Abstract
Unlike most bacteria, Streptococcus pneumoniae (pneumococcus) has two evolutionarily distinct ABC transporters (Pst1 and Pst2) for inorganic phosphate (Pi) uptake. The genes encoding a two-component regulator (PnpRS) are located immediately upstream of the pst1 operon. Both the pst1 and pst2 operons encode putative PhoU-family regulators (PhoU1 and PhoU2) at their ends. This study addresses why S. pneumoniae contains dual Pi uptake systems and the regulation and contribution of the Pst1 and Pst2 systems in conditions of high (mM) Pi amount and low (μM) Pi amount. We show that in unencapsulated mutants, both pst1 and pst2 can be deleted, and Pi is taken up by a third Na+/Pi co-transporter, designated as NptA. In contrast, either pst1 or pst2 is unexpectedly required for the growth of capsule producing strains. We used a combination of mutational analysis, transcript level determinations by qRT-PCR and RNA-Seq, assays for cellular PnpR~P amounts by SDS-PAGE, and pulse-Pi uptake experiments to study the regulation of Pi uptake. In high Pi medium, PhoU2 serves as the master negative regulator of Pst2 transporter function and PnpR~P levels (post-transcriptionally). ΔphoU2 mutants have high PnpR~P levels and induction of the pst1 operon, poor growth, and sensitivity to antibiotics, possibly due to high Pi accumulation. In low Pi medium, Pst2 is still active, but PnpR~P amount and pst1 operon levels increase. Together, these results support a model in which pneumococcus maintains high Pi transport in high and low Pi conditions that is required for optimal capsule biosynthesis.
Keywords: PhoU, PnpRS two-component system (TCS), PnpR~P phosphorylation, Pst1 and Pst2 Pi ABC transporters, NptA Na+/Pi co-transporter
Introduction
Phosphorus is an essential element in all cells because of its structural and metabolic roles in nearly all biological processes, including the composition of nucleic acids, phospholipids, and energy intermediates. A preferred source of phosphorous for bacterial cells is environmental inorganic orthophosphate (; Pi). The mechanism of extracellular Pi uptake has been studied intensively in Escherichia coli and Bacillus subtilis as model organisms (Hulett, 1993; Takemaru et al., 1996; Wanner, 1996; Qi et al., 1997; Lamarche et al., 2008; Hsieh and Wanner, 2010; Botella et al., 2011, 2014), and recently in other bacterial species (Braibant et al., 1996; Gonin et al., 2000; Zaborina et al., 2008; Rifat et al., 2009; Shi and Zhang, 2010; Burut-Archanai et al., 2011; Cheng et al., 2012; Wang et al., 2013; de Almeida et al., 2015; Lubin et al., 2016). Generally, bacterial high-affinity Pi uptake systems consist of an ATP-binding cassette (ABC) transporter, designated as Pst (for phosphate-specific transporter), which contains at least four component subunits: an extracellular Pi binding protein (PstS), two transmembrane channel proteins (PstCA), and a cytoplasmic ATPase (PstB) (see Figure 1; Hsieh and Wanner, 2010). The expression of most bacterial Pst transporters is regulated at the transcriptional level by a two-component regulatory system (TCS), which has different designations in different bacteria (Hulett, 1993; Novak et al., 1999; Throup et al., 2000; Howell et al., 2006; Glover et al., 2007). Many bacteria also regulate Pi uptake by an ancillary negative regulatory protein, designated PhoU (Steed and Wanner, 1993; Botella et al., 2011, 2014; de Almeida et al., 2015; Lubin et al., 2016).
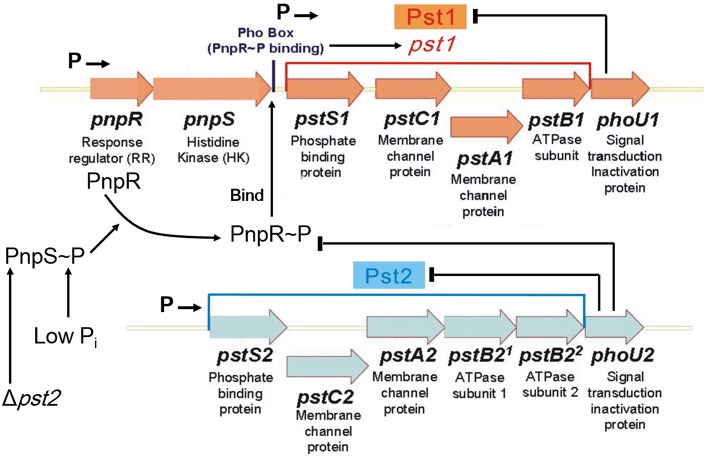
Figure 1.
Model for regulation of the dual Pst1 and Pst2 ABC Pi transporters in Streptococcus pneumoniae D39. Genomic sequencing and transcriptome analyses show that pnpRS, pst1-phoU1, and pst2-phoU2 are organized into three distinct operons that are transcribed from separate promoters, indicated by Ps followed by arrows (see text). The Pst2 ABC Pi transporter is constitutively expressed in both high and low Pi conditions. PhoU2 inhibits phosphorylation of the PnpR response regulator (RR) and negatively regulates the transport activity of Pst2. In the absence of PhoU2, PnpR is phosphorylated by the PnpS histidine kinase (HK). PnpR~P binds to the Pho box upstream of the pst1-phoU1 operon and activates transcription and expression of the genes encoding the Pst1 transporter and PhoU1 regulator; however, PnpR~P does not autoregulate its own (pnpRS) transcription. In contrast to PhoU2, PhoU1 negatively regulates the transport activity of Pst1, but does not regulate phosphorylation of PnpR. In the absence of the Pst2 transporter or in low Pi conditions, the PnpRS two component system (TCS) activates the expression of the genes encoding the Pst1 transporter and PhoU1 regulator. Either the Pst1 or Pst2 Pi transporter is required in encapsulated cells. Therefore, the regulated Pst1 and constitutive Pst2 Pi transport systems may act as a redundant failsafe to ensure that capsule biosynthesis is maintained during variations in Pi conditions. See the text for additional details.
In E. coli and related enterobacteria, the histidine kinase (HK) and response regulator (RR) that mediate Pi transport are designated as PhoR and PhoB, respectively, and the regulation of Pi uptake involves a PhoU regulator (Hsieh and Wanner, 2010; Gardner et al., 2014, 2015). Briefly, when [Pi] > 4.0 μM, the expression of the phoB-phoR regulator and pst transporter operons is inhibited by PhoU by a mechanism described below (Hsieh and Wanner, 2010). When the [Pi] is depleted to < 4.0 μM, PhoU releases inhibition of the PhoR HK and the PstB subunit of the transporter, allowing autophosphorylation of the PhoR HK, phosphoryl transfer to the PhoB RR, and activation of transcription by PhoB~P of operons in the phosphate (pho) regulon, including the pst transporter, the phoB-phoR regulator, and other operons involved in the uptake and assimilation of phosphorous-containing compounds (Wanner, 1996; Hsieh and Wanner, 2010). PhoB~P activates transcription by binding to the Pho box sequence upstream from the promoters of the regulon operons, including phoB-phoR, which provides autoregulation of the TCS proteins (Wanner, 1996; Martin, 2004; Lubin et al., 2016). Usually, the sequence of the consensus Pho box is different between Gram-negative (e.g., E. coli) and Gram-positive (e.g., B. subtilis) bacteria (Martin, 2004). Since the Pst transporter is not activated in E. coli at high Pi concentrations, this system is considered as a high-affinity transporter that works predominantly at low Pi concentrations (Wanner, 1996). PhoB/R, the Pst transporter, and members of the Pho regulon are important for virulence in many pathogenic Gram-negative bacteria, including E. coli, Vibrio cholerae, Proteus mirabilis, and Pseudomonas aeruginosa (Jacobsen et al., 2008; Lamarche et al., 2008; Zaborina et al., 2008; Pratt et al., 2010; Chekabab et al., 2014a,b). In P. aeruginosa, the PstS Pi binding protein also plays roles in adhesion and a Pi-independent role in biofilm formation (Zaborina et al., 2008; Neznansky et al., 2014; Shah et al., 2014).
PhoU is a negative regulator of Pho regulon expression in E. coli and many other bacteria (Muda et al., 1992; Wanner, 1996; Hsieh and Wanner, 2010; Gardner et al., 2014; de Almeida et al., 2015). Although, PhoU is an important regulator in many bacteria, it is notably absent from certain Gram-positive bacteria, including B. subtilis (Qi et al., 1997; Moreno-Letelier et al., 2011). phoU deletion in E. coli, P. aeruginosa, and Mycobacterium marinum leads to growth defects (Steed and Wanner, 1993; Wanner, 1996; Rice et al., 2009; Wang et al., 2013; de Almeida et al., 2015). In E. coli, this growth defect is reversed by deletion of the pst transporter operon or the phoBR TCS operon (Steed and Wanner, 1993; Wanner, 1996). These observations suggest that the growth defect of phoU mutants is caused by unregulated function of the Pst transport system, leading to excess Pi accumulation (Wanner, 1996; Rice et al., 2009). ΔphoU mutants also accumulate increased amounts of poly-orthophosphate (poly-Pi) in E. coli, M. marinum, P. aeruginosa, and Caulobacter crescentus (Morohoshi et al., 2002; Hirota et al., 2013; Wang et al., 2013; de Almeida et al., 2015; Lubin et al., 2016). Poly-Pi also accumulates in E. coli K-12 cells in stationary phase in high Pi medium, and this accumulation is correlated with inhibition of biofilm formation mediated by PhoB~P RR phosphorylation with acetyl-phosphate acting as donor (Grillo-Puertas et al., 2016). The rate of Pi uptake was reported to increase in an E. coli ΔphoU in one study (Rice et al., 2009), but not in another (Steed and Wanner, 1993). Besides defective growth, ΔphoU mutants exhibit higher sensitivity to a diverse range of antibiotics in E. coli, Mycobacterium tuberculosis, M. marinum, and P. aeruginosa (Li and Zhang, 2007; Shi and Zhang, 2010; Wang et al., 2013; de Almeida et al., 2015) and a defect in mutagenic DNA break repair in E. coli (Gibson et al., 2015).
Recent papers demonstrate that E. coli PhoU interacts with the PAS domain of the PhoR HK and with the PstB ATPase protein, in support of the dual inhibition of Pho regulon transcription and Pst mediated transport in E. coli (Gardner et al., 2014, 2015). Three crystal structures of PhoU-like proteins have been reported from Aquifex aeolicus, Thermotoga maritima, and P. aeruginosa (Liu et al., 2005; Oganesyan et al., 2005; Lee et al., 2014), showing that PhoU consists of two symmetric, three-alpha-helix bundles. However, these PhoU proteins showed several quaternary structures in crystals, including monomer, dimer, or hexamer packing (Liu et al., 2005; Oganesyan et al., 2005; Lee et al., 2014). Gel filtration shows that purified E. coli PhoU forms a dimer in solution (Gardner et al., 2014). In addition, divalent cation binding of magnesium and manganese is required for E. coli PhoU binding to the cytoplasmic side of the inner membrane and may play a role in formation of a ternary regulatory complex containing PhoU, PhoR, and PstB (Gardner et al., 2014). On the other hand, a recent study suggests that a different paradigm operates in C. crescentus, where PhoU does not modulate PhoR HK activity directly (Lubin et al., 2016). Instead, PhoU may negatively regulate the activity of the Pst transporter in response to Pi availability in C. crescentus (Lubin et al., 2016).
Besides Pst ABC transporters, Pi is taken up by symporter secondary transport systems. In E. coli, two additional Pi transporters, PitA and PitB, have been identified that are symporters of divalent cations, such as Mg2+ and Ca2+ (van Veen et al., 1994; Wanner, 1996; Harris et al., 2001; Jackson et al., 2008). PitA and PitB have been considered as low-affinity Pi transporters that predominantly function in high Pi environments (Wanner, 1996; Hsieh and Wanner, 2010). Moreover, expression of pitA is induced by Zn2+ addition (Jackson et al., 2008), suggesting that PitA may act primarily as a metal ion transporter instead of a Pi transporter (Beard et al., 2000).
Unlike E. coli, B. subtilis, or C. crescentus, which contain only one Pst transporter, Streptococcus pneumoniae (pneumococcus) encodes two evolutionarily distinct Pi ABC pump transporters, Pst1 and Pst2 (Figure 1; Lanie et al., 2007; Moreno-Letelier et al., 2011). The multigene pst1 and pst2 operons are located at different locations in the pneumococcus chromosome (Lanie et al., 2007). Only one phoBR-like TCS, pnpRS, encoding the PnpR RR and the PnpS HK, is encoded upstream of the pst1 operon, and both the pst1 and pst2 transporter operons encode separate PhoU-family regulators, designated PhoU1 and PhoU2 (Figure 1). The PnpRS TCS, Pst1 transporter, and PhoU1 were initially studied in unencapsulated S. pneumoniae laboratory strain R6x (Novak et al., 1999), before the discovery of the second Pst2 transporter and PhoU2 regulator. This work indicated that pnpRS operon expression was not regulated by Pi amount, and that mutants deficient in the PstB1 ATPase subunit seemed to show decreased Pi uptake in certain growth media (Novak et al., 1999). Subsequent work suggests that upregulation of pst1 operon expression is correlated with increased β-lactam antibiotic resistance in low-affinity pbp2x mutants and some clinical isolates of S. pneumoniae (Soualhine et al., 2005; Engel et al., 2014).
