Abstract
Enterobacter aerogenes (Enterobacteriaceae) is an important opportunistic pathogen that causes hospital-acquired pneumonia, bacteremia, and urinary tract infections. Recently, multidrug-resistant E. aerogenes have been a public health problem. To develop an effective antimicrobial agent, bacteriophage phiEap-2 was isolated from sewage and its genome was sequenced because of its ability to lyse the multidrug-resistant clinical E. aerogenes strain 3-SP. Morphological observations suggested that the phage belongs to the Siphoviridae family. Comparative genome analysis revealed that phage phiEap-2 is related to the Salmonella phage FSL SP-031 (KC139518). All of the structural gene products (except capsid protein) encoded by phiEap-2 had orthologous gene products in FSL SP-031 and Serratia phage Eta (KC460990). Here, we report the complete genome sequence of phiEap-2 and major findings from the genomic analysis. Knowledge of this phage might be helpful for developing therapeutic strategies against E. aerogenes.
Enterobacter aerogenes is a gram-negative bacterium of the Enterobacteriaceae family1. This bacterium is widely found in the human gastrointestinal tract and environment2. E. aerogenes has been reported to be an important opportunistic pathogen for humans3, and it is resistant to multiple antibiotics that are normally used to treat infections caused by Enterobacter4. It causes hospital-acquired infections such as pneumonia, bacteremia, urinary tract infection, surgical site infection, and meningitis2. Phages by their very nature would seem to be good candidates for antibacterial therapy5. Although the use of phages for therapeutic purposes has raised concerns over the potential for immunogenicity, restriction/modification, rapid toxin release by lytic action, development of bacteria resistance and so on5,6,7,8, phage therapy was still actively pursued. For example, a Siphoviridae phage 1535 against K. pneumoniae has been reported to have great potential for treating pneumonia and other infections caused by K. pneumoniae9, and the application of the Yersinia phage PY100 for the control of Y. enterocolitica at the post-harvest level seems to be promising10. To date, there are only two reported E. aerogenes phages, F20 (JN672684)4 which is a member of the Siphoviridae family of T1-like viruses, and an unclassified phage UZ111. To expand the repertoire of phages available for targeting clinically relevant E. aerogenes, a novel phage (phiEap-2) against E. aerogenes was isolated from hospital sewage, and the biology and genomics of the phage were characterized. This phage has specific lytic activity against a carbapenem-nonsusceptible E. aerogenes strain, 3-SP, which contains an NDM-1 carbapenemase-producing plasmid designated as p3SP-NDM and is resistant to multiple β-lactam antibiotics including imipenem and meropenem2. The phage is a member of the Siphoviridae family; the morphology, one-step growth curve, stability studies and complete genome sequence of the phage were determined.
Results and Discussion
Morphology
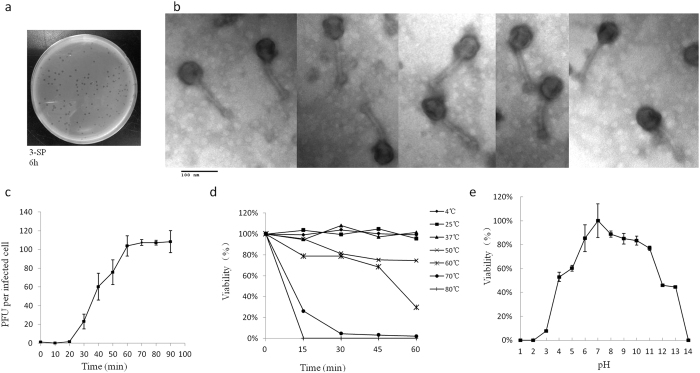
Phage phiEap-2 produced large, clear, round plaques of 1–2 mm in diameter on a lawn of E. aerogenes 3-SP (Fig. 1a). The phage was purified and examined by TEM after negative staining (Fig. 1b). The phage had a capsid that was 55 nm in diameter and a non-contractile tail that was about 117 nm long and 10 nm in diameter; these morphological features indicate that this virus belongs to the Siphoviridae family. Host range tests suggested that phiEap-2 was specific for E. aerogenes (Table 1).
Figure 1. Isolated enterobacteria phage phiEap-2.
(a) Plaques of phage phiEap-2 on E. aerogenes 3-SP. (b) Transmission electron micrograph (TEM) of phage phiEap-2 at × 150 000. The bar indicates 100 nm. (c) One-step growth curve of phiEap-2. Phages were grown in an exponential phase culture of E. aerogenes. (d) Stability of phage phiEap-2 at different temperatures. (e) Stability of phage phiEap-2 at different pH.
