Summary
Introduction
The discovery of disease-associated loci through genome-wide association studies (GWAS) is the leading approach to the identification of novel biological pathways for human disease. To date, GWAS have had been limited by relatively small sample sizes and yielded relatively few loci associated with ischemic stroke The National Institute of Neurological Disorders Stroke Genetics Network (NINDS-SiGN) is an international consortium that has taken a systematic approach to phenotyping and produced the largest ischemic stroke GWAS to date.
Methods
In order to identify genetic loci associated with ischemic stroke, we performed a two-stage genome-wide association study. The first stage consisted of 16,851 cases with state-of-the-art phenotyping and 32,473 stroke-free controls. Cases were aged 16 to 104 years, recruited between 1989 and 2012, and subtyped by centrally trained and certified investigators using the web-based protocol, Causative Classification of Stroke (CCS). We constructed case-control strata by identify samples genotyped on (nearly) identical arrays and of similar genetic ancestral background. Data was cleaned and imputed using dense imputation reference panels generated from whole-genome sequence data. Genome-wide testing was performed within each stratum for each available phenotype, and summary level results were combined using inverse variance-weighted fixed effects meta-analysis. The second stage consisted of in silico look-ups of 1,372 SNPs in 20,941 cases and 364,736 stroke-free controls, with cases previously subtyped using the TOAST classification system according to local standards. The two stages were then jointly analyzed in a final meta-analysis.
Findings
We identified a novel locus at 1p13.2 near TSPAN2 associated with large artery atherosclerosis (LAA)-related stroke (stage I OR for the G allele at rs12122341 = 1·21, p = 4.50 × 10−8; stage II OR = 1·19, p = 1·30 × 10−9). We also confirmed four loci robustly associated with ischemic stroke and reported in prior studies, including PITX2 and ZFHX3 for cardioembolic stroke, and HDAC9 for LAA stroke. The 12q24 locus near ALDH2, originally associated with all ischemic stroke but not with any specific subtype, exceeded genome-wide significance in the meta-analysis of small artery stroke. Other loci, including NINJ2, were not confirmed.
Interpretation
Our results identify a novel LAA-stroke susceptibility gene and now indicate that all loci implicated by GWAS to date are subtype specific. Follow-up studies will be necessary to determine whether the locus near TSPAN2 yields a novel therapeutic approach to stroke prevention. Given the subtype-specificity of these associations, the rich phenotyping available in SiGN is likely to prove vital for further genetic discovery in ischemic stroke.
Funding
National Institute of Neurological Disorders and Stroke (NINDS), National Institutes of Health (NIH).
Introduction
Worldwide, stroke is the second leading cause of death and a major contributor to dementia and age-related cognitive decline. Globally, approximately 15 million people suffer a stroke each year, with an increasing number of stroke deaths annually.1 Most survivors are left with permanent disability, making stroke the world’s leading cause of adult incapacity as well.2 Strokes result from the sudden occlusion or rupture of a blood vessel supplying the brain, and are accordingly categorized as ischemic (vessel occlusion) or hemorrhagic (vessel rupture) on the basis of neuroimaging. Ischemic cases account for up to 85% of all strokes.
Although hypertension, atrial fibrillation, diabetes mellitus, and cigarette smoking are known risk factors for stroke,3 a substantial proportion of risk remains unexplained and may be due to inherited genetic variation. Discovering genetic variants predisposing to stroke is a vital first step toward the development of improved diagnostics and novel therapies that offer the hope of reducing the disease burden. Genome-wide association studies (GWAS) have thus far yielded only a handful of confirmed loci,4–7 which together account for a small proportion of the heritable risk.8
Ischemic stroke occurs when blood flow to a brain region is interrupted due to blockage of a blood vessel. Because vessel occlusion can occur through a variety of mechanisms, ischemic stroke can be classified based on presumed mechanism into specific subtypes: large artery atherosclerosis (LAA), cardioembolism (CE) and small artery occlusion (SAO). All but one GWAS association for ischemic stroke have been subtype-specific, indicating the need for studies better powered to detect subtype-specific associations. The National Institute of Neurological Disorders and Stroke (NINDS) Stroke Genetics Network (NINDS-SiGN)9 is the largest and most comprehensive GWAS of stroke and its subtypes to date. We sought to detect newly associated polymorphisms and to confirm previously reported associations with risk of ischemic stroke and its subtypes.
Methods
We performed a two-stage joint association analysis of ischemic stroke and its subtypes. Stage I consisted of a GWAS, followed by an in silico association analysis of top SNPs in independent samples in stage II; both stages were then jointly analyzed to identify loci exceeding genome-wide significance. Compared to separate discovery and replication analyses, this approach improves power for discovery while maintaining equivalent type I error.10
Study sample
For stage I, ischemic stroke cases with consistent neuroimaging and adequate clinical data to allow phenotypic classification were included from 31 existing collections. The ischemic stroke cases in stage II met similar requirements except existing TOAST subtyping was used for phenotypic classification. Details for each collection, including funding information and study design, can be found in the Supplementary Note.
For each collection, approval for inclusion in the SiGN analysis complied with local ethical standards and with local institutional review board/ethics committee oversight. All cases and controls provided informed consent for genetic studies either directly or through surrogate authorization.
Stroke subtype classification
The NINDS-SiGN9 utilized two subtyping systems: the recently developed Causative Classification of Stroke (CCS) system, a standardized web-based subtype classification system,11 and the more widely used Trial of Org 10172 in Acute Stroke Treatment (TOAST) subtype classification system12,13. Both of these subtyping systems are based on a similar conceptual framework but are operationalized differently. The TOAST subtyping system is based on application of written rules requiring clinician judgment, and patients with conflicting potential etiologies are placed into an undetermined category. The CCS subtyping system classifies patients algorithmically based on inputs to a web-based form and has two different approaches to classifying patients with conflicting potential etiologies. The CCS system generates both causative (CCSc) and phenotypic (CCSp) subtype categories. CCSc categorization utilizes historical, examination, and test data from each ischemic stroke subject to assign the most probable cause in the presence of competing etiologies; CCSp categorization utilizes abnormal test findings to assign each case into one or more major etiologic groups without using rules to determine the most likely etiology.