S. pneumoniae is a common commensal bacterium that primarily colonizes the human nasopharynx (Donkor, 2013; Chao et al., 2014; Hakansson et al., 2015; Short and Diavatopoulos, 2015), but can become an opportunistic pathogen, causing several serious respiratory and invasive diseases (Henriques-Normark and Tuomanen, 2013; Vernatter and Pirofski, 2013; Ferreira and Gordon, 2015; Gratz et al., 2015; Oliver and Swords, 2015). Therefore, the Pst1 and Pst2 transporters must mediate Pi acquisition from several niches with vastly different Pi concentrations in human hosts (Orihuela et al., 2001; Wilson, 2005, 2008). Signature-tagged mutagenesis (STM) screens and a study of the role of the PnpR RR (also called RR04) indicated that the pnpRS, pst1, and pst2 operons are all required for full pneumococcal virulence (Polissi et al., 1998; Throup et al., 2000; Hava and Camilli, 2002; McCluskey et al., 2004; Paterson et al., 2006; Trihn et al., 2013). Consistent with these earlier studies, a recent Tn-Seq study showed that PhoU1 is important for nasopharynx colonization, whereas PhoU2 is important for lung infection (van Opijnen and Camilli, 2012).
In this report, we studied the transcriptional and functional regulation of the pneumococcal Pst1 and Pst2 Pi transporters under growth conditions containing high (≈18 mM) or low (≈100 μM) concentrations of Pi. Our results show that pst2 operon transcription is constitutive, but Pst2 transporter activity is negatively regulated by PhoU2. In addition, PhoU2 negatively regulates PnpR RR phosphorylation and transcription of the pst1 operon at high concentrations of Pi. Therefore, PhoU2 resembles E. coli PhoU in that it functions in regulating the level of RR phosphorylation, besides modulating Pst2 transporter activity. In contrast, PhoU1 resembles C. crescentus PhoU in that it only regulates Pst1 transporter activity and does not modulate PnpRS TCS function. Our results also indicate that encapsulated S. pneumoniae requires the function of Pst1 or Pst2 for growth, whereas a symporter, named NptA, can provide sufficient Pi to allow the growth of unencapsulated mutants in high Pi conditions.
Materials and methods
Bacterial strains and growth conditions
Strains used in this study are listed in Table S1. Encapsulated strains were derived from parent strain IU1781 (D39 cps+ rpsL1), and unencapsulated strains were derived from parent strains IU1945 (D39 Δcps), IU1824 (D39 Δcps rpsL1), and IU3309 (D39 Δcps2E rpsL1), which are derivatives of serotype 2 S. pneumoniae strain D39 IU1690 (Lanie et al., 2007). Strains containing antibiotic markers were constructed by transformation of competent pneumococcal cells with linear DNA amplicons synthesized by overlapping fusion PCR (Ramos-Montanez et al., 2008; Tsui et al., 2010). Strains containing markerless alleles in native chromosomal loci were constructed using allele replacement via the Pc-[kan-rpsL+] (Janus cassette; Sung et al., 2001). Primers used to synthesize different amplicons are listed in Table S2. All constructs were confirmed by DNA sequencing of chromosomal regions corresponding to the amplicon region used for transformation. Bacteria were grown on plates containing trypticase soy agar II (modified; Becton-Dickinson) and 5% (vol/vol) defibrinated sheep blood (TSAII-BA). Plates were incubated at 37°C in an atmosphere of 5% CO2. For selections of transformants, TSAII-BA plates contained 250 μg/mL kanamycin, 0.3 μg/mL erythromycin, or 250 μg/mL streptomycin. Strains were cultured statically in Becton-Dickinson brain heart infusion (BHI) broth at 37°C in an atmosphere of 5% CO2, and growth was monitored by OD620 as described before (Tsui et al., 2016). Transformants were single-colony-isolated on TSAII-BA plates containing antibiotics twice before growth in antibiotic-containing BHI broth for storage (Tsui et al., 2016). All mutant constructs were confirmed by DNA sequencing of chromosomal regions corresponding to the amplicon region used for transformation.
Static growth in BHI broth, which contains ≈18 mM Pi, was used as a high Pi condition. For growth curves, strains were inoculated into 3 mL of BHI broth, serially diluted, and grown overnight. The next day, cultures with OD620 = 0.1–0.3 were diluted into 5 mL of fresh BHI broth to OD620 ≈ 0.002, and growth was monitored hourly. C+Y medium (Lacks and Hotchkiss, 1960) was used for studies of moderately low Pi condition. We determined that C+Y broth (no added Pi) already contains ≈1.5 mM Pi (see Results). A modified chemically defined medium (mCDM) (Carvalho et al., 2013) was used for moderate and low Pi conditions. To optimize growth, the concentrations of choline-HCl and all amino acids amounts were increased by 1000-fold and tyrosine was added to 100 mg/L compared to the CDM recipe in Carvalho et al. (2013). In addition, 40 mM MOPS buffer was added to mCDM, which was adjusted to a final pH = 7.4 with 10 M NaOH. mCDM contains 36.4 mM Pi(Carvalho et al., 2013). mCDM with no Pi was made by omitting KH2PO4 and K2HPO4 and adding KCl to 50.8 mM. mCDM media with 2 mM or 1 mM Pi was made by mixing mCDM and mCDM lacking Pi in a ratio of 2–34.4 or 1–35.4, respectively. mCDM with 1 mM Pi was diluted 10 or 100-fold with mCDM lacking Pi make mCDM with 100 μM and 10 μM Pi, respectively. For growth in mCDM, 3 mL overnight cultures were grown as described above, and the next day, cultures with OD620 = 0.1–0.3 were centrifuged (5125 × g, 5 min, 25°C), washed with 3 mL mCDM lacking Pi twice, and resuspended in 3 mL mCDM lacking Pi. Cells were then diluted in 5 mL of mCDM with 2, 1 mM, 100, 10 μM, or no Pi to OD≈0.005 and growth of static cultures in an atmosphere of 5% CO2 was monitored hourly at OD620.
Antibiotic disk diffusion assays
Overnight cultures were diluted and grown in 5 mL of BHI to OD620 ≈ 0.1. 100 μL of cultures were mixed with 3 mL of nutrient-broth soft agar [0.8% (w/v) nutrient broth and 0.7% (w/v) Bacto Agar (Difco)] and poured onto TSAII-BA plates. After 15 min, antibiotics disks were placed at the middle of plates, which were incubated at 37°C in an atmosphere of 5% CO2 overnight for 16 h. Diameters of zones of growth inhibition were measured with a ruler, and P-values were calculated by unpaired t-test in GraphPad Prism. Antibiotic disks were from Becton, Dickinson Co.: cefotaxime (30 μg); cefazolin (30 μg); cefamandole (30 μg); ceftazidime (30 μg); amdinocillin (10 μg); vancomycin (30 μg); gentamicin (120 μg); and tetracycline (30 μg).
RNA preparation, qRT-PCR, and RNA-seq analyses
To study high Pi conditions, overnight cultures were diluted and grown in 5 mL of BHI to OD ≈0.15. Cells were collected by centrifuging at 16,000 × g for 5 min at 4°C. 1 mL of RNApro solution (MP Biomedicals) was added to resuspend cell pellets. The suspension was transferred to a Lysing Matrix B tube (MP Biomedicals), which was shaken 3X in FastPrep homogenizer (6.0 M/s for 40 s each). Cell debris and lysing matrix were removed by centrifugation at 16,000 × g for 5 min at 4°C. 700 μL of supernates was transferred to a new microcentrifuge tube and incubated at room temperature for 5 min. 300 μL of chloroform was then added followed by incubation at room temperature for 5 min. Mixtures were centrifuged at 16,000 × g for 5 min at 4°C. 280 μL of the upper, aqueous phase was collected and mixed with 140 μL of 100% Ethanol in a new microcentrifuge tube for RNA precipitation. RNA purification was done using miRNeasy minikit (Qiagen), including on-column treatment with DNase I (Qiagen), following the manufacturer's instructions. 5 μg of purified RNA was treated by DNase from a DNA-free DNA removal kit (Ambion). 125 ng of treated RNA was used to synthesize cDNA by a qScript Felex cDNA synthesis kit (Quanta Biosciences). Synthesized cDNA was diluted 1:6 in water and then serially diluted 1:5 in water three more times. qRT-PCR reactions contained 10 μL of 2 × Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent), 2 μL of each 2 μM primers (Table S3), 0.3 μL of a 1:500 dilution of ROX reference dye, and 6 μL of diluted cDNA. Samples were run in an MX3000P thermocycler (Stratagene) with Program MxPro v. 3.0. Transcript amounts were normalized to gyrA mRNA amount and compared with transcript amounts of the wild-type parent strain by unpaired t-test in GraphPad Prism (Kazmierczak et al., 2009).
To study low Pi conditions, bacteria from overnight BHI broth cultures were washed, diluted, and grown in 5 mL of mCDM medium containing 36.4 mM Pi to OD620 ≈ 0.15. Cells were collected by centrifugation, washed twice with 5 mL mCDM lacking Pi, and resuspended in mCDM containing 36.4 mM Pi or 10 μM Pi. Cultures were incubated at 37°C in an atmosphere of 5% CO2 for 30 min. Lysis, RNA extraction, purification, and qRT-PCR reactions were performed as described above, except that 16S rRNA was used to normalize transcript amounts, because gyrA was down-regulated under low Pi condition. All transcript amounts were compared with the wild-type parent strain grown in mCDM containing 36.4 mM.
RNA samples for RNA-Seq analyses were prepared from 30 mL cultures as described previously (Hoover et al., 2015) cDNA library construction, single-end, 100 bp-sequencing on a HiSeq 2000 sequencer (Illumina), and bioinformatic analyses were performed as described in Hoover et al. (2015). False-discovery rates (FDR) were calculated using Benjamini and Hochberg's algorithm (Benjamini and Hochberg, 1995) and a gene or region was defined as differentially expressed if it had an up- or down-fold change of 1.8 with a FDR < 0.05. RNA-Seq data were deposited in the NCBI GEO database under accession number GSE80637.
Phos-tag SDS-PAGE and western blot
Phos-tag SDS-PAGE and standard Western blotting were carried out as described previously (Wayne et al., 2010, 2012; Tsui et al., 2014). To study high Pi conditions, overnight BHI broth cultures were diluted and grown up to OD620 ≈ 0.2 in 30 mL of BHI. Cells expressing PnpR-L-FLAG3 were lysed using a FastPrep homogenizer, and cell lysates were resolved by Phos-tag SDS-PAGE at 4°C (Wayne et al., 2012). Cells expressing PstS2-HA were lysed by the same method, but resolved by standard SDS-PAGE (Tsui et al., 2014). PnpR-L-FLAG3 and PstS2-HA were detected by Western blotting as described previously (Tsui et al., 2014) using anti-FLAG or anti-HA antibody as primary antibody. Chemiluminescent signal in protein bands was quantitated by using an IVIS imaging system as described in Wayne et al. (2010).
To study low Pi conditions, strains were grown in 30 mL of mCDM to OD620 ≈ 0.2 as described above. Cells were collected by centrifugation and washed twice with 30 mL of mCDM lacking Pi. Cell pellets were resuspended in 30 mL of mCDM containing 36.4 mM Pi or lacking Pi and incubated statically at 37°C in an atmosphere of 5% CO2 for 40 min. Proteins samples were extracted and Phos-tag SDS-PAGE was performed as described above.
Qualitative quellung assay for capsule
Overnight cultures were diluted and grown in 5 mL of BHI to OD≈0.15. 1 μL of culture was mixed with 1 μL of Type 2 pneumococcal antiserum (Statens Serum Institut) on a glass slide. A cover slip was placed on top of the mixture, which was viewed immediately with a 100X objective by a phase-contrast microscope. Cells surrounded by capsule appear enlarged or swollen.
