Abstract
Map4k4, originally identified in siRNA screens and characterized by tissue specific gene deletions, is emerging as a regulator of glucose homeostasis and cardiovascular health. Recent studies have shown that Map4k4 gene ablation or inhibition of its kinase activity attenuates hyperglycemia and plaque formation in mouse models of insulin resistance and atherosclerosis, and suggest roles for Map4k4 in multiple signaling systems, including NFκB activation, small GTPase regulation, the Hippo cascade, and regulation of cell dynamics by FERM domain proteins. This new and promising area of inquiry raises key questions that need to be addressed in the future, such as defining which of the above or other effectors mediate Map4k4 control of metabolic and vascular functions, and identifying upstream activators of Map4k4.
Keywords: diabetes, insulin resistance, atherosclerosis, Sterile 20 kinases
Map4k4 as a novel target in metabolic and cardiovascular disease
An exciting aspect of investigations into the underlying mechanisms of major diseases is uncovering unexpected proteins that may serve as targets for new therapeutics. The Sterile 20-related kinase mitogen activated kinase kinase kinase kinase-4 (Map4k4) has emerged as such a potential therapeutic target in recent studies on animal models of obesity, insulin resistance, type 2 diabetes and atherosclerosis. Concurrently, Map4k4 was surprisingly shown to participate in pathways distinct from the canonical Map kinase signaling pathway suggested by its name and the function of its yeast ortholog. We review here the role of Map4k4 in these pathways that regulate energy metabolism, inflammation and cell dynamics, and address the connections of these cellular functions to whole-body phenotypes relevant to metabolic and cardiovascular diseases.
Sterile 20 kinases and Map4ks: Emerging roles in cell and animal function
The Saccharomyces cerevisiae sterile 20 gene was originally identified as a mutant allele in the haploid yeast mating pathway, and its product Ste20p was found to be a serine kinase functioning as a mitogen activated kinase kinase kinase kinase (Map4k) upstream of the Fus3p Map kinase [1, 2]. This Map kinase cascade regulates responses to pheromones and other environmental signals in yeast. In mammals, kinases with catalytic domains related to Ste20p have diversified into a group of over 30 members divisible into two major subgroups: the p21-activated kinase (PAK) and germinal center kinase (GCK) families, the latter comprising the mammalian Map4ks including Map4k4 [3].
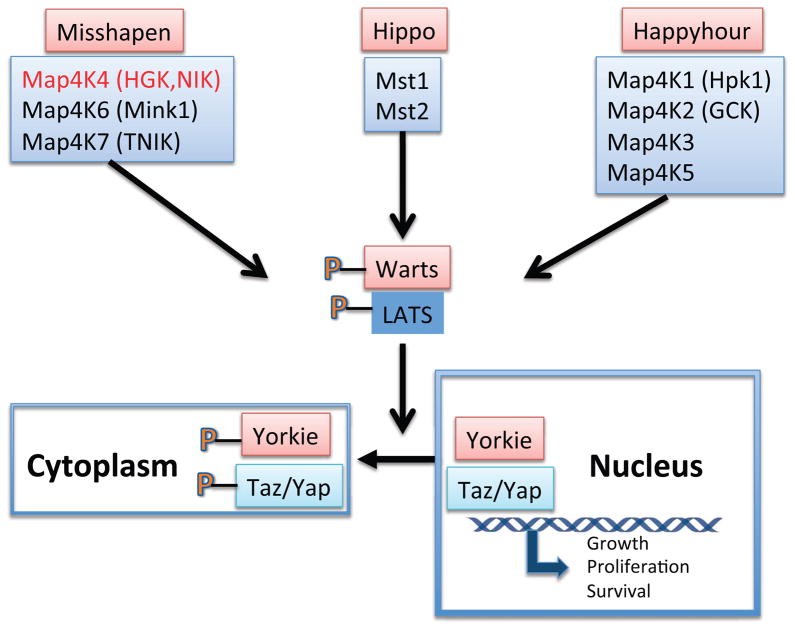
Increasing recent interest in Ste20 kinase function has been spurred in part by genetic studies in Drosophila melanogaster that identified Ste20 kinases as key regulators of development and organ size, most notably members of the Hippo signaling pathway (Figure 1). The Hippo mutation in Drosophila leads to dramatic overgrowth of multiple tissues as a result of greater cell number and cell size. The Hippo gene encodes a Ste20 kinase (most closely related to mammalian Ste20 kinases Mst1/2) that phosphorylates and activates whose mammalian homolog is LATS (Warts). Warts in turn phosphorylates and promotes cytoplasmic retention of the transcription factor Yorkie, thus downregulating Yorkie-dependent genes [4]. Recent work elaborating a parallel Hippo pathway in mammalian cells has demonstrated that a variety of Ste20 kinases can function as upstream activators by phosphorylation of LATS [5–7], among them multiple members of the Map4 family including Map4k4, Map4k1/HPK1, Map4k2/GCK, Map4k3/GLK, Map4k5/KHS Map4k6/MINK1, Map4k7/TNIK. In some cellular contexts, the specific contribution of Map4k4 to LATS phosphorylation appears minor, and is detectable only in the background of multiple deletion of other LATS kinases [6]. However, evidence for a direct interaction of Map4k4 with LATS and a strong effect of Map4k4 deletion on latrunculin-stimulated LATS phosphorylation in mouse embryo fibroblasts [5] supports the view that at least in some situations Map4k4 may be a major contributor to Hippo signaling. The Drosophila Ste20 kinase Tao-1 has also been shown to phosphorylate Warts [8], but a similar function of three mammalian homologs Tao 1, 2 and 3 has not been reported to date.