Table 1. Host range infection of the phage phiEap-2 and efficiency of plating.
| Species | ID | Infection | Efficiency of plating (EOP) |
|---|---|---|---|
| E. aerogenes | 3-SP | + | 1.00 |
| E. aerogenes | 201316724 | + | 1.12 |
| E. aerogenes | 2015-301 | + | 0.84 |
| E. aerogenes | 13208 | − | 0 |
| E. aerogenes | A29864 | − | 0 |
| E. aerogenes | A36179 | − | 0 |
| E. aerogenes | AH10 | − | 0 |
| E. aerogenes | AH12 | + | 0.97 |
| E. aerogenes | AH13 | + | 0.85 |
| E. aerogenes | AH14 | + | 0.60 |
| E. aerogenes | AH15 | + | 0.20 |
| E. aerogenes | AH17 | + | 0.51 |
| E. aerogenes | AH18 | + | 1.03 |
| E. aerogenes | AH2 | − | 0 |
| E. aerogenes | AH20 | + | 0.18 |
| E. aerogenes | AH21 | + | 1.01 |
| E. aerogenes | AH22 | + | 0.59 |
| E. aerogenes | AH24 | − | 0 |
| E. aerogenes | AH25 | + | 0.03 |
| E. aerogenes | AH28 | + | 1.12 |
| E. aerogenes | AH29 | + | 0.63 |
| E. aerogenes | AH3 | + | 0.23 |
| E. aerogenes | AH30 | + | 0.29 |
| E. aerogenes | AH32 | + | 0.80 |
| E. aerogenes | AH33 | − | 0 |
| E. aerogenes | AH34 | + | 0.61 |
| E. aerogenes | AH36 | + | 0.75 |
| E. cloacae | T5282 | − | 0 |
| E. cloacae | TI3 | − | 0 |
| E. sakazakii | 45401 | − | 0 |
| E. sakazakii | 45402 | − | 0 |
| Serratia marcescens | wk2050 | − | 0 |
| S. marcescens | 201315732 | − | 0 |
| S. marcescens | wj-1 | − | 0 |
| S. marcescens | wj-2 | − | 0 |
| S. marcescens | wj-3 | − | 0 |
| Escherichia coli | ATCC 25922 | − | 0 |
| Klebsiella pneumoniae | ATCC BAA-1706 | − | 0 |
| Achromobacter xylosoxidans | A22732 | − | 0 |
| Leclercia adcarboxglata | P10164 | − | 0 |
| Raoultella ornithinolytica | YNKP001 | − | 0 |
| Stenotrophomonas maltophilia | 9665 | − | 0 |
| Citrobacter freundii | P10159 | − | 0 |
| Vibrio parahaemolyticus | J5421 | − | 0 |
| Pseudomonas aeruginosa | PA01 | − | 0 |
| Acinetobacter baumannii | N1 | − | 0 |
| Shigella sonnei | #1083 | − | 0 |
−absent; +present.
Life cycle parameters
Multiplication parameters of phage phiEap-2 were determined using one-step growth curve conditions (Fig. 1c). The latent period, defined as the time interval between the adsorption and the beginning of the first burst, was about 25 min. A burst time of 60 min and average burst size of 100 plaque-forming units (pfu)/cell, which was calculated as the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells during the latent period, were observed. The thermal stability of phage phiEap-2 was determined at different temperatures from 4 to 80 °C (Fig. 1d). No significant loss in the phage titer was observed from 4 to 37 °C. Phage phiEap-2 had a titer reduction at 50 °C, and the titers dramatically decreased at 60 °C. Stability of phage phiEap-2 with different pH were also conducted (Fig. 1e). The phage was stable over a broad range of pH from 6 to 11. A significant reduction in phage titre was observed when it was extremely acidic (pH 3) or basic (pH 14). It was noticed that still nearly half of the phages existed at pH 4 or pH 13. The results suggested that the phage appears to tolerate better basic than acidic conditions.
General features of the phiEap-2 genome
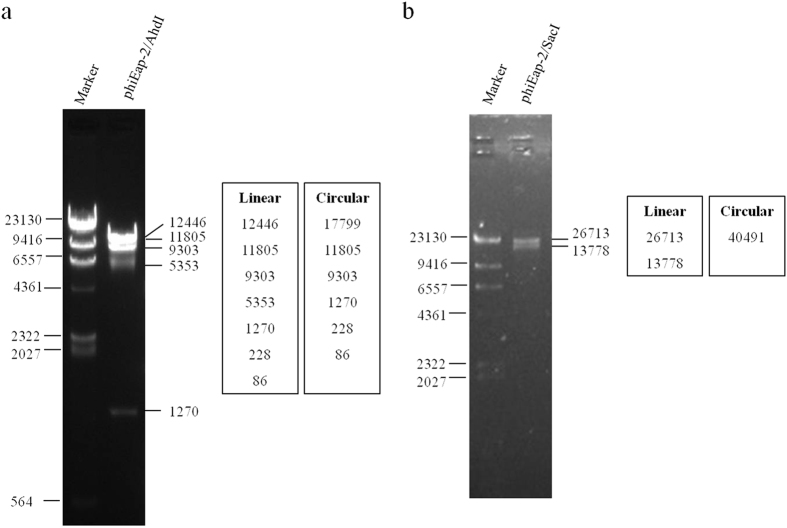
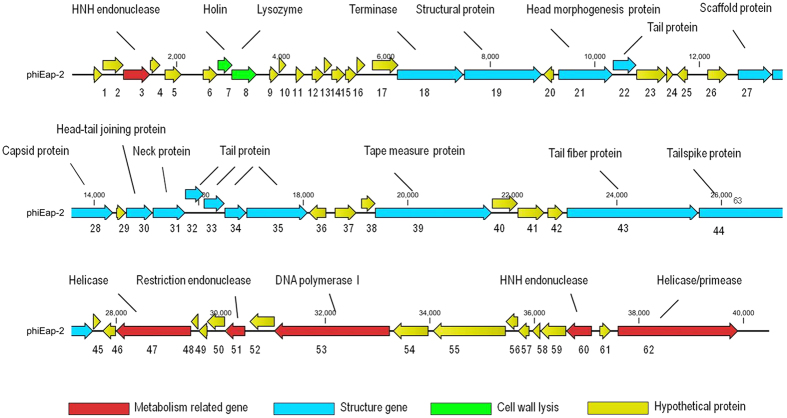