Pi concentration determination
The Pi amount in C+Y broth was determined by the colorimetric method described in Katewa and Katyare (2003). Briefly, standards were prepared by dilution of a KH2PO4 stock to give final Pi concentrations of 2.5, 5.0, 10.0, 20.0, and 40.0 μM Pi in 2.4 mL of water in glass tubes. 0.8 mL of 3N H2SO4 was added to each standard tube. 0.4 mL of 2.5% (w/v) ammonium molybdate (prepared in 3N H2SO4) was added in each tube. Last, 0.4 mL of reducing agents (20 mg of hydrazine sulfate and 20 mg of ascorbic acid dissolved in 1 mL of 0.1N H2SO4) was added to each tube. After 2 h at room temperature, A820 was determined and plotted to generate a standard curve. C+Y broth (no added Pi) was diluted 100X with water, Pi content was assayed as described above, and Pi concentration was determined from the standard curve.
Pi uptake assays
To study high Pi conditions, strains were grown in 5 mL of BHI broth to OD600 ≈ 0.2. Cells were centrifuged at 5125 × g for 5 min at room temperature, washed twice with 5 mL of mCDM lacking Pi, and resuspended at room temperature in 5 mL of mCDM lacking Pi. K2H32PO4 (107–108 dpm; 8500–9120 Ci/mmole; Perkin Elmer) was added to a final concentration of 1 mM at t=0, and 100 μL of cells was collected by vacuum filtration (0.22 μm GSWP; 13 mm diameter; Millipore) at 1, 2, 4, 6, 10 min after addition of 32P. Filters were washed 3X with 3 mL of room-temperature 1 × PBS (Ambion). Washed filters were transferred individually into 20-mL glass scintillation vials to which 5 mL of a biodegradable counting cocktail was added. Dpm of each sample was determined using a TRI-CARB 2100TR Liquid Scintillation Counter (Perkin Elmer), Pi amount incorporated at each time point was calculated.
To study low Pi condition, strains were grown to OD600 ≈ 0.2 in mCDM containing 36.4 mM Pi as described above. Cells were centrifuged at 5125 × g for 5 min at room temperature, and washed twice with 5 mL of mCDM lacking Pi. Cell pellets were resuspended in 5 mL of mCDM lacking Pi and incubated 1 h at 37°C in an atmosphere of 5% CO2. 200 μM of K2H32PO4 (107–108 dpm) was added at t = 0, and samples were withdrawn, filtered, and counted as described above.
Results
ΔphoU2 mutants show a growth defect and increased sensitivity to a range of antibiotics that is reversed by inactivation of Pst2 transport
The Pst transporter is not needed for growth of E. coli at high Pi concentrations >4 μM, and ΔphoU mutations lead to a severe growth defect that is reversed by inactivation of the Pst transporter (Steed and Wanner, 1993; Rice et al., 2009). To determine the roles of the Pst1 and Pst2 transporters and their regulation, we constructed a series of markerless deletion mutants [ΔphoU2 (IU6375); ΔphoU1 (IU6377); ΔpnpRS (IU6381); Δpst2 (IU6610); Δpst1 (IU6638)] in S. pneumoniae serotype 2 strain D39, which is encapsulated and virulent (Lanie et al., 2007; Figure 1, Table S1). Mutants were first grown in BHI broth, which contains a high Pi concentration (≈18 mM). Only the ΔphoU2 mutant showed a significant decrease in growth yield compared to the parent and other mutants (Figure 2A, Figure S1A, Table S4). Growth yield was restored when the ΔphoU2 mutation was complemented by an ectopic copy of the phoU2+ gene expressed from the PftsA promoter at the bgaA site (IU6397) (Figure 2A, Table S4). Similar results were obtained in mutants in unencapsulated D39 derivative strain K579 and E579 (data not shown).
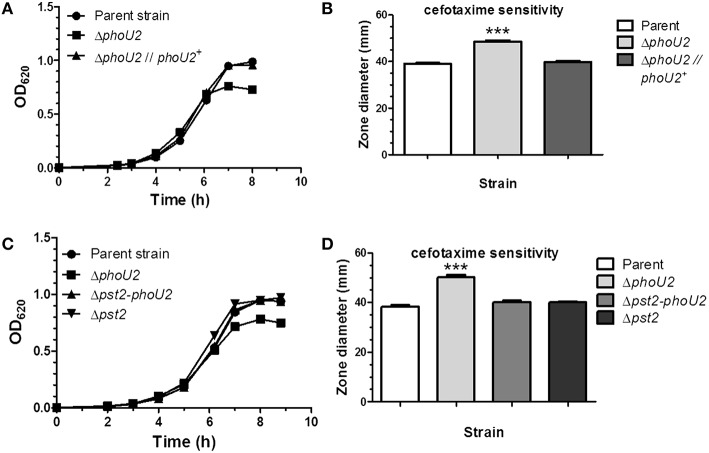
Figure 2.
Deletion of phoU2 leads to a lower growth yield and increased β-lactam antibiotic sensitivity that are reversed by a Δpst2 mutation. (A) Representative growth curves in BHI broth (≈18 mM Pi) of encapsulated parent strain (IU1781), a ΔphoU2 mutant (IU6375), and a ΔphoU2 mutant complemented by ectopic expression of PhoU2+ (IU6397). Strains were grown as described in Materials and Methods. A linear scale for OD620 is used to emphasize differences in growth yields. Growth yields and rates are quantitated for multiple determinations in Table S4. (B) Cefotaxime sensitivity assays of encapsulated parent strain (IU1781), a ΔphoU2 mutant (IU6375), and a PhoU2+-complemented ΔphoU2 mutant (IU6397). Cefotaxime disk sensitivity assays of bacteria grown in BHI broth were performed as described in Material and Methods. P-values were calculated by unpaired t-tests relative to the parent strain using GraphPad Prism; (n ≥ 3); ***P < 0.001. Increased sensitivity to other β-lactam antibiotics, vancomycin, gentamicin, and tetracycline of a ΔphoU2 mutant compared to its isogenic parent strain is shown in Table S5. (C) Representative growth curves of encapsulated parent strain (IU1781) and ΔphoU2 (IU6375), Δpst2-phoU2 (IU6550), and Δpst2 (IU6610) mutants in BHI broth. (D) Cefotaxime sensitivity assays for encapsulated parent strain (IU1781), and ΔphoU2 (IU6375), Δpst2-phoU2 (IU6550), and Δpst2 (IU6610) mutants. (n ≥ 3); ***P < 0.001.
Inactivation of phoU in E. coli generates higher sensitivity to various kinds of antibiotics than the parent strain (Li and Zhang, 2007). Since the pneumococcal ΔphoU2 mutant shows a similar growth defect as the ΔphoU mutant in E. coli (Figure 2A, Figure S1A), we tested antibiotic sensitivity to several antibiotics. Of the Pho regulon mutants tested in the encapsulated strain, only the ΔphoU2 mutant showed increased sensitivity to β-lactams and other classes of antibiotics, including glycopeptides and protein synthesis inhibitors, on plates that contain relatively high Pi content (Figure 2B, Figure S1B, and Table S5).
Previous work in E. coli (Steed and Wanner, 1993; Wanner, 1996; Rice et al., 2009; Hsieh and Wanner, 2010) and C. crescentus, in which PhoU is essential for growth (Lubin et al., 2016), indicated that the reduced growth of phoU deletion or depletion mutants could be reversed by inactivation of the Pst pump. This reversal was interpreted to mean that PhoU negatively regulates the Pst transporter itself, and in its absence, deleterious phosphate compounds accumulate that disrupt growth and metabolism. In the pneumococcal encapsulated D39 genetic background, a Δpst2-phoU2 deletion mutant lacking the Pst2 transporter and PhoU2 regulator grew similar to the parent strain and did not show increased sensitivity to antibiotics caused by the absence of PhoU2 alone (Figures 2C,D, Table S4). Likewise, single deletions of genes encoding each component of the Pst2 transporter restored growth yield of a ΔphoU2 mutant back to wild-type (data not shown). Western blot analysis showed that the ΔphoU2 mutation did not change the cellular amount of the PstS2 transporter subunit fused to the HA epitope tag (Figure S3), and by inference the amount of the Pst2 transporter. We conclude that the decreased growth yield and antibiotic sensitivity of encapsulated pneumococcus ΔphoU2 mutants in high Pi conditions are dependent on function of the Pst2 transporter, consistent with negative regulation of the Pst2 transporter by PhoU2.
PhoU2 negatively regulates transcription activation of the pst1 operon by the PnpRS TCS in high Pi conditions
As noted in the Introduction, PhoU negatively regulates the PhoBR TCS in E. coli, but not in C. crescentus (Hsieh and Wanner, 2010; Lubin et al., 2016). Consequently, we tested whether ΔphoU1 or ΔphoU2 deletions affected transcript levels of the pst1 or pst2 operons under high Pi conditions (see Figure 1). We first performed RNA-Seq analyses of ΔphoU2::kanrpsL+ and ΔphoU2::kanrpsL+ ΔphoU1::Pc-erm mutants growing in early-middle exponential phase in BHI broth, which contains a high (18 mM) concentration of Pi (Table S6). In both strains, only transcript amounts of the pst1 transporter operon, including phoU1 in the single mutant, were strongly induced (≈22X). The transcript amounts of the pnpRS regulator and pst2 transporter operons were not induced, and the number of other genes in the Pho regulon appears to be limited in S. pneumoniae D39 (Table S6). Notably, pneumococcus encodes neither an alkaline phosphatase (phoA) nor a pathway for synthesis of teichuronic acids lacking phosphate (Wanner, 1996; Botella et al., 2011, 2014). Besides the strong induction of pst1 operon transcription, there were small (2-4X) changes in the relative amounts of only a handful of other transcripts, including some corresponding to metabolic and stress-responsive genes, possibly reflecting the defective growth of these phoU2 mutants. In both mutants, one of the stronger responses was a decrease in the relative transcript amounts of the genes encoding the glycerol facilitator (GlpF) and glycerol kinase (GlpK). A putative, somewhat degenerate Pho box is located -125 bp upstream of the glpK reading frame. Together, these results suggested that PhoU2 negatively regulates pst1 operon expression under high Pi conditions, whereas the pnpRS and pst2 operons are constitutively expressed.
These conclusions were confirmed by qRT-PCR analysis of combinations of markerless deletion mutations in the pst and pnpRS genes (Table 1). RNA-Seq transcriptome analysis indicates that the pnpRS, pst1-phoU1, and pst2-phoU2 operons are separately transcribed (Table S6, Figure 1, Figures S2A,B). Hence, we quantitated the relative amounts of the pnpR, pstS1, and pstS2 transcripts normalized to gyrA transcript amount by qRT-PCR to represent pnpRS, pst1-phoU1, and pst2-phoU2 operon expression (see Materials and Methods; Wayne et al., 2012). Consistent with the RNA-Seq results, the ΔphoU2 mutations caused ≈16X increase in pst1-phoU1 operon transcript amount, but no change in expression of pst2 or pnpRS operon (Table 1, line 2). In contrast, a ΔphoU1 mutation did not cause a significant change in the relative amounts of transcript from any of the three operons (Table 1, line 3). No increase in pst1-phoU1 transcript amount in the ΔphoU2 mutant was detected in complementation experiments in which a wild-type copy of phoU2+ was expressed from an ectopic site (Table 1, line 4).
Table 1.
Relative transcript amounts of the pst1, pst2, and pnpRS operons in regulatory mutantsa.
| Strainsb | Relative transcript amount of pstS1c | Relative transcript amount of pstS2d | Relative transcript amount of pnpRe |
|---|---|---|---|
| 1. Parent strain (IU1781) | ≡1 | ≡1 | ≡1 |
| 2. ΔphoU2 (IU6375) | +16.0 ± 1.8 (n = 4) (***)f | +1.2 ± 0.6 (n = 2) (ns) | −1.2 (n = 1) |
| 3.ΔphoU1 (IU6377) | +1.6 ± 0.4 (n = 4) (ns) | +1.3 ± 0.4 (n = 2) (ns) | +1.1 (n = 1) |
| 4. ΔphoU2//phoU2+ (IU6397) | +1.3 ± 0.3 (n = 3) (ns) | NDg | ND |
| 4. ΔpnpR (IU6379) | −4.8±0.2 (n = 2) (**) | −1.1±0.1 (n = 2) (ns) | ND |
| 6. ΔpnpS (IU6496) | −2.4±0.4 (n = 2) (**) | −1.4±0.1 (n = 2) (ns) | ND |
| 7. ΔpnpRS (IU6381) | −2.0 ± 0.6 (n = 2) (*) | −1.1±0.2 (n = 2) (ns) | ND |
| 8. ΔphoU2 ΔpnpR (IU6573) | −4.4±1.1 (n = 4) (***) | ND | ND |
| 9. ΔphoU2 ΔpnpS (IU6595) | −2.1±0.3 (n = 4) (**) | ND | ND |
| 10. ΔphoU2 ΔpnpRS (IU6575) | −2.5±0.6 (n = 5) (**) | ND | ND |
| 11. ΔphoU2 ΔphoU1 (IU6499) | +19.6 ± 1.6 (n = 5) (***) | −1.4±0.1(n = 2) (ns) | +1.0 (n = 1) |
| 12. Δpst2-phoU2 (IU6550) | +43.8 ± 4.2 (n = 4) (***) | − | +1.1 ± 0.0 (n = 3) (ns) |
| 13. Δpst2 (IU6610) | +44.8 ± 1.5 (n = 4) (***) | − | ND |
| 14. Δpst2-phoU2 ΔphoU1 (IU6612) | +44.8 ± 5.5 (n = 3) (**) | − | ND |
| 15. Δpst1 (IU6638) | − | +1.0 (n = 1) | ND |
RNA preparation and qRT-PCR were performed as described in Materials and Methods.
