in many cellular processes [1]. It contains a transactivation domain at the N-terminus and a DNA-binding domain at the C-terminus which consists of three zinc fingers (ZF). Sp1-ZF binds to GC-rich boxes with the consensus sequence 5’-G/T-GGGCGG-G/A [2]. The Sp1-ZF domain contains three contiguous classic Cys2-His2 motifs with the consensus sequence Cys-X2–4-Cys-X11–12-His-X3-His. Each Cys2-His2 ZF is organized by two β-sheets and an α-helix on which two Cys and two His residues are respectively located and are coordinately bound by one Zn+2 in the tetrahedral geometry. This geometry orients the fingers for site-specific recognition of GC boxes on targeted DNA [3].
Several metal ions can affect Sp1’s transcriptional activities through interfering Sp1 ZF-DNA interactions. Sp1 regulates the expression of transporters for these metal ions, making Sp1 an important cellular regulator of metal ion homeostasis. We previously demonstrated that cadmium Cd(II) suppressed Sp1’s transcription activity that regulates Cd transporter, Zip8, expression [4].
Sp1 plays a critical role in regulating copper (Cu) homeostasis. Cu is an essential micronutrient but is toxic when in excess, therefore, cellular bioavailable Cu has to be properly regulated We previously demonstrated that under Cu deficiency, Sp1 expression is transcriptionally self-upregulated by binding to its own promoter where 10 copies of GC-boxes are located [5]. Elevated Sp1 then transcriptionally upregulates expression of the high-affinity human copper transporter 1 (hCtr1) to transport Cu from the extracellular milieu. When cellular Cu levels reach the upper limit, Sp1 expression is silenced, thereby downregulating hCtr1 to prevent Cu overload [6, 7].
Importantly, hCtr1 can also transport cisplatin (cis-diamminedichloroplatinum (II), cDDP), a well-recognized antitumor agent in cancer chemotherapy [8]. cDDP functions as a competitor for hCtr1-mediated Cu(I) transport, causing reduced cellular Cu levels, resulting in increased Sp1 and hCtr1 expression [9]. These results demonstrated that Sp1 can transcriptionally up- and down-regulate Sp1 and hCtr1 in response to Cu deficiency and affluence, respectively. However, the underlying mechanisms are unknown.
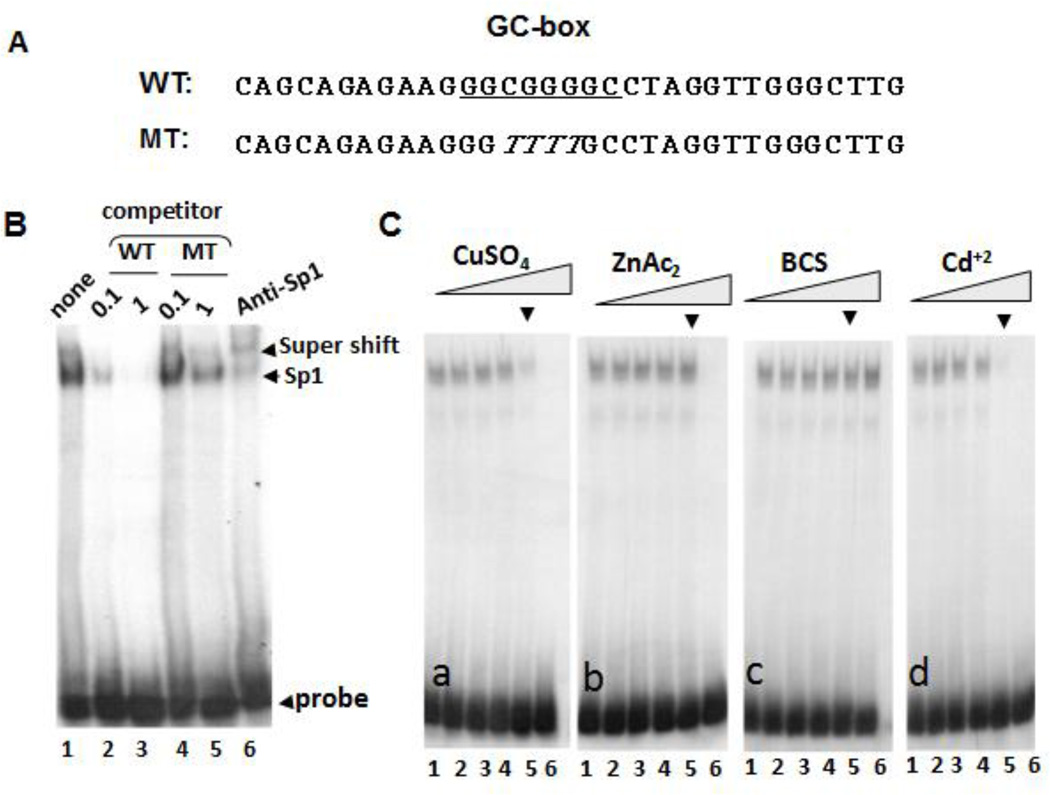
An important rate-limiting step in Sp1-mediated transcriptional regulation involves Sp1 binding to GC box sequences at the promoters of target genes. In this communication, we employed a gel electrophoretic mobility shift (GEMS) approach to investigate the effects of Cu(II) concentrations (replete and deplete conditions) and cDDP on the stability of Sp1-DNA binding. GEMS assay used 32P-labeled double-stranded DNA