The genome of phage phiEap-2 is 40,491 bp in length with a 51.95% GC content. When the original sequencing was completed, the assembly of the random library of sequences yielded a closed, circular genome. However, based on the genome sequence analysis and the length of the fragments generated by digestion with AhdI (Fig. 2a) and SacI (Fig. 2b), the genome is linear rather than circular. Sixty-two open reading frames (ORFs) were predicted in the phage genome. Twenty-three ORFs were functionally annotated, and thirty-nine ORFs were annotated as hypothetical proteins. The orientation of genome annotation was chosen so that most genes (71%) were on the plus strand. The capsid and tail genes were identified, as well as representatives of the DNA replication, packaging, and lysis proteins. Comparative genome analysis of phiEap-2 with existing phages supports the hypothesis that phiEap-2 has no similarity to the previously published E. aerogenes phage F204, but it is related to the previously sequenced Salmonella phage FSL SP-031 (KC139518)12 (Fig. 3). Both of phiEap-2 and FSL SP-031 belong to the Siphoviridae family of viruses. This family is characterized by having a double-stranded DNA genome, an isometric head, and a long, non-contractile tail. FSL SP-031 encoded 59 predicted proteins despite a genome of 42, 215 bp, which was larger than that of phiEap-2. For comparison, the GC content of FSL SP-031 is 51.3%. PhiEap-2 and FSL SP-031 shared thirty-five orthologous genes; the amino acid identities for these genes are shown in Table 2. The phiEap-2 phage has a similar gene arrangement that was observed in other Siphoviridae phages13: genes for head and tail assembly were arranged together with the head genes 5′ to the tail genes. phiEap-2 genes were categorized into three functional groups according to the homology search-based annotation of functional genes (Fig. 4). ORF information, such as the position of genes, protein length, directions of transcription, size, function, and homology between phiEap-2 genes and other phage-related genes, is shown in Table 2.
Figure 2. Restriction fragment length polymorphism analysis of phiEap-2 DNA.
Genomic DNA from phage phiEap-2 was digested with the enzymes indicated (AhdI and SacI) and run on an agarose gel (0.7%). The length of fragments generated by digestion of the linear genome or the circular genome was showed on the right side of the electrophoresis map.
Figure 3. Phylogenetic tree based on large terminase subunits of selected bacteriophages.
The large terminase subunits were compared using the ClustalW program, and the phylogenetic tree was generated using the neighbour-joining method and 1000 bootstrap replicates. FSL SP-031, GenBank accession no. NC_021775; phiEap-2, GenBank accession no. KT287080; Eta, GenBank accession no. KC460990; FSL SP-101, GenBank accession no. KC139511; K1-dep(1), GenBank accession no. GU196278; SS3e, GenBank accession no. NC_006940; T1, GenBank accession no. AY216660.
Table 2. Phage phiEap-2 gene annotations.
| ORFs | Strand | Nucleotide position | Length (aa)a | Conserved Protein Domain Family | Best match (%) | Vs FSL SP-031 | Vs Eta | Function |
|---|---|---|---|---|---|---|---|---|
| ORF1 | + | 409–582 | 58 | |||||
| ORF2 | + | 582–983 | 133 | FSL SP-031 orf18 (54%) | orf18 (54%) | |||
| ORF3 | + | 976–1494 | 172 | pfam13392; pfam00847; PHA00280 | SETP7 orf42 (49%) | HNH endonuclease | ||
| ORF4 | + | 1491–1688 | 65 | FSL SP-031 orf19 (89%) | orf19 (89%) | |||
| ORF5 | + | 1770–2096 | 108 | pfam11753 | FSL SP-031 orf20 (82%) | orf20 (82%) | ||
| ORF6 | + | 2500–2781 | 93 | SETP7 orf57 (53%) | orf22 (53%) | |||
| ORF7 | + | 2783–3067 | 94 | SETP7 orf58 (57%) | orf23 (45%) | Class I holin | ||
| ORF8 | + | 3048–3533 | 161 | cd00737; COG3772; pfam00959 | FSL SP-031 orf24 (80%) | orf24 (80%) | Lysozyme | |
| ORF9 | + | 3772–3957 | 62 | |||||