Strains were markerless deletion mutants derived from encapsulated parent strain IU1781.
Relative pstS1 gene transcript amount was used to represent pst1 operon expression.
Relative pstS2 gene transcript amount was used to represent pst2 operon expression.
Relative pnpR gene transcript amount was used to represent pnpRS operon expression.
***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant. P-values were calculated by an unpaired t-test in GraphPad Prism. P-value is not available when n = 1.
ND, not determined.
We next determined that the increased transcription of the pst1-phoU1 operon in the ΔphoU2 mutant is mediated by the PnpRS TCS through increased phosphorylation of the PnpR~P RR. ΔpnpR, ΔpnpS, or ΔpnpRS mutants showed slightly reduced pst1-phoU1 operon transcript amounts compared to the parent strain, with no change in pst2-phoU2 operon expression (Table 1, lines 5–7). Likewise, ΔpnpR ΔphoU2, ΔpnpS ΔphoU2, and ΔpnpRS ΔphoU2 double mutants showed reduced pst1-phoU1 operon relative transcript amounts (Table 1, lines 8–10), instead of the sizable increase observed for the ΔphoU2 single mutant containing an active PnpRS TCS (Table 1, line 2). To confirm directly that PhoU2 acts as a negative regulator of PnpRS function under high Pi conditions, we performed Phos-tag SDS PAGE analysis to determine PnpR~P RR phosphorylation levels (see Materials and Methods; Figure 3). In these experiments, we fused three tandem copies of the FLAG epitope tag to the C-terminus of the PnpR RR regulator expressed from its normal chromosomal locus (Figure 3, Table S1). The PnpR-L-FLAG3 RR induced pst1-phoU1 transcription to a similar extent as wild-type (untagged) PnpR+ in a ΔphoU2 mutant (data not shown). In the phoU2+ strain, essentially no PnpR~P (< 1%) was detected in cells growing exponentially in high-Pi BHI broth (Figure 3). In contrast, the ΔphoU2 mutant contained ≈45% PnpR~P, which accounts for the high (≈16X) increase in pst1-phoU1 transcript detected (Table 1, line 2). We conclude that PhoU2 negatively regulates PnpR~P amounts and transcription of the pst1-phoU1 operon, but does not regulate transcription of the pnpRS or pst2-phoU2 operon, which are constitutively expressed.
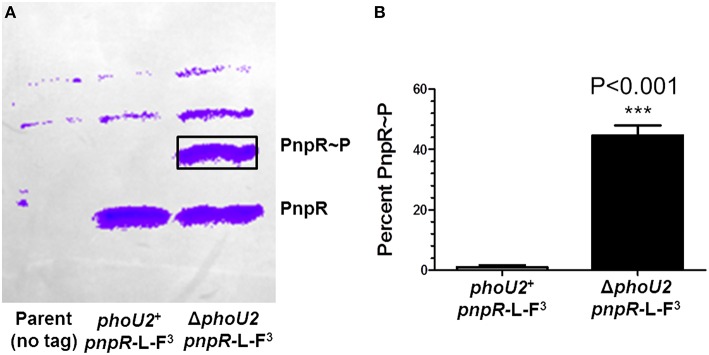
Figure 3.
Approximately 45% of PnpR is phosphorylated (PnpR~P) in a ΔphoU2 mutant. (A) Representative Phos-tag SDS-PAGE of the encapsulated parent strain not expressing a FLAG-tagged protein (IU1781, lane 1) and of the pnpR-L-FLAG3 (IU6689, lane 2), and ΔphoU2 pnpR-L-FLAG3 (IU6687, lane 3) mutants growing exponentially in BHI broth. The gel was Western blotted with anti-FLAG antibody as described in Materials and Methods. The top two bands in all lanes are nonspecific. The upper anti-FLAG-specific band corresponds to phosphorylated PnpR~P, and the lower band corresponds to unphosphorylated PnpR. Control experiments show that the upper band is heat-sensitive, as expected for PnpR~P (Figure S4). (B) Quantification of 3 independent Phos-tag SDS-PAGE experiments. Less than 1% of PnpR was phosphorylated in the phoU2+ strain, whereas ≈45% of PnpR is phosphorylated (PnpR~P) in the ΔphoU2 mutant. P-value was determined by an unpaired t-test in GraphPad Prism; ***P < 0.001.
Consistent with this interpretation, a putative PnpR~P binding site (Pho-box) sequence (TTTACACAATCTTTACA; Martin, 2004) is located 92 bp upstream of pstS1 reading frame gene (Figure 1), but no recognizable Pho-box sequences can be found upstream of the pnpRS or pst2-phoU2 operon. Finally, we tested whether induction of pst1-phoU1 operon expression contributes to the growth defect and antibiotic sensitivity of a ΔphoU2 mutant (Figure 2). A ΔphoU2 Δpst1 or ΔphoU2 Δpst1-phoU1 double mutant showed the same decrease in growth yield (Figure 4A, Table S4) and antibiotic sensitivity (Figure 4B) as the ΔphoU2 single mutant, indicating that these defects were caused primarily by misregulation of the Pst2 transporter in high Pi conditions, instead of induced expression of the Pst1 transporter.
Figure 4.
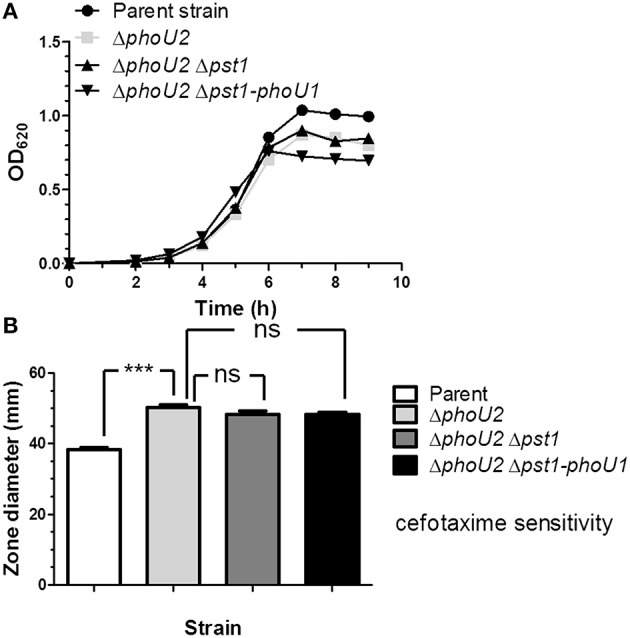
Deletion of pst1 or pst1-phoU1 in a ΔphoU2 mutant does not recover growth yield or antibiotic sensitivity. (A) Representative growth curves of encapsulated parent strain (IU1781), and ΔphoU2 (IU6375), ΔphoU2 Δpst1 (IU6664), and ΔphoU2 Δpst1-phoU1 (IU6854) mutants in BHI broth. Growth yields and rates are quantitated in Table S4. (B) Cefotaxime sensitivity assays for encapsulated parent strain (IU1781), and ΔphoU2 (IU6375), ΔphoU2 Δpst1 (IU6664), and ΔphoU2 Δpst1-phoU1 (IU6854) mutants. (n ≥ 3); ***P < 0.001. See Materials and Methods, the legend to Figure 2, and the text for additional details.
Transcription of the pst1 operon is further induced by the absence of the Pst2 transporter under high Pi conditions
We next tested whether absence of the Pst2 transporter affects pst1 operon expression in bacteria growing in high Pi-BHI broth. Surprisingly, relative transcript amounts from the pst1 operon increased by ≈44 fold in the Δpst2 phoU2+, Δpst2-phoU2, and Δpst2-phoU2 ΔphoU1 mutant compared to the parent strain (Table 1, lines 12, 13, and 14). By contrast, pst2 operon transcription is unchanged in a Δpst1 phoU1+ mutant compared to the parent (Table 1, line 15). The similar induction in the Δpst2 phoU2+ and Δpst2-phoU2 mutants (Table 1, lines 12 and 13) implies that negative regulation of the PnpRS TCS by PhoU2 depends on a functional Pst2 transporter system. Moreover, the similarity of pst1 induction in the Δpst2-phoU2 ΔphoU1 and other mutants implies that PhoU1 does not directly regulate pst1 transcription. This conclusion was supported by the similar induction of pst1 operon transcription in the ΔphoU2 phoU1+ and ΔphoU2 ΔphoU1 mutants (Table 1, lines 2 and 11). Instead, the growth characteristics of these mutants imply that PhoU1 negatively regulates the activity of the Pst1 transporter, in parallel to the negative regulation of the activity of Pst2 by PhoU2 (Figure 5A, Table S4). In BHI broth, the parent, ΔphoU1, and Δpst2-phoU2 mutant show similar growth and antibiotic sensitivity (Figure 5; Table S4). In contrast, the ΔphoU2 and ΔphoU1 Δpst2-phoU2 mutants showed reduced growth yield and increased antibiotic sensitivity (Figure 5, Table S4), consistent with increased Pi accumulation caused by misregulation of the Pst2 and Pst1 transporters, respectively. This interpretation was further supported by the reduced growth rate and yield and increased antibiotic sensitivity of the ΔphoU2 ΔphoU1 mutant compared to the ΔphoU2 and Δpst2-phoU2 ΔphoU1 mutants (Figure 5; Table S4).
Figure 5.
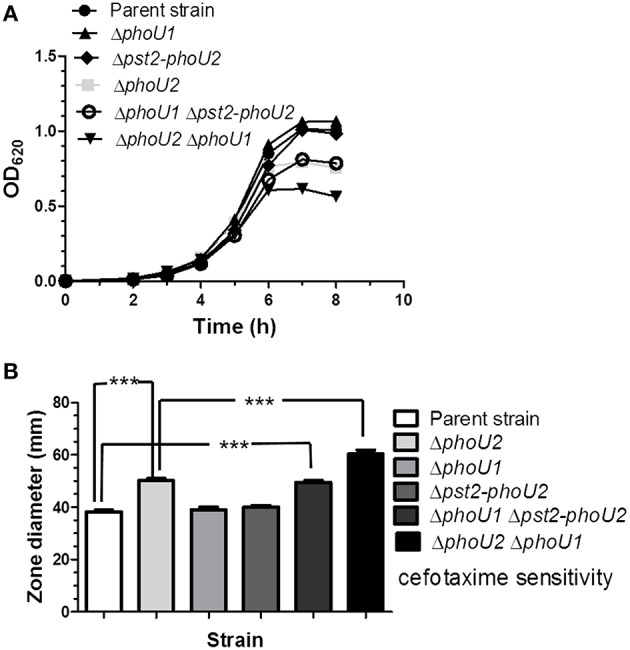
A ΔphoU1 Δpst2-phoU2 double mutant phenocopies a ΔphoU2 single mutant for decreased growth yield and increased cefotaxime sensitivity, which are even greater in a ΔphoU1 ΔphoU2 double mutant. (A) Representative growth curves of encapsulated parent strain (IU1781) and ΔphoU2 (IU6375), ΔphoU1 (IU6377), Δpst2-phoU2 (IU6550), ΔphoU1 Δpst2-phoU2 (IU6612), and ΔphoU2 ΔphoU1 (IU6499) mutants in BHI broth. (n ≥ 3) Growth yields and rates are quantitated in Table S4. (B) Cefotaxime sensitivity assays for encapsulated parent strain (IU1781) and ΔphoU2 (IU6375), ΔphoU1 (IU6377), Δpst2-phoU2 (IU6550), ΔphoU1 Δpst2-phoU2 (IU6612), and ΔphoU2 ΔphoU1 (IU6499) mutants. (n ≥ 3); ***P < 0.001. See Materials and Methods, the legend to Figure 2, and the text for additional details.
Either the Pst1 or Pst2 transporter is required in the encapsulated, but not in the unencapsulated, D39 strain
E. coli encodes an alternate low-affinity Pi uptake (Pit) system that functions in the absence of the high-affinity Pst transporter in high Pi conditions (Wanner, 1996; Harris et al., 2001; Hsieh and Wanner, 2010). However, we were unable to construct Δpst2 Δpst1 mutants in the encapsulated strain of D39 (Table 2, top two sections), where the ΔnptA amplicon was used as a positive control in the transformations. Likewise, we were unable to construct a Δpst2 ΔpnpRS double mutant, where induction of the Pst1 transporter was negated by the absence of the PnpRS TCS (Table 1, top section; Figure 1). As expected, the cps+ ΔpnpRS-pst1 mutant could be constructed (IU6133), where the Pst2 transporter is functional.
