Abstract
Commensal microbiota are critical for the development of local immune responses. Here, we show that gut microbiota can regulate CD4 T cell polarization during pulmonary fungal infections. Vancomycin drinking water significantly decreased lung Th17 cell numbers during acute infection, demonstrating that Gram-positive commensals contribute to systemic inflammation. We next tested a role for RegIIIγ, an IL-22 inducible anti-microbial protein with specificity for Gram-positive bacteria. Following infection, increased accumulation of Th17 cells in the lungs of RegIIIγ−/− and Il22−/− mice was associated with intestinal segmented filamentous bacteria (SFB) colonization. Although gastrointestinal delivery of recombinant RegIIIγ decreased lung inflammatory gene expression and protected Il22−/− mice from weight-loss during infection, RegIIIγ had no direct effect on SFB colonization, fungal clearance or lung Th17 immunity. We further show that vancomycin only decreased lung IL-17 production in mice colonized with SFB. To determine the link between gut microbiota and lung immunity, serum transfer experiments revealed that IL-1 receptor ligands increase the accumulation of lung Th17 cells. These data suggest that intestinal microbiota including SFB can regulate pulmonary adaptive immune responses.
Introduction
CD4+ helper T cells (Th) orchestrate pulmonary host defense by differentiating into specialized subsets (1). Interleukin-12 induces the generation of IFN-γ-secreting Th1 cells, while the combination of IL-6 plus TGF-β induces IL-17-producing Th17 cells. Although the cytokines that program Th differentiation are fairly well defined, less is known regarding the regulation of Th immunity in vivo. Studies on opportunistic fungal infections have illustrated complementary functions for effector Th subsets. The mechanisms through which CD4 T cells control pulmonary infections include stimulating antimicrobial protein release from epithelium, macrophage activation and neutrophil recruitment (1).
Aspergillus fumigatus is a ubiquitous spore-forming soil fungus that people inhale every day (2). Once conidia reach the alveoli, they are usually destroyed by macrophages and neutrophils within 24 to 48 hours. Immunocompromised patients are susceptible to invasive aspergillosis, during which conidia germinate into hyphae and disseminate to other tissues such as the brain, kidneys or heart (2). Th1 cells are protective against aspergillosis (3), while Th17 cells are critical for vaccine-induced anti-fungal immunity (4). These powerful inflammatory responses to fungi can also cause pathology. For instance, chronic exposure to conidia may lead to allergic bronchopulmonary aspergillosis in cystic fibrosis or asthma patients, corresponding to strong Th2 responses (1, 2). Aspergillus fumigatus is the most prevalent cause of severe pulmonary allergic disease (5). In chronic granulomatous disease, defective superoxide generation in neutrophils leads to increased IL-17 production, which exacerbates acute lung injury during A. fumigatus infection (6). Regulatory T cells (Tregs) are induced by the tryptophan metabolite kynurenine, reducing lung damage during aspergillosis but also potentially increasing fungal persistence (6). Understanding the mechanisms that regulate inflammatory T cell cytokine production towards A. fumigatus has substantial therapeutic value in both invasive disease as well as fungal allergy.
Commensal bacteria residing in the intestinal lumen have considerable effects on host defense. Studies in germ-free (GF) mice revealed that commensals protect against pathogenic infections through competitive growth inhibition or stimulation of innate immune receptors (7). Colonization of the gut is critical for the maturation of lymphoid tissues such as Peyer's patches, facilitating interactions among B cells and T cells (8). Although helpful for controlling infections, activated T cells in the gut also have the potential for driving colitis (9, 10). These studies demonstrated that microbial products as well as T cell-intrinsic properties influence intestinal inflammation. Microbiota also affect immune responses in peripheral tissues (11), and contribute to systemic anti-viral immunity (12, 13, 14). Loss of microbial diversity has also been implicated in allergic inflammation (15). Altogether, this suggests host factors that modulate microbial composition in the gut may also regulate peripheral T cell activation during infectious challenges.
The intestinal mucosa provides a physical barrier from microbes, maintained by the generation of a mucus layer, antimicrobial proteins and IgA (16). Paneth cells shape the composition of intestinal microbiota by producing antimicrobial peptides. For instance, alpha-defensin 5 increases the ratio of Bacteroidetes to Firmicutes in the distal small intestine (17). RegIIIγ is produced by Paneth cells in response to IL-22 and has bactericidal activity against Gram-positive organisms (18, 19). RegIIIγ contributes to maintaining the spatial separation between microbiota and the intestinal cell wall (20). In the absence of RegIIIγ, the microbiota is enriched for segmented filamentous bacteria (SFB), which are sufficient to induce Th17 cells (21, 22). Although RegIIIγ protects against Gram-positive pathogens in the gut (19, 23), its impact on pulmonary immunity to fungal pathogens is unknown.
Here, we investigated a role for commensal bacteria in lung immunity to A. fumigatus. We found that vancomycin-sensitive bacteria amplify lung IL-17 production early following fungal infection. Endogenous expression of RegIIIγ or Il22 was linked to decreased intestinal SFB colonization, leading to decreased IL-17 production in the lungs and small intestine. The phenotype of expanded CD4+ IL-17+ cells in Il22−/− mice was transferrable either with intestinal microbiota or serum to wild-type (WT) mice. Pre-incubation of serum with an IL-1 receptor antagonist blunted the IL-17 response, demonstrating that commensals induce serum IL-1 receptor ligands which contribute to lung IL-17 production. Finally, reconstituting Il22−/− mice with local RegIIIγ protected them from weight-loss during A. fumigatus infection and reduced inflammatory cytokines in lung tissue. Thus, factors that shape the balance of intestinal commensal species regulate lung inflammatory responses including Th17 cell priming.
Materials and Methods
Mice
C57BL/6 mice (B6) from Jackson Laboratory (Figs. 2, 3, 5D; Bar Harbor, ME) or Taconic Farms (Figs. 1, 4, 5A-C, 5E, 6; Germantown, NY or Cambridge City, IN) were used. OT-II mice were purchased from Jackson Laboratory. Il22−/− mice (24) were bred at Taconic Farms (Cambridge City, Indiana). Mice with a targeted deletion for all six exons of RegIIIγ were generated as described below, and backcrossed to B6 for ten generations. Mice were housed at Louisiana State University Health Sciences Center and the University of Pittsburgh under specific pathogen-free conditions, and were handled in accordance to NIH federal guidelines.
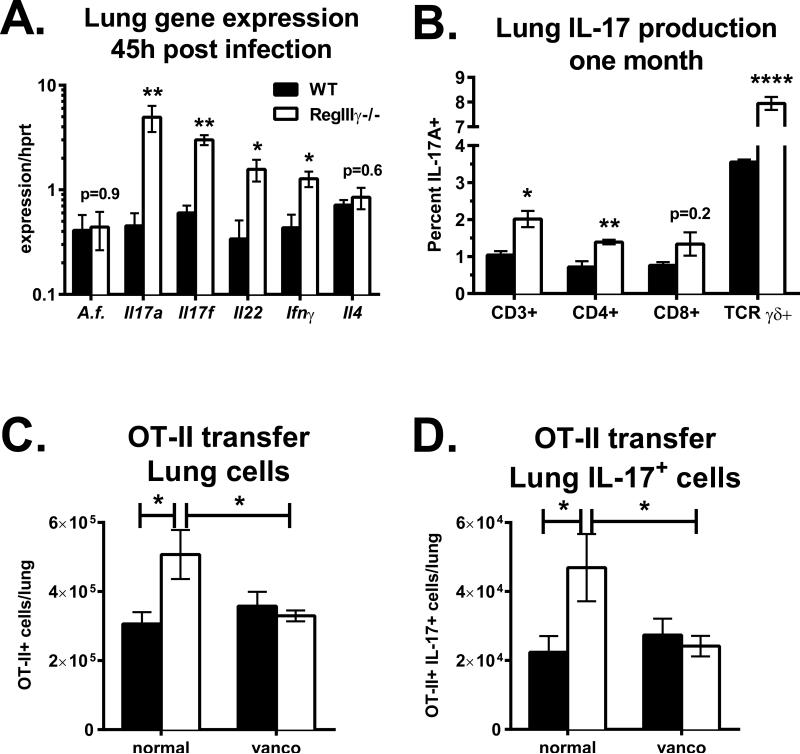
Figure 2. RegIIIγ inhibits pulmonary type 17 immunity by modulating intestinal microbiota.