For stage I, each site assigned stroke subtypes using the CCS system (Supplementary Note). For stage II, we identified additional sites having subtyped stroke cases with GWAS data. Since all available CCS cases were included in stage I, we used the corresponding subtype categories from TOAST in stage II.
For both CCS and TOAST, each case was categorized according to five ischemic stroke subtypes: cardioembolic (CE), large artery atherosclerosis (LAA), small artery occlusion (SAO), undetermined (UNDETER), and other. The “other” classification was also available but ultimately not analyzed due to low sample counts and limited power. For semantic convenience, we use the term “undetermined” in this manuscript to describe similar categories in both CCS and TOAST. However, in CCS we are referring to cryptogenic cases in which no cause was identified after adequate evaluation, while TOAST undetermined cases included those with incomplete evaluation, multiple causes, and the truly cryptogenic.
Quality control
The full details of genotyping and quality control (QC) are provided in the Supplementary Note and outlined in Supplementary Figure 1. Briefly, newly-genotyped cases and a small number of controls were genotyped on the Illumina 5M array for inclusion in stage I analyses. All other cases had been previously genotyped on various Illumina platforms (Supplementary Note). Publicly-available external controls were selected to match cases based on ancestral background and genotyping array.
Cases and controls newly genotyped together formed separate analysis groups (Krakow and Leuven, Table 1). The remaining cases and controls were matched based on genotyping platform to maximize SNP content and pool samples from the same cohort or geographic region (Table 1). Merged cases and controls were assigned ancestry-specific analysis strata in two steps (Supplementary Note). Samples were projected onto HapMap 314 data using PCA to establish a group of European-ancestry samples (EUR). Then, a hyperellipsoid clustering technique was implemented on the basis of PCs within self-reported groups of non-Hispanic black and Asian participants. The hyperellipsoid analysis established a group of non-Hispanic black (AFR) and one of Asian participants. Samples not grouped as EUR, AFR, or Asian, formed the Hispanic (HIS) stratum. Asian-ancestry samples were excluded from further analysis due to small sample size. After establishing the ancestry-based composite groups, we performed PCA again to confirm ancestral homogeneity within each case-control strata. Case-control strata then underwent extensive QC (Supplementary Note). Finally, each stratum was prephased15 and imputed. EUR samples were imputed using a merged reference panel comprised of the 1000 Genomes Project (1KG) Phase I16 and the Genome of the Netherlands17; AFR and HIS samples were imputed using the 1KG Phase I reference panel only. Summary-level imputed data from one additional cohort (VISP) was added to the stage I meta-analysis.
Table 1. Case and control cohorts included in the two-stage NINDS SiGN stage I and stage II.
Stage I case cohorts were matched to external controls based on genotyping array, cohort, and ancestry. Alternating shading indicates separate analysis case-control groups constructed for stage I analyses from contributing cohorts (which were primarily case- or control-only cohorts). Hispanic samples were an exception and are not shown as a separate group here, as limited samples required that we pool all available Hispanic samples into a single analysis stratum. Stage II consisted of in silico SNP look-ups of summary-level results in (previously analyzed) case-control sets.
| Cohort | Location | Array | Ancestry | Cases | Controls |
|---|---|---|---|---|---|
| Stage I | |||||
| BRAINS | U.K. | 650Q | EUR | 267 | – |
| MGH-GASROS | U.S.A. | 610 | EUR | 111 | – |
| ISGS | U.S.A. | 610 | EUR | 351 | – |
| SWISS | U.S.A. | 610 | EUR | 25 | – |
| HABC | U.S.A. | 1M | EUR | – | 1,586 |
| EDIN | U.K. | 660 | EUR | 566 | – |
| MUNICH | U.K. | 660 | EUR | 1,131 | – |
| OXVASC | U.K. | 660 | EUR | 457 | – |
| STGEORGE | U.K. | 660 | EUR | 418 | – |
| KORA | Germany | 550 | EUR | – | 804 |
| WTCCC | U.K. | 660 | EUR | – | 5,150 |
| GEOS | U.S.A. | 1M | AFR, EUR | 843 | 880 |
| BRAINS | U.K. | 5M | EUR, HIS | 110 | – |
| MGH-GASROS | U.S.A. | 5M | AFR, EUR, HIS | 456 | – |
| GCNKSS | U.S.A. | 5M | AFR, EUR, HIS | 482 | – |
| ISGS | U.S.A. | 5M | AFR, EUR, HIS | 178 | – |
| MCISS | U.S.A. | 5M | AFR, EUR, HIS | 619 | – |
| MIAMISR | U.S.A. | 5M | AFR, EUR, HIS | 294 | – |
| NHS | U.S.A. | 5M | EUR, HIS | 314 | – |
| NOMAS | U.S.A. | 5M | AFR, EUR, HIS | 358 | – |
| REGARDS | U.S.A. | 5M | AFR, EUR, HIS | 304 | – |
| SPS3 | The Americas, Spain | 5M | AFR, EUR, HIS | 949 | – |
| SWISS | U.S.A. | 5M | AFR, EUR, HIS | 181 | – |
| WHI | U.S.A. | 5M | AFR, EUR, HIS | 454 | – |