Table 2.
The Δpst1 Δpst2 double mutant cannot be constructed in encapsulated strain D39a.
| Recipient strainb | Ampliconc | Number of colonies on transformation plates after ≈20 hd |
|---|---|---|
| Δpst2 cps+ (encapsulated) | ΔnptA (control) | 100–150 (n = 3) |
| ΔpnpRS | 0–10 (n = 3) | |
| Δpst1 | 0–4 (n = 3) | |
| Δpst1-phoU1 | 0 (n = 3) | |
| No DNA (control) | 0 (n = 3) | |
| Δpst1 cps+ (encapsulated) | ΔnptA (control) | 100–150 (n = 3) |
| Δpst2 | 0 (n = 3) | |
| Δpst2-phoU2 | 0 (n = 3) | |
| No DNA (control) | 0 (n = 3) | |
| Δpst2 cps mutants (unencapsulated) | ΔnptA (control) | 100–150 (n = 3) |
| ΔpnpRS | 250–300 (n = 3) | |
| Δpst1 | 100–150 (n = 3) | |
| Δpst1-phoU1 | 250–300 (n = 3) | |
| No DNA (control) | 0 (n = 3) | |
| Δpst1 cps mutants (unencapsulated) | ΔnptA (control) | 100–150 (n = 3) |
| Δpst2 | 100–150 (n = 3) | |
| Δpst2-phoU2 | 150–200 (n = 3) | |
| No DNA (control) | 0 (n = 3) |
Transformations were performed as described in Materials and Methods.
Transformations were performed into multiple cps+ strains [IU1690 (D39); IU1781 (D39 rpsL1)] and cps mutants [IU1824 (D39 Δcps rpsL1); IU1945 (D39 Δcps); IU3309 (D39 Δcps2E rpsL1)] with similar results. D39 Δcps2E rpsL1 Δpst1 Δpst2 mutants could not be repaired back to cps+ (data not shown).
Amplicons were synthesized as described in Materials and Methods (see Table S2). Amplicons used for transformations contained the Pc-kanrpsL+ or Pc-erm antibiotic cassette for selection. Transformations with the ΔnptA amplicon or without DNA were the positive or negative control, respectively.
≤ 10 colonies on plates indicates accumulation of unencapsulated suppressor mutants (see text).
In all Δpst2 Δpst1 cps+ transformations, several colonies (< 10) appeared upon prolonged incubation (Table 2, top two sections). These suppressor colonies had a rougher appearance than the smooth colonies of D39 cps+ strains (data not shown), suggesting that capsule production was lost or reduced in these Δpst2 Δpst1 transformants. The Quellung test for serotype 2 capsule (see Materials and Methods) confirmed that a suppressor strain (IU6413) had lost its capsule (data not shown). This result suggested that unlike in cps+ strains, we would be able to construct Δcps Δpst1 Δpst2 mutants in the D39 genetic background. This hypothesis was confirmed by transformation experiments (Table 2, bottom two sections), and growth experiments showing that Δcps Δpst1 Δpst2 triple mutants grew comparably to the Δcps single mutant in BHI broth (Figure S5). Thus, we conclude that either the Pst1 or Pst2 Pi transporter must be functional in encapsulated D39 strains and that the Δpst1 Δpst2 cps+ double mutant is not viable.
NptA is the third Pi uptake system that functions in Δcps Δpst1 Δpst2 mutants
Normal growth of the Δcps Δpst1 Δpst2 mutant in high Pi medium (Figure 5) implies that sufficient Pi is being taken up by a third uptake system. BLAST searches did not reveal a close pneumococcal homolog of the Pit symporters of E. coli and B. subtilis. During these searches, we found another candidate gene, spd_0443, which encodes a putative Na+/Pi-cotransporter II-like protein. Spd_0443 homologs have been shown to act as Pi transporters in mammalian intestines and kidneys (Katai et al., 1999) and in certain bacterial species, such as V. cholerae and Vibrio vulnificus (Lebens et al., 2002; Staley and Harwood, 2014). Consistent with a role in Pi uptake, we could not delete spd_0443 in a Δcps Δpst1 Δpst2 mutant in high Pi medium (Table 3), but we could delete spd_0443 in the Δpst1 or Δpst2 single mutant (Table 2). Thus, the Spd_0443 Na+/Pi-cotransporter likely acts as a third Pi uptake system in S. pneumoniae. Because the iron transporter in S. pneumoniae is already named “Pit,” we renamed Spd_0443 as NptA (Na+-dependent phosphate transporter A), similar to Vibrio species (Lebens et al., 2002).
Table 3.
NptA (Na+/Pi co-transporter) is a third Pi uptake system.
| Recipient straina | Ampliconb | Number of colonies on transformation plates after ≈20 h |
|---|---|---|
| Δpst1 cps | ΔpnpR (control) | 150–200 (n = 3) |
| ΔnptA | 100–150 (n = 3) | |
| No DNA (control) | 0 (n = 3) | |
| Δpst2 cps | ΔpnpR (control) | 150–200 (n = 3) |
| ΔnptA | 100–150 (n = 3) | |
| No DNA (control) | 0 (n = 3) | |
| Δpst1 Δpst2 cps | ΔpnpR (control) | 150–200 (n = 3) |
| ΔnptA | 0 (n = 3) | |
| No DNA (control) | 0 (n = 3) |
Transformations were performed as described in Materials and Methods. Transformations were performed into two cps mutants [IU1824 (D39 Δcps rpsL1); IU3309 (D39 Δcps2E rpsL1)] with similar results.
Amplicons were synthesized as described in Materials and Methods (see Table S2). Amplicons used for transformations contained the Pc-kanrpsL+ or Pc-erm antibiotic cassette for selection. Transformations with the ΔpnpR amplicon or without DNA were the positive and negative control, respectively.
Δpst1-phoU1 or Δpst2-phoU2 deletion has no effect on growth under low Pi conditions
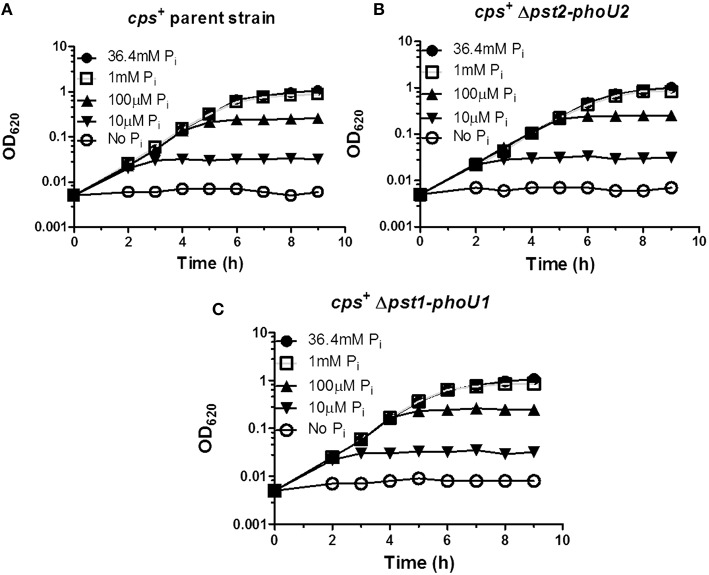
We next examined the roles of the Pst1 and Pst2 transporters and their regulators (Figure 1) under low Pi culture conditions. A previous study used the semi-defined C+Y medium as a low Pi condition (Novak et al., 1999). However, encapsulated strains grew in C+Y broth without Pi addition (Figure S6), and direct chemical assay (see Materials and Methods; Katewa and Katyare, 2003) showed that C+Y broth (with no added Pi) contains ≈1.5 mM Pi (Figure S7). Therefore, a modified chemically defined medium (mCDM) was used to study low Pi culture conditions (see Materials and Methods; Carvalho et al., 2013). The growth yield of the wild-type encapsulated strain was dependent on Pi concentration below 1 mM Pi, with growth detectable down to ≈10 μM and no growth without Pi addition (Figure 6A). Growth rates and yields of the Δpst2-phoU2 and Δpst1-phoU1 mutants were similar to those of the parent strain (Figures 6B,C), implying that a functional Pst1 or Pst2 transporter is sufficient for optimal growth under low Pi conditions.
Figure 6.
Deletion of the pst1-phoU1 or pst2-phoU2 operon does not affect growth under low Pi conditions in mCDM media. (A) Representative growth curves of encapsulated parent strain (IU1781) in mCDM with 36.4, 1 mM, 100, 10 μM, and no Pi. (B) Representative growth curves of a Δpst2-phoU2 mutant (IU6550) in mCDM with 36.4, 1 mM, 100, 10 μM, and no Pi. (C) Representative growth curves of a Δpst1-phoU1 mutant (IU6830) in mCDM with 36.4, 1 mM, 100, 10 μM, and no Pi. mCDM with different concentrations of Pi was prepared as described in Materials and Methods. Growth curves were determined at least 3X independently.
We used unencapsulated (Δcps) mutants to determine the effects of Pi concentration when both the Pst1 and Pst2 transporters were absent. Similar to the encapsulated strains (Figure 6), growth yield of the parent, Δpst1-phoU1, and Δpst2-ΔphoU2 single mutants decreased below 1 mM Pi and was still detectable at 10 μM Pi (Figures 7A–C). By contrast, the growth of the Δpst2-phoU2 Δpst1-phoU1 double mutant was not fully supported even by 2 mM Pi, and growth yield showed apparent autolysis in 1 mM Pi and no growth with ≥100 μM Pi (Figure 7D). We conclude that in low Pi conditions, the NptA transporter alone is not sufficient for growth of D39-derived S. pneumoniae, consistent with NptA acting as a low-affinity Pi transporter compared to Pst1 or Pst2.
Figure 7.
The NptA Na+/Pi co-transporter alone cannot provide sufficient Pi for long-term growth in mCDM containing moderately high (1 mM) and lower Pi concentrations. (A) Representative growth curves of unencapsulated parent strain Δcps (IU1945) in mCDM with 36.4, 2, 1 mM, 100, 10 μM, and no Pi. (B) Representative growth curves of unencapsulated Δcps Δpst2-phoU2 mutant (K583) in mCDM with 36.4, 2, 1 mM, 100, 10 μM, and no Pi. (C) Representative growth curves of unencapsulated Δcps Δpst1-phoU1 mutant (E595) in mCDM with 36.4, 2, 1 mM, 100, 10 μM, and no Pi. (D) Representative growth curves of unencapsulated Δcps Δpst2-phoU2 Δpst1-phoU1 mutant (IU5774), which only contains the NptA Na+/Pi transporter, in mCDM with 36.4, 2, 1 mM, 100, 10 μM, and no Pi. Growth curves were determined at least 3X independently.
Low Pi induces pst1 operon transcript amount >100X via phosphorylation of the PnpR~P RR
We determined the effect of low Pi concentration on pst1 operon expression in the encapsulated D39 strain. Strains were grown in high-Pi (36.4 mM) mCDM, washed, and resuspended in mCDM containing high (36.4 mM) or low (10 μM) Pi (see Materials and Methods). After 30 min, samples were taken for qRT-PCR using 16S rRNA as the normalization standard (see Materials and Methods). In all strains tested, Pi limitation to 10 μM induced relative pst1 transcript amount by >100X compared to the wild-type parent strain in high (36.4 mM) Pi (Table 4). Similar to strains grown in BHI (Table 1), ΔphoU2 and ΔphoU2 ΔphoU1 mutants showed ≈25X induction of pst1 operon transcript amount when grown in mCDM containing high Pi (Table 4), whereas a ΔphoU1 mutant showed no increase (Table 4). In these mutants, reduction of Pi concentration to 10 μM induced pst1 transcript amounts by an additional ≈5–100X (Table 4). Pi limitation of a ΔpnpRS TCS mutant showed no increase in relative pst1 operon transcript amounts (data not shown), indicating a dependence on the PnpRS TCS for induction of pst1 operon transcription in low Pi media. This conclusion was confirmed by Phos-tag SDS PAGE, which showed that phosphorylation of the PnpR RR (PnpR~P) went from < 1% in mCDM containing 36.4 mM Pi to ≈80% following a shift to mCDM lacking Pi for 40 min (Figure S8A). We conclude that transcription of pneumococcal pst1 transporter operon is strongly induced by the PnpR~P RR in low Pi conditions.
Table 4.