B6 Jackson (black bars) or RegIIIγ−/− mice (white bars) were infected with A. fumigatus via o.p. aspiration. A. Lung gene expression on day 2 normalized to Hprt (n=5). B. The percent of IL-17 producing cells within individual T cell populations one month following infection (n=3-4). C and D. Mice on vancomycin drinking water for one month or normal water were adoptively transferred with OT-II cells, followed by o.p. immunization with OVA plus cholera toxin. One week later, lung cells were restimulated and stained for intracellular IL-17. C. Total numbers of lung-resident OT-II T cells (CD4+ TCR Vα2+). D. Total numbers of IL-17 producing OT-II CD4 T cells. Data in C and D are combined from two experiments with n=6-8. In one experiment, vancomycin was combined with neomycin, metronidazole and ampicillin.
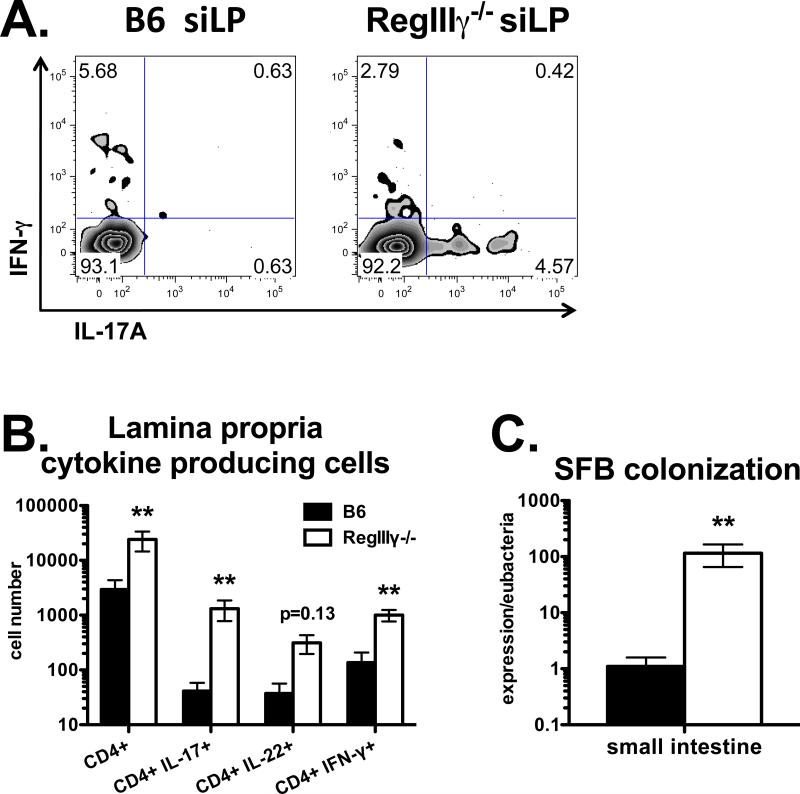
Figure 3. RegIIIγ suppresses intestinal Th17 cell priming.
Mice were injected i.p. with OVA plus LPS. Three weeks later, siLP lymphocytes were stimulated with PMA plus ionomycin. A. Flow cytometry plots of IL-17 vs. IFN-γ staining, gated on CD4 T cells. B. Total numbers of siLP CD4 T cells producing IL-17, IL-22 or IFN-γ. Data in A and B are combined from two experiments with n=6. C. SFB colonization determined by PCR from small intestinal cDNA, normalized to Eubacteria. Data in C are combined from 4 experiments with n=8-9.
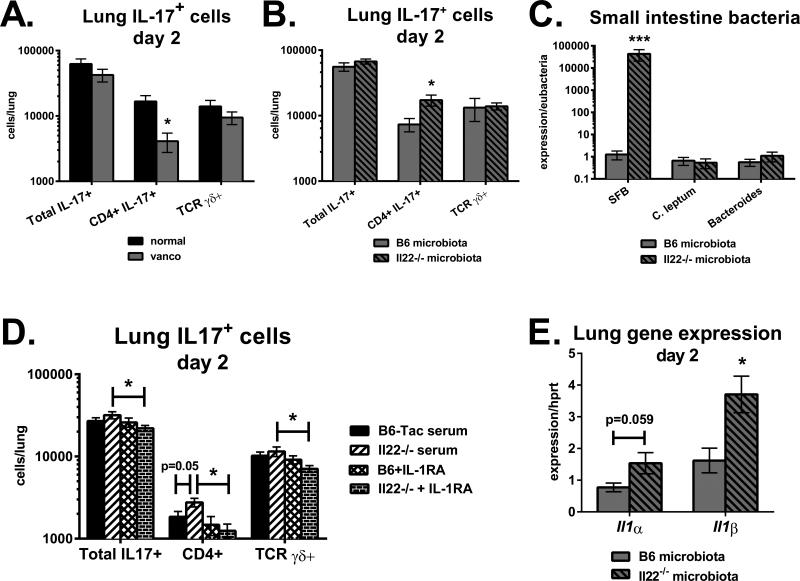
Figure 5. Interleukin-22 suppresses pulmonary Th17 cell accumulation through serum IL-1 receptor ligands.
A. Vancomycin-treated B6 Taconic mice and normal water controls were infected with A. fumigatus via o.p. aspiration. Two days following infection, lung cells were stimulated with PMA plus ionomycin and stained for intracellular IL-17. Shown are total numbers of IL-17+ cells, CD4+ IL-17+ cells and TCR γδ+ IL-17+ cells. Data are combined from two experiments with n=5. B, C and E: Vancomycin-treated B6 mice were placed on normal water and given a fecal transplant from B6 Taconic or Il22−/− donors. Fifteen days following gavage, mice were neutrophil-depleted and infected with A. fumigatus. B. Total numbers of lung IL-17-producing cells two days following infection. C. Distal small intestine cDNA was analyzed for bacterial species-specific 16S rRNA primers, normalized to Eubacteria. E. Expression of Il1α and Il1β in lung homogenates on day 2. Data in B, C and E are combined from two experiments with n=6-8. D. B6 Jackson mice were neutrophil-depleted and infected. Seven hours later, serum from naïve B6 Taconic or Il22−/− mice was transferred i.v. Additional groups received serum pre-mixed with IL-1RA (anakinra). Shown are the total numbers of IL-17+ cells in lungs on day 2. Data in D are combined from 2-3 experiments with n=8-12.
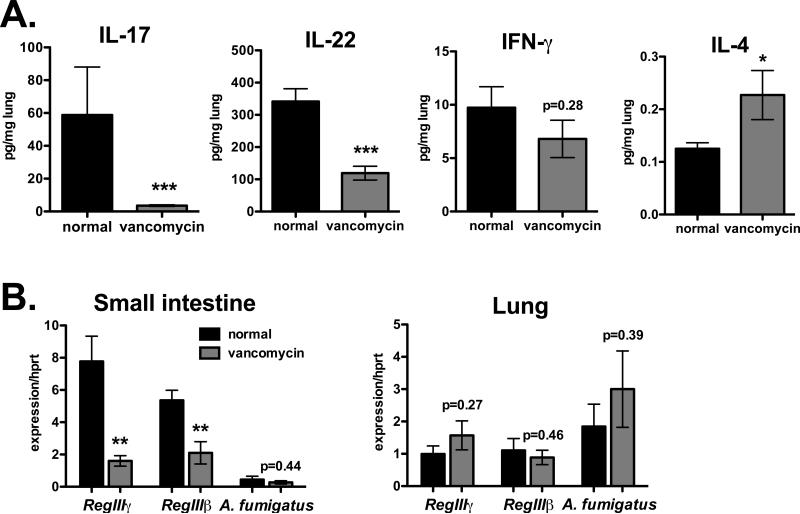
Figure 1. Commensal bacteria amplify lung IL-17 production during fungal infection.
B6 mice on vancomycin drinking water for one month (gray bars), or maintained on normal water (black bars) were injected i.p. with the neutrophil-depleting antibody 1A8, followed by o.p. infection with A. fumigatus the next day. Tissues were collected on day 2. A. Cytokine levels in lung homogenates normalized to total protein. B. Gene expression in distal small intestine (left) or lung tissue (right), normalized to Hprt. Data are combined from two experiments with n=10.
Figure 4. Interleukin-22 suppresses lung IL-17 levels during fungal infection.