| ORF10 | + | 3954–4097 | 47 | FSL SP-031 orf25 (93%) | orf25 (93%) | |||
| ORF11 | + | 4275–4445 | 56 | pfam10930 | FSL SP-031 orf26 (87%) | orf26 (87%) | ||
| ORF12 | + | 4585–4821 | 78 | FSL SP-031 orf27 (49%) | orf27 (49%) | |||
| ORF13 | + | 4818–4973 | 52 | FSL SP-031 orf28 (50%) | orf28 (50%) | |||
| ORF14 | + | 4960–5220 | 86 | pYD38-A orf74 (74%) | ||||
| ORF15 | + | 5223–5444 | 73 | pfam06322 | FSL SP-031 orf29 (72%) | orf29 (72%) | ||
| ORF16 | + | 5441–5608 | 56 | |||||
| ORF17 | + | 5738–6241 | 167 | FSL SP-031 orf31 (60%) | orf31 (60%) | |||
| ORF18 | + | 6216–7487 | 423 | TIGR01547; pfam03237 | FSL SP-031 orf32 (89%) | orf32 (89%) | orf40 (57%) | Terminase large subunit |
| ORF19 | + | 7500–8987 | 495 | pfam13264 | K1-dep(1) orf2 (80%) | orf33 (77%) | orf42 (65%) | Structural protein |
| ORF20 | − | 9014–9217 | 68 | |||||
| ORF21 | + | 9302–10345 | 347 | TIGR01641; pfam04233; COG2369 | FSL SP-031 orf34 (91%) | orf34 (91%) | orf43 (66%) | Head morphogenesis protein |
| ORF22 | + | 10345–10794 | 149 | pfam07679 | FSL SP-031 orf35 (67%) | orf35 (67%) | orf44 (55%) | Tail protein |
| ORF23 | + | 10794–11363 | 189 | FSL SP-031 orf35 (35%) | orf35 (35%) | orf44 (35%) | ||
| ORF24 | + | 11367–11501 | 44 | BA3 orf20 (66%) | ||||
| ORF25 | − | 11567–11779 | 71 | Acj61 p098 (39%) | ||||
| ORF26 | + | 12153–12542 | 129 | FSL SP-031 orf37 (61%) | orf37 (61%) | |||
| ORF27 | + | 12736–13386 | 216 | Eta orf50 (64%) | orf40 (54%) | orf50 (64%) | Scaffold protein | |
| ORF28 | + | 13393–14361 | 322 | K1-dep(1) orf8 (93%) | Major capsid protein | |||
| ORF29 | + | 14422–14607 | 61 | K1-dep(1) orf 9 (56%) | ||||
| ORF30 | + | 14611–15117 | 168 | PRK00007 | FSL SP-031 orf45 (75%) | orf45 (75%) | orf54 (70%) | Head-tail joining protein |
| ORF31 | + | 15120–15743 | 207 | FSL SP-031 orf46 (73%) | orf46 (73%) | orf55 (72%) | Neck protein | |
| ORF32 | + | 15740–16099 | 119 | FSL SP-031 orf47 (82%) | orf47 (82%) | orf56 (77%) | Tail protein | |
| ORF33 | + | 16096–16494 | 132 | pfam04883 | FSL SP-031 orf48 (58%) | orf48 (58%) | orf57 (51%) | Tail protein |
| ORF34 | + | 16494–16910 | 138 | pfam13554 | FSL SP-031 orf49 (90%) | orf49 (90%) | orf58 (81%) | Tail protein |
| ORF35 | + | 16913–18085 | 390 | FSL SP-031 orf50 (86%) | orf50 (86%) | orf59 (77%) | Tail protein | |
| ORF36 | − | 18108–18440 | 111 | FSL SP-031 orf51 (41%) | orf51 (41%) | |||
| ORF37 | + | 18602–19018 | 138 | FSL SP-031 orf52 (82%) | orf52 (82%) | orf61 (57%) | ||
| ORF38 | + | 19108–19380 | 90 | FSL SP-031 orf53 (81%) | orf53 (81%) | orf62 (53%) | ||
| ORF39 | + | 19373–21607 | 744 | pfam06791; PTZ00121 | FSL SP-031 orf54 (65%) | orf54 (65%) | orf63 (53%) | Tail tape measure protein |
| ORF40 | + | 21610–22104 | 164 | Eta orf64 (75%) | orf55 (64%) | orf64 (75%) | ||
| ORF41 | + | 22101–22613 | 170 | pfam08875 | Eta orf66 (78%) | orf56 (69%) | orf66 (78%) | |
| ORF42 | + | 22673–22975 | 100 | Eta orf67 (64%) | orf57 (58%) | orf67 (64%) | ||
| ORF43 | + | 23041–25557 | 838 | COG4733 | FSL SP-031 orf58 (60%) | orf58 (60%) | orf68 (60%) | Tail fiber protein |
| ORF44 | + | 25570–27558 | 662 | pfam12708; pfam13229; TIGR04247 | Eta orf69 (37%) | orf59 (24%) | orf69 (37%) | Tailspike protein |
| ORF45 | + | 27555–27707 | 50 | |||||
| ORF46 | − | 27736–27993 | 86 | pfam13600; cd14812 | FSL SP-101 orf1 (72%) | orf1 (68%) | ||
| ORF47 | − | 27996–29432 | 478 | cd00046; pfam00271; pfam00176; smart00487; COG0553 | FSL SP-031 orf2 (96%) | orf2 (96%) | Helicase | |
| ORF48 | − | 29429–29572 | 48 | |||||