Figure 1. Sterile 20 kinases in Hippo signaling.
Drosophila kinases (orange boxes) and their mammalian counterparts (blue boxes) phosphorylate and activate the Warts or LATS (mammalian) protein kinases. These in turn phosphorylate the Yorkie or Taz/Yap transcription factor, resulting in exclusion from the nucleus and downregulation of growth-promoting genes (see references 4 and 6).
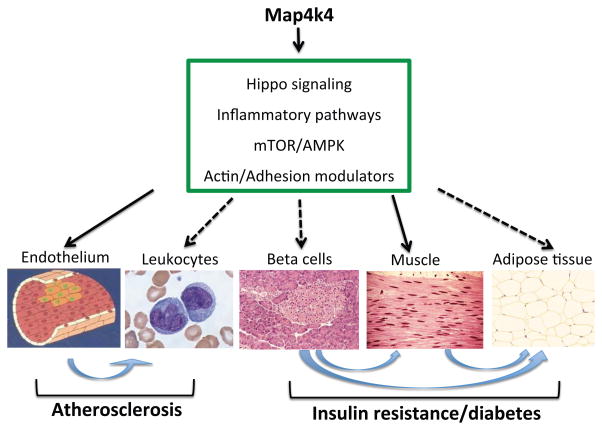
In addition to this recently identified association with the Hippo pathway, Map4ks and other Ste20 kinases have been independently identified in a variety of gene association studies, loss or gain of function screens, and protein-protein interaction studies [9–16]. Most notably, Map4k4 has repeatedly emerged as a gene involved in the promotion of cancer invasion and metastasis, and associated with poor prognosis in advanced and metastatic cancers [17–21]. We discuss this interesting aspect below in the context of cellular pathways modulated by Map4k4, however, in this review we focus on Map4k4 function in metabolic and cardiovascular disease (Figure 2, Key Figure).
Figure 2. Signaling and tissue targets of Map4k4.
Studies in cellular models have implicated the involvement of Map4k4 as a signaling node in a variety of regulatory pathways (within box) relevant to downstream nutrient responses, cell growth and dynamics, and inflammatory activation. Animal models have demonstrated significant effects of Map4k4 in muscle and endothelium in insulin resistance and atherosclerosis, respectively (solid arrows). Contributions of Map4k4 in leukocytes, pancreatic beta cells and adipose tissue are uncertain (dashed arrows), but Map4k4 action may modulate inter-organ communication relevant to metabolic and cardiovascular diseases (blue arrows).
Map4k4 in metabolic disease models
Interest in a potential role of Map4k4 in metabolism emerged from results of an siRNA-mediated loss of function screen of protein kinases expressed in differentiated 3T3-L1 adipocytes [22]. This screen was designed to identify novel components of a signaling pathway connecting insulin receptor activation to increased glucose uptake and other metabolic responses in adipocytes. Among the protein kinases assessed for their contribution to insulin-stimulated glucose uptake and insulin signaling in these studies, several targets were identified that upon silencing resulted in enhanced glucose uptake, suggesting they function as negative regulators of the pathway [22]. Most prominent among these was Map4k4. Map4k4 was originally identified in mammalian cells as Nck-interacting kinase [23] in a yeast two-hybrid screen using the Nck SH3 domain as bait. It was independently identified as HPK/GCK like kinase (Hgk) in studies that reported a function for this kinase in the JNK signaling pathway [16], suggesting a parallel function to the yeast Ste20p. Silencing of Map4k4 expression in differentiated 3T3-L1 adipocytes resulted in increased expression of the key adipogenic transcription factors C/EBPα, C/EBPβ, and PPARγ and of the insulin-regulated glucose transporter GLUT4, resulting in increased adipogenesis, glucose uptake, and triglyceride (TG) depletion [22]. Further studies in adipocytes demonstrated these effects of Map4k4 depletion are mediated in part through mTOR-dependent regulation of translation resulting in increased levels of PPARγ protein [24]. Based on the association of Map4k4 with signaling pathways associated with inflammation, it was hypothesized that Map4k4 could be a mechanistic connection between obesity-associated inflammation and dysfunction of adipose tissue associated with insulin resistance and type 2 diabetes (T2D) [25]. Increased expression of Map4k4 in adipocytes treated with TNFα [26] gave further credence to this idea.