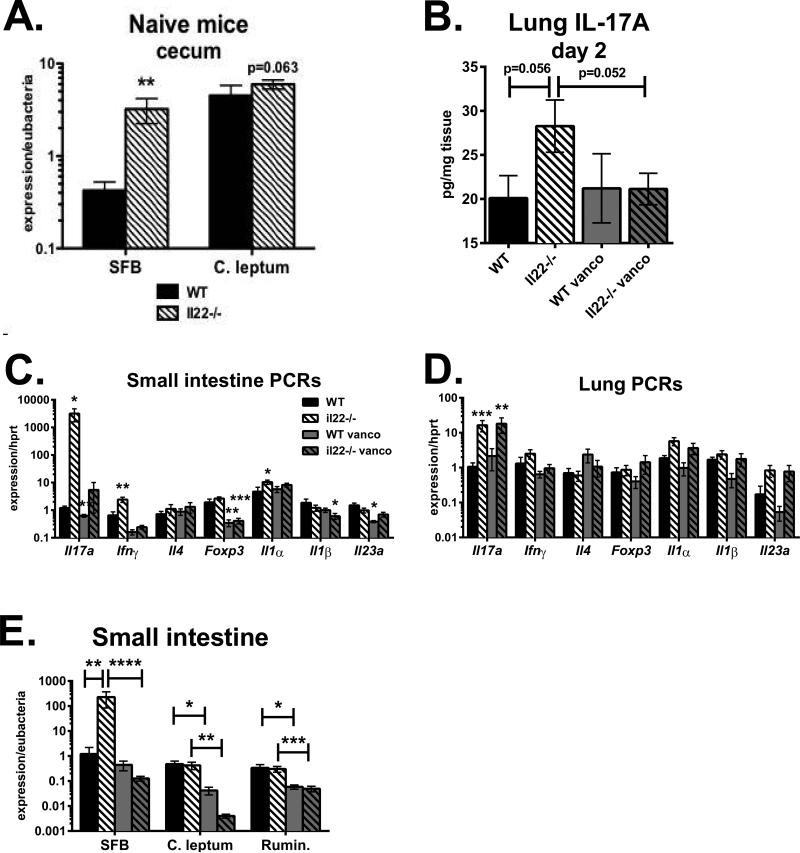
A. SFB and C. leptum expression from cecum stool DNA of untreated WT-Taconic (closed bars) and Il22−/− mice (striped bars), normalized to Eubacteria. Data are combined from four experiments with n=17 and shown as mean +/− SEM. B-E: WT and Il22−/− mice were placed on vancomycin drinking water for one month (gray bars) or maintained on normal water. Then, all mice were placed on normal water and injected i.p. with 1A8 antibody. The next day, mice were infected with A. fumigatus. B. Interleukin-17 levels in lung homogenate on day 2, normalized to total protein. C-D: Real-time PCR for indicated genes in distal small intestine (C) or lung (D) on day 2, normalized to Hprt. E. Real-time PCR on small intestine cDNA for SFB, C. leptum or Ruminococcaceae 16S rRNA, normalized to Eubacteria. Data in B-E are combined from two experiments with n=7-9.
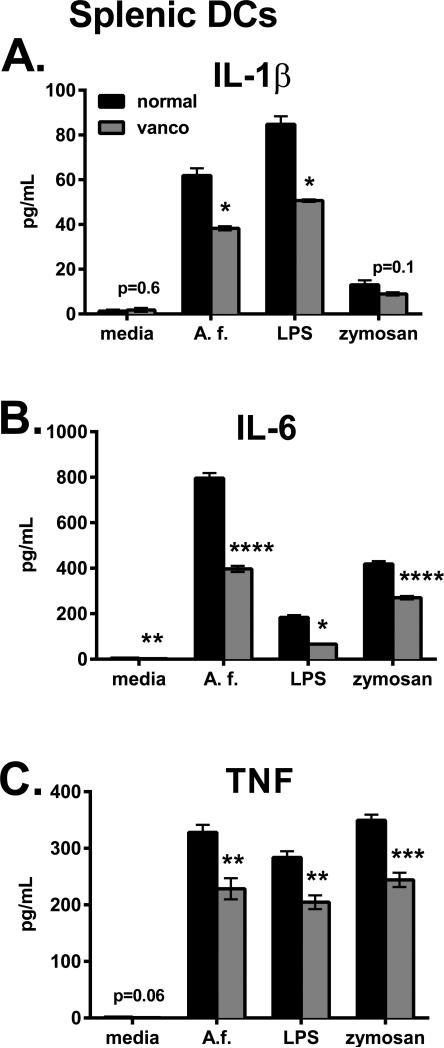
Figure 6. Microbiota condition splenic DCs for anti-fungal cytokine responses.
Splenic CD11c+ cells were purified and stimulated overnight with heat-killed swollen A. fumigatus (A.f.), LPS or zymosan, and cytokine levels were measured in supernatants. Data are from one representative experiment of two performed and shown as mean +/− SEM (4-5 wells).
Generation of RegIIIγ−/− mice
Regions from the mouse RegIIIγ locus were amplified from genomic DNA by PCR and cloned into the pDelboy 3X targeting vector. The fragments included a 3.5kb 5’ flanking region ending at the RegIIIγ transcription start site, a 2.5 kb fragment cloned between two lox p sites and comprising all 6 exons of the RegIIIγ gene, and a 3.4 kb 3’ flanking region. The targeting vector also contains a neomycin resistance gene, a thymidylate kinase gene, and a beta-gal gene. Cre mediated recombination between the loxp sites results in deletion of all coding portions of the RegIIIγ gene and brings the beta-gal gene immediately downstream of the RegIIIγ 5’ promoter.
The vector was linearized, electroporated into ES cells and clones underwent positive and negative selection with G418 and ganciclovir. Successfully targeted clones were analyzed by PCR and Southern blot and three successfully targeted clones were identified. One clone was microinjected into blastocysts using standard techniques, and the resulting chimeric mice were bred to generate heterozygous mutant offspring. Male RegIIIγfl/+ heterozygotes were crossed with Tie2 Cre females to generate RegIIIγ+/− offspring. These were interbred to generate RegIIIγ−/− offspring and were also backcrossed onto the C57BL/6 background for 10 generations.
By quantitative PCR, RegIIIγ−/− mice showed mean 240-fold lower expression of RegIIIγ mRNA in the trachea compared to control mice. As these mice have no detectable RegIIIγ coding region we believe that the residual RegIIIγ signal is from transcripts of other highly related Reg family genes including RegIIIβ.
Infections and treatments
Mice were infected with A. fumigatus via o.p. aspiration (107 organisms). For some experiments (Figs. 1, 4B-E, 5B-E, 7), mice were neutrophil-depleted by i.p. injection of antibody 1A8 (0.6mg; Bio X Cell, West Lebanon, NH) one day prior to infection. Serum was given i.v. (0.25mL) in the presence of anakinra (Kineret, Swedish Orphan Biovitrum; Stockholm, Sweden) or vehicle 7h following infection. Vancomycin was administered in drinking water at 0.5g/L for at least 4 weeks, then mice were placed on normal water one day prior to infection. Il22−/− mice were gavaged with recombinant RegIIIγ protein (40ug (25)) or vehicle for every other day beginning on day -9 prior to infection for a total of six treatments. For fecal transplant experiments, stool pellets were homogenized in water and poured over a 100uM cell strainer. Recipient mice were then gavaged with 0.4mL of fecal matter.
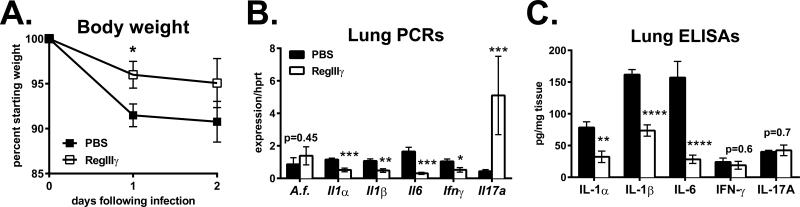
Figure 7. Intestinal RegIIIγ delivery is anti-inflammatory during pulmonary fungal infection.
A. Il22−/− mice were gavaged with recombinant mouse RegIIIγ (40ug; open symbols) or vehicle control (closed symbols), as described in Materials and Methods. Mice were then neutrophil-depleted and infected. A. Percent body weight relative to day 0 as mean +/− SEM. B. Lung gene expression on day 2, normalized to Hprt. C. Lung cytokines measured by ELISA on day 2, normalized to total protein. Data are combined from two experiments with n=8-10.
To examine pulmonary Th17 cell priming, TCR transgenic OT-II T cells were negatively selected using a CD4 T cell isolation kit (Miltenyi Biotec; Bergisch Gladbach, Germany), and 0.5 million cells were adoptively transferred into mice via retro-orbital injection. The next day, mice were inoculated via o.p. aspiration with OVA (100ug; Sigma Aldrich; St. Louis, MO) plus cholera toxin (1ug; List Biological Laboratories, Inc.; Campbell, CA). Lungs were collected on day 7. To examine intestinal Th17 cell priming, mice were injected i.p. with OVA (1mg) at time 0 and E. coli-derived LPS (50-60ug; Sigma-Aldrich) 18 hours later. Small intestinal lamina propria lymphocytes were isolated 3 weeks later.
Tissue processing
To analyze gene expression, lungs and distal small intestine were placed in TRIzol (Life Technologies, Carlsbad, CA), homogenized and processed according to the manufacturer's protocol. One microgram of RNA was used to synthesize cDNA (iScript, Bio-Rad; Hercules, CA). Real-time PCR primers were purchased from Applied Biosystems (Foster City, CA) and used with Taqman Universal PCR Master Mix (Applied Biosytems) or SsoFast Probes Supermix (Bio-Rad). PCRs were run on a Bio-Rad CFX96 Real Time System.