Relative transcript amounts of the pst1 operon in different mutants in mCDM containing high and low Pi concentrationsa.
| Strainsb | Pi concentration in mCDM | Relative transcript amount of pstS1 operonc |
|---|---|---|
| Parent strain (IU1781) | 36.4 mM | ≡1 |
| 10 μM | 209.7 ± 37.9 (n = 6) (***)d | |
| ΔphoU2 (IU6375) | 36.4 mM | 24.1 ± 3.9 (n = 3) (***) |
| 10 μM | 201.6 ± 47.6 (n = 3) (***) | |
| ΔphoU1 (IU6377) | 36.4 mM | 1.4 ± 0.1 (n = 3) (ns) |
| 10 μM | 145.4 ± 25.3 (n = 4) (***) | |
| ΔphoU2 ΔphoU1 (IU6499) | 36.4 mM | 28.3 ± 0.5 (n = 3) (***) |
| 10 μM | 136.3 ± 12.6 (n = 3) (***) |
RNA preparation and qRT-PCR from strains in mCDM containing high and low Pi concentrations were performed as described in Materials and Methods.
Strains were markerless deletion mutants derived from encapsulated parent strain IU1781 (see Table S1).
Relative pstS1 gene transcript amount was used to represent pst1 operon expression.
***P < 0.001; ns, not significant. P-values were calculated by an unpaired t-test in GraphPad Prism.
Deletion of pst1-phoU1 and pst2-phoU2 reduces the rate of Pi uptake by ≈50% in an unencapsulated strain
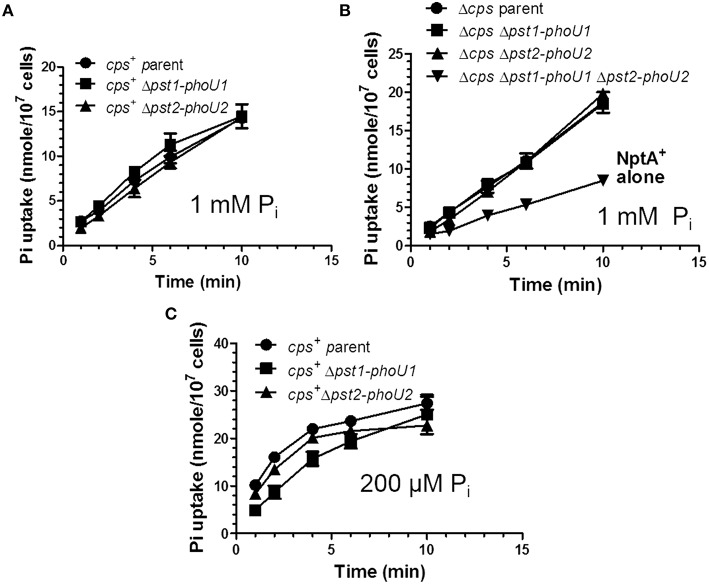
Previously, the relative rate of Pi uptake was reported to be reduced by ≈30% in a ΔpstB1 mutant of laboratory strain R6x grown in C+Y medium (no added Pi) (Novak et al., 1999), which turns out to contain a relatively high (1.5 mM) Pi content (above; Figure S7). To the contrary, 32Pi uptake experiments in encapsulated and unencapsulated D39 mutants lacking the Pst1 (Δpst1-phoU1) or Pst2 (Δpst2-phoU2) transporter showed linear rates of 32Pi uptake for at least 10 min that were similar to those of the wild-type parent strains in mCDM containing 1 mM total Pi (Figures 8A,B, Table S7). R6x is a derivative of R6 (Tiraby and Fox, 1973), which is an unencapsulated derivative of D39 (Lanie et al., 2007). The difference between this and the previous result (Novak et al., 1999) may partly reflect the large number of mutations in the R6 laboratory strain compared to the D39 strain progenitor genetic background (Lanie et al., 2007). In contrast to the single mutants, the unencapsulated Δpst1-phoU1 Δpst2-phoU2 double mutant showed a significant drop (≈50%) in the rate of Pi uptake compared to the wild-type parent strain or the single mutants in mCDM containing 1 mM Pi (Figure 8B, Table S7) or in C+Y broth (data not shown). This result implicates both the Pst1 and Pst2 transporters in Pi uptake in S. pneumoniae D39.
Figure 8.
The Pst1 or Pst2 transporter is sufficient for Pi uptake under moderately high Pi conditions (1 mM), but the Pst1 transporter is required for optimal Pi uptake under low Pi conditions (200 μM) following Pi limitation. (A) Pi uptake assays of encapsulated parent strain (IU1781) and Δpst2-phoU2 (IU6550) and Δpst1-phoU1 (IU6830) mutants in 1 mM total 32Pi in mCDM (n = 3). Pi uptake was determined as the initial rate of 32Pi incorporation into cells in mCDM in pulse-labeling experiments described in Materials and Methods. See Tables S7 for initial rates of Pi uptake (n = 3). (B) Pi uptake assays of unencapsulated parent strain Δcps (IU1945) and Δcps Δpst2-phoU2 (K583), Δcps Δpst1-phoU1 (E595), and Δcps Δpst1-phoU1 Δpst2-phoU2 (IU5774) mutants in 1 mM Pi mCDM. See Tables S7 for initial rates of Pi uptake (n = 3). (C) Pi uptake assay of encapsulated parent strain (IU1781) and Δpst2-phoU2 (IU6550) and Δpst1-phoU1 (IU6830) mutants in 200 μM total Pi mCDM following Pi starvation for 1 h as described in Materials and Methods (n = 3). Initial rates of Pi uptake were determined with the first min of labeling and are listed in Table S8 (n = 3).
Pi uptake is reduced in a Δpst1-phoU1 mutant limited for total Pi
To determine Pi uptake under low Pi conditions, we incubated encapsulated parent, Δpst1-phoU1, or Δpst2-phoU2 mutants in mCDM lacking Pi for 1 h, and then added 32Pi (total [Pi] = 200 μM), and sampled Pi uptake with time (Figure 8C). Unlike the constant rate of 32P uptake observed at 1 mM total Pi (Figures 8A,B), Pi uptake leveled off after about 5 min in 200 μM Pi (Figure 8C). The initial rate of Pi uptake by the encapsulated parent strain was markedly greater (≈8X) in 200 μM Pi than in 1 mM Pi (Figures 8A,C, Tables S7, S8). The initial rate of Pi uptake was comparable for the parent strain and the Δpst2-phoU2 mutant (Figure 8C, Table S8). In contrast, the initial rate of Pi uptake was reduced by ≈2X in the Δpst1-phoU1 mutant compared to the parent or Δpst2-phoU2 mutant (Figure 8C, Table S8). These results support a primary role for the Pst1 transporter in low-Pi conditions.
Discussion
In this paper we demonstrate for the first time that regulation of the two evolutionarily distinct Pst Pi transport systems is linked in several ways in the human commensal and pathogen, S. pneumoniae. Niches encountered by pneumococcus in human hosts contain different Pi concentrations, including nasal fluid (≈5 mM), saliva (≈1 mM), and serum (≈1 mM) (Bansal, 1990; Wilson, 2005, 2008). Several aspects of this regulatory network are unusual, compared to the usual negative regulation of Pi uptake even at moderate Pi concentrations in other bacterial species (Figure 1; Hulett, 1993; Hsieh and Wanner, 2010; Botella et al., 2011). The pst1-phoU1 and pst2-phoU2 operons are completely separated in the pneumococcal chromosome, and although upstream of the pst1-phoU1 operon, the pnpRS operon, which encodes the PnpRS TCS, is independently expressed and not autoregulated (Table 1, Figure 1). Deletion of gene spd_1614, which encodes a third putative PhoU-like protein, did not lead to growth phenotypes and was not studied further (data not shown).
Expression of the Pst2 transporter is constitutive under the conditions tested here, including media containing high Pi concentrations (Table 1, Table S6, and Figure S3). However, Pst2 uptake of Pi is likely negatively regulated by the PhoU2 protein, whose absence causes a drop in growth yield and increased antibiotic sensitivity in mutants growing in high Pi medium (Tables S4, S5, Figures 1, 2, and Figure S3). In addition, PhoU2 negatively regulates the transcription activation of the pst1-phoU1 operon by the phosphorylated PnpR~P RR, such that a ΔphoU2 deletion mutation leads to a substantial increase in pst1-phoU1 transcript amounts (Table 1, Figures 1, 3, and Figure S4). The PhoU1 protein, which shares only 34% amino acid identities with PhoU2, does not play a reciprocal role in negatively regulating pst1-phoU1 transcription (Table 1, lines 3, 11, and 14), but PhoU1 likely negatively regulates Pi uptake by the Pst1, but not the Pst2, Pi transporter (Figures 1, 4, 5). Thus, the PhoU2 protein can regulate both PnpR phosphorylation level and Pst2 transporter function, similar to the PhoU homolog in E. coli (Hsieh and Wanner, 2010; Gardner et al., 2014, 2015). In contrast, PhoU1 function is restricted to regulating transporter but not TCS function, similar to the PhoU in C. crescentus (Lubin et al., 2016). E. coli PhoU interacts with the PAS domain of the PhoR HK (Gardner et al., 2014, 2015). However, the pneumococcal PnpS HK lacks a recognizable PAS domain, and it remains to be determined whether PhoU2 regulation of PnpR~P levels is through direct interactions with the PnpS HK. Likewise, it is unknown whether PhoU2 interacts with the PstB1 subunit of the Pst1 transporter to exert control over pst1-phoU1 operon transcription, by analogy to control in E. coli (Gardner et al., 2014, 2015). Interactions of PhoU1 or PhoU2 with subunits of the Pst1 or Pst2 transporters also remain to be determined.
The requirement of a functional Pst1 or Pst2 for growth of the encapsulated serotype 2 strain provides a biological rationale for the constitutive expression and function of the Pst2 transporter at high Pi concentrations (Table 2) and why S. pneumoniae maintains dual Pi uptake systems. Capsule is one of the most important factors required for pneumococcal colonization, carriage, and virulence in its human host (Briles et al., 1992; Morona et al., 2004, 2006; Bentley et al., 2006; Hyams et al., 2010). Expression of the Pst1 transporter is strongly induced by low Pi conditions (Table 4), and lack of PhoU1 does not change pst1-phoU1 operon expression in high Pi media (Tables 2, 4). These results, and a reduced rate of Pi uptake by mutants lacking Pst1 (Figure 8C, Table S8) indicate that Pst1 functions mainly at low Pi concentrations. However, encapsulated S. pneumoniae strains require high-affinity Pi transport even in high-Pi media (Table 2), and this Pi uptake cannot be provided by the low-affinity NptA Na+/Pi cotransporter that replaces Pst1 and Pst2 in unencapsulated strains (Figure 8B, Table S7). The reason underlying this link between Pst-mediated Pi transport and the maintenance of capsule is not currently clear. A recent report suggests that low Pi conditions induce capsule biosynthesis in M. tuberculosis (van de Weerd et al., 2016). Overexpression of capsule could alter the metabolism of S. pneumoniae thereby inhibiting the growth of cps+ Δpst1 Δpst2 mutants and leading to the appearance of spontaneous cps mutants (Table 2). However, starvation of the wild-type D39 cps+ strain for Pi for 1 h did not reveal a qualitative change in capsule amount in the Quellung reaction (data not shown). Moreover, pneumococcal capsule biosynthesis is positively regulated by phosphorylation of regulatory protein CpsD (Yother, 2011), and this protein phosphorylation would likely be reduced during Pi limitation.
The regulatory pathways that mediate capsule induction in M. tuberculosis involve sigma factors and poly-Pi kinases (van de Weerd et al., 2016) that are absent from S. pneumoniae. As mentioned in the Introduction, ΔphoU mutants of other bacteria accumulate poly-Pi (Morohoshi et al., 2002; Hirota et al., 2013; Wang et al., 2013; de Almeida et al., 2015; Lubin et al., 2016), possibly because high intracellular Pi concentration disrupt metabolic homeostasis leading to defects in growth accompanied by general antibiotic sensitivity. RNA-Seq results reported here indicate that the pneumococcal Pho regulon contains a limited number of recognizable genes involved in Pi accumulation (Table S6). Notably, S. pneumoniae serotype 2 strain D39 as well as some other serotype strains, such as TIGR4, lack recognizable homologs of poly-Pi kinases Ppk1 and Ppk2 (Zhang et al., 2002) and Mg2+-dependent poly-Pi exopolyphosphatase Ppx (Akiyama et al., 1993), but do encode a degradative Mn2+-dependent, inorganic pyrophosphatase (PpaC) (Lanie et al., 2007); therefore, it is unclear whether these S. pneumoniae strains produces poly-Pi. DAPI staining experiments that revealed poly-Pi accumulation in C. crescentus ΔphoU mutants (Lubin et al., 2016) were inconclusive and did not indicate any differences in staining of the phoU2+ parent and ΔphoU2 mutant in high-Pi BHI broth (data not shown). Similarly, DAPI-based assays of extracts did not show any difference between the parent and ΔphoU2 mutant indicative of different poly-Pi amounts (data not shown).