| WUSTL | U.S.A. | 5M | AFR, EUR, HIS | 449 | – |
| HRS | U.S.A. | 2.5M | AFR, EUR, HIS | – | 11,174 |
| OAI | U.S.A. | 2.5M | AFR, EUR | – | 3,882 |
| HCHS/SOL | U.S.A. | 2.5M | HIS | – | 1,214 |
| KRAKOW | Poland | 5M | EUR, HIS | 880 | 717 |
| LEUVEN | Belgium | 5M | EUR, HIS | 460 | 453 |
| BASICMAR | Spain | 5M | EUR, HIS | 890 | – |
| ADHD | Spain | 1M | EUR | – | 411 |
| INMA | Spain | 1M | EUR | – | 807 |
| GRAZ | Austria | 610 | EUR | – | 815 |
| GRAZ | Austria | 5M | EUR | 609 | – |
| SAHLSIS | Sweden | 5M | EUR, HIS | 783 | – |
| LUND | Sweden | 5M | EUR, HIS | 613 | – |
| MDC1 | Sweden | 610 | EUR, HIS | 211 | 1,362 |
| ASGC | Australia | 610 | EUR | 1,109 | 1,200 |
| VISP | U.S.A., Canada, Scotland | 1M | AFR, EUR | 1,979 | – |
| Melanoma Study | U.S.A. | 1M | EUR | – | 1,047 |
| HANDLs | U.S.A. | 1M | AFR | – | 971 |
| Total | – | – | – | 16,851 | 32,473 |
| Stage II | |||||
| ARIC | U.S.A. | Affy 6.0 | AFR | 263 | 2,466 |
| CADISP | Multi-cohort | Illumina 610 | EUR | 555 | 9,259 |
| CHARGE2 | Multi-cohort | Multi-chip | EUR | 3,100 | 75,530 |
| CHS | U.S.A. | Illumina Omni 1M | AFR | 110 | 623 |
| deCODE | Iceland | Multi-chip | EUR | 5,291 | 228,512 |
| GLASGOW | U.K. | ImmunoChip | EUR | 599 | 1,775 |
| HVH | U.S.A. | Illumina 370CNV | EUR | 577 | 1,330 |
| INTERSTROKE | Multi-cohort | Cardio-metabochip | AFR, EAS, EUR, HIS | 1,771 | 2,103 |
| LUND | Sweden | 635 | EUR | 546 | 528 |
| MDC | Sweden | 5M | EUR | 1,304 | 3,504 |
| METASTROKE2 | Multi-cohort | Multi-chip | EUR | 1,729 | 7,925 |
| RACE | Pakistan | 660 | SAS | 2,385 | 5,193 |
| SAHLSIS | Sweden | 750 | EUR | 299 | 596 |
| SIFAP | Germany | 2.5M | EUR | 981 | 1,825 |
| SIGNET-REGARDS | U.S.A. | Affy 6.0 | AFR | 258 | 2,094 |
| SWISS/ISGS | U.S.A. | Illumina 610 or 660 | AFR | 173 | 389 |
| UTRECHT | The Netherlands | ImmunoChip | EUR | 556 | 1,145 |
| VHIR-FMT-BARCELONA | Spain | HumanCore ExomeChip | EUR | 545 | 320 |
| WGHS3 | U.S.A. | Human Hap300 and custom array | EUR | 440 | 22,725 |
| Total | – | – | – | 21,482 | 367,842 |
| Joint | – | – | – | 38,333 | 400,315 |
Only TOAST subtypes available for stage I
Contributing cohorts are described in the Supplementary Note
Not included in the ischemic stroke and CE analyses due to overlap with CHARGE
EUR, European ancestry; AFR, African ancestry; HIS, Hispanic; EAS, East Asian; SAS = South Asian. Cohort abbreviations: BRAINS, Biorepository of DNA in Stroke; MGH-GASROS, Massachusetts General Hospital – Genes Affecting Stroke Risk and Outcome Study; ISGS, Ischemic Stroke Genetics Study; SWISS, Siblings with Ischemic Stroke Study; HABC, Health ABC; EDIN, Edinburgh Stroke Stoke; OXVASC, Oxford Vascular Study; STGEORGE, St. George’s Hospital; KORA, MONICA/KORA Ausburg Study; WTCCC, Wellcome Trust Case Control Consortium; GEOS, Genetics of Early Onset Stroke; GCNKSS, Greater Cincinnati/Northern Kentucky Stroke Study; MCISS, Middlesex County Ischemic Stroke Study; MIAMISR, Miami Stroke Registry and Biorepository; NHS, Nurses’ Health Study; NOMAS, Northern Manhattan Study (NOMAS); REGARDS, Reasons for Geographic and Racial Differences in Stroke; SPS3, Secondary Prevention of Small Subcortical Strokes; WHI, Women’s Health Initiative; WUSTL, Washington University St. Louis; HRS, Health and Retirement Study; OAI, Osteoarthritis Initiative; HCHS/SOL, The Hispanic Community Health Study/Study of Latinos; LEUVEN, Leuven Stroke Genetics Study; BASICMAR, Base de Datos de Ictus del Hospital del Mar; ADHD, Attention-deficit Hyperactivity Disorder; INMA, Infancia y medio ambiente; SAHLSIS, Sahlgrenska Academy Study of Ischemic Stroke; LUND, Lund Stroke Registry; MDC, Malmo Diet and Cancer Study; ASGC, Australian Stroke Genetics Collaborative; VISP, Vitamin Intervention for Stroke Prevention; HANDLs, Health/Aging in Neighborhoods of Diversity across the Lifespan Study; ARIC, Atherosclerosis Risk in Communities Study; CADISP, Cervical Artery Dissection and Ischemic Stroke Patients; CHARGE, Cohorts for Aging and Research in Genetic Epidemiology; CHS, Cardiovascular Health Study; HVH, Heart and Vascular Health Study; GLASGOW, Glasgow ImmunoChip Study; RACE, Risk Assessment of Cardiovascular Events; SIFAP, Stroke in Young Fabry Patients; SIGNET, The Sea Island Genetics Network; UTRECHT, Utrecht ImmunoChip Study/PROMISe Study; WGHS, Women’s Genome Health Study.
Stage I genome-wide association analysis
After QC and imputation, 16,851 cases and 32,473 controls across 15 ancestry-specific groups were available for genome-wide testing (Table 1, Supplementary Note). Within each stratum, we analyzed the all ischemic stroke phenotype, as well as four primary subtypes (CE, LAA, SAO and UNDETER) as determined by CCSc, CCSp, and TOAST, which was available in 12,612 (74.8%) cases. All GWAS were adjusted for sex and the top ten principal components; genome-wide testing was uncorrected for age, as age information was missing for the bulk of controls.