For ex vivo cytokine production, tissues were placed in cell lysis buffer (Cell Signaling Technology; Danvers, MA) with protease inhibitor cocktail tablets (Roche Applied Science; Penzberg, Germany), homogenized and centrifuged at 10,000 × g. Cytokine levels in supernatants were measured by ELISA (eBioscience; San Diego, CA; BioLegend; San Diego, CA) or Milliplex Mouse Cytokine/Chemokine Immunoassay (EMD Millipore; Billerica, MA) and normalized to total protein levels (Pierce BCA Protein Assay Kit; Thermo Fisher Scientific; Waltham, MA).
For flow cytometry, lungs were digested with collagenase D (3mg/mL; Roche Applied Science) in Dulbecco's Modified Eagle Medium for 1h at 37C, crushed through 100uM cell strainers (BioExpress; Kaysville, UT) and treated with ammonium chloride to lyse red blood cells. Small intestinal lamina propria lymphocytes were isolated as previously described (26), with some modifications. Briefly, small intestinal tissue was flushed, sliced open longitudinally, cut into ~1 cm pieces and washed with Ca/Mg-free BSS. Tissue was stirred at 37°C in Ca/Mg-free BSS containing 5 mM EDTA and 0.15 mg/mL dithioerythritol, with the supernatant removed. Then, tissue was incubated at 37°C in BSS containing 1mM CaCl2, 1mM MgCl2 and 1mg/mL collagenase D (Roche Applied Science). Cells were fractionated on a 44% and 67% percoll gradient (Amersham Biosciences; Piscataway, NJ), with lymphocytes partitioning at the interface. Single cell suspensions were incubated in IMDM with 10% fetal bovine serum, phorbol 12-myristate 13-acetate (50ng/mL, Sigma-Aldrich), ionomycin (750ng/mL, Sigma-Aldrich) and golgiplug (Becton Dickinson Pharmingen; Franklin Lakes, NJ) for 4h at 37C. Cells were stained with antibodies directed against CD3ε, CD4, CD8, TCR Vα2, TCR γδ, IL-17A, IL-22 or IFN-γ (eBioscience, BD Pharmingen).
For in vitro cytokine production, splenic DCs were purified using CD11c microbeads (Miltenyi Biotec). Cells were stimulated overnight with heat-killed swollen A. fumigatus (10:1 organisms:DC), LPS (50ng/mL) or zymosan (100ug/mL; InvivoGen). Cytokine levels were determined by Milliplex Mouse Cytokine/Chemokine Immunoassay (EMD Millipore).
Microbiota analysis
Real-time PCR
DNA from small intestinal stool or cecum was isolated using QIAamp DNA Stool Mini Kit (Qiagen; Venlo, Netherlands), and 90-180ng was used as a template for PCR. In some experiments, cDNA from distal small intestine was used as a template. Primers for Eubacteria, SFB (27), Bacteroides, C. leptum (28) and Ruminococcaceae were purchased from Integrated DNA Technologies (Coralville, IA). Samples were diluted with RNase/DNase-free water and mixed with iQ SYBR Green Supermix (Bio-Rad). PCRs were performed on a CFX96 Real-Time System (Bio-Rad). Relative gene expression for bacteria species (SFB, Bacteroides, C. leptum) was normalized to Eubacteria.
16S Next Generation Sequencing
DNA from cecal contents was prepared for sequencing as previously described (29). Briefly, DNA was PCR amplified using 515F and 806R bacterial 16S PCR primers containing sample-specific barcodes and sequence adaptors. PCR products were pooled in equimolar amounts and sequenced on an Illumina MiSeq using a 300 cycle V2 sequencing kit. Sequences were processed in the QIIME pipeline (30). Sequences were split and trimmed to a minimum quality score of 20, aligned to the reference greengenes database (31) using uclust (32) and a reference OTU threshold of 97%, and analyzed for taxonomic and alpha diversity. Data are publicly available on MG-RAST, accession 4541676.3 (URL: http://metagenomics.anl.gov/metagenomics.cgi?page=MetagenomeOverview&metagenome=4541676.3).
Histopathology
B6-Taconic mice on normal water, vancomycin water and Il22−/− mice were injected i.p. with a neutrophil-depleting antibody (1A8; 0.6mg) one day prior to o.p. infection with A. fumigatus. Two days later, lungs were inflated with formalin and stained with hematoxylin and eosin. The severity of disease was scored by a pathologist (DAP) blinded to the groups. Individual fields were scored at ×200 magnification, with a minimum of 20 fields per slide. Scoring in each field was judged on the percentage of inflammation according to the following scale: 0= no inflammation; 1= 1-25% inflammation; 2= 26-50% inflammation; 3= 51-75% inflammation; 4= 76-100% inflammation.
Dendritic cell/T cell co-cultures
Naïve CD4 T cells (CD4+ CD62L+) and CD11c+ cells were isolated from spleens and lungs, respectively, using magnetic bead selection (Miltenyi Biotec). T cells and DCs were mixed at 2.4:1 (Fig. S3A) or 3:1 (Fig. S3B) in IMDM plus 10 percent FBS for three days. Cultures were treated with heat-killed swollen A. fumigatus, anti-CD3 (1ug/mL), A. fumigatus hyphae or left unstimulated.
Statistical analysis
A two-tailed Student's t-test was used when variance between the groups was similar as determined by an F test. For comparing groups with an unequal variance, a two-tailed Mann-Whitney test was performed. In figures, statistical significance is indicated by the p-value if p>0.05, or * (p<0.05), ** (p<0.01), *** (p<0.001) or **** (p<0.0001).
Results
Commensal bacteria amplify IL-17 production in lungs during fungal infection
To study the role of commensal bacteria in pulmonary anti-fungal immunity, C57BL/6 (B6) mice from Taconic Farms, known to be colonized with SFB (22), were placed on drinking water containing vancomycin (0.5g/L) for at least 4 weeks, and then infected with A. fumigatus (107 organisms) via oropharyngeal (o.p.) aspiration. Aspergillus fumigatus is an opportunistic pathogen that colonizes immunocompromised patients. To mimic this situation, mice were injected with the neutrophil-depleting antibody 1A8 one day prior to infection. Although vancomycin treatment did not affect A. fumigatus burden in the lungs at early time points, there was a substantial reduction in lung IL-17 and IL-22 levels during this acute infection (Fig. 1). In contrast, IFN-γ was not affected by vancomycin whereas IL-4 was significantly increased. This suggested that commensal bacteria in the intestine preferentially increase the production of cytokines associated with Type 17 immunity in the lung, and decrease Type 2 responses. Overall, vancomycin treatment did not affect disease severity determined by H&E stained lung sections at the 48 hour time point (Fig. S1A), possibly due to neutrophil depletion. Intestinal colonization induces production of antimicrobial proteins including the RegIII family (18). Accordingly, vancomycin diminished the expression of RegIIIβ and RegIIIγ in small intestinal tissue but not in the lungs (Fig. 1B). Therefore, vancomycin-sensitive commensals, i.e. Gram-positive bacteria, influence the expression of local anti-microbial factors in the gut as well as type 17 cytokines in the lung.
Endogenous RegIIIγ suppresses lung IL-17 production during fungal infection
The bactericidal activity of RegIIIγ is attributed to its binding of cell wall peptidoglycan on Gram-positive bacteria (18). Since oral vancomycin treatment potently decreased IL-17 and IL-22 production in lung tissue (Fig. 1A), and RegIIIγ−/− mice have been reported to have increased colonization with SFB (20), we hypothesized that the endogenous antimicrobial factor RegIIIγ may similarly regulate pulmonary Th17 cytokines. Of note, our line of RegIIIγ−/− mice was colonized with SFB (Fig. 3C), as previously reported for an independent line (20). To test our hypothesis, SFB-positive RegIIIγ−/− and B6 Jackson mice that lacked SFB were infected with A. fumigatus via o.p. aspiration. Lungs were harvested two days later and analyzed for gene expression by real-time PCR. Pulmonary fungal burden was similar between WT and RegIIIγ−/− mice, suggesting that RegIIIγ was dispensable for anti-fungal host defense (Fig. 2A, A.f.). However, the expression of the Th17-related genes Il17a, Il17f and Il22 were elevated in the lungs of RegIIIγ−/− mice, indicating more activated T cells. To determine which T cell subsets contribute to IL-17 production, lymphocytes were isolated from lungs one month following infection, restimulated in vitro with PMA plus ionomycin and analyzed by flow cytometry. Approximately one percent of total CD3+ T cells in WT mice produced IL-17A (Fig. 2B). In RegIIIγ−/− mice, approximately 2 percent of total T cells produced IL-17, indicating that SFB colonization in RegIIIγ−/− mice was linked to increased pulmonary Th17 cell priming and/or maintenance (Fig. 2B). This trend was consistent when analyzing the individual CD4+, CD8+ and TCR γδ+ T cell subsets. Thus, lung-resident T cells from infected RegIIIγ−/− mice had a 2-fold increase in IL-17 potential compared to T cells isolated from WT mice.