Further studies by several laboratories provided additional weight to an association of Map4k4 with obesity and metabolic disease in both human subjects and mouse models. Preadipocytes and primary mature adipocytes from obese individuals displayed increased expression of Map4k4 compared to cells from non-obese individuals [27]. Another study of a large cohort of human subjects who were characterized for parameters of insulin sensitivity and glucose tolerance revealed a set of common polymorphisms in the Map4k4 locus that associated with T2D and insulin resistance [28]. Further indication of relevance to human metabolic disease came from a study indicating that Map4k4 contributes to inflammatory signals that impair insulin sensitivity in human skeletal muscle. Silencing of Map4k4 in human primary skeletal muscle cells prevents TNFα-induced insulin resistance, while siRNA against Map4k4 introduced into myotubes from insulin-resistant T2D patients restored insulin sensitivity and glucose uptake to normal healthy levels [29]. Recently, a large-scale human study identified increased Map4k4 gene expression in atherosclerosis [30] which was also observed in a smaller number of samples from human atherosclerotic lesions [31].
Based on these indications of a potential role for Map4k4 in promoting metabolic disease, mouse models have been developed for inducible and tissue-specific ablation of Map4k4 expression to explore functions of Map4k4 in vivo (Table 1). Since whole-body Map4k4-knockout (KO) animals die at E9.5 due to failure of mesodermal cells to migrate away from the primitive streak [32], a mouse line with loxP sites flanking exon7 of Map4k4 and a tamoxifen inducible, ubiquitously expressed Cre was generated (iMap4k4-KO mice, [33]). This enabled ablation of Map4k4 expression in adult animals, bypassing any developmental effects of Map4k4 loss at the embryonic stage. Treatment of these animals with tamoxifen caused efficient deletion of Map4k4 in all tissues examined. The floxed Map4k4 allele was also employed to effect tissue-specific deletion when crossed with appropriate tissue-specific promoter driven Cre expressing lines to delineate Map4k4 functions in many relevant tissues.
Table 1.
Metabolic or cardiovascular phenotypes of Map4k4 knockout models.
| Cre/tissue(s) deleted | Metabolic or cardiovascular phenotype | Ref |
|---|---|---|
| Ubiquitin-ERT2/whole body (inducible) | ↑insulin sensitivity ↓insulin (HFD fed animals) |
[33] |
| Adiponectin/white and brown adipose tissue | none | [33] |
| Albumin/liver | none | [33] |
| Myf5/skeletal muscle, brown adipose tissue | ↑glucose tolerance ↑insulin sensitivity (HFD fed animals) |
[33] |
| CD4/T-cells | ↑glucose ↑insulin ↓glucose tolerance ↓insulin sensitivity (chow fed, lean animals) |
[46] |
| Cdh5(PAC)-ERT2/endothelial cells (inducible) | ↓plaque formation (Western diet, ApoE−/− mice) | [31] |
Map4k4 contributes to obesity-associated insulin resistance and hyperglycemia
The systemic metabolic effects of tamoxifen-inducible global Map4k4 depletion were tested in iMap4k4-KO mice or floxed controls, on a normal chow or high fat diet (HFD), an established model of diet-induced obesity (DIO) and insulin resistance. Despite equivalent weight gain on HFD, iMap4k4-KO mice maintain significantly lower fasting blood glucose compared to floxed controls. This is associated with greatly improved insulin response in insulin tolerance tests, enhanced insulin signaling to Akt in adipose tissue and liver, and decreased markers of inflammation in adipose tissue. Interestingly, despite increased insulin sensitivity and lower fasting blood glucose, iMap4k4-KO mice on HFD exhibit delayed glucose clearance in a glucose tolerance test (GTT). This is probably a result of lower basal and glucose-stimulated insulin levels in these animals [33], which may indicate an additional role of Map4k4 in regulating beta cell function and insulin secretion.
Notably, a recent study describing a novel small molecule Map4k4 kinase inhibitor showed dramatically improved glycemic control in the ob/ob (leptin deficient and hyperphagic) mouse model of T2D, following treatment with the compound [34]. This study did not identify the relevant target tissue for Map4k4 inhibition, but confirms the potential of Map4k4 as a therapeutic target for T2D.
The simplest explanation for improved insulin response upon whole body deletion or inhibition of Map4k4 would be direct improvements in insulin sensitivity in adipose tissue and/or liver, possibly via improved resistance to obesity-associated inflammation in adipose tissue or resistance to hepatic steatosis. However, selective deletion of Map4k4 in adipose tissue (via adipocyte expressed adiponectin-Cre) or liver (albumin-Cre) had no effect on insulin sensitivity or glucose homeostasis in either lean or obese, HFD mice. Furthermore, adipose tissue showed similar levels of obesity-associated inflammation, while hepatic steatosis was unchanged in the liver KO model.