Post GWAS, SNPs with frequency < 1% showed excessive genomic inflation and were consequently removed. Imputed SNPs were checked for consistent frequency with the continental populations represented in 1KG Phase I; SNPs with a frequency difference >30% were removed. After post-GWAS QC, 9·3M – 15·4M SNPs were available across the study strata for meta-analysis. We performed inverse variance-weighted fixed effects meta-analysis using MANTEL18 in each of the 15 traits. Lambda of the 15 meta-analyses ranged from 0·936 – 1·005 (Supplementary Figure 2).
The first stage GWAS revealed 1,372 SNPs in 268 loci associated with ischemic stroke or a specific subtype in any of the CCS or TOAST traits at p < 1 × 10−6.
Genetic correlation among CCSc, CCSp, and TOAST subtypes
We then extracted the z-scores (SNP betas divided by their respective standard error) from each of the stage I GWAS phenotypes and calculated correlation (Pearson’s r) between pairings of z-scores, calculating the correlation for all possible trait pairings. The analysis revealed moderate to strong genetic correlation (Figure 1) between the standardized SNP effects in CCSc, CCSp, and TOAST, despite previously noted modest phenotypic correlation.19 The observed genetic correlation indicated that TOAST subtyping was appropriate for inclusion in the second stage of analysis.
Figure 1. Genetic and phenotypic correlation of CCS Causative, CCS Phenotypic, and TOAST subtyping methods in stage I analyses.
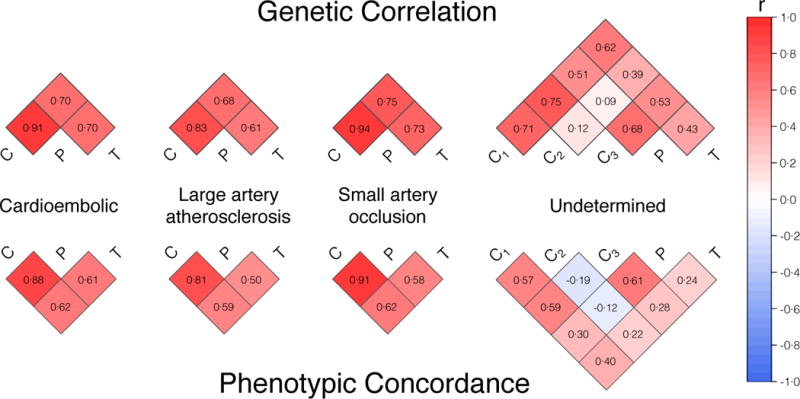
All cases with an available CCS subtype were included in stage I analyses. Genome-wide z-scores from the CCS Causative (C), CCS Phenotypic (P), and TOAST (T) GWAS were checked for correlation between each possible pair of traits. The moderate to strong genetic correlation within subtypes indicated that additional TOAST-subtyped cases were suitable for follow-up analyses. Phenotypic correlations were also strong within subtype-specific clusters. (Top) Pearson’s r correlation coefficients (mathematically equivalent in this scenario to the Lin’s concordance correlation coefficient) are printed within each square to indicate genetic correlation. (Bottom) Cohen’s kappas are printed within each square to indicate phenotypic agreement. C1, all undetermined (CCS Causative); C2, incomplete and unclassified (CCS Causative); C3, cryptogenic and cardioembolic minor (CCS Causative). The C2 and C3 classifications are mutually exclusive.
Stage II analysis
Stage II consisted of an in silico look-up of association results for the stage I nominally significant in 18 independent studies, totaling 20,941 TOAST-subtyped cases and 364,736 controls (Table 1 and Supplementary Table 1). The SNPs selected for stage II for each subtype were aggregated such that, e.g., SNPs with p < 1 × 10−6 from the three CE GWAS (CCSc, CCSp, and TOAST) were all selected for lookup in the independent CE TOAST cases and matched controls. This process was repeated for the other subtypes.
Joint analysis
Results from the in silico lookups from stage II were meta-analyzed with the results from stage I. Genome-wide significance in joint analysis was set at p < 1 × 10−8, after correcting for testing five subtypes. Lambda in the ischemic stroke joint analysis was 1·005 and ranged from 0·936 – 0·998 in the subtype analyses (Supplementary Figure 3).
The SiGN study was a cooperative agreement with the United States National Institute of Neurological Disorders and Stroke (NINDS). Although the NINDS participated in the design of the study, the study investigators were solely responsible for the data collection, analysis, and interpretation. The analysis team had full access to all data included in the study.
Results
After extensive data QC (Supplementary Figure 1, Supplementary Note), 16,851 stroke cases and 32,473 controls comprised the stage I sample; an additional independent set of 20,941 cases and 364,736 controls comprised stage II, enabling joint analysis of a combined 37,893 cases and 400,315 controls across five primary (independent) traits (IS, and the subtypes CE, LAA, SAO, and UNDETER).
In the joint analysis of CCS (stage I) and TOAST (stage II) results, SNPs in two novel loci exceeded genome-wide significance (p < 1 × 10−8 after correcting for five independent traits). Four common SNPs in LD (r2 > 0·57, 1KG European-ancestry (EUR) samples) near TSPAN2 were genome-wide significant for LAA (rs12122341, CCSp (stage I) and TOAST (stage II): OR for the G allele = 1·19, p = 1·30 × 10−9; Figure 2a, Table 2).
Figure 2. Forest plot and regional association plot of rs12122341.