Microbiota in RegIIIγ-deficient mice regulate pulmonary Th17 cell priming
To test if intestinal Gram-positive bacteria drive pulmonary Th17 cell priming in RegIIIγ−/− mice, they were placed on vancomycin drinking water for at least 4 weeks. In one of two experiments, vancomycin was combined with ampicillin, metronidazole and neomycin, while control mice were maintained on normal water. Following antibiotic treatment, mice were adoptively transferred with TCR transgenic OT-II CD4 T cells. One day later, mice were immunized with OVA plus the mucosal adjuvant cholera toxin (33) via o.p. aspiration. On day 7, lung single cell suspensions were stimulated with PMA plus ionomycin and analyzed for IL-17 production. The total number of OT-II CD4+ T cells recovered from the lungs of RegIIIγ−/− mice on normal water was 1.8-fold higher compared to WT Jackson mice (Fig. 2C). Vancomycin drinking water significantly reduced lung T cell accumulation in RegIIIγ−/− mice to similar numbers as WT mice, demonstrating the gut microbiota in RegIIIγ−/− mice contributes to pulmonary Th17 cell accumulation. In contrast, vancomycin did not significantly affect Th17 cells in WT mice from Jackson Labs that were not colonized with SFB, suggesting their microbiota did not influence Th17 cell accumulation in the lungs (Fig. 2D). These data demonstrate that gut microbiota in RegIIIγ−/− mice inhibits pulmonary Th17 cell priming in a T cell-extrinsic manner.
To further test the link between SFB colonization and pulmonary cytokines, we purchased SFB-negative RegIIIγ−/− mice from Jackson Labs. Following neutrophil depletion and infection, lung IL-17A, IL-22, IFN-γ and IL-10 were comparable to B6 Jax mice (Fig. S2, top). Fungal burdens in RegIIIγ−/− Jax mice were slightly elevated compared to B6 Jax, but not statistically different (Fig. S2, middle). Thus, elevated IL-17 production in RegIIIγ−/− mice (Fig. 2) was linked to SFB colonization.
RegIIIγ suppresses intestinal Th17 cell priming
Since RegIIIγ is highly expressed by intestinal epithelium (18), we next determined its effect on intestinal T helper cell priming to a model antigen ovalbumin (OVA). Mice were immunized intraperitoneally (i.p.) with OVA plus LPS. Three weeks later, lymphocytes from small intestinal lamina propria (siLP) were restimulated in vitro with PMA plus ionomycin. The total numbers of CD4+ T cells isolated from siLP were 8-fold higher in RegIIIγ−/− mice compared to WT Jackson mice (Fig. 3B), suggesting that endogenous RegIIIγ suppresses T cell priming in the intestinal mucosa. This was accompanied by increased total numbers of CD4+ T cells producing the effector cytokines IL-17, IL-22 or IFN-γ. Notably, immunized RegIIIγ−/− mice had 32-fold more Th17 cells than WT. As noted above, RegIIIγ limits colonization with SFB by maintaining the spatial separation between commensal microbes and mucosal epithelia (20). Our RegIIIγ−/− mice were also colonized with SFB prior to (not shown) and after immunization (Fig. 3C), correlating with increased percentages of CD4+ T cells producing IL-17 (Fig. 3A). Unimmunized RegIIIγ−/− mice contained 2.5 to 3-fold more Th17 cells in the siLP and spleen, respectively, compared to unimmunized WT mice (not shown). Altogether these data demonstrate an association between SFB colonization in RegIIIγ−/− mice and preferential induction of the Th17 pathway in mucosal tissues.
Endogenous Il22 expression suppresses lung IL-17 levels during fungal infection
RegIIIβ and RegIIIγ expression in intestinal epithelium is regulated by IL-22 during gastrointestinal infections (19). In the steady-state, we observed that some naïve Il22−/− mice expressed lower levels of RegIII family members in the distal small intestine compared to WT Taconic controls, although this was not true for all mice examined. However, on a consistent basis we found that Il22−/− mice had increased levels of cecal SFB colonization compared to WT Taconic mice (Fig. 4A). If SFB influences pulmonary IL-17 responses, we reasoned that SFB-colonized Il22−/− mice should have a similar phenotype as the RegIIIγ−/− strain (Figs. 2, 3). To test this, WTTaconic and Il22−/− mice were infected with A. fumigatus via o.p. aspiration. One day prior to infection, mice were neutrophil-depleted by i.p. injection of antibody 1A8. To determine if any potential differences between the mouse strains were influenced by gut microbiota, some mice were placed on vancomycin-drinking water for one month prior to the experiment. On day 2, Il22−/− mice had a 1.5-fold increase in lung IL-17 compared to WT Taconic (Fig. 4B), consistent with results observed in RegIIIγ−/− mice colonized with SFB (Fig. 2). Vancomycin drinking water decreased IL-17 levels by 25 percent in Il22−/− mice, suggesting that IL-22-mediated alterations in intestinal flora regulate pulmonary IL-17 production. In support, vancomycin water significantly decreased SFB colonization in Il22−/− mice (Fig. 4E). In B6 control mice, vancomycin decreased lung IL-17 levels in only one of two experiments (Fig. 4B). Further analysis revealed that B6 Taconic mice were SFB-negative in one of the experiments. Thus, vancomycin only decreased lung IL-17 levels in SFB-colonized mice. Levels of C. leptum and Ruminococcaceae were similar between WT and Il22−/− mice, and decreased by vancomycin. This is consistent with a study demonstrating that several Clostridium species including Ruminococcus are sensitive to vancomycin (34). Therefore, lung IL-17 levels correlated with intestinal SFB colonization. When analyzing cytokines in the small intestine, Il22−/− mice on normal water had a dramatic 3000-fold increase in Il17a expression compared to WT controls, which was reduced by 99.8 percent upon pretreatment with vancomycin drinking water (Fig. 4C). There was also a 3.7-fold increase in Ifnγ expression in the small intestine of Il22−/− mice, which was decreased by vancomycin. Expression of Il4 and Foxp3 were similar between WT Taconic and Il22−/− mice in both the intestine and lung (Figs. 4C, 4D), suggesting that endogenous IL-22 did not affect Th2 or Treg differentiation. Lung Il17a expression correlated with Il1α in the lungs and small intestine of Il22−/− mice (Figs. 4C, 4D). Therefore, the absence of IL-22 creates a permissive environment for SFB growth, augmenting mucosal IL-17 production in both the gut and lung.
To examine the pulmonary T cell subsets affected by microbiota, B6 Taconic mice on vancomycin water versus normal water were infected with A. fumigatus via o.p. aspiration. Two days later, lung single cell suspensions were stimulated with PMA plus ionomycin and stained for intracellular IL-17. Vancomycin significantly decreased the number of lung CD4+ cells producing IL-17, but did not change the number of γδ T cells making IL-17 (Fig. 5A). Next, the impact of IL-22-conditioned microbiota was assessed by returning vancomycin-treated mice to normal water, followed by a fecal transplant from B6 Taconic (low SFB) or Il22−/− donor mice (high SFB). Fifteen days following the microbiota transfer, mice were neutrophil-depleted and then infected with A. fumigatus the next day. Recipients of microbiota from SFB-high colonized Il22−/− mice had significantly greater CD4+ IL-17+ cells, while numbers of γδ+ IL-17+ cells were not affected (Fig. 5B). Notably, the number of lung CD4+ IL-17+ cells directly correlated with SFB colonization in the small intestine (Fig. 5C) and the cecum (data not shown). A previous study found that microbiota-induced IL-1β increases intestinal Th17 cell numbers (35). We found that SFB-high microbiota from Il22−/− donors increased Il1α/β gene expression in the lungs following fungal infection (Fig. 5E). Our data demonstrate that vancomycin-sensitive bacteria in the gut augment IL-17 production by CD4+ T cells during pulmonary fungal infection, and that increased Th17 responses observed in Il22−/− mice are transferrable with SFB-containing microbiota.
