Abstract
Successful gene therapy of neurological disorders is predicated on achieving widespread and uniform transgene expression throughout the affected disease area in the brain. However, conventional gene vectors preferentially travel through low-resistance perivascular spaces and/or are confined to the administration site even with the aid of a pressure-driven flow provided by convection-enhanced delivery. Biodegradable DNA nanoparticles offer a safe gene delivery platform devoid of adverse effects associated with virus-based or synthetic non-biodegradable systems. Using a state-of-the-art biodegradable polymer, poly(β-amino ester), we engineered colloidally stable sub-100 nm DNA nanoparticles coated with a non-adhesive polyethylene glycol corona that are able to avoid the adhesive and steric hindrances imposed by the extracellular matrix. Following convection enhanced delivery, these brain-penetrating nanoparticles were able to homogeneously distribute throughout the rodent striatum and mediate widespread and high-level transgene expression. These nanoparticles provide a biodegradable DNA nanoparticle platform enabling uniform transgene expression patterns in vivo and hold promise for the treatment of neurological diseases.
Keywords: non-viral gene delivery, poly(β-amino esters), brain gene therapy, convection-enhanced delivery, extracellular matrix
1. INTRODUCTION
The continuing discovery of numerous genetic targets as well as emerging technologies for nucleic acid manipulation have renewed the interest in gene therapy as a therapeutic approach for the treatment of debilitating central nervous system (CNS) diseases.1 Synthetic DNA nanoparticles (DNA-NP) offer a cost-effective way to deliver therapeutic nucleic acids without the risk of insertional mutagenesis or immunogenic reactions often observed with virus-based delivery vehicles.1a, 2 Amongst those, poly(β-amino esters) (PBAE) have demonstrated the ability to condense a large number of plasmids3 and mediate transgene expression in a variety of hard-to-transfect cells in vitro with efficiencies surpassing those of leading non-viral gene delivery systems and/or commercially available transfection agents.4 Moreover, their biodegradable nature allows efficient gene delivery without the undesired cytotoxic or inflammatory responses associated with the use of non-biodegradable or slowly degrading synthetic DNA-NP.4j, 5 The highly tailorable nature of PBAE allows the development of polymer libraries that can be rapidly screened to pinpoint polymer structures imparting highly efficient in vitro gene transfer in a cell-specific manner.4a-i, 4k In particular, specific PBAE polymers have shown to provide selective transfection of human brain tumor cells4g and in vivo gene transfer to an orthotopic brain tumor without signs of toxicity.6 However, similar to gold-standard viral and non-viral gene vectors,17 clinical use of conventional PBAE-based DNA-NP, formulated by a previously reported method 4a, b, (PBAE-CP hereafter) is likely limited by their inability to achieve widespread distribution throughout the brain parenchyma. Given that most neurological disorders are characterized by highly disseminated disease areas,7 it is critical to design gene vectors capable of traveling unhindered in the brain, and thus, achieve therapeutically effective gene transfer uniformly throughout the brain.8
Following interstitial administration, gene vector distribution in the brain predominantly takes place through the narrow, tortuous extracellular space (ECS) between cells and blood vessels.9 The brain extracellular matrix (ECM), the primary structural element of the ECS, imposes an adhesive and steric barrier to the movement of conventional gene vectors through the ECS.10 Similar to other non-viral gene vectors formulated with cationic polymers or lipids,11 the positive surface charges of PBAE-CP facilitate their aggregation in physiological ionic environments and lead to adhesive interactions with negatively charged ECM macromolecules.12 As a result, PBAE-CP administered in the brain are most likely trapped within the ECS.13 Even with the aid of a pressure-driven bulk flow provided by convection-enhanced delivery (CED), conventional DNA-NP cannot overcome steric and adhesive interactions, resulting in their confinement to the immediate vicinity of the infusion site11 or the fluid filled perivascular spaces.14 This distribution pattern precludes DNA-NP from reaching and transfecting the widely dispersed target cells regardless of administration route.8b, 11b, 11d We thus, have designed brain-penetrating DNA-NP, based on PBAE polymers, that provide widespread and uniform transgene expression throughout the brain parenchyma following intracranial infusion by CED.