Taken together, our results suggest that S. pneumoniae may maintain the regulated Pst1 and constitutive Pst2 Pi transport systems as a failsafe to ensure capsule biosynthesis is maintained during variations in Pi conditions. Coordination between the two Pst Pi transport systems is coordinated by the PhoU2 protein that modulates transcription of the pst1-phoU1 operon by the PnpRS TCS and separately regulates the function of the Pst2 transporter. Most bacteria encode a single Pst-type Pi transporter as part of a Pho regulon that encode numerous phosphate accumulation and sparing proteins (Hulett, 1993; Wanner, 1996; Hsieh and Wanner, 2010). The limited Pho regulon of S. pneumoniae may reflect restriction to the human host, where Pi is the primary source of phosphorus (Wilson, 2008). Even among the Streptococcus, dual Pst systems are limited to only a few species, including S. pneumoniae, S. pseudopneumoniae, S. dysgalactiae, S. equi (Group C), S. porcinus, S. agalactiae (Group B), and S. equinus, and are absent from the major Viridans and A Groups. Outside of the Streptococcus, multiple Pst Pi transporters have only been reported in a very limited number of bacteria, including Synechocystis sp. PCC 6803 and M. tuberculosis (Braibant et al., 1996; Suzuki et al., 2004; Burut-Archanai et al., 2011; Tischler et al., 2013). In Synechocystis, low Pi conditions activate expression of both of the Pst Pi transporters that are present (Suzuki et al., 2004), whereas in M. tuberculosis one Pst transporter is activated by low Pi and at least one other Pst transporter seems to be constitutively expressed, like Pst2 in S. pneumoniae (Tischler et al., 2013).
Author contributions
JZ, DS, KW, MW contributed to the conception and design of the work; JZ, DS carried out acquisition and analysis of data; JZ, DS, KW, MW interpreted data for the work; JZ, MW drafted and wrote the final version of the paper; JZ, DS, KW, MW approved the final version to be published.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kevin Bruce, Tiffany Tsui, and other laboratory members for methods and critical discussions of this work, and James Ford, Kurt Zimmer, and Doug Rusch for assistance with Illumina DNA sequencing and RNA-Seq analyses. This work was supported by grants R21AI095814 and R01GM114315 (to MW).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00063
References
- Akiyama M., Crooke E., Kornberg A. (1993). An exopolyphosphatase of Escherichia coli. J. Biol. Chem. 268:633–639. [PubMed] [Google Scholar]
- Bansal V. K. (1990). Serum Inorganic Phosphorus, in Clinical Methods: The History, Physical, and Laboratory Examinations, eds Walker H. K., Hall W. D., Hurst J. W. (Boston, MA: Butterworths Butterworth Publishers, a division of Reed Publishing; ), 895–899. [PubMed] [Google Scholar]
- Beard S. J., Hashim R., Wu G., Binet M. R., Hughes M. N., Poole R. K. (2000). Evidence for the transport of zinc(II) ions via the Pit inorganic phosphate transport system in Escherichia coli. FEMS Microbiol. Lett. 184, 231–235. 10.1111/j.1574-6968.2000.tb09019.x [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. Met. 57, 289–300. [Google Scholar]
- Bentley S. D., Aanensen D. M., Mavroidi A., Saunders D., Rabbinowitsch E., Collins M., et al. (2006). Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. 10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella E., Devine S. K., Hubner S., Salzberg L. I., Gale R. T., Brown E. D., et al. (2014). PhoR autokinase activity is controlled by an intermediate in wall teichoic acid metabolism that is sensed by the intracellular PAS domain during the PhoPR-mediated phosphate limitation response of Bacillus subtilis. Mol. Microbiol. 94, 1242–1259. 10.1111/mmi.12833 [DOI] [PubMed] [Google Scholar]
- Botella E., Hübner S., Hokamp K., Hansen A., Bisicchia P., Noone D., et al. (2011). Cell envelope gene expression in phosphate-limited Bacillus subtilis cells. Microbiology 157, 2470–2484. 10.1099/mic.0.049205-0 [DOI] [PubMed] [Google Scholar]
- Braibant M., Lefèvre P., de Wit L., Ooms J., Peirs P., Huygen K., et al. (1996). Identification of a second Mycobacterium tuberculosis gene cluster encoding proteins of an ABC phosphate transporter. FEBS Lett. 394, 206–212. 10.1016/0014-5793(96)00953-2 [DOI] [PubMed] [Google Scholar]
- Briles D. E., Crain M. J., Gray B. M., Forman C., Yother J. (1992). Strong association between capsular type and virulence for mice among human isolates of Streptococcus pneumoniae. Infect. Immun. 60, 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burut-Archanai S., Eaton-Rye J. J., Incharoensakdi A. (2011). Na+-stimulated phosphate uptake system in Synechocystis sp. PCC 6803 with Pst1 as a main transporter. BMC Microbiol. 11:225. 10.1186/1471-2180-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S. M., Kuipers O. P., Neves A. R. (2013). Environmental and nutritional factors that affect growth and metabolism of the pneumococcal serotype 2 strain D39 and its nonencapsulated derivative strain R6. PLoS ONE 8:e58492. 10.1371/journal.pone.0058492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y., Marks L. R., Pettigrew M. M., Hakansson A. P. (2014). Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front. Cell. Infect. Microbiol. 4:194. 10.3389/fcimb.2014.00194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekabab S. M., Harel J., Dozois C. M. (2014a). Interplay between genetic regulation of phosphate homeostasis and bacterial virulence. Virulence 5, 786–793. 10.4161/viru.29307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekabab S. M., Jubelin G., Dozois C. M., Harel J. (2014b). PhoB activates Escherichia coli O157:H7 virulence factors in response to inorganic phosphate limitation. PLoS ONE 9:e94285. 10.1371/journal.pone.0094285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Wakefield M. J., Yang J., Tauschek M., Robins-Browne R. M. (2012). Genome-wide analysis of the Pho regulon in a pstCA mutant of Citrobacter rodentium. PLoS ONE 7:e50682 10.1371/journal.pone.0050682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida L. G., Ortiz J. H., Schneider R. P., Spira B. (2015). phoU inactivation in Pseudomonas aeruginosa enhances accumulation of ppGpp and polyphosphate. Appl. Environ. Microbiol. 81, 3006–3015. 10.1128/AEM.04168-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor E. S. (2013). Understanding the pneumococcus: transmission and evolution. Front. Cell. Infect. Microbiol. 3:7. 10.3389/fcimb.2013.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel H., Mika M., Denapaite D., Hakenbeck R., Mühlemann K., Heller M., et al. (2014). A low-affinity penicillin-binding protein 2x variant is required for heteroresistance in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 58, 3934–3941. 10.1128/AAC.02547-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D. M., Gordon S. B. (2015). Mechanisms Causing the Inflammatory Response to Streptococcus pneumoniae, in Streptococcus pneumoniae Molecular Mechanisms of Host-Pathogen Interactions, eds Brown J. M., Hammerschmidt S., Orihuela C. (London: Academic Press; ), 383–400. [Google Scholar]
- Gardner S. G., Johns K. D., Tanner R., McCleary W. R. (2014). The PhoU protein from Escherichia coli interacts with PhoR, PstB, and metals to form a phosphate-signaling complex at the membrane. J. Bacteriol. 196, 1741–1752. 10.1128/JB.00029-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. G., Miller J. B., Dean T., Robinson T., Erickson M., Ridge P. G., et al. (2015). Genetic analysis, structural modeling, and direct coupling analysis suggest a mechanism for phosphate signaling in Escherichia coli. BMC Genet. 16 (Suppl. 2):S2. 10.1186/1471-2156-16-S2-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson J. L., Lombardo M. J., Aponyi I., Vera Cruz D., Ray M. P., Rosenberg S. M. (2015). Atypical role for PhoU in mutagenic break repair under stress in Escherichia coli. PLoS ONE 10:e0123315. 10.1371/journal.pone.0123315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover R. T., Kriakov J., Garforth S. J., Baughn A. D., Jacobs W. R., Jr. (2007). The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189, 5495–5503. 10.1128/JB.00190-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonin M., Quardokus E. M., O'Donnol D., Maddock J., Brun Y. V. (2000). Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 182, 337–347. 10.1128/JB.182.2.337-347.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz N., Loh L. N., Tuomanen E. (2015). Pneumococcal invasion: development of Bacteremia and Meningitis, in Streptococcus pneumoniae Molecular Mechanisms of Host-Pathogen Interactions, eds Brown J. M., Hammerschmidt S., Orihuela C. (London: Academic Press; ), 433–451. [Google Scholar]
- Grillo-Puertas M., Rintoul M. R., Rapisarda V. A. (2016). PhoB activation in non-limiting phosphate condition by the maintenance of high polyphosphate levels in stationary phase inhibits biofilm formation in Escherichia coli. Microbiology. 10.1099/mic.0.000281. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Hakansson A. P., Marks L. R., Roche-Hakansson H. (2015). Pneumococcal genetic transformation during colonization and biofilm formation, in Streptococcus pneumoniae Molecular Mechanisms of Host-Pathogen Interactions, eds Brown J. M., Hammerschmidt S., Orihuela C. (London: Academic Press; ), 129–142. [Google Scholar]
- Harris R. M., Webb D. C., Howitt S. M., Cox G. B. (2001). Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 183, 5008–5014. 10.1128/JB.183.17.5008-5014.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava D. L., Camilli A. (2002). Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45, 1389–1406. 10.1046/j.1365-2958.2002.03106.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques-Normark B., Tuomanen E. I. (2013). The pneumococcus: epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 3:a010215. 10.1101/cshperspect.a010215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota R., Motomura K., Nakai S., Handa T., Ikeda T., Kuroda A. (2013). Stable polyphosphate accumulation by a pseudo-revertant of an Escherichia coli phoU mutant. Biotechnol. Lett. 35, 695–701. 10.1007/s10529-012-1133-y [DOI] [PubMed] [Google Scholar]
- Hoover S. E., Perez A. J., Tsui H.-C. T., Sinha D., Smiley D. L., DiMarchi R. D., et al. (2015). A new quorum sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol. Microbiol. 97, 229–243. 10.1111/mmi.13029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A., Dubrac S., Noone D., Varughese K. I., Devine K. (2006). Interactions between the YycFG and PhoPR two-component systems in Bacillus subtilis: the PhoR kinase phosphorylates the non-cognate YycF response regulator upon phosphate limitation. Mol. Microbiol. 59, 1199–1215. 10.1111/j.1365-2958.2005.05017.x [DOI] [PubMed] [Google Scholar]
- Hsieh Y. J., Wanner B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203. 10.1016/j.mib.2010.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulett F. M. (1993). Regulation of phosphorus metabolism, in Bacillus Subtilis and Other Gram-Positive Bacteria Biochemistry, Physiology, and Molecular Genetics, eds Sonenshein A. L., Hoch J. A., Losick R. (Washington, DC: ASM Press; ), 229–235. [Google Scholar]
- Hyams C., Camberlein E., Cohen J. M., Bax K., Brown J. S. (2010). The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect. Immun. 78, 704–715. 10.1128/IAI.00881-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Binet M. R., Lee L. J., Ma R., Graham A. I., McLeod C. W., et al. (2008). Expression of the PitA phosphate/metal transporter of Escherichia coli is responsive to zinc and inorganic phosphate levels. FEMS Microbiol. Lett. 289, 219–224. 10.1111/j.1574-6968.2008.01386.x [DOI] [PubMed] [Google Scholar]
- Jacobsen S. M., Lane M. C., Harro J. M., Shirtliff M. E., Mobley H. L. (2008). The high-affinity phosphate transporter Pst is a virulence factor for Proteus mirabilis during complicated urinary tract infection. FEMS Immunol. Med. Microbiol. 52, 180–193. 10.1111/j.1574-695X.2007.00358.x [DOI] [PubMed] [Google Scholar]
- Katai K., Miyamoto K., Kishida S., Segawa H., Nii T., Tanaka H., et al. (1999). Regulation of intestinal Na+-dependent phosphate co-transporters by a low-phosphate diet and 1,25-dihydroxyvitamin D3. Biochem. J. 343, 705–712. 10.1042/bj3430705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katewa S. D., Katyare S. S. (2003). A simplified method for inorganic phosphate determination and its application for phosphate analysis in enzyme assays. Anal. Biochem. 323, 180–187. 10.1016/j.ab.2003.08.024 [DOI] [PubMed] [Google Scholar]
- Kazmierczak K. M., Wayne K. J., Rechtsteiner A., Winkler M. E. (2009). Roles of rel(Spn) in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72, 590–611. 10.1111/j.1365-2958.2009.06669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S., Hotchkiss R. D. (1960). A study of the genetic material determining an enzyme in Pneumococcus. Biochim. Biophys. Acta 39, 508–518. 10.1016/0006-3002(60)90205-5 [DOI] [PubMed] [Google Scholar]