| ORF49 | − | 29572–29745 | 58 | |||||
| ORF50 | − | 29736–30080 | 114 | PHA00527 | Marshall orf108 (47%) | |||
| ORF51 | − | 30077–30469 | 130 | pfam08774 | FSL SP-031 orf3 (65%) | orf3 (65%) | Restriction endonuclease | |
| ORF52 | − | 30552–31031 | 159 | pfam13392; pfam00847; PHA00280 | ||||
| ORF53 | − | 31021–33234 | 737 | cd08642; smart00482; pfam00476; pfam01612; PRK14975; TIGR00593; COG0749 | FSL SP-031 orf5 (74%) | orf5 (74%) | DNA polymerase I | |
| ORF54 | − | 33293–33973 | 226 | pfam10991 | FSL SP-031 orf7 (85%) | orf7 (85%) | ||
| ORF55 | − | 34058–35452 | 464 | pfam10926 | FSL SP-031 orf9 (72%) | orf9 (72%) | ||
| ORF56 | − | 35452–35688 | 78 | FSL SP-031 orf11 (58%) | orf11 (58%) | |||
| ORF57 | − | 35685–35906 | 74 | |||||
| ORF58 | − | 35955–36113 | 53 | |||||
| ORF59 | − | 36117–36608 | 163 | PRK13108; TIGR00457 | FSL SP-031 orf14 (42%) | orf14 (42%) | ||
| ORF60 | − | 36611–37099 | 162 | pfam13392; pfam00847; PHA00280 | SETP7 orf42 (67%) | HNH endonuclease | ||
| ORF61 | + | 37244–37465 | 73 | cd00093; pfam13443; smart00530; COG3655; PRK09706; COG1396 | FSL SP-031 orf15 (39%) | orf15 (39%) | ||
| ORF62 | + | 37594–39897 | 767 | pfam13481; cd01125 | FSL SP-031 orf16 (75%) | orf16 (75%) | Replicative helicase/primease |
aamino acids.
Figure 4. Genomic structure of phiEap-2.
The genome map was performed using the CLC Main Workbench, version 6.1.1 (CLC bio, Aarhus, Denmark). Arrows represent predicted ORFs, the direction of the arrow represents the direction of transcription. Different colors denote different functional groups of bacteriophage genes.
DNA metabolism
At least six genes in the phiEap-2 genome that play a role in nucleotide metabolism were identified, including a helicase, a replicative helicase-primase, a restriction endonuclease, a DNA polymerase, and two alleles of HNH endonuclease. gp47 encodes a helicase consisting of an SNF2 family N-terminal domain (pfam00176) and a helicase-conserved C-terminal domain (pfam00271). A BLASTP search of the helicase revealed significant identity with the FSL SP-031 orf2 (96%). gp62, which encodes a replicative helicase-primase containing an AAA_25 (pfam13481) domain and a hexameric replicative helicase RepA region, showed 75% identity with FSL SP-031 orf16. The restrictive endonuclease encoded by gp51 of phiEap-2 showed homology to FSL SP-031 orf3 (65%) and contained a VRR-NUC domain (pfam08774). gp53, which encodes a DNA polymerase I containing a 3′-5′ exonuclease domain (pfam01612) and a DNA polymerase family A domain (pfam00476), showed similarity to FSL SP-031 orf5 (74%). Proteins containing an HNH motif bind to nucleic acids and possess endonuclease activity14. The two HNH endonucleases encoded by gp60 and gp3 were homologous except for some amino acid changes; both of the proteins consist of an AP2 domain (pfam00847) and an HNH endonuclease domain (pfam13392), and they shared 67% and 49% identity with Salmonella phage SETP7 (NC_022754) gp42, respectively. The HNH endonucleases are unique to phiEap-2 compared with FSL SP-031.
Cell wall lysis-related genes
PhiEap-2 has a holin-encoding gene (gp7) immediately upstream of the lysozyme gene (gp8), suggesting that it may use a holin-dependent lytic mechanism. The lysis genes are located to the left of the terminase gene. As in many other double-stranded DNA phages, gp7, which encodes a predicted holin composed of 94 amino acids, and gp8, which encodes a lysozyme with 80% homology to the FSL SP-031 orf24, seemed to be involved in the holin-endolysin system.