probe containing a Sp1-binding sequence. We chose the GC-box sequence (Fig. 1A) located at site 10 of the Sp1 promoter which was responsive to Cu concentration-dependent Sp1 expression [5]. The probe was mixed with a nuclear extract prepared from non-small cell lung cancer (NSCLC) cells and nonspecific competitor poly(deoxyinosine-deoxycytidine) (poly(dI-dC)). The mixture was separated by gel electrophoresis as previously described [4]. We identified Sp1-DNA complex formation using the non-radioactively labeled wild-type GC-box oligonucleotides vs. mutant oligonucleotides as competitors (Fig. 1B, lanes 2 – 5). The Sp1-DNA complex was then confirmed using anti-Sp1 antibody in a super-shift assay (Fig. 1B, lane 6). We then included various concentrations of Cu+2, Zn+2, bathocuproine sulfonate (BCS, a Cu chelator), or Cd+2 in the reaction mixture to determine their effects on the stability of the Sp1-DNA complex. We found that at the concentration at which Cu+2 ions caused destabilization of the Sp1-DNA complex (Fig 1C [a], arrow head), no destabilization of the complex was found with Zn+2 ions at the same concentration (Fig, 1C [b]), although higher concentration of Zn+2 caused destabilization (Fig. 1C [b] lane 6). These results suggest that Cu+2 was more effective in reducing Sp1-DNA stability than Zn+2.
Fig. 1.
Effects of CuSO4, ZnCl2, BCS, and CdCl2 on Sp1-probe binding. [A] Sequences of wild-type (WT) used as the probe and a mutant (MT) used as competitor, The box contains GC-rich Sp1-binding site. [B] Identification of Sp1-probe complex by GEMS. [C] Competition of Sp1-probe binding by increased concentrations of different metal salts and BCS as indicated (lanes 1 to 6 correspondingly represent 0, 1, 10, 50, 100, and 200 µM, in each panel, using 100 µM as reference a point, arrow).
Interestingly, including BCS in the reaction mixture increased Sp1-DNA complex stabilization (Fig. 1C [c]). Because it has been demonstrated that BCS treatment does not change the available cellular Zn2+ pool [10], these results demonstrated that Cu chelation enhances Sp1-DNA stability. On the other hand, Cd(II), like Cu(II), induced destabilization of Sp1-DNA complex (Fig. 1C [d]).
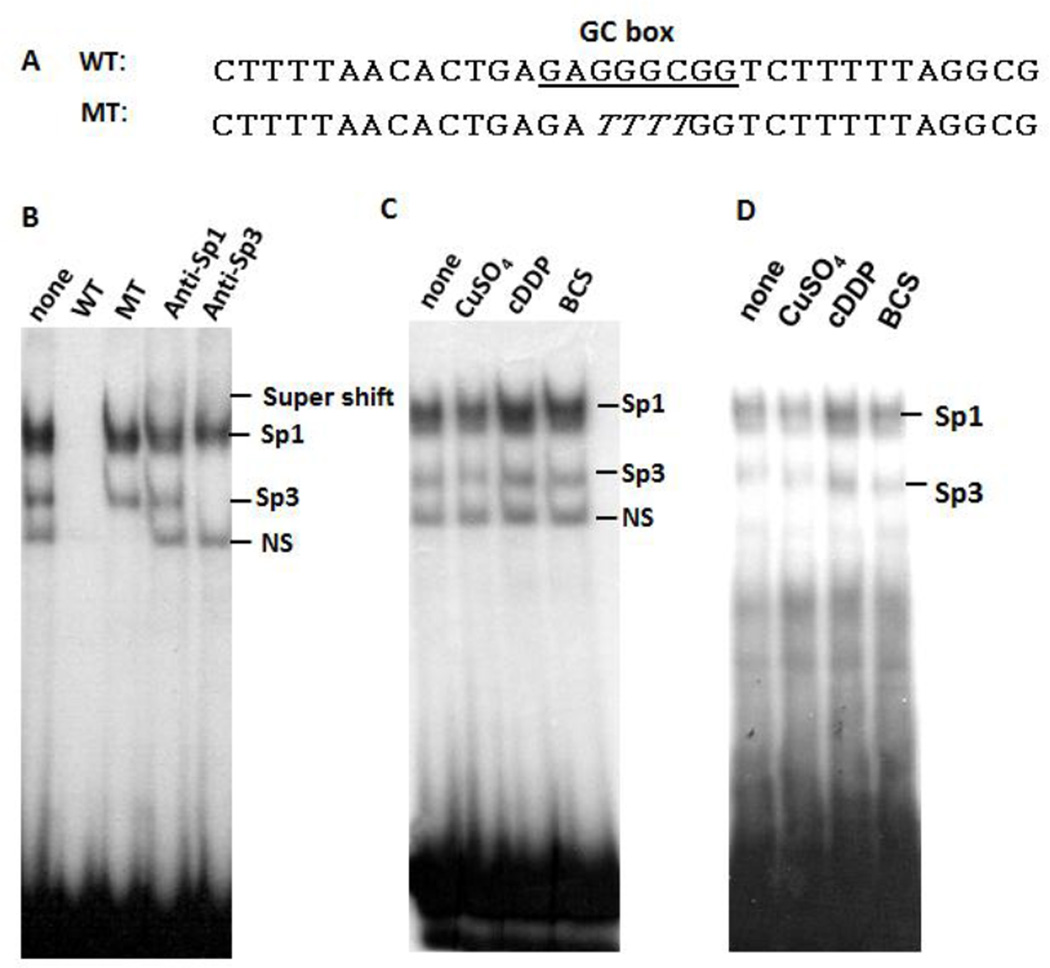
To investigate the generality of these observations, we performed similar experiments using nuclear extracts prepared from HEK293 embryonic kidney cells and a 32P-labeled oligonucleotide probe from the sequence located at Site 8 at the Sp1 promoter [5] (Fig. 2A). In this experiment, we identified both Sp1-DNA and Sp3-DNA complexes by GMES using anti-Sp1 and anti-Sp3 antibodies in super-shift assay (Fig. 2B). We found that including CuSO4 in the reaction mixture destabilized both Sp1-DNA and Sp3-DNA complexes, whereas cDDP or BCS enhanced stabilization of both complexes (Figs. 2C & 2D). Given the fact that we previously found that cDDP and BCS can also coordinately upregulate of Sp1 and hCtr1 in an animal tumor model [6], we believe that these results are likely to be applicable to in vivo situations.