2. RESULTS AND DISCUSSION
2.1 Engineering and characterization of PBAE-based brain-penetrating DNA nanoparticles
In our recent proof-of-principle study,11d we have demonstrated that non-biodegradable DNA-NP, possessing ~50 nm particle diameter and a non-adhesive polyethylene glycol (PEG) surface coating, provided widespread transgene expression in brain parenchyma following CED.11d Here, we sought to engineer similarly sized and PEG-shielded PBAE-based DNA-NP capable of rapidly penetrating brain tissue (PBAE-BPN hereafter). We first formulated DNA-NP solely derived from PEGylated PBAE (PBAE-PEG) polymers; however, the DNA-NP exhibited particle diameters larger than 100 nm (i.e. 107 ± 10 nm) and a high polydispersity index (PDI = 0.36) (Table S1). We thus employed our method of using a mixture of non-PEGylated PBAE and PBAE-PEG polymers (Figures S1 and S2) at an optimal ratio in order to condense plasmid DNA.11d, 15 Using this method, we produced PBAE-BPN with dense PEG coatings, possessing the physicochemical properties required for rapid penetration through the brain parenchyma:13b sub-100 nm hydrodynamic diameter and near neutral ζ-potential (Table S.1, Table 1). We next engineered PBAE-BPN based on PBAE polymers with varying chemical structures (Figure S1), and confirmed that all formulations possessed similar “brain-penetrating” physicochemical properties (Table S2). This finding suggests that our approach is highly tailorable and PBAE-BPN can be formulated using different PBAE core polymers that may provide enhanced intracellular gene delivery in a cell-specific manner.4 In agreement with previous findings, non-PEGylated PBAE-CP displayed larger particle diameter (120 ± 3.6 nm) and positive ζ-potentials (35 ± 1.6 mV) (Figure 1B-D and Table 1).16 The biodegradable nature of PBAE and dense PEG surface shielding rendered PBAE-BPN non-toxic to neuronal cells, allowing for safe delivery of plasmid DNA even at concentrations associated with cytotoxicity when cells were treated with conventional gene vectors (Figure S3).
Table 1.
DNA-NP characterization
| Hydrodynamic Diameter ± SEM (nm)a) | ζ-potential ± SEM (mV)b) | PDIa) | |
|---|---|---|---|
| PBAE-CP | 120 ± 3.6 | 35.3 ± 1.6 | 0.1 |
| PBAE-BPN | 50 ± 2.7 | 2.5 ± 5.8 | 0.1 |
Hydrodynamic diameter and PDI were measured by DLS in 10 mM NaCl at pH 7.0. Data represents the mean ± SEM (N = 3 measurements).
ζ-potential was measured by laser Doppler anemometry in 10 mM NaCl at pH 7.0. Data represents the mean ± SEM (N = 3 measurements).
Figure 1. Physicochemical characterization of DNA-NP.
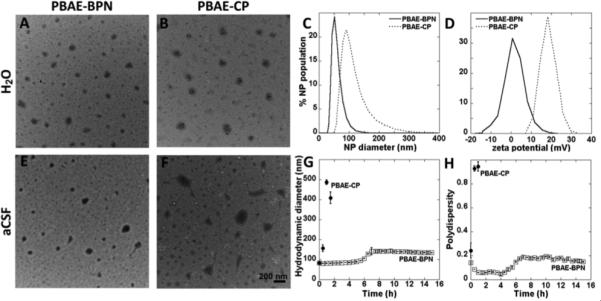
(A,B) Transmission electron micrographs of (A) PBAE-BPN and (B) PBAE-CP in ultrapure water, Scale bar = 200 nm. (C,D) Distribution of (C) hydrodynamic diameter and (D) ζ-potential as characterized in 10 mM NaCl at pH 7.0, by dynamic light scattering (DLS) and laser Doppler anemometry, respectively. (E-H) Colloidal stability of DNA-NP in aCSF at 37°C as monitored by (E,F) transmission electron micrographs immediately after incubation with aCSF (Scale bar = 200 nm) as well as (G) hydrodynamic diameter and (H) PDI over 15 h of incubation in aCSF or until PDI > 0.5. Data represents the mean ± SEM.