Structural proteins
The arrangement of genes encoding phiEap-2 structure assembly proteins followed the conserved synteny and gene orders of Siphovirus13. Entire structural gene products encoded by phiEap-2, except for capsid protein, had orthologous gene products in FSL SP-031 and Eta (Table 2). The observation revealed a significant evolutionary relationship between FSL SP-031 and Eta. BLASTP analysis of gp18 of phiEap-2 showed significant homology to the C-terminal sequence of the Eta terminase large subunit. Different from Eta, whose terminase small subunit was located immediately upstream of the terminase large subunit, the phiEap-2 phage has only one terminase subunit. It is necessary to point out that gp28, especially the region that encodes the major capsid protein of phiEap-2, revealed extensive conservation with Escherichia phage K1-dep(1) and had no sequence similarity with FSL SP-031 or Eta, although coat proteins of FSL SP-031 and Eta were located in the same position of the genome. Five ORFs (gp22 and gp32–35) were annotated as tail proteins, and they had amino acid identities with FSL SP-031 orthologous genes that ranged from 58% to 90%. Tail fibers and tailspikes are appendages in the phage tail that facilitate the initial binding of the phage to the bacterial host and have a role in host specificity12. The tail fiber protein encoded by gp43 shares 60% amino acid identity with FSL SP-031 orf58. The tailspike protein encoded by gp44 has a mosaic nature: the N terminus displayed similarity (59%) to FSL SP-031 orf59, and the C-terminus more closely resembles a hypothetical protein of E. aerogenes. This observation is consistent with previous reports that these genes often show diversity due to recombination12 and also may suggest that these genes are linked to the evolution of host specificity12. phiEap-2 gp39, which encodes a tape measure protein (TMP) and is the largest gene in the genome, contains a TMP_2 domain (pfam06791) in the N-terminal end. TMP is important for the assembly of phage tails, is involved in tail-length determination, and corresponds to the length of the phage tail15,16. Comparisons of the phiEap-2 and FSL SP-031 proteins revealed 65% identity, and the N and C termini were highly similar.
Concluding remarks
The emergence of multiple antibiotic resistant E. aerogenes strains has limited the use of antibiotics to control this pathogen. The phiEap-2 genome does not encode any phage lysogeny factors, toxins, antibiotic resistance genes or pathogen-related genes, indicating that phiEap-2 may be considered a virulent phage with no side effects. Stability is the primary requirement when considering phage for commercial use4. Here, we isolated a phage from sewage, reported its sequence analysis, and presented data relating to its initial characterization. TEM showed that the phage belonged to the Siphoviridae family; these phage possess isometric heads and long, non-contractile tails. The genome of the phage was found to be double-stranded DNA and showed sequence homology to Salmonella phage FSL SP-031. The structural gene module showed a degree of sequence conservation with FSL SP-031 and Serratia phage Eta. The evolution of phage is thought to involve the exchange of functional modules via the loss or acquisition of genetic material by recombination between phage and also between phage and their hosts. The evolutionary advantage of this genetic recombination is thought to assist phage in their permanent adaptation to changing environmental conditions or in their quest to infect new hosts17. Although there is significant diversity among phages18, the structural gene module was found at an equivalent location in the genomes of the three members of the Siphoviridae (phiEap-2, FSL SP-031 and Eta), indicating an evolutionary connection between these phage. Characterization of phage phiEap-2 will assist in its exploitation as a therapeutic candidate against E. aerogenes and as a biocontrol agent to prevent contamination by E. aerogenes. However, considering the development of bacterial resistance to phage and the fact that relatively few candidate phage that lyse E. aerogenes have been properly characterized, there is demand for the isolation of novel E. aerogenes phage to expand the repertoire of phage available for targeting clinically significant E. aerogenes. Furthermore, an increased repertoire of available phage may allow for the development of multi-phage cocktails that may be broadly effective against a wide range of bacterial targets. In our future work, this possibility will be explored. In this study, the phenotypic features and genetic properties of the phage were examined, providing the basis for future therapeutic work. The sequenced phage might also be used in investigations of phage–bacterium interactions.
Methods
Phage isolation
E. aerogenes strains were grown in Luria–Bertani (LB) broth medium at 37 °C. Multidrug-resistant E. aerogenes strain 3-SP was used as a host strain for phage isolation from Bejing hospital sewage. The sewage samples were centrifuged at 12,000 × g for 10 min to remove the solid impurities. The supernatants were filtered through a 0.22-μm pore-size membrane filter to remove bacterial debris. Filtrate (300 μL) was added to 5 ml LB broth medium and mixed with 200 μL E. aerogenes culture (optical density at 600 nm, OD600 = 0.6) to enrich the phage at 37 °C for 8 h. Then, the culture was centrifuged at 12,000 × g for 10 min, and the supernatant was filtered with a 0.22-μm pore-size membrane filter to remove the residual bacterial cells. Filtrate diluted into medium (100 μL) was mixed with 300 μL E. aerogenes in LB culture (OD600 = 0.6) and 3 ml molten top soft nutrient agar (0.7% agar), which was then overlaid on solidified base nutrient agar (1.5% agar)19. Following incubation for 8 h at 37 °C, clear phage plaques were picked from the plate. The phage titer was determined using the double-layered method.
Purification of the phage
To prepare phiEap-2 for transmission electron microscopy (TEM) studies, cell debris from 400 ml E. aerogenes strain 3-SP infected with phiEap-2 was pelleted by low-speed centrifugation (8,500 × g, 20 min, 4 °C). Phage particles were precipitated with 1 M NaCl and 10% polyethylene glycol (PEG) 8000 at 4 °C with stirring for 60 min. The precipitated phage particles were harvested by low-speed centrifugation (8,500 × g, 20 min, 4 °C). Phage particles were resuspended in TM buffer (50 mM Tris-HCl [pH 7.8], 10 mM MgSO4) and extracted with an equal volume of chloroform. After low-speed centrifugation (3,000 × g, 15 min, 4 °C), the aqueous phase was sedimented at about 25,000 × g for 60 min20.