Fig. 2.
Effects of CuSO4, cDDP and BCS on Sp1-probe binding. [A] Nucleotide sequence of WT used as probe and a MT sequence used as competitor. Sp1-binding GC-rich sequence is indicated by box. [B] Identification of Sp1-probe and Sp3-probe complexes as indicated. [C] Effects of CuSO4, cDDP, and BCS (100 µM each) on Sp1-probe binding stability using nuclear extract prepared from HEK293 cells. [D] Similar experiment as described in [C] except nuclear extract was prepared from SCLC cells.
Our in vitro GEMS assays have the following important implications for the mechanistic regulation of Cu homeostasis and platinum drug transport: (i) We found that BCS and cDDP enhanced Sp1-DNA binding stability, consistent with their ability in inducing Sp1/hCtr1 transcriptional activities (Fig. 3A). In contrast, Cu(II) destabilizes Sp1-DNA interaction, also consistent with its suppressive effect on Sp1 regulated Sp1/hCtr1 expression (Fig. 3B). Thus, mechanisms by which these agents can transcriptionally upregulate and downregulate Sp1/hCtr1 expression can be explained by their effects on the stability of Sp1 binding to their respective promoters. The negative effect of Cu on Sp1-DNA stability may be explained by the displacement of Zn(II) in the geometry in the Sp1-ZF domain by Cu(II). Although Sp1-DNA complex formation primarily relies on DNA base and Sp1-ZF contact, interactions between His ligands and DNA phosphate backbone are also important [3]. Cu(II) may also neutralize negative changes on the DNA phosphate backbone, thus destabilizing its interactions with the His ligands of ZF.
Fig. 3.
Diagram depicting the relationships between Cu chelators and cDDP [A], and Cu [B], in the regulation of Sp1 and hCtr1 expression.
The observation that Cu chelation induces Sp1-DNA stabilization thereby increased hCtr1 expression is intriguing. This approach has been taken to clinical trial using copper chelator in combination of Pt-based drug to improve the efficacy of cancer chemotherapy by enhanced Pt drug transport [7, 11]. While the precise mechanism for enhanced Sp1-DNA stability by Cu chelation remains to be critically elucidated, we speculate that this may be due to enhanced interaction between DNA phosphate groups and His ligands under reduced Cu(II) conditions. Further studies are needed to clarify this possibility.