We have previously reported that particles as small as 200 nm are immobilized in the brain parenchyma via steric obstruction imposed by the brain ECM, regardless of non-adhesive surface coatings.13b Thus, it is imperative that DNA-NP do not agglomerate beyond this size in physiological fluids present in the brain ECS. Following incubation in artificial cerebrospinal fluid (aCSF) at 37°C, PBAE-BPN retained sub-100 nm diameters for 5 h and then doubled in size to ~140 nm, which was further retained for at least the next 10 h. We also confirmed that PBAE-BPN formulated with varying PBAE polymers retained their particle diameter in aCSF (Table S2). On the contrary, PBAE-CP aggregated immediately upon incubation in aCSF, reaching diameters of ~150 nm and ~490 nm after 30 min and 1 h, respectively (Figure 1E-G). Within 2 h of incubation in aCSF, the PDI of PBAE-CP increased to 1, underscoring complete loss of colloidal stability, whereas PDI of PBAE-BPN was well preserved (≤ 0.2) throughout the study up to 16 h (Figure 1H). These findings suggest that PBAE-BPN are most likely to retain the brain-penetrating property in vivo, while PBAE-CP are unlikely to diffuse within the brain parenchyma due not only to the highly positive surface charge, but also to their tendency towards aggregating to sizes larger than the mesh spacings of brain ECM.
2.2 PBAE-BPN rapidly diffuse in healthy brain tissue ex vivo
We next investigated whether the surface PEG coatings allow PBAE-BPN to efficiently avoid adhesive interactions with ECM components and, thus, to rapidly penetrate through brain parenchyma. Using multiple particle tracking (MPT), we assessed the diffusion of fluorescently labeled DNA-NP in rat brain tissue ex vivo.11d, 13b As evidenced by the highly constrained, non-Brownian trajectories, PBAE-CP were immobilized in the brain parenchyma (Figure 2A). In contrast, PBAE-BPN demonstrated relatively unhindered diffusion, leading to trajectories that span several microns in distance. Quantitatively, the ensemble-averaged mean square displacement (<MSD>) of PBAE-BPN was ~50-fold greater than that of PBAE-CP at a timescale of 1 s (Figure 2B). Compared to their theoretical diffusion rates in cerebrospinal fluid, PBAE-BPN were slowed only by 150-fold, but PBAE-CP were slowed by over 3000-fold. To avoid any biased interpretation that may be caused by outliers, we analyzed the distributions of individual DNA-NP mean square displacements (MSD). The histogram distribution of logarithms of individual MSD (log10MSD) revealed a unimodal distribution of PBAE-BPN with the majority of NP (> 95%) demonstrating log10MSD greater than −2 (Figure 2C). In comparison, PBAE-CP exhibited a wider range of MSD values, with 50% displaying log10MSD smaller than −2. Overall, the diffusion of PBAE-BPN, due to the surface PEG shielding and resistance to aggregation, was markedly improved compared to that of conventional uncoated PBAE-CP. This finding is in good agreement with our previous reuslts that dense surface coverage of PEG corona provided enhanced diffusion of non-biodegradable DNA-NP through brain tissues from healthy rats.11d Of note, we achieved similarly favorable physicochemical properties, stability in aCSF, and diffusion in brain parenchyma irrespective of core cationic polymer,11d suggesting that our method of formulating brain-penetrating DNA-NP is highly generalizable.