Electron microscopy
Phage particles were negatively stained with 2% (wt/vol) phosphotungstic acid, pH 7. Stained particles were observed in a Philips EM 300 electron microscope operated at 80 kV. Dimensions were measured on photographic prints at a final magnification of 150,000×21.
One-step growth curve
A mid-exponential-phase culture (10 ml) of E. aerogenes strain 3-SP (OD600 = 0.4 to 0.6) was harvested by centrifugation and resuspended in 0.25 volume of fresh LB (ca. 109 colony-forming units/ml). 106–107 pfu/ml phage was added at a multiplicity of infection of 0.1 and allowed to adsorb for 5 min at room temperature. The mixture was then centrifuged at 10,000 rpm for 10 min at room temperature, pelleted cells were resuspended in 10 ml LB, and incubation was continued at 37 °C. Samples were taken at 10-min intervals for 80 min. The samples were immediately diluted and plated for phage titration22. Burst size is calculated as the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells during the latent period. Measurement of phage’s latent-period duration was accomplished by detecting the delay between phage adsorption of a bacterium and the liberation of phage virions23,24,25.
Stability
A temperature-controlled incubator or water bath was used to determine the stabilities at different temperatures or pH. Briefly, a 1.5-ml tube containing equal volumes of phage (2.5 × 108 pfu/ml) were incubated at a specified temperature or pH. After treatment, the tube was cooled slowly and placed in an ice water bath; samples were assayed to determine surviving phages. The results were expressed as a percentage of the initial viral counts. Each assay was performed as three repetitions and the values represented are the means.
Host range determination
The lytic activity of phiEap-2 was tested against 14 species as determined by standard spot tests26. The strains to be tested were grown overnight in LB. Briefly, 10 μL purified phage suspension containing approximately 108 pfu/ml were spotted in the middle of a lawn of bacteria and left to dry before overnight incubation. Bacterial sensitivity to a bacteriophage was established by bacterial lysis at the spot where the phage was deposited. Each strain was tested three times at 37 °C. Efficiency of plating (EOP), the ratio of pfu/ml obtained with an assay host to the pfu/ml obtained with the isolation host, was calculated using the double layer plaque method. Assay host refers to the tested E. aerogenes clinical isolates, and isolation host refers to the E. aerogenes isolate 3-SP with which we initially isolated the phage.
Preparation of phage DNA
The precipitated phage were resuspended in SM buffer27 and purified by Caesium chloride (CsCl) gradient ultracentrifugation based on a method previously described28. The gradients were prepared in Beckman SW32.1 tubes by subsequently underlaying 1.5 ml of each 1.33, 1.45, 1.6 and 1.7 g/cm3 CsCl solution. Phages were gently added on top of the 1.33 g/cm3 CsCl. The tubes were centrifuged at 140,000 g for 3 h at 4 °C. The opalescent phage band was collected using a glass pasteur pipette and dialysed (1000 kDa MWCO), twice for 2 h and once overnight, against 250 volumes (500 ml) of SM buffer to remove CsCl. Phages were concentrated and the titre was determined. Purified phage titres were 1 × 1011 pfu/ml and stored in the dark at 4 °C 28. DNA of high titer suspensions (1010 pfu/ml) of filtered phage lysate was extracted with the phenol-chloroform (24:1, vol/vol) method and precipitated with ethanol. The samples were analyzed on 0.7 to 1.0% agarose gels.
Genome sequencing and computational analysis
The purified phage phiEap-2 genomic DNA was sequenced using an Illumina HiSeq 2500 sequencer. The preparation of the library was done using a KAPA Hyper Prep Kit Illumina platforms following the manufacturer’s instructions. The assembly used 7,907,550 reads, or 988.4 MB, of raw data to give a 24410 × coverage of the genome. The reads were assembled using SSAKE (v3.8) assembly software. The final assembled sequence was searched against the current protein and nucleotide databases (http://www.ncbi.nlm.nih.gov/) using the basic local alignment search tool (BLAST)29. Protein BLAST (BLASTP) (http://www.ncbi.nlm.nih.gov/BLAST/) was used to identify putative homologies and proteins sharing similarities with predicted phage proteins. The CLC Main Workbench, version 6.1.1 (CLC bio, Aarhus, Denmark), was used for genome annotation. Simulation of the restriction enzyme mapping of the phiEap-2 genome sequence was carried out using the software package DNAStar. The phiEap-2 DNA was digested by selected restriction endonucleases (AhdI and SacI, purchased from New England Biolabs, Ipswich, MA, USA). For a reaction system of 20 μL, 10 units of the restriction endonuclease and 200 ng of phiEap-2 DNA were used. The mixture was incubated at 37 °C for 120 min and then used to perform agarose gel electrophoresis. Agarose gel electrophoresis was subsequently performed to separate the restriction fragments. Phylogenetic analysis with the published genome sequences of related phages was performed using ClustalW. Multiple sequence alignment was carried out using Mauve software.