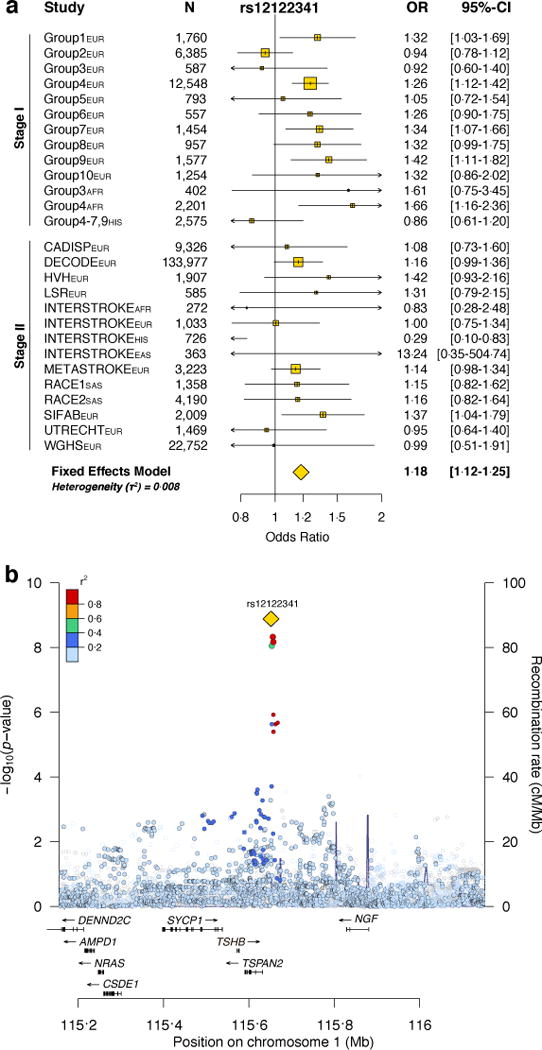
(a) Rs12122341 was associated to large artery atherosclerosis (LAA) subtype in joint analysis of CCS Phenotypic cases and controls (stage I) and TOAST-subtyped LAA cases and their matched controls (stage II). (b) Rs12122341 lies on chromosome 1 near the TSPAN2 locus. EUR, European-ancestry; AFR, African-ancestry; HIS, Hispanic; EAS, East Asian ancestry; SAS, South Asian ancestry.
Table 2. Novel and previously-identified loci implicated in ischemic stroke and its subtypes through genome-wide testing.
For subtype-specific loci, ORs and their corresponding p-values are reported for the CCS Causative (C), CCS Phenotypic (P), and TOAST (T) subtypes. Risk allele frequency (RAF) was computed using 1000 Genomes (Phase I) European-ancestry samples (EUR), African-ancestry samples (AFR), and samples from the Americas (AMR). Association results were looked up in TOAST-subtyped cases and their matched controls and meta-analyzed with stage I results from CCS Causative, CCS Phenotypic, and TOAST cases. IS, ischemic stroke; LAA, large artery atherosclerosis; UND, undetermined; CE, cardioembolic; SAO, small artery occlusion.
| Trait | SNP | Chr | Risk allele | RAF (%) | Nearest Gene | Stage I | Stage II | Joint analysis | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EUR | AFR | AMR | Cases-C | OR [95% CI] | P-value | Cases-T | OR [95% CI] | P-value | Cases-C | OR [95% CI] | P | |||||
| Cases-P | Cases-P | P | ||||||||||||
| Cases-T | Cases-T | P | ||||||||||||
| Novel loci | ||||||||||||||
| LAA | rs12122341 | 1 | G | 25·7 | 8·8 | 19·5 | TSPAN2 | 2,454 | 1·20 [1·12 – 1·29] | 3·38 × 10−7 | 2,249 | 1·15 [1·04 – 1·26] | 5·25 × 10−3 | 4,703 | 1·18 [1·12 – 1·25] | 8·32 × 10−9 |
| 2,715 | 1·21 [1·13 – 1·30] | 4·50 × 10−8 | 4,964 | 1·19 [1·12 – 1·26] | 1·30 × 10−9 | |||||||||
| 2,346 | 1·15 [1·07 – 1·24] | 1·61 × 10−4 | 4,595 | 1·15 [1·08 – 1·22] | 2·70 × 10−6 | |||||||||
| UND | rs74475935 | 16 | G | 0·2 | 1·8 | 0·6 | ABCC1 | 2,392* | 5·17 [2·99 – 8·92] | 3·69 × 10−9 | 3,469 | 1·87 [0·55 – 6·41] | 3·16 × 10−1 | 5,861 | 4·63 [2·77 – 7·72] | 4·70 × 10−11 |
| 1,062* | 8·68 [4·55 – 16·58] | 5·94 × 10−11 | 4,531 | 6·89 [3·80 – 12·47] | 1·85 × 10−10 | |||||||||
| 3,593 | 2·18 [1·16 – 4·10] | 1·58 × 10−2 | 7,062 | 2·11 [1·20 – 3·70] | 9·22 × 10−3 | |||||||||
| Previously-identified loci, stage I p < 1 × 10−6 | ||||||||||||||
| IS | rs10744777 | 12 | T | 66·7 | 4·5 | 5·2 | ALDH2 | 16,851 | 1·10 [1·06 – 1·13] | 3·07 × 10−8 | 21,042 | 1·05 [1·01 – 1·08] | 6·55 × 10−3 | 37,893 | 1·07 [1·5 – 1·09] | 4·20 × 10−9 |
| IS | rs2634074 | 4 | T | 2·1 | 4·8 | 4·1 | PITX2 | 16,851 | 1·09 [1·06 – 1·13] | 2·56 × 10−7 | 21,042 | 1·10 [1·07 – 1·14] | 2·00 × 10−8 | 37,893 | 1·10 [1·07 – 1·12] | 2·68 × 10−14 |
| IS | rs2107595 | 7 | A | 15·7 | 2·2 | 2·2 | HDAC9 | 16,851 | 1·10 [1·06 – 1·14] | 7·74 × 10−7 | 21,042 | 1·07 [1·03 – 1·11] | 1·70 × 10−4 | 37,893 | 1·09 [1·06 – 1·12] | 8·60 × 10−10 |
| CE | rs2200733 | 4 | T | 12·0 | 2·2 | 2·6 | PITX2 | 3,071 | 1·39 [1·28 – 1·50] | 1·24 × 10−16 | 3,991 | 1·36 [1·26 – 1·46] | 1·21 × 10−16 | 7,062 | 1·37 [1·30 – 1·45] | 1·04 × 10−29 |