Interleukin-22 suppresses pulmonary Th17 cell accumulation through serum IL-1 receptor ligands
We next addressed a potential role for SFB containing microbiota to regulate lung DC function. In vitro, lung CD11c+ cells from B6 Taconic mice on normal water versus vancomycin were nearly equivalent at inducing IL-17 from polyclonal naïve CD4 T cells in the presence of heat-killed A. fumigatus (Fig. S1B). This suggested that lung DCs from vancomycin-treated mice can mount a normal response to A. fumigatus in vitro. In support, overnight stimulation of lung CD11c+ cells from B6 Taconic mice on normal water versus vancomycin resulted in similar secretion of IL-1α, IL-1β and IL-6 (Fig. S1D). This contrasted with what we observed for splenic DCs, in which vancomycin-sensitive bacteria were required for optimal cytokine production in response to heat-killed A. fumigatus, zymosan or LPS (Fig. 6). In vivo, we found that infection increased the number of lung CD11c+ CD11b+ cells similarly in B6 Taconic, B6 plus vancomycin and Il22−/− mice (Fig. S1E). Further, SFB colonization did not impact IL-1α or IL-1β production by lung APCs, measured by intracellular cytokine staining. If SFB colonization activates lung DCs, then Il22−/− DCs would be more potent than B6 at inducing Th17 differentiation. Co-culture of lung CD11c+ cells with A. fumigatus-specific naïve T cells revealed that IL-17 secretion in vitro did not correlate with SFB colonization in DC donor mice (Fig. S1C). Overall, these data suggest that lung DCs were not directly influenced by gastrointestinal microbiota and additional factors contribute to IL-17 production following pulmonary fungal infection.
To determine the role of serum factors on pulmonary Th17 cell accumulation, SFB-negative B6 Jackson mice were infected with A. fumigatus and then adoptively transferred with serum from naïve B6 Taconic or Il22−/− SFB-colonized mice 7 hours later. Some groups of mice received serum pre-mixed with an IL-1 receptor antagonist (IL-1RA; anakinra), as Il1α expression was upregulated in the gut of Il22−/− mice infected with A. fumigatus (Fig. 4C). Two days following infection, lung single cell suspensions were restimulated with PMA plus ionomycin and stained for intracellular IL-17A. Mice receiving sera from SFB colonized Il22−/− mice had increased numbers of CD4+ IL-17+ T cells (Fig. 5D). This was dependent on IL-1 receptor signaling, as pre-incubation of Il22−/− serum with IL-1RA significantly decreased Th17 cell numbers as well as TCR γδ+ IL-17+ cells. Notably, donor serum from Il22−/− mice contained elevated levels of IL-1α protein (Il22−/− serum: 958 +/− 198pg/mL IL-1α; B6 Taconic serum: 501 +/− 35pg/mL IL-1α), while IL-1β protein levels were similar to B6 Taconic (Il22−/− serum: 99 +/− 4pg/mL IL-1β; B6 serum: 107 +/− 3pg/mL IL-1β). In contrast to the role of IL-1R signaling, systemic IL-23 administration did not significantly affect IL-17 levels in the lung on day 2 (data not shown). Overall, these data demonstrate that IL-22 modulates serum IL-1 receptor ligands that augment lung Th17 cell accumulation during fungal infection.
Intestinal RegIIIγ delivery is anti-inflammatory during pulmonary fungal infection
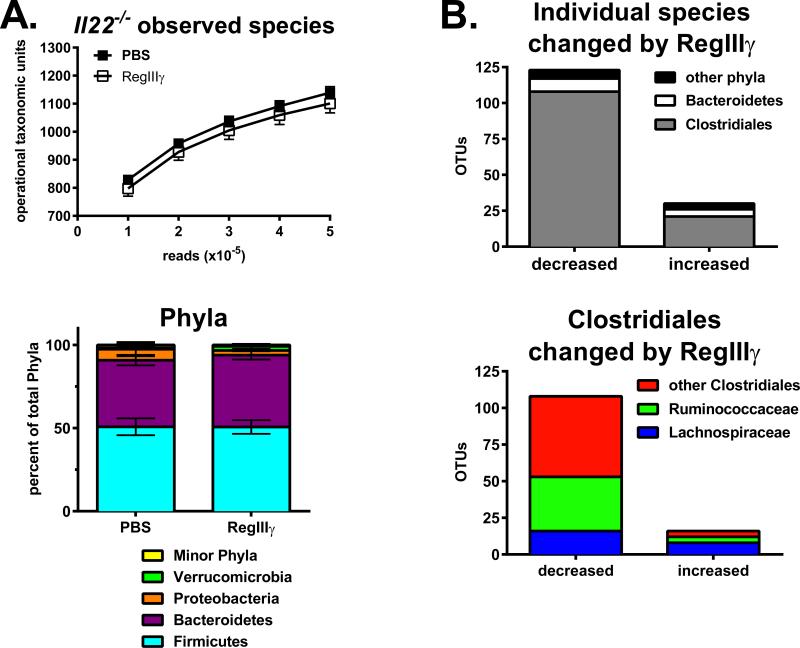
Next we tested if recombinant RegIIIγ can rescue the phenotype in Il22−/− mice, as RegIIIγ is an IL-22 inducible gene. To directly deliver RegIIIγ to the gut, mice were gavaged with the recombinant protein at 48-hour intervals from one to nine days prior to infection. A total of five treatments were given, and tissues were collected two days following infection. RegIIIγ gavage protected mice from weight-loss, losing only 4 percent body mass on day 1 while the group receiving PBS vehicle lost 9 percent (Fig. 7A). Aspergillus fumigatus burden was not significantly different between the two groups (Fig. 7B, A.f.), suggesting that rather than having a direct anti-fungal effect, RegIIIγ was anti-inflammatory in this model. Supporting this, expression of Il1α, Il1β, Il6 and Ifnγ were significantly reduced in lungs of mice receiving recombinant RegIIIγ (Fig. 7B), and the cytokines IL-1α, IL-1β and IL-6 were also decreased (Fig. 7C). Surprisingly, IL-17 protein levels were comparable between RegIIIγ-treated and control mice. Analysis of the small intestine and cecum revealed that RegIIIγ had no effect on SFB colonization (data not shown). Fluorescence activated cell sorting was used to sort RegIIIγ+ and RegIIIγ− fractions. This was followed by PCR which revealed that endogenous RegIIIγ does not preferentially bind to SFB-enriched microbiota (Kumar, unpublished observations). Overall, these findings complement our data demonstrating that SFB colonization correlates with lung Th17 responses. To characterize bacteria species that may contribute to weight loss during fungal infection, 16S rDNA sequencing was performed on cecal contents. Exogenous RegIIIγ preferentially decreased species in the Clostridiales order and had no discernable effect on Bacteroidetes (Fig. 8). Many of the species affected were non-Lachnospiraceae including Ruminococcaceae. Our data demonstrates that RegIIIγ delivery to the gut reduces the expression of lung inflammatory genes during fungal infection.
Figure 8. Selective depletion of Clostridiales Ruminococcaceae by RegIIIγ treatment.
DNA from cecal contents was isolated from Il22−/− mice used in Figure 7, and used as a template for bacterial 16S rDNA PCR analysis, as described previously (63). Amplicons were sequenced using Illumina MiSeq, and analyzed for operational taxonomic units (OTUs) at the phylum, class, order and family levels. A. Total number of OTUs defined as species in PBS versus RegIIIγ treatment groups (top), and the distribution of major phyla (bottom). B. Total number of individual species that were changed by ≥2-fold in response to RegIIIγ treatment categorized by phyla or order as indicated (top), and total number of individual species within Clostridiales that were changed by RegIIIγ treatment categorized by family, as indicated (bottom). Data are combined from two experiments with n=8-10.
SFB colonization augments Th17 immunity during pulmonary fungal infection
Our data suggests that colonization with SFB, rather than host genotype for RegIIIγ or Il22, determines the strength of pulmonary Th17 cell priming. To address this, we compared B6 mice that were SFB-high versus SFB-low. Routine screening revealed a cohort of Taconic mice that were SFB-negative upon arrival, in contrast to previous experiments using this vendor. This cohort was gavaged with SFB-high fecal suspensions from other Taconic donors in house and rested for two weeks on normal or vancomycin water. For comparison, B6 mice from Jackson Labs (SFB-low) were also used. One day prior to infection, mice were neutrophil-depleted. Results demonstrated that vancomycin water significantly decreased lung Th17 cell numbers in SFB-high Taconic mice, but not SFB-low Jackson, mice (Fig. S3). Accordingly, vancomycin significantly decreased SFB colonization in Taconic mice. Although C. leptum was also decreased by vancomycin, there were no differences in C. leptum colonization between Taconic and Jackson mice (Fig. S3). This provides further evidence that intestinal SFB colonization promotes lung Th17 immunity.