Skeletal muscle also contributes significantly to insulin-mediated glucose disposal. Based on previous data showing a significant role of Map4k4 in promoting differentiation and myotube formation in C2C12 muscle cells [35], the effect of Map4k4 deletion in early muscle development by employing a Myf5-Cre to delete Map4k4 in the skeletal muscle lineage was assessed [33]. These mice do in fact display protection from obesity-associated insulin resistance and hyperglycemia. Surprisingly this protection is associated with a dramatic improvement in insulin signaling to Akt in visceral adipose depot, a tissue where Map4k4 is intact. However, no change in insulin responsiveness in either liver or muscle could be detected in these animals. Thus, Map4k4 may mediate the expression of a signal emanating from a Myf5 lineage cell likely muscle that is delivered to adipose depots and enhances adipose tissue insulin sensitivity (Figure 2). The nature of this signal is unknown. One caveat to the interpretation of these results is that the Myf5 lineage also includes precursors of brown adipocytes and a limited subset of white adipocytes in specific depots [36]. However, since the adiponectin-Cre drives gene deletion in mature brown adipose cells as well as all white adipose depots, the effect of Myf5-Cre-driven deletion of Map4k4 on adipocytes is likely mediated through cells other than adipocytes.
Endothelial Map4k4 promotes atherosclerosis
Map4k4 is highly expressed in endothelial cells, which line the circulatory and lymphatic vasculature. Since Map4k4 plays a role in inflammation and lipid pathways, it is reasonable to think that it might function in the development of atherosclerosis, which is triggered by an inflammatory response to lipid-mediated vascular damage. Furthermore, Map4k4 has been reported to promote endothelial permeability [37], which could contribute to atherosclerosis by enabling leucocyte extravasation as well as transport of macromolecules including oxidized lipids to plaques (Figure 2). When Map4k4 was deleted in endothelial cells of adult Apoe−/− mice using a tamoxifen-inducible Ve-cadherin-Cre, mice on a high fat/high cholesterol Western diet (to initiate atherosclerosis), exhibited greater than 50% decreases in atherosclerotic lesion area and lipid accumulation in the aortic root [31]. This was associated with decreased macrophage accumulation in plaque areas. Furthermore, homing of intravenously injected GFP-expressing macrophages to plaque areas was substantially reduced in Map4k4 endothelial KO mice. Dramatically reduced atherosclerotic lesion area was also observed in both Apoe−/− and Ldlr−/− mice treated with a selective small molecule MAP4K4 inhibitor.
These effects of Map4k4 in endothelial function as well as its role in angiogenesis [38, 39] could also contribute to vascular functions that influence insulin sensitivity and glucose homeostasis including delivery of insulin to skeletal muscle [40], as well as modulation of inflammation in adipose tissue [41]. Endothelial cell Map4k4 has also been implicated in lymphatic vessel formation and function [42]. Thus effects of whole body deletion or systemic inhibition on insulin resistance and glucose homeostasis mediated by endothelial cells cannot be ruled out.
Signaling pathways of Map4k4: activators and effectors
A significant challenge in research on Map4k4 functions relates to the plethora of signaling pathways by which it has been reported to operate (see below). It is possible that Map4k4 represents a key node in each of these reported pathways, but more information is required for firm conclusions to be drawn. Another confounding aspect of this field is that many of the Ste20-like protein kinases in mammalian tissues may overlap in their substrates and exhibit functional redundancies. Recent studies delineating Map4k4 function in the Hippo pathway have indeed indicated a high degree of overlap and redundancy [7]. Based on these studies, all of the mammalian homologs of the fly proteins Hippo, Misshapen and Happyhour, are upstream of LATS and Yap/Taz (Figure 1). Numerous inputs influence the Hippo pathway, but specific signals and activators of the Ste20 kinases relevant to Hippo regulation in various cell types have not been well established.
Inflammatory pathways: Cytokine signaling to Jnk and NfKb
Substantial evidence exists for involvement of Map4k4 in regulating some physiological functions through Hippo-independent cellular signaling pathways. Since Ste20p in yeast functions as a Map4k in a classical Map kinase cascade analogous to such pathways in mammalian cells, a reasonable initial hypothesis was that Map4k4 functions in the canonical Jnk pathway and transmits stress signals through a Map kinase cascade. Indeed, early studies seemed to confirm this supposition. Several publications reported the activation of JNK by Map4k4, but in each case the conclusion was based on ectopic cotransfection and high expression of Map4k4 and JNK [16, 23]. While these results could be replicated, neither siRNA mediated depletion nor adenovirus mediated overexpression of Map4k4 had any effect on endogenous JNK activation by TNFα in 293 cells [43]. Moreover, siRNA knockdown of JNK decreased TG deposition in adipocytes, while Map4k4 knockdown increased lipogenesis and TG accumulation. Furthermore, TNFα activation of JNK is unaffected by knockdown of Map4k4 in human umbilical vein endothelial cells (HUVECs), yet increased adhesion molecule expression following TNFα treatment (a well established phenomenon that promotes leukocyte adhesion and extravasation) was reduced by Map4k4 knockdown in the same cells [31].