| 3,695 | 1·39 [1·29 – 1·49] | 3·26 × 10−19 | 7,686 | 1·37 [1·30 – 1·45] | 2·79 × 10−32 | |||||||||
| 3,427 | 1·37 [1·27 – 1·48] | 1·02 × 10−16 | 7,418 | 1·36 [1·29 – 1·44] | 8·05 × 10−30 | |||||||||
| CE | rs7193343 | 16 | T | 17·4 | 2·4 | 18·9 | ZFHX3 | 3,071 | 1·17 [1·09 – 1·26] | 1·12 × 10−5 | 3,991 | 1·15 [1·07 – 1·23] | 7·93 × 10−5 | 7,062 | 1·17 [1·10 – 1·22] | 7·28 × 10−9 |
| 3,695 | 1·19 [1·11 – 1·27] | 2·93 × 10−7 | 7,686 | 1·17 [1·11 – 1·23] | 2·29 × 10−10 | |||||||||
| 3,427 | 1·17 [1·09 – 1·25] | 1·45 × 10−5 | 7,418 | 1·16 [1·10 – 1·22] | 8·88 × 10−9 | |||||||||
| LAA | rs11984041 | 7 | T | 9·3 | 2·2 | 6·7 | HDAC9 | 2,454 | 1·30 [1·18 – 1·42] | 8·46 × 10−8 | 2,249 | 1·15 [1·03 – 1·29] | 1·16 × 10−2 | 4,703 | 1·23 [1·15 – 1·33] | 1·10 × 10−8 |
| 2,715 | 1·29 [1·18 – 1·42] | 3·50 × 10−8 | 4,964 | 1·24 [1·15 – 1·33] | 4·52 × 10−9 | |||||||||
| 2,346 | 1·30 [1·17 – 1·43] | 3·62 × 10−7 | 4,595 | 1·23 [1·14 – 1·33] | 4·48 × 10−8 | |||||||||
| SAO | rs10744777 | 12 | T | 66·7 | 4·5 | 5·2 | ALDH2 | 2,736 | 1·19 [1·11 – 1·27] | 9·10 × 10−7 | 2,426 | 1·12 [1·03 – 1·21] | 4·66 × 10−3 | 5,162 | 1·16 [1·10 – 1·22] | 2·77 × 10−8 |
| 2,734 | 1·20 [1·12 – 1·28] | 6·82 × 10−8 | 5,160 | 1·17 [1·11 – 1·23] | 2·92 × 10−9 | |||||||||
| 3,147 | 1·13 [1·06 – 1·21] | 1·05 × 10−4 | 5,573 | 1·13 [1·07 – 1·18] | 1·62 × 10−6 | |||||||||
| Previously-identified loci, stage I p > 1 × 10−6 | ||||||||||||||
| IS | rs34166160 | 12 | A | 0·9 | 0·0 | 0·3 | NINJ2 | 16,851 | 1·20 [0·96 – 1·48] | 1·06 × 10−1 | ||||||
| IS | rs11833579 | 12 | G | 75·8 | 79·4 | 68·0 | NINJ2 | 16,851 | 1·02 [0·95 – 1·01] | 2·15 × 10−1 | ||||||
| IS | rs505922 | 9 | C | 35·1 | 32·6 | 23·5 | ABO | 16,851 | 1·07 [1·04 – 1·10] | 2·03 × 10−5 | ||||||
| CE | rs505922 | 9 | C | 35·1 | 32·6 | 23·5 | ABO | 3,071 | 1·04 [0·98 – 1·10] | 1·88 × 10−1 | ||||||
| 3,695 | 1·04 [0·98 – 1·10] | 1·62 × 10−1 | ||||||||||||
| 3,427 | 1·08 [1·02 – 1·15] | 5·66 × 10−3 | ||||||||||||
| LAA | rs505922 | 9 | C | 35·1 | 32·6 | 23·5 | ABO | 2,454 | 1·09 [1·02 – 1·17] | 6·93 × 10−3 | ||||||
| 2,715 | 1·11 [1·04 – 1·18] | 1·29 × 10−3 | ||||||||||||
| 2,346 | 1·14 [1·06 – 1·21] | 2·15 × 10−4 | ||||||||||||
| LAA | rs556621 | 6 | T | 29·1 | 8·1 | 40·7 | 6p21 | 2,454 | 1·04 [0·97 – 1·11] | 3·18 × 10−1 | ||||||
| 2,715 | 1·02 [0·95 – 1·19] | 6·36 × 10−1 | ||||||||||||
| 2,346 | 1·11 [1·04 – 1·19] | 2·55 × 10−3 | ||||||||||||
| LAA | rs2383207 | 9 | G | 49·9 | 4·5 | 41·3 | CDKN2B-AS1 | 2,454 | 1·12 [1·05 – 1·19] | 4·34 × 10−4 | ||||||
| 2,715 | 1·11 [1·05 – 1·19] | 7·93 × 10−4 | ||||||||||||
| 2,346 | 1·09 [1·02 – 1·17] | 8·13 × 10−3 | ||||||||||||
Results from the CCS cryptogenic phenotype
A second locus emerged as genome-wide significant, but only in samples of African ancestry, and thus must be interpreted with marked caution given the small sample sizes in which it was found. Rs74475935 in ABCC1 on chromosome 16 was associated with the undetermined phenotype (Table 2, Supplementary Figure 5), driven by a variant with rare frequency (MAF ~0·01%) in European-ancestry samples and low frequency (MAF ~1·5%) in African-ancestry samples. This result requires further replication in larger samples.