Another important finding in this communication is that it appears the “free Cu(II) ions” are sufficient for regulating Sp1-DNA interaction. It has been suggested that virtually all cellular Cu is tightly bound, and intracellular Cu trafficking is carried out by Cu chaperones to different cellular compartments, i.e, CCS to superoxide dismutase in the cytoplasm [12], Cox17 to mitochondrial cytochrome C oxidase, and Atox1 to two trans-Golgi P1B-type ATPases (ATP7A and ATP7B) [13]. It has been reported that ZF domain is involved in nuclear targeting of Sp1 [14]. However, our results from a cell-free system demonstrating that free Cu can affect Sp1-DNA binding suggest that regulation of Sp1-mediated transcriptional regulation by Cu may not need a carrier protein. Lastly, while hCtr1 transports Cu(I) but not Cu(II), our current results demonstrate that free Cu(II) can affect Sp1-DNA stability. Cu(I) can be converted to Cu(II) by Fenton reaction under oxidative stress conditions, whereas Cu(II) can be converted back to Cu(I) by metal reductase [7], suggesting that cellular Cu(I) and Cu(II) are interconvertable.
In conclusion, this is the first demonstration describing the effects of Cu, Cu chelator, and cDDP on Sp1-DNA stability. The results mirror the effects of these agents on Sp1-regulation of Sp1 and hCtr1 expression. Our findings provide insights into the mechanisms of Cu homeostasis regulation and hCtr1-regulated platinum drug transport.
Hightlights.
The human high-affinity copper transporter (hCtr1) transports both Cu and cisplatin
hCtr1 is regulated by Cu availability and cisplatin via Specific protein 1 (Sp1) factor
Cu(II) de-stabilizes Sp1-DNA binding
Cu chelator and cisplatin increase Sp1-DNA binding
Effects of Sp1-DNA binding by Cu(II)/cDDP mirror their effects on hCtr1/Sp1 expression
Acknowledgments
This study was supported by the NCI grant (R01-CA149260 to MTK), and the National Science Council, Taiwan (NSC102-2628-006-014-MY3 to HHWC).
Table of Abbreviations
- ATP7A and ATP7B
two trans-Golgi P1B-type ATPases
- BCS
bathocuproine sulfonate
- cDDP
cisplatin
- GEMS
gel-electrohoretic mobility shift
- hCtr1
high-affinity copper transporter
- NSCLC
non-small cell lung cancer
- Poly (dI-dC)
poly(deoxyinosine-deoxycytidine)
- Sp1
Specific protein 1
- ZF
Zinc finger
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC, Hung JJ. Oncogene. 2012;31:3973–3988. doi: 10.1038/onc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wierstra I. Biochemical and biophysical research communications. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 3.Pavletich NP, Pabo CO. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 4.Aiba I, Hossain A, Kuo MT. Mol Pharmacol. 2008;74:823–833. doi: 10.1124/mol.108.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang ZD, Tsai WB, Lee MY, Savaraj N, Kuo MT. Molecular pharmacology. 2012;81:455–464. doi: 10.1124/mol.111.076422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang ZD, Long Y, Tsai WB, Fu S, Kurzrock R, Gagea-Iurascu M, Zhang F, Chen HH, Hennessy BT, Mills GB, Savaraj N, Kuo MT. Molecular cancer therapeutics. 2012;11:2483–2494. doi: 10.1158/1535-7163.MCT-12-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen HH, Chen WC, Liang ZD, Tsai WB, Long Y, Aiba I, Fu S, Broaddus R, Liu J, Feun LG, Savaraj N, Kuo MT. Expert opinion on therapeutic targets. 2015;19:1307–1317. doi: 10.1517/14728222.2015.1043269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo MT, Fu S, Savaraj N, Chen HH. Cancer Res. 2012;72:4616–4621. doi: 10.1158/0008-5472.CAN-12-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang ZD, Long Y, Chen HH, Savaraj N, Kuo MT. Journal of biological inorganic chemistry : JBIC : a publication of the Society of Biological Inorganic Chemistry. 2014;19:17–27. doi: 10.1007/s00775-013-1051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su W, Jackson S, Tjian R, Echols H. Genes & development. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 11.Fu S, Hou MM, Wheler J, Hong D, Naing A, Tsimberidou A, Janku F, Zinner R, Piha-Paul S, Falchook G, Kuo MT, Kurzrock R. Investigational new drugs. 2014;32:465–472. doi: 10.1007/s10637-013-0051-8. [DOI] [PubMed] [Google Scholar]
- 12.Pope CR, De Feo CJ, Unger VM. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20491–20496. doi: 10.1073/pnas.1309820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores AG, Unger VM. The Journal of membrane biology. 2013;246:903–913. doi: 10.1007/s00232-013-9592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito T, Kitamura H, Uwatoko C, Azumano M, Itoh K, Kuwahara J. Biochemical and biophysical research communications. 2010;403:161–166. doi: 10.1016/j.bbrc.2010.10.036. [DOI] [PubMed] [Google Scholar]