More recent findings indicate the NfkB pathway, independent of JNK signaling, is employed by Map4k4 to increase expression of adhesion molecules Icam-1 and Vcam-1 in response to TNFα. Canonical NfkB activation occurs by phosphorylation and degradation of IkB, permitting the nuclear translocation of the p65 transcriptional activator within the NfkB complex [44]. Silencing of Map4k4 expression did not affect IkB phosphorylation or degradation, nonetheless TNF-α-induced NFκB transcriptional activity as assessed by a luciferase reporter containing NFκB binding elements that was attenuated when Map4k4 was silenced [31]. Furthermore, using p65 chromatin immunoprecipitation (ChIP) in HUVECs, TNF-α-induced p65 binding to the VCAM-1, E-selectin, and IKBα promoters was reduced in Map4k4-silenced cells, suggesting that Map4k4 may regulate NFκB target gene expression via a distinct mechanism.
Taken together with other studies showing association of Map4k4 with inflammation pathways [26, 29, 45], these findings suggest a role for Map4k4 signaling independent of canonical Map kinase cascades or classical NfKb activation mechanisms. Notably the small molecule inhibitor of Map4k4 activity also dramatically blunts TNFα upregulation in a lipopolysaccharide challenge model [34], consistent with earlier findings with siRNA-mediated knockdown of Map4k4 [45]. We note that one cell-specific model of Map4k4 KO contradicts this general scheme: T-cell specific KO of Map4k4 (CD4-Cre) has been reported to promote Th17 differentiation and accumulation of Th17 cells in adipose tissue, leading to insulin resistance [46]. However, another recent study reported that CD4-Cre mediated KO of Map4k4 led to inhibition of CD4 T-cell proliferation and enhanced regulatory T-cell differentiation [47], thus potentially complex roles of Map4k4 in T-cell function have yet to be established.
AMP Kinase and mTOR: Map4k4 influence on cultured adipocyte metabolism
Interest in Map4k4 as a metabolic regulator was initially based on improved glucose transport and TG synthesis upon knockdown of Map4k4 in cultured adipocytes [22] and involvement of the mTOR pathway in mediating translation of key adipogenic regulators [24]. However, mTOR also transmits signals of nutrient sufficiency to mediate increased lipogenesis through SREBP-1, a transcriptional regulator of numerous genes involved in lipid synthesis [48]. Conversely, AMP kinase (AMPK) signaling in response to energy depletion inhibits the major anabolic process of lipogenesis by downregulation of SREBP-1, and also by inhibitory phosphorylation of acetyl-CoA carboxylase (Acc), a key enzyme in the de novo lipogenesis pathway. Interestingly, Map4k4 was also found to regulate SREBP-1 and lipogenesis in an mTOR-dependent manner [43]. Surprisingly, it was discovered that Map4k4 is required for full activation of AMPK in response to nutrient stress signals. Silencing of Map4k4 expression blunted AMPK phosphorylation of substrates including Acc1, providing a hypothesis on how depletion of Map4k4 enhances lipogenesis. However, adipose specific KO of Map4k4 has no detectable metabolic phenotype in the HFD-fed obese mouse model, indicating these effects, if they do occur in vivo, are masked by more complex regulation or redundancy in the adipose tissue environment.
Map4k4 function in cytoskeletal dynamics and cell adhesion
Although a comprehensive set of direct Map4k4 substrates is not yet available, several studies have identified Map4k4 as an upstream regulator of proteins involved in cytoskeletal dynamics or adhesion including Arp2 [49], Farp1 [50], moesin [39], IQSEC [51] and Pyk2 [10]. Deletion of Map4k4 in endothelial cells altered membrane dynamics resulting in reduced endothelial cell migration and impaired angiogenesis [39]. Map4k4 has also been reported to alter endothelial barrier function and permeability through Rap GTPases [37]. These alterations in membrane dynamics, adhesion and cell-cell interactions are likely correlated with the observations of Map4k4 involvement in tumor metastasis and invasiveness [18–21, 52]; however, as noted earlier, altered endothelial function is also associated with metabolic dysfunctions in insulin resistance as well as atherosclerosis.
Concluding remarks and future perspectives
The studies described here include evidence for significant roles of Map4k4 in the development and progression of metabolic and cardiovascular dysfunctions. Based on data generated in vitro with cultured adipocytes, it is surprising that selective deletion of Map4k4 in adipocytes or in liver is without detectable metabolic phenotype. Strikingly, however, Map4k4 appears to be involved in a large number of vascular functions, perhaps most notably endothelial mobility, adhesion properties and permeability, which combine to drive atherosclerosis and glucose intolerance. The dramatic attenuation of atherosclerosis in endothelial-specific Map4k4 KO mice, and the replication of this effect by treating mice with a selective Map4k4 inhibitor, suggest major signaling pathways are at play. However, revealing the connections of these physiological phenotypes to such signaling pathways of Map4k4 has just begun and will remain a fruitful area of investigation. Among the remaining challenges is systematic identification of upstream activators as well as effectors of Map4k4 function (see Outstanding Questions). Several substrates of Map4k4 have been reported [39, 49, 50, 53, 54] and phosphopeptides dependent on Map4k4 activity identified in HepG2 cells [50] but further identification and validation of Map4k4 targets in tissues of interest should be informative.