We also confirmed previously published loci PITX24 and ZFHX35 for CE stroke, and HDAC96 for LAA stroke, all exceeding genome-wide significance in these samples (Table 2). The 12q24·12 locus near ALDH2, previously shown to be associated with all ischemic stroke but not with any specific subtype,7 exceeded genome-wide significance in the joint analysis of all ischemic stroke (OR for the T allele = 1·07, p = 4·20 × 10−9). However, the association was even stronger for SAO in CCSp (stage I) and TOAST (stage II) (OR = 1·17, p = 2·92 × 10−9) and was nearly genome-wide significant for SAO in the joint analysis of CCSc (stage I) and TOAST (stage II) (OR = 1·16, p = 2·77 × 10−8). Evidence for association was markedly reduced with other subtypes in our study (OR < 1·1 and p > 4 × 10−3 for CE, LAA, and undetermined in the combined CCSp and TOAST analysis. Systematic testing accounting for shared controls (Supplementary Note) revealed a nominally significant difference in the magnitude of the OR between SAO and the combined non-SAO subtypes (p = 0·048, Supplementary Figure 6), suggesting that the effect of 12q24·12 may be specific for SAO.
In contrast, we failed to show even nominal evidence for association to NINJ2, previously implicated in ischemic stroke (rs34166160, OR for the A allele = 1·20, p = 0·106), though we had 100% power in our sample size to detect a nominal association (p < 0.05) at the locus. In the full stage I analysis, nominal evidence for association was observed for both the 6p2120 and CDKN2B-AS121 loci in LAA, and for the ABO22 locus in IS, LAA, and CE (Table 2). When restricting our analysis to only those samples not used for initial discovery, CDKN2B-AS1 was nominally associated with LAA (OR for the G allele = 1·09, p = 0·009) and ABO was nominally associated to ischemic stroke (OR for the C allele = 1·07, p = 2·5 × 10−4), LAA (OR = 1·15, p = 2·5 × 10−4) and CE (OR = 1·09, p = 0·007; Supplementary Table 2). For 6p21, however, we observed no evidence for association (OR for the T allele = 1·04, p = 0·304).
Discussion
We performed the largest GWAS of ischemic stroke and stroke subtypes to date. Our results reveal a novel association with LAA. The lead SNP, rs12122341, is located in an intergenic region 23.6kb upstream of TSPAN2, the gene encoding tetraspanin-2 (Figure 2b). The lead SNP is in LD with intronic and UTR variants in TSPAN2 (r2 > 0·3, 1KG EUR) but it is located in a DNA sequence immediately adjacent to TSPAN2 that can be bound by several transcription factor proteins, including CTCF. In fact, this sequence is a promotor and enhancer site that is marked by histone modification and DNase hypersensitivity according to ENDCODE and ROADMAP Epigenomics experimental data (Supplementary Figure 7)23,24, suggesting a potential role for this SNP in gene regulation. An intergenic SNP near rs12122341 was recently reported to be associated with migraine25, but the two SNPs are not in LD (r2 = 0·03, 1KG EUR).
TSPAN2, the gene closest to rs12122341, is a member of the transmembrane 4 (tetraspanin) superfamily. It mediates signal transduction to regulate cell development, activation, growth and motility. TSPAN2 knock-out mice exhibit increased neuroinflammation, indicated by activation of microglia and astrocytes with no effect on myelination and axon integrity.26 Notably, TSPAN is highly expressed in artery and whole blood cells (Supplementary Figure 7), which aligns with our observed association of TSPAN2 with LAA stroke.
Whether the association of rs12122341 arises to the locus’ regulation of TSPAN2 or other nearby genes will require further functional evaluation.
The additional association in undetermined stroke (rs74475935) is in a gene-rich region with LD-paired SNPs (r2 > 0·1, 1KG African-ancestry samples) stretching up to 4 Mb. Due to the low sample size for rs74475935 (610 cases) and the dearth of African-ancestry samples available, studies that explicitly interrogate large samples of African descent are necessary to fully evaluate the robustness of this signal.
Thus far, only four loci – PITX24, ZFHX35, HDAC96, and 12q24·127 – have been repeatedly identified in GWAS of ischemic stroke, all subtype specific except for 12q24·12. Although the 12q locus association was originally discovered for IS, our analysis indicates it is likely specific to SAO. These findings suggest that ischemic stroke subtypes carry distinct genetic signatures. Our analysis of genetic correlation across the traits, however, also revealed that the subtypes share subtle genetic relationships (Supplementary Figure 8, Supplementary Table 3a), an observation supported by a recent study that identified genetic overlap between the LAA and SAO subtypes.27 Future efforts will help dissect both the shared and unique genetic architectures within and between subtypes.
To date, GWAS of ischemic stroke, subtypes (all associations thus far have been subtype-specific), have utilized far smaller sample sizes than studies performed in other complex traits. The SiGN study, the largest to date, was well powered (75·1%) to find common SNP subtype-specific associations of larger effect (MAF = 25%, OR = 1·2, in 3,000 cases and 30,000 controls) but markedly less well powered to find lower-frequency or lower-effect SNPs (MAF 10% and OR 1·2: 13·8% power; MAF 25% and OR 1·1: 1·1% power). Because of the quasi-linear relationship between sample size and discovered loci,28 and because large-scale GWAS in other complex traits have yielded hundreds of SNP-disease associations,29–31 studying larger samples in ischemic stroke subtypes will likely yield additional associated common variants. Furthermore, the implementation of whole genome sequencing studies in stroke will begin to test whether rare alleles in the population account for a substantial proportion of disease heritability.