To confirm that SFB-enriched microbiota augments lung Th17 immunity, vancomycin-treated mice were gavaged with fecal suspensions from Taconic donors (SFB-high) or Jackson donors (SFB-low). Three weeks following microbiota transfer, mice were neutrophil depleted and infected, as described in Materials and Methods. Transfer of SFB-enriched microbiota into Jackson recipients significantly increased total numbers of IL-17+ and CD4+ IL-17+ cells in the lungs (Fig. S4A). In contrast, transferring SFB-low microbiota from Jackson donors into Taconic recipients did not significantly impact Th17 cell numbers (Fig. S4B). Altogether, our data demonstrate a link between intestinal SFB colonization and pulmonary Th17 immunity.
Discussion
Commensal microbiota support the development of immune responses by stimulating pattern recognition receptors. By targeting growth pathways that are shared among classes of bacteria, antibiotics decrease intestinal species diversity, which modulates the immune tone of the gut (36). For instance, metronidazole depletes Clostridium species which induce the development of colonic Tregs (28, 37). On the other hand, vancomycin depletes Gram-positive bacteria and decreases the population of small intestinal Th17 cells (38). Here, we show that oral vancomycin has a significant effect on lung IL-17 production during an acute fungal infection (Fig. 1). Antibiotics have also been shown to inhibit Th17-mediated asthma (39). These data demonstrate that antibiotics, while required for treating intestinal infections, have systemic anti-inflammatory effects that can inhibit responses to secondary infections. In support, oral vancomycin renders mice susceptible to vancomycin-resistant Enterococci through the downregulation of antimicrobial proteins including RegIIIγ (40). We demonstrate that another consequence of RegIIIγ depletion may lead to increased accumulation of Th17 cells following pulmonary challenges (Fig. 2). This was attributed to commensal bacteria including SFB, as the phenotype observed in RegIIIγ−/− mice was restored with vancomycin drinking water. Thus, our data show that modulating the balance of intestinal bacteria species significantly affects T cell polarization at non-local sites during infections.
Beneficial effects of commensal bacteria include stimulating the development of intestinal lymphoid tissue, anti-microbial protein secretion and lymphocyte activation. It is not surprising that GF or commensal-depleted animals are susceptible to intestinal infections; however, they also succumb to systemic, skin and respiratory pathogens (12, 13, 41, 42). Two mechanisms have been proposed to explain the role of commensals in systemic inflammation (43). First, innate immune agonists such as bacterial cell wall products may directly provide an adjuvant effect. Second, chronic low-level stimulation of APCs by commensal products, termed tonic signaling, epigenetically programs them to respond robustly to a future infection (12, 13). We observed that vancomycin-sensitive commensals condition splenic DCs to produce increased levels of inflammatory cytokines in response to fungal products (Fig. 6), providing evidence for epigenetic programming. Further, Quintin, et al. demonstrated that a primary fungal infection trains monocytes for increased cytokine production during a secondary infection (44). In our model, lung DCs from vancomycin-treated mice were not hyporesponsive to A. fumigatus (Fig. S1D), and it remains possible that commensals provide an acute adjuvant effect. Future studies will be aimed to decipher the mechanism of how Gram-positive commensals amplify lung IL-17 production.
Secretion of RegIIIγ into the small intestine promotes the physical separation of luminal bacteria from epithelium (20). Therefore, RegIIIγ may block DC stimulation with commensal products or microbial translocation across the GI tract. SFB directly adhere to intestinal villi and provide an adjuvant effect to restore small intestinal Th17 cells in GF mice (21, 22, 45). Since RegIIIγ was found to inhibit mucosa-associated SFB colonization (20), this may explain why small intestinal CD4 T cells, including Th17 and Th1 cells, are significantly increased in RegIIIγ−/− mice following i.p. immunization (Fig. 3). It is not known if RegIIIγ has direct bactericidal activity against SFB or if it inhibits colonization indirectly. Nevertheless, several inflammatory diseases are associated with a compromised intestinal epithelial barrier. Microbial translocation is increased during inflammatory bowel disease, chronic HIV infection, hepatitis B or C infection and pancreatitis, contributing to systemic immune activation (46). Interleukin-22 is critical for epithelial repair following injury (19, 47), and production of IL-22 by innate lymphoid cells prevents microbial translocation in the steady-state (48). This suggests that intestinal IL-22 activity may have systemic anti-inflammatory effects. We found that Il22−/− mice have increased IL-17 in the lungs and small intestine during acute fungal infection, compared to WT mice (Fig. 4). This phenotype was vancomycin-sensitive, correlated with SFB colonization, and was transferrable with intestinal microbiota (Fig. 5B), demonstrating that IL-22 was modulating microbial composition in the gut. Overall, cumulative evidence points to a model suggesting that IL-22 helps to prevent systemic immune activation by inhibiting SFB colonization, blocking microbial translocation and decreasing pulmonary Th17 cell polarization towards fungal antigens.
The cellular sources of IL-17 during infections may influence its physiological effects. Deletion of integrin αvβ8 on CD11c+ cells inhibits Ag-specific Th17 responses and protects against airway hyperresponsiveness independently of airway inflammation (49). Interleukin-17 produced by CD4 T cells, not γδ T cells, enhances airway smooth muscle contraction. In our study, SFB colonization led to preferential accumulation of Th17 cells into lung tissue, and had no effect on the number of γδ T cells making IL-17 (Figs. 5A, 5B). Further studies are necessary to define the role of SFB colonization on airway hyperresponsiveness.
The regulation and function of RegIIIγ in vivo is compartmentalized. In Paneth cells, IL-22 upregulates RegIIIγ mRNA (19, 48, 50) while stimulation of the MyD88 pathway induces RegIIIγ release from secretory granules in an inactive pro-form (51, 52). Following its secretion into the intestine, trypsin cleaves 11 amino acids from the N-terminus of RegIIIγ, resulting in a 15 kD isoform with potent bactericidal activity (52). In lung epithelium, IL-6 upregulates RegIIIγ expression during Staphylococcus aureus infection (53). We showed that RegIIIγ neutralization delays S. aureus clearance, while recombinant RegIIIγ delivery to the lungs accelerates clearance. Although the cleaved 15 kD isoform is detectable in bronchoalveolar lavage fluid, the enzyme responsible for RegIIIγ cleavage in lungs is not known. Thus, IL-22 and IL-6 control RegIIIγ expression in the gut and lungs, respectively. Since both of these cytokines activate the STAT3 transcription factor, the compartmentalized regulation of RegIIIγ expression may be determined by the local release of STAT3-activating cytokines and/or epithelial receptor expression. The regulation of RegIIIγ at multiple checkpoints is one way the innate immune system controls bacteria colonization on mucosal surfaces.
The role of RegIIIγ in host defense is pathogen-specific. Although RegIIIγ binds to the surface of A. fumigatus, it does not significantly affect fungal viability at pH 7 (data not shown). This is similar to what has been reported for other fungi (18). Following infection, RegIIIγ did not significantly affect pulmonary fungal clearance (Figs. 2A, 7B, S2). Our finding that vancomycin drinking water decreases lung Th17 cell numbers in RegIIIγ−/− mice demonstrates that RegIIIγ expression is not sufficient to regulate Th17 immunity (Fig. 2D). The increased Th17 cell numbers in RegIIIγ−/− mice compared to B6 controls was due to microbiota differences (Fig. 2). Thus, when using SFB-negative strains, fungal clearance and IL-17 levels were comparable in B6 and RegIIIγ−/− mice (Fig. S2). Our data suggest that RegIIIγ did not play a direct role in pulmonary anti-fungal immunity.
Here we demonstrated that differences in microbiota between strains of mice can impact pulmonary immunity. Certain bacteria including SFB may stimulate the systemic release of IL-1 receptor ligands, augmenting lung Th17 cell accumulation during fungal infections. The source of IL-1 receptor ligands remains an interesting question. We observed that SFB-enriched microbiota increased the expression of Il1β in lung tissue (Fig. 5E), but did not affect the number of lung CD11c+ CD11b+ cells producing IL-1α or IL-1β (Fig. S1E). This suggests that non-DCs may provide a source of IL-1 receptor ligands during infection. It also remains possible that T cell responsiveness to IL-1 receptor ligands might be regulated by microbiota. Our work complements a study showing that increased activity of the NLRP3 inflammasome in response to a high-fat diet leads to IL-17-dependent airway hyperreactivity (54). The authors found IL-1β produced by macrophages drives the accumulation of ILC3 cells that mediate disease through IL-17. Our findings suggest that microbiota can influence susceptibility to autoimmune disease. Animal studies have identified commensals that provoke or prevent autoimmunity by modulating the balance of Th17 cells and Tregs (55, 56). Intestinal Tregs recognize microbial antigens (57, 58), and increase the ratio of Firmicutes to Bacteroidetes species (59), suggesting that T cells can in turn influence microbial composition. Thus, commensal organisms have integral roles in the development of local and systemic immune responses.