Outstanding Questions.
What are the cellular and intracellular signals and upstream activators that modulate Map4k4 activity? Are these distinct from other activators of Hippo signaling?
What are the direct substrates and downstream effectors of Map4k4, especially in Hippo-independent pathways? Which of these are specific vs. shared with other Map4ks, or other Ste20 kinases?
What is the Map4k4-modulated signal from Myf5-lineage cells that improves insulin sensitivity in adipose tissue? What are the specific functions of Map4k4 in beta-cell function and insulin secretion?
Given the apparent involvement of Map4k4 in endothelial cell function, what other roles does it play in physiological or developmental regulation of vasculature (both blood and lymphatic)?
Does Map4k4 integrate nutrient or inflammatory signals with other regulators of Hippo signaling such as cell tension?
None of the animal models of Map4k4 depletion developed to date display significant changes in organ size that might be predicted from dysregulation of the Hippo pathway as have been observed in other KO models of LATS kinases [55, 56]. However, this could be explained by redundancy or overlapping functions of closely related Ste20/Map4 kinases, like Map4k6/MINK1 and Map4k7/TNIK, which converge on LATS phosphorylation to regulate Hippo signaling. The relation of Hippo pathway components to processes that may be relevant to metabolic phenotypes such as adipocyte proliferation and differentiation [57], and to PI3K-mTOR signaling pathway [58], should be investigated further in this context. Nonetheless the profound effects of Map4k4 deletion, particularly in endothelial and Myf5-lineage cells, imply Hippo-independent functions that are not redundant with other Map4k’s/LATS kinases. Another open question is the role of Map4k4 in pancreatic beta cell function that is suggested by the reduced insulin secretion in iMap4k4KO mice [33].
It will also be of great interest to understand how Map4k4 might play a role in integrating its associated signaling pathways. Inhibition of mTOR, activation of AMPK and activation of the Hippo pathway are conceptually consistent with a role for Map4k4 in restricting anabolic functions and cell growth in response to nutrient, endocrine, inflammatory or other signals. Furthermore, it will be interesting to determine whether Map4k4 functions in coordinating these pathways with cellular dynamics mediated by regulators of cytoskeletal function or adhesion. In this regard it is of interest that AMPK mediates energy stress signaling to the Hippo pathway by phosphorylation of angiomotin-like 1 (AMOTL1) [59], which sequesters Yap/TAZ in the cytosol [60]. It is intriguing that angiomotin family members are also associated with regulation of endothelial cell shape, migration and cell-cell connections [61–63], suggesting a possible common signaling nexus involving Map4k4, which is also consistent with observations of a major role of Map4k4 in endothelial function.
Collectively, a picture is starting to emerge implicating Map4k4 in various cellular pathways, including responses to nutrient availability, inflammation and cell growth, as well as having key roles in insulin resistance and atherosclerosis. Future research to further decipher Map4k4 signaling nodes in metabolic and cardiovascular diseases is warranted.
Trends.
Mediators of energy metabolism and inflammation can contribute to dysfunction in metabolic diseases such as T2D and atherosclerosis, and may be valid targets for pharmaceutical therapies.
Map4k4 contributes to the development of insulin resistance associated with obesity and atherosclerosis. Genetic knockdown and small molecule inhibition of Map4k4 are protective in these pathologies in mouse models, especially in attenuating plaque formation in atherosclerosis.
Originally associated with canonical Map kinase signaling in yeast and mammalian cells, Map4k4 now appears to participate in the Hippo signaling pathway, inflammatory cascades, energy regulatory pathways (mTOR, AMPK), and cytoskeletal dynamics.
Understanding the role of Map4k4 in connections between these cellular functions to disease phenotypes will provide new insights into the pathology of metabolic and cardiovascular diseases.