Despite its overall large sample size, the SiGN study has several limitations (in addition to the power considerations discussed above). First, sample inclusion is heavily biased towards individuals of European descent; inclusion of non-European populations will improve power for locus discovery32 and be especially informative for future fine-mapping efforts.33 Second, the inclusion of TOAST-based stage II samples likely added phenotypic heterogeneity (Figure 1, Supplementary Table 3b), potentially reducing power.19 Third, many of the participating studies within SiGN (and in particular the publicly-available controls) had limited or no stroke-specific risk factor data available. Such data is key to disentangling potential gene-environment interactions. Future genetic studies of stroke will continue to face additional challenges related to the phenotype, including high prevalence of the disease (lifetime risk ~20%), its late onset (primarily > 65 years), the contribution of other cardiovascular disease and environment to its etiology, and difficulties subtyping (in SiGN 12.6 – 22.3% of all cases analyzed were ultimately classified as undetermined by CCS or TOAST).
Our use of CCS enabled identification of candidate SNPs that did not reach significance for stage II follow-up in TOAST, including those SNPs at the TSPAN2 locus. This refinement may reflect a reduction in phenotypic heterogeneity that CCS introduces through its capture of clinical stroke features, completeness of diagnostic investigations, and, where possible, classification of cases with multiple potential etiologies into the most probable causes. The association signal of the CCS-discovered TSPAN2 locus was, however, improved upon inclusion of TOAST samples, suggesting that leveraging the genetic correlation underlying the subtyping methods and allowing for broader inclusion of cases, regardless of subtyping system, can yield discovery of more susceptibility loci. Further studies will determine whether the rich repository of individual-level data created through the use of the CCS will uncover novel phenotypes, revealing biological mechanisms and broadening our understanding of stroke’s genetic architecture.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization. WHO | The top 10 causes of death. 2014 http://www.who.int/mediacentre/factsheets/fs310/en/. (accessed April 23, 2015)
- 2.Mozaffarian D, Benjamin EJ, Go AS, et al. Heart Disease and Stroke Statistics–2015 Update: A Report From the American Heart Association. Circulation. 2014;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 3.Meschia JF, Bushnell C, Boden-Albala B, et al. Guidelines for the Primary Prevention of Stroke: A Statement for Healthcare Professionals From the American Heart Association/American Stroke Association. Stroke. 2014;45:3754–832. doi: 10.1161/STR.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gretarsdottir S, Thorleifsson G, Manolescu A, et al. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–9. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellenguez C, Bevan S, Gschwendtner A, et al. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–33. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kilarski LL, Achterberg S, Devan WJ, et al. Meta-analysis in more than 17,900 cases of ischemic stroke reveals a novel association at 12q24.12. Neurology. 2014;83:678–85. doi: 10.1212/WNL.0000000000000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bevan S, Traylor M, Adib-Samii P, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43:3161–7. doi: 10.1161/STROKEAHA.112.665760. [DOI] [PubMed] [Google Scholar]
- 9.Meschia JF, Arnett DK, Ay H, et al. Stroke Genetics Network (SiGN) study: design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke. 2013;44:2694–702. doi: 10.1161/STROKEAHA.113.001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 11.Ay H, Benner T, Arsava EM, et al. A computerized algorithm for etiologic classification of ischemic stroke: The causative classification of stroke system. Stroke. 2007;38:2979–84. doi: 10.1161/STROKEAHA.107.490896. [DOI] [PubMed] [Google Scholar]
- 12.Adams H, Bendixen B, Kappelle L, et al. Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 13.Kolominsky-Rabas PL, Weber M, Gefeller O, Neundoerfer B, Heuschmann PU. Epidemiology of ischemic stroke subtypes according to TOAST criteria: incidence, recurrence, and long-term survival in ischemic stroke subtypes: a population-based study. Stroke. 2001;32:2735–40. doi: 10.1161/hs1201.100209. [DOI] [PubMed] [Google Scholar]
- 14.Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.1000 Genomes Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Francioli LC, Menelaou A, Pulit SL, et al. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 18.de Bakker PIW, Ferreira MAR, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122–8. doi: 10.1093/hmg/ddn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McArdle PF, Kittner SJ, Ay H, et al. Agreement between TOAST and CCS ischemic stroke classification: the NINDS SiGN study. Neurology. 2014;83:1653–60. doi: 10.1212/WNL.0000000000000942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holliday EG, Maguire JM, Evans T-J, et al. Common variants at 6p21.1 are associated with large artery atherosclerotic stroke. Nat Genet. 2012;44:1147–51. doi: 10.1038/ng.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith JG, Melander O, Lövkvist H, et al. Common genetic variants on chromosome 9p21 confers risk of ischemic stroke a large-scale genetic association study. Circ Cardiovasc Genet. 2009;2:159–64. doi: 10.1161/CIRCGENETICS.108.835173. [DOI] [PubMed] [Google Scholar]
- 22.Williams FMK, Carter AM, Hysi PG, et al. Ischemic stroke is associated with the ABO locus: the EuroCLOT study. Ann Neurol. 2013;73:16–31. doi: 10.1002/ana.23838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward LD, Kellis M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esserlind AL, Christensen AF, Le H, et al. Replication and meta-analysis of common variants identifies a genome-wide significant locus in migraine. Eur J Neurol. 2013;20:765–72. doi: 10.1111/ene.12055. [DOI] [PubMed] [Google Scholar]
- 26.De Monasterio-Schrader P, Patzig J, Möbius W, et al. Uncoupling of neuroinflammation from axonal degeneration in mice lacking the myelin protein tetraspanin-2. Glia. 2013;61:1832–47. doi: 10.1002/glia.22561. [DOI] [PubMed] [Google Scholar]
- 27.Holliday EG, Traylor M, Malik R, et al. Genetic overlap between diagnostic subtypes of ischemic stroke. Stroke. 2015;46:615–9. doi: 10.1161/STROKEAHA.114.007930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood AR, Esko T, Yang J, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–86. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulit SL, Voight BF, de Bakker PIW. Multiethnic genetic association studies improve power for locus discovery. PLoS One. 2010;5:e12600. doi: 10.1371/journal.pone.0012600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaitlen N, Paşaniuc B, Gur T, Ziv E, Halperin E. Leveraging Genetic Variability across Populations for the Identification of Causal Variants. Am J Hum Genet. 2010;86:23–33. doi: 10.1016/j.ajhg.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