Intestinal SFB colonization may regulate pulmonary host defense through various mechanisms, including anti-microbial protein secretion, immune cell activation and recruitment. A recent study found that SFB protects against acute Staphylococcus aureus pneumonia (60). SFB-colonized mice had significantly increased levels of IL-6 and IL-22, as well as neutrophil cell numbers, in bronchoalveolar lavage. In their study, IL-22 action in the lung was critical for host defense against S. aureus (60). Our data shows that IL-22 action in the gut may also regulate lung immunity by modulating the microbiota. This is consistent with other studies measuring the impact of IL-22 on gut microbiota (61, 62). Although we provide evidence that systemic IL-1R ligands contribute to pulmonary Th17 immunity (Fig. 5D), more studies are necessary to fully define the link between SFB colonization and host defense.
Supplementary Material
Footnotes
Research reported in this publication was supported by the NIAID NIH Award Number R37-HL079142 to J.K.K., R01 DK070855 to L.V.H. and J.P.M. was partially supported by F32AI096772. L.V.H. is also supported by the Howard Hughes Medical Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations: B6, C57BL/6; DC, dendritic cell; GF, germ-free; o.p., oropharyngeal; RegIIIγ, Regenerating islet-derived III-gamma; SFB, segmented filamentous bacteria; siLP, small intestinal lamina propria; Treg, regulatory T cell; WT, wild-type.
Disclosures
W.O. is an employee of Genentech; the authors have no additional financial interests.
References
- 1.Chen K, Kolls JK. T cell-mediated host immune defenses in the lung. Annu. Rev. Immunol. 2013;31:605–633. doi: 10.1146/annurev-immunol-032712-100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Latge JP. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cenci E, Mencacci A, Bacci A, Bistoni F, Kurup VP, Romani L. T cell vaccination in mice with invasive pulmonary aspergillosis. J. Immunol. 2000;165:381–388. doi: 10.4049/jimmunol.165.1.381. [DOI] [PubMed] [Google Scholar]
- 4.Wuthrich M, Gern B, Hung CY, Ersland K, Rocco N, Pick-Jacobs J, Galles K, Filutowicz H, Warner T, Evans M, Cole G, Klein B. Vaccine-induced protection against 3 systemic mycoses endemic to North America requires Th17 cells in mice. J. Clin. Invest. 2011;121:554–568. doi: 10.1172/JCI43984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhary N, Marr KA. Impact of Aspergillus fumigatus in allergic airway diseases. Clin. Transl. Allergy. 2011;1:4. doi: 10.1186/2045-7022-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romani L, Fallarino F, De Luca A, Montagnoli C, D'Angelo C, Zelante T, Vacca C, Bistoni F, Fioretti MC, Grohmann U, Segal BH, Puccetti P. Defective tryptophan catabolism underlies inflammation in mouse chronic granulomatous disease. Nature. 2008;451:211–215. doi: 10.1038/nature06471. [DOI] [PubMed] [Google Scholar]
- 7.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr. Opin. Immunol. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 9.Morrissey PJ, Charrier K, Braddy S, Liggitt D, Watson JD. CD4+ T cells that express high levels of CD45RB induce wasting disease when transferred into congenic severe combined immunodeficient mice. Disease development is prevented by cotransfer of purified CD4+ T cells. J. Exp. Med. 1993;178:237–244. doi: 10.1084/jem.178.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int. Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 11.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat. Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West CE. Gut microbiota and allergic disease: new findings. Curr Opin Clin. Nutr. Metab. Care. 2014;17:261–266. doi: 10.1097/MCO.0000000000000044. [DOI] [PubMed] [Google Scholar]
- 16.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat. Rev. Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 17.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 20.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 22.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 2007;204:1891–1900. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, Danilenko DM, Valdez P, Kasman I, Eastham-Anderson J, Wu J, Ouyang W. Interleukin-22, a T(H)17 cytokine, mediates IL-23-induced dermal inflammation and acanthosis. Nature. 2007;445:648–651. doi: 10.1038/nature05505. [DOI] [PubMed] [Google Scholar]
- 25.Cash HL, Whitham CV, Hooper LV. Refolding, purification, and characterization of human and murine RegIII proteins expressed in Escherichia coli. Protein Expr. Purif. 2006;48:151–159. doi: 10.1016/j.pep.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat. Protoc. 2007;2:2307–2311. doi: 10.1038/nprot.2007.315. [DOI] [PubMed] [Google Scholar]
- 27.Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect. Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov, Umesaki Y, Itoh K, Honda K. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 33.Lee JB, Jang JE, Song MK, Chang J. Intranasal delivery of cholera toxin induces th17-dominated T-cell response to bystander antigens. PLoS One. 2009;4:e5190. doi: 10.1371/journal.pone.0005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narushima S, Sugiura Y, Oshima K, Atarashi K, Hattori M, Suematsu M, Honda K. Characterization of the 17 strains of regulatory T cell-inducing human-derived Clostridia. Gut microbes. 2014;5:333–339. doi: 10.4161/gmic.28572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med. 2012;209:251–258. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012;33:459–466. doi: 10.1016/j.it.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, Finlay BB, Littman DR. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemaire MM, Dumoutier L, Warnier G, Uyttenhove C, Van Snick J, de Heusch M, Stevens M, Renauld JC. Dual TCR expression biases lung inflammation in DO11.10 transgenic mice and promotes neutrophilia via microbiota-induced Th17 differentiation. J. Immunol. 2011;187:3530–3537. doi: 10.4049/jimmunol.1101720. [DOI] [PubMed] [Google Scholar]
- 40.Brandl K, Plitas G, Mihu CN, Ubeda C, Jia T, Fleisher M, Schnabl B, DeMatteo RP, Pamer EG. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U S A. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abt MC, Artis D. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr. Opin. Microbiol. 2013;16:4–9. doi: 10.1016/j.mib.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quintin J, Saeed S, Martens JH, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C, Joosten LA, Xavier RJ, van der Meer JW, Stunnenberg HG, Netea MG. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis CP, Savage DC. Habitat, succession, attachment, and morphology of segmented, filamentous microbes indigenous to the murine gastrointestinal tract. Infect. Immun. 1974;10:948–956. doi: 10.1128/iai.10.4.948-956.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu. Rev. Immunol. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 2008;14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, Shibata N, Grunberg S, Sinha R, Zahm AM, Tardif MR, Sathaliyawala T, Kubota M, Farber DL, Collman RG, Shaked A, Fouser LA, Weiner DB, Tessier PA, Friedman JR, Kiyono H, Bushman FD, Chang KM, Artis D. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kudo M, Melton AC, Chen C, Engler MB, Huang KE, Ren X, Wang Y, Bernstein X, Li JT, Atabai K, Huang X, Sheppard D. IL-17A produced by alphabeta T cells drives airway hyper-responsiveness in mice and enhances mouse and human airway smooth muscle contraction. Nat. Med. 2012;18:547–554. doi: 10.1038/nm.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukherjee S, Partch CL, Lehotzky RE, Whitham CV, Chu H, Bevins CL, Gardner KH, Hooper LV. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J. Biol. Chem. 2009;284:4881–4888. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi SM, McAleer JP, Zheng M, Pociask DA, Kaplan MH, Qin S, Reinhart TA, Kolls JK. Innate Stat3-mediated induction of the antimicrobial protein Reg3gamma is required for host defense against MRSA pneumonia. J. Exp. Med. 2013;210:551–561. doi: 10.1084/jem.20120260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HY, Lee HJ, Chang YJ, Pichavant M, Shore SA, Fitzgerald KA, Iwakura Y, Israel E, Bolger K, Faul J, Dekruyff RH, Umetsu DT. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat. Med. 2014;20:54–61. doi: 10.1038/nm.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochoa-Reparaz J, Mielcarz DW, Wang Y, Begum-Haque S, Dasgupta S, Kasper DL, Kasper LH. A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol. 2010;3:487–495. doi: 10.1038/mi.2010.29. [DOI] [PubMed] [Google Scholar]
- 56.Wu HJ, Ivanov, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32:815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cebula A, Seweryn M, Rempala GA, Pabla SS, McIndoe RA, Denning TL, Bry L, Kraj P, Kisielow P, Ignatowicz L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, Santacruz N, Peterson DA, Stappenbeck TS, Hsieh CS. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Josefowicz SZ, Niec RE, Kim HY, Treuting P, Chinen T, Zheng Y, Umetsu DT, Rudensky AY. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gauguet S, D'Ortona S, Ahnger-Pier K, Duan B, Surana NK, Lu R, Cywes-Bentley C, Gadjeva M, Shan Q, Priebe GP, Pier GB. Intestinal Microbiota of Mice Influences Resistance to Staphylococcus aureus Pneumonia. Infect. immun. 2015;83:4003–4014. doi: 10.1128/IAI.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, Fu YX, Zhou L. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J. Immunol. 2013;190:5306–5312. doi: 10.4049/jimmunol.1300016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.