Acknowledgments
The authors thank Dr. Laura Danai, Dr. Rachel Roth Flach, and members of the Czech laboratory for helpful comments and suggestions. Work on Map4k4 in the authors’ lab is supported by grant R37-DK030898 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Leberer E, et al. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. Embo j. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramer SW, Davis RW. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawat SJ, Chernoff J. Regulation of mammalian Ste20 (Mst) kinases. Trends Biochem Sci. 2015;40:149–156. doi: 10.1016/j.tibs.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng Z, et al. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, et al. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meng Z, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Y, et al. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev Cell. 2015;34:642–655. doi: 10.1016/j.devcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boggiano JC, et al. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins CS, et al. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci U S A. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loftus JC, et al. A Novel Interaction between Pyk2 and MAP4K4 Is Integrated with Glioma Cell Migration. J Signal Transduct. 2013;2013:956580. doi: 10.1155/2013/956580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh SH, et al. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008;15:283–298. doi: 10.1038/sj.cdd.4402258. [DOI] [PubMed] [Google Scholar]
- 12.Machida N, et al. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem. 2004;279:15711–15714. doi: 10.1074/jbc.C300542200. [DOI] [PubMed] [Google Scholar]
- 13.Mack KD, et al. Functional identification of kinases essential for T-cell activation through a genetic suppression screen. Immunol Lett. 2005;96:129–145. doi: 10.1016/j.imlet.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Poinat P, et al. A conserved interaction between beta1 integrin/PAT-3 and Nck-interacting kinase/MIG-15 that mediates commissural axon navigation in C. elegans. Curr Biol. 2002;12:622–631. doi: 10.1016/s0960-9822(02)00764-9. [DOI] [PubMed] [Google Scholar]
- 15.Ramnarain DB, et al. Differential gene expression analysis reveals generation of an autocrine loop by a mutant epidermal growth factor receptor in glioma cells. Cancer Res. 2006;66:867–874. doi: 10.1158/0008-5472.CAN-05-2753. [DOI] [PubMed] [Google Scholar]
- 16.Yao Z, et al. A novel human STE20-related protein kinase, HGK, that specifically activates the c-Jun N-terminal kinase signaling pathway. J Biol Chem. 1999;274:2118–2125. doi: 10.1074/jbc.274.4.2118. [DOI] [PubMed] [Google Scholar]
- 17.Han SX, et al. Lowered HGK expression inhibits cell invasion and adhesion in hepatocellular carcinoma cell line HepG2. World J Gastroenterol. 2010;16:4541–4548. doi: 10.3748/wjg.v16.i36.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao JM, et al. A five-gene signature as a potential predictor of metastasis and survival in colorectal cancer. J Pathol. 2010;220:475–489. doi: 10.1002/path.2668. [DOI] [PubMed] [Google Scholar]
- 19.Liu AW, et al. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res. 2011;17:710–720. doi: 10.1158/1078-0432.CCR-10-0331. [DOI] [PubMed] [Google Scholar]
- 20.Qiu MH, et al. Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol Res Pract. 2012;208:541–548. doi: 10.1016/j.prp.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Wright JH, et al. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol. 2003;23:2068–2082. doi: 10.1128/MCB.23.6.2068-2082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X, et al. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc Natl Acad Sci U S A. 2006;103:2087–2092. doi: 10.1073/pnas.0507660103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su YC, et al. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. Embo j. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guntur KV, et al. Map4k4 negatively regulates peroxisome proliferator-activated receptor (PPAR) gamma protein translation by suppressing the mammalian target of rapamycin (mTOR) signaling pathway in cultured adipocytes. J Biol Chem. 285:6595–6603. doi: 10.1074/jbc.M109.068502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilherme A, et al. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tesz GJ, et al. Tumor necrosis factor alpha (TNFalpha) stimulates Map4k4 expression through TNFalpha receptor 1 signaling to c-Jun and activating transcription factor 2. J Biol Chem. 2007;282:19302–19312. doi: 10.1074/jbc.M700665200. [DOI] [PubMed] [Google Scholar]
- 27.Isakson P, et al. Impaired preadipocyte differentiation in human abdominal obesity: role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes. 2009;58:1550–1557. doi: 10.2337/db08-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sartorius T, et al. Association of common genetic variants in the MAP4K4 locus with prediabetic traits in humans. PLoS One. 2012;7:e47647. doi: 10.1371/journal.pone.0047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouzakri K, Zierath JR. MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J Biol Chem. 2007;282:7783–7789. doi: 10.1074/jbc.M608602200. [DOI] [PubMed] [Google Scholar]
- 30.Ali ZA, et al. Oxido-reductive regulation of vascular remodeling by receptor tyrosine kinase ROS1. J Clin Invest. 2014;124:5159–5174. doi: 10.1172/JCI77484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roth Flach RJ, et al. Endothelial protein kinase MAP4K4 promotes vascular inflammation and atherosclerosis. Nat Commun. 2015;6:8995. doi: 10.1038/ncomms9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Y, et al. Mesodermal patterning defect in mice lacking the Ste20 NCK interacting kinase (NIK) Development. 2001;128:1559–1572. doi: 10.1242/dev.128.9.1559. [DOI] [PubMed] [Google Scholar]