- Lamarche M. G., Wanner B. L., Crépin S., Harel J. (2008). The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473. 10.1111/j.1574-6976.2008.00101.x [DOI] [PubMed] [Google Scholar]
- Lanie J. A., Ng W. L., Kazmierczak K. M., Andrzejewski T. M., Davidsen T. M., Wayne K. J., et al. (2007). Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189, 38–51. 10.1128/JB.01148-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebens M., Lundquist P., Söderlund L., Todorovic M., Carlin N. I. (2002). The nptA gene of Vibrio cholerae encodes a functional sodium-dependent phosphate cotransporter homologous to the type II cotransporters of eukaryotes. J. Bacteriol. 184, 4466–4474. 10.1128/JB.184.16.4466-4474.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. J., Park Y. S., Kim S. J., Lee B. J., Suh S. W. (2014). Crystal structure of PhoU from Pseudomonas aeruginosa, a negative regulator of the Pho regulon. J. Struct. Biol. 188, 22–29. 10.1016/j.jsb.2014.08.010 [DOI] [PubMed] [Google Scholar]
- Li Y., Zhang Y. (2007). PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51, 2092–2099. 10.1128/AAC.00052-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lou Y., Yokota H., Adams P. D., Kim R., Kim S. H. (2005). Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters. J. Biol. Chem. 280, 15960–15966. 10.1074/jbc.M414117200 [DOI] [PubMed] [Google Scholar]
- Lubin E. A., Henry J. T., Fiebig A., Crosson S., Laub M. T. (2016). Identification of the PhoB regulon and role of PhoU in the phosphate starvation response of Caulobacter crescentus. J. Bacteriol. 198, 187–200. 10.1128/JB.00658-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín J. F. (2004). Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J. Bacteriol. 186, 5197–5201. 10.1128/JB.186.16.5197-5201.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluskey J., Hinds J., Husain S., Witney A., Mitchell T. J. (2004). A two-component system that controls the expression of pneumococcal surface antigen A (PsaA) and regulates virulence and resistance to oxidative stress in Streptococcus pneumoniae. Mol. Microbiol. 51, 1661–1675. 10.1111/j.1365-2958.2003.03917.x [DOI] [PubMed] [Google Scholar]
- Moreno-Letelier A., Olmedo G., Eguiarte L. E., Martinez-Castilla L., Souza V. (2011). Parallel evolution and horizontal gene transfer of the pst operon in Firmicutes from oligotrophic environments. Int. J. Evol. Biol. 2011:781642 10.4061/2011/781642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohoshi T., Maruo T., Shirai Y., Kato J., Ikeda T., Takiguchi N., et al. (2002). Accumulation of inorganic polyphosphate in phoU mutants of Escherichia coli and Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 68, 4107–4110. 10.1128/AEM.68.8.4107-4110.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona J. K., Miller D. C., Morona R., Paton J. C. (2004). The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189, 1905–1913. 10.1086/383352 [DOI] [PubMed] [Google Scholar]
- Morona J. K., Morona R., Paton J. C. (2006). Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc. Natl. Acad. Sci. U.S.A. 103, 8505–8510. 10.1073/pnas.0602148103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muda M., Rao N. N., Torriani A. (1992). Role of PhoU in phosphate transport and alkaline phosphatase regulation. J. Bacteriol. 174, 8057–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neznansky A., Blus-Kadosh I., Yerushalmi G., Banin E., Opatowsky Y. (2014). The Pseudomonas aeruginosa phosphate transport protein PstS plays a phosphate-independent role in biofilm formation. FASEB J. 28, 5223–5233. 10.1096/fj.14-258293 [DOI] [PubMed] [Google Scholar]
- Novak R., Cauwels A., Charpentier E., Tuomanen E. (1999). Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J. Bacteriol. 181, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan V., Oganesyan N., Adams P. D., Jancarik J., Yokota H. A., Kim R., et al. (2005). Crystal structure of the “PhoU-like” phosphate uptake regulator from Aquifex aeolicus. J. Bacteriol. 187, 4238–4244. 10.1128/JB.187.12.4238-4244.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver M. B., Swords W. E. (2015). Pneumococcal biofilms and bacterial persistence during otitis media infections, in Streptococcus pneumoniae Molecular Mechanisms of Host-Pathogen Interactions, eds Brown J. M., Hammerschmidt S., Orihuela C. (London: Academic Press; ), 293–308. [Google Scholar]
- Orihuela C. J., Mills J., Robb C. W., Wilson C. J., Watson D. A., Niesel D. W. (2001). Streptococcus pneumoniae PstS production is phosphate responsive and enhanced during growth in the murine peritoneal cavity. Infect. Immun. 69, 7565–7571. 10.1128/IAI.69.12.7565-7571.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson G. K., Blue C. E., Mitchell T. J. (2006). Role of two-component systems in the virulence of Streptococcus pneumoniae. J. Med. Microbiol. 55, 355–363. 10.1099/jmm.0.46423-0 [DOI] [PubMed] [Google Scholar]
- Polissi A., Pontiggia A., Feger G., Altieri M., Mottl H., Ferrari L., et al. (1998). Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66, 5620–5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J. T., Ismail A. M., Camilli A. (2010). PhoB regulates both environmental and virulence gene expression in Vibrio cholerae. Mol. Microbiol. 77, 1595–1605. 10.1111/j.1365-2958.2010.07310.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Kobayashi Y., Hulett F. M. (1997). The pst operon of Bacillus subtilis has a phosphate-regulated promoter and is involved in phosphate transport but not in regulation of the pho regulon. J. Bacteriol. 179, 2534–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Montañez S., Tsui H. C., Wayne K. J., Morris J. L., Peters L. E., Zhang F., et al. (2008). Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol. Microbiol. 67, 729–746. 10.1111/j.1365-2958.2007.06082.x [DOI] [PubMed] [Google Scholar]
- Rice C. D., Pollard J. E., Lewis Z. T., McCleary W. R. (2009). Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75, 573–582. 10.1128/AEM.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifat D., Bishai W. R., Karakousis P. C. (2009). Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 200, 1126–1135. 10.1086/605700 [DOI] [PubMed] [Google Scholar]
- Shah M., Zaborin A., Alverdy J. C., Scott K., Zaborina O. (2014). Localization of DING proteins on PstS-containing outer-surface appendages of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 352, 54–61. 10.1111/1574-6968.12368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Zhang Y. (2010). PhoY2 but not PhoY1 is the PhoU homologue involved in persisters in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 65, 1237–1242. 10.1093/jac/dkq103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K. R., Diavatopoulos D. A. (2015). Nasopharyngeal Colonization with Streptococcus pneumoniae, in Streptococcus pneumoniae Molecular Mechanisms of Host-Pathogen Interactions, eds. Brown J. M., Hammerschmidt S., Orihuela C. (London: Academic Press; ), 279–291. [Google Scholar]
- Soualhine H., Brochu V., Ménard F., Papadopoulou B., Weiss K., Bergeron M. G., et al. (2005). A proteomic analysis of penicillin resistance in Streptococcus pneumoniae reveals a novel role for PstS, a subunit of the phosphate ABC transporter. Mol. Microbiol. 58, 1430–1440. 10.1111/j.1365-2958.2005.04914.x [DOI] [PubMed] [Google Scholar]
- Staley C., Harwood V. J. (2014). Differential expression of a sodium-phosphate cotransporter among Vibrio vulnificus strains. Microb. Ecol. 67, 24–33. 10.1007/s00248-013-0300-6 [DOI] [PubMed] [Google Scholar]
- Steed P. M., Wanner B. L. (1993). Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175, 6797–6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C. K., Li H., Claverys J. P., Morrison D. A. (2001). An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl. Environ. Microbiol. 67, 5190–5196. 10.1128/AEM.67.11.5190-5196.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Ferjani A., Suzuki I., Murata N. (2004). The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in synechocystis. J. Biol. Chem. 279, 13234–13240. 10.1074/jbc.M313358200 [DOI] [PubMed] [Google Scholar]
- Takemaru K., Mizuno M., Kobayashi Y. (1996). A Bacillus subtilis gene cluster similar to the Escherichia coli phosphate-specific transport (pst) operon: evidence for a tandemly arranged pstB gene. Microbiology 142, 2017–2020. 10.1099/13500872-142-8-2017 [DOI] [PubMed] [Google Scholar]
- Throup J. P., Koretke K. K., Bryant A. P., Ingraham K. A., Chalker A. F., Ge Y., et al. (2000). A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35, 566–576. 10.1046/j.1365-2958.2000.01725.x [DOI] [PubMed] [Google Scholar]
- Tiraby J. G., Fox M. S. (1973). Marker discrimination in transformation and mutation of pneumococcus. Proc. Natl. Acad. Sci. U.S.A. 70, 3541–3545. 10.1073/pnas.70.12.3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler A. D., Leistikow R. L., Kirksey M. A., Voskuil M. I., McKinney J. D. (2013). Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect. Immun. 81, 317–328. 10.1128/IAI.01136-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trihn M., Ge X., Dobson A., Kitten T., Munro C. L., Xu P. (2013). Two-component system response regulators involved in virulence of Streptococcus pneumoniae TIGR4 in infective endocarditis. PLoS ONE 8:e54320. 10.1371/journal.pone.0054320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H. C., Boersma M. J., Vella S. A., Kocaoglu O., Kuru E., Peceny J. K., et al. (2014). Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol. Microbiol. 94, 21–40. 10.1111/mmi.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H. C., Mukherjee D., Ray V. A., Sham L. T., Feig A. L., Winkler M. E. (2010). Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192, 264–279. 10.1128/JB.01204-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui H. T., Zheng J. J., Magallon A. N., Ryan J. D., Yunck R., Rued B. E., et al. (2016). Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol. Microbiol. 100, 1039–1065. 10.1111/mmi.13366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Weerd R., Boot M., Maaskant J., Sparrius M., Verboom T., van Leeuwen L. M., et al. (2016). Inorganic phosphate limitation modulates capsular polysaccharide composition in Mycobacteria. J. Biol. Chem. 291, 11787–11799. 10.1074/jbc.M116.722454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T., Camilli A. (2012). A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 22, 2541–2551. 10.1101/gr.137430.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen H. W., Abee T., Kortstee G. J., Konings W. N., Zehnder A. J. (1994). Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry 33, 1766–1770. 10.1021/bi00173a020 [DOI] [PubMed] [Google Scholar]
- Vernatter J., Pirofski L. A. (2013). Current concepts in host-microbe interaction leading to pneumococcal pneumonia. Curr. Opin. Infect. Dis. 26, 277–283. 10.1097/QCO.0b013e3283608419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Mao Y., Yu J., Zhu L., Li M., Wang D., et al. (2013). PhoY2 of Mycobacteria is required for metabolic homeostasis and stress response. J. Bacteriol. 195, 243–252. 10.1128/JB.01556-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. (1996). Phosphorus assimilation and control of the phosphate regulon, in Escherichia coli and Salmonella typhimurium cellular and molecular biology, Vol. 2, eds Neidhardt F. C., Curtis R., III., Ingraham J. L., Lin E. C. C., Low K. B., Magasanik B., Reznikoff W. S., Riley M., Schaechter M., Umbarger H. E. (Washington, DC: ASM Press; ), 1357–1381. [Google Scholar]
- Wayne K. J., Li S., Kazmierczak K. M., Tsui H. C., Winkler M. E. (2012). Involvement of WalK (VicK) phosphatase activity in setting WalR (VicR) response regulator phosphorylation level and limiting cross-talk in Streptococcus pneumoniae D39 cells. Mol. Microbiol. 86, 645–660. 10.1111/mmi.12006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne K. J., Sham L. T., Tsui H. C., Gutu A. D., Barendt S. M., Keen S. K., et al. (2010). Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae. J. Bacteriol. 192, 4388–4394. 10.1128/JB.00578-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M. (2005). Microbial Inhabitants of Humans: Their Ecology and Role in Health and Disease. Cambridge: Cambridge University Press. [Google Scholar]
- Wilson M. (2008). Bacteriology of Humans an Ecological Perspective. Malen, MA: Blackwell Publishing. [Google Scholar]
- Yother J. (2011). Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu. Rev. Microbiol. 65, 563–581. 10.1146/annurev.micro.62.081307.162944 [DOI] [PubMed] [Google Scholar]
- Zaborina O., Holbrook C., Chen Y., Long J., Zaborin A., Morozova I., et al. (2008). Structure-function aspects of PstS in multi-drug-resistant Pseudomonas aeruginosa. PLoS Pathog. 4:e43. 10.1371/journal.ppat.0040043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ishige K., Kornberg A. (2002). A polyphosphate kinase (PPK2) widely conserved in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16678–16683. 10.1073/pnas.262655199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.