Additional Information
Accession code: The annotated genome sequence for the phage phiEap-2 was deposited in the NCBI nucleotide database under the accession number KT287080.
How to cite this article: Li, E. et al. Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci. Rep. 6, 28338; doi: 10.1038/srep28338 (2016).
Acknowledgments
This work received support from National Natural Science Foundation of China Grant (31200137).
Footnotes
Author Contributions E.L., X.W. and Y.M. did the experiments and contributed equally to this study as joint first authors. Z.Y., H.L., W.L. and X.W. analyzed the data. C.L., Z.S., R.Z., H.Y., A.J. and W.Y. provided the bacterial strains. J.Y. and X.Z. designed the experiments. X.Z. managed the project. X.Z. wrote the article.
References
- Eevers N. et al. Draft Genome Sequence of Enterobacter aerogenes, a DDE-Degrading and Plant Growth-Promoting Strain Isolated from Cucurbita pepo. Genome Announc 3, doi: 10.1128/genomeA.00317-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. et al. NDM-1 encoded by a pNDM-BJ01-like plasmid p3SP-NDM in clinical Enterobacter aerogenes. Front Microbiol 6, 294 doi: 10.3389/fmicb.2015.00294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin-Regli A. & Pages J. M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6, 392 doi: 10.3389/fmicb.2015.00392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra C. K., Choi T. J. & Kang S. C. Isolation and characterization of a bacteriophage F20 virulent to Enterobacter aerogenes. J Gen Virol 93, 2310–2314 (2012). [DOI] [PubMed] [Google Scholar]
- Summers W. C. Bacteriophage therapy. Annu Rev Microbiol 55, 437–451 (2001). [DOI] [PubMed] [Google Scholar]
- Paul V. D. et al. Lysis-deficient phages as novel therapeutic agents for controlling bacterial infection. BMC Microbiol 11, 195 doi: 10.1186/1471-2180-11-195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I., Manor M., Kiro R. & Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA 112, 7267–7272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong M. & Ryu S. Bacteriophage PBC1 and its endolysin as an antimicrobial agent against Bacillus cereus. Appl Environ Microbiol 81, 2274–2283 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F. et al. Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Biomed Res Int 2015, 752930 doi: 10.1155/2015/752930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orquera S. et al. Control of Campylobacter spp. and Yersinia enterocolitica by virulent bacteriophages. J Mol Genet Med 6, 273–278 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verthe K. et al. Stability and activity of an Enterobacter aerogenes-specific bacteriophage under simulated gastro-intestinal conditions. Appl Microbiol Biotechnol 65, 465–472 (2004). [DOI] [PubMed] [Google Scholar]
- Moreno Switt A. I. et al. Genomic characterization provides new insight into Salmonella phage diversity. BMC Genomics 14, 481 doi: 10.1186/1471-2164-14-481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull G. F. Bacteriophage genomics. Curr Opin Microbiol 11, 447–453 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kala S. et al. HNH proteins are a widespread component of phage DNA packaging machines. Proc Natl Acad Sci USA 111, 6022–6027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Hendrix R. W. & Duda R. L. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell 16, 11–21 (2004). [DOI] [PubMed] [Google Scholar]
- Katsura I. & Hendrix R. W. Length determination in bacteriophage lambda tails. Cell 39, 691–698 (1984). [DOI] [PubMed] [Google Scholar]
- Veesler D. & Cambillau C. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev 75, 423–433, first page of table of contents (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost L. S., Leplae R., Summers A. O. & Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3, 722–732 (2005). [DOI] [PubMed] [Google Scholar]
- Germida J. J. & Casida L. E. Ensifer adhaerens Predatory Activity Against Other Bacteria in Soil, as Monitored by Indirect Phage Analysis. Appl Environ Microbiol 45, 1380–1388 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. et al. Characterization of phiCFP-1, a virulent bacteriophage specific for Citrobacter freundii. J Med Virol 88, 895–905 (2016). [DOI] [PubMed] [Google Scholar]
- Li E. et al. Characterization of a novel Achromobacter xylosoxidans specific siphoviruse: phiAxp-1. Sci Rep 6, 21943 doi: 10.1038/srep21943 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajunen M., Kiljunen S. & Skurnik M. Bacteriophage phiYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J Bacteriol 182, 5114–5120 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedon S. T. Lysis of lysis-inhibited bacteriophage T4-infected cells. J Bacteriol 174, 8073–8080 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbruck M. The Growth of Bacteriophage and Lysis of the Host. J Gen Physiol 23, 643–660 (1940). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundling A., Manson M. D. & Young R. Holins kill without warning. Proc Natl Acad Sci USA 98, 9348–9352 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. Phage host range and efficiency of plating. Methods Mol Biol 501, 141–149 (2009). [DOI] [PubMed] [Google Scholar]
- Summer E. J. Preparation of a phage DNA fragment library for whole genome shotgun sequencing. Methods Mol Biol 502, 27–46 (2009). [DOI] [PubMed] [Google Scholar]
- Frampton R. A. et al. Genome, Proteome and Structure of a T7-Like Bacteriophage of the Kiwifruit Canker Phytopathogen Pseudomonas syringae pv. actinidiae. Viruses 7, 3361–3379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]