- 33.Danai LV, et al. Inducible Deletion of Protein Kinase Map4k4 in Obese Mice Improves Insulin Sensitivity in Liver and Adipose Tissues. Mol Cell Biol. 2015;35:2356–2365. doi: 10.1128/MCB.00150-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ammirati M, et al. Discovery of an in Vivo Tool to Establish Proof-of-Concept for MAP4K4-Based Antidiabetic Treatment. ACS Med Chem Lett. 2015;6:1128–1133. doi: 10.1021/acsmedchemlett.5b00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, et al. Identification of Map4k4 as a novel suppressor of skeletal muscle differentiation. Mol Cell Biol. 2013;33:678–687. doi: 10.1128/MCB.00618-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez-Gurmaches J, et al. PTEN loss in the Myf5 lineage redistributes body fat and reveals subsets of white adipocytes that arise from Myf5 precursors. Cell Metab. 2012;16:348–362. doi: 10.1016/j.cmet.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pannekoek WJ, et al. Rap1 and Rap2 antagonistically control endothelial barrier resistance. PLoS One. 2013;8:e57903. doi: 10.1371/journal.pone.0057903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ndubaku CO, et al. Structure-Based Design of GNE-495, a Potent and Selective MAP4K4 Inhibitor with Efficacy in Retinal Angiogenesis. ACS Med Chem Lett. 2015;6:913–918. doi: 10.1021/acsmedchemlett.5b00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vitorino P, et al. MAP4K4 regulates integrin-FERM binding to control endothelial cell motility. Nature. 2015;519:425–430. doi: 10.1038/nature14323. [DOI] [PubMed] [Google Scholar]
- 40.Kubota T, et al. The role of endothelial insulin signaling in the regulation of glucose metabolism. Rev Endocr Metab Disord. 2013;14:207–216. doi: 10.1007/s11154-013-9242-z. [DOI] [PubMed] [Google Scholar]
- 41.Sengenes C, et al. The role of endothelial cells in inflamed adipose tissue. J Intern Med. 2007;262:415–421. doi: 10.1111/j.1365-2796.2007.01853.x. [DOI] [PubMed] [Google Scholar]
- 42.Roth Flach RJ, et al. Endothelial MAP4K4 is critical for lymphatic vascular development and function. Mol Cell Biol. 2016 doi: 10.1128/MCB.01121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danai LV, et al. Map4k4 suppresses Srebp-1 and adipocyte lipogenesis independent of JNK signaling. J Lipid Res. 2013;54:2697–2707. doi: 10.1194/jlr.M038802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 45.Aouadi M, et al. Orally delivered siRNA targeting macrophage Map4k4 suppresses systemic inflammation. Nature. 2009;458:1180–1184. doi: 10.1038/nature07774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang HC, et al. HGK/MAP4K4 deficiency induces TRAF2 stabilization and Th17 differentiation leading to insulin resistance. Nat Commun. 2014;5:4602. doi: 10.1038/ncomms5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang H, et al. MAP4K4 deletion inhibits proliferation and activation of CD4(+) T cell and promotes T regulatory cell generation in vitro. Cell Immunol. 2014;289:15–20. doi: 10.1016/j.cellimm.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Shao W, Espenshade PJ. Expanding roles for SREBP in metabolism. Cell Metab. 2012;16:414–419. doi: 10.1016/j.cmet.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LeClaire LL, et al. The Nck-interacting kinase NIK increases Arp2/3 complex activity by phosphorylating the Arp2 subunit. J Cell Biol. 2015;208:161–170. doi: 10.1083/jcb.201404095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwaid AG, et al. MAP4K4 Is a Threonine Kinase That Phosphorylates FARP1. ACS Chem Biol. 2015 doi: 10.1021/acschembio.5b00679. [DOI] [PubMed] [Google Scholar]
- 51.Yue J, et al. Microtubules regulate focal adhesion dynamics through MAP4K4. Dev Cell. 2014;31:572–585. doi: 10.1016/j.devcel.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang N, et al. Silencing SOX2 Expression by RNA Interference Inhibits Proliferation, Invasion and Metastasis, and Induces Apoptosis through MAP4K4/JNK Signaling Pathway in Human Laryngeal Cancer TU212 Cells. J Histochem Cytochem. 2015;63:721–733. doi: 10.1369/0022155415590829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgartner M, et al. The Nck-interacting kinase phosphorylates ERM proteins for formation of lamellipodium by growth factors. Proc Natl Acad Sci U S A. 2006;103:13391–13396. doi: 10.1073/pnas.0605950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan W, et al. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J Biol Chem. 2001;276:31349–31356. doi: 10.1074/jbc.M102679200. [DOI] [PubMed] [Google Scholar]
- 55.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song H, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An Y, et al. Lats2 modulates adipocyte proliferation and differentiation via hippo signaling. PLoS One. 2013;8:e72042. doi: 10.1371/journal.pone.0072042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tumaneng K, et al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeRan M, et al. Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 2014;9:495–503. doi: 10.1016/j.celrep.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aase K, et al. Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 2007;21:2055–2068. doi: 10.1101/gad.432007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bratt A, et al. Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem. 2005;280:34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- 63.Ernkvist M, et al. p130-angiomotin associates to actin and controls endothelial cell shape. Febs j. 2006;273:2000–2011. doi: 10.1111/j.1742-4658.2006.05216.x. [DOI] [PubMed] [Google Scholar]