Figure 2. Diffusion of DNA-NP in rat brain parenchyma ex vivo.
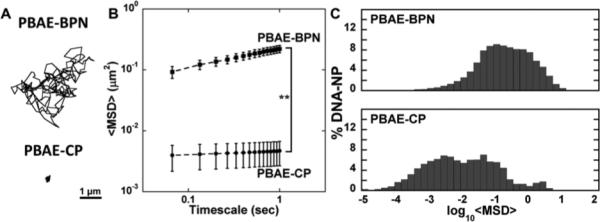
(A) Representative DNA-NP trajectories over 20 s in rat brain. Scale bar = 1 μm. (B) Ensemble-averaged mean square displacements (<MSD>) of DNA-NP as a function of timescale in rat brain tissue. Data represents the mean ± SEM of 8 experiments, with N ≥ 500 DNA-NP tracked for each experiment. **p < 0.001 (C) Histograms describing the distribution of individual log10MSD of respective DNA-NP at a timescale (τ) of 1 s.
2.3 PBAE-BPN provide widespread distribution in healthy brain following CED
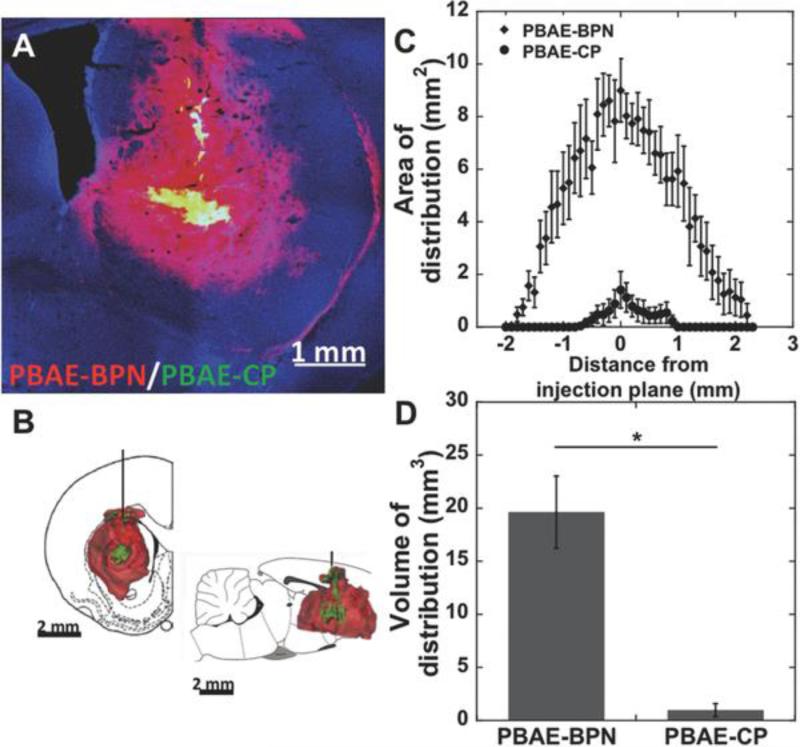
CED provides a pressure gradient during intracranial administration of therapeutics, enabling greater distribution away from the point of infusion.7 However, the distribution of therapeutic nanoparticles following CED is highly dependent on their intrinsic abilities to penetrate brain parenchyma.11a-c CED does not significantly improve particle distribution if the convective force cannot overcome the hindrances that lead to particle entrapment. We thus investigated whether the efficient ex vivo brain penetration of PBAE-BPN (Figure 2) would translate to widespread and uniform distribution following CED. Following co-administration of Cy3-labeled PBAE-CP and Cy5-labeled PBAE-BPN, we discovered that PBAE-CP were confined to the immediate vicinity of the infusion site, while PBAE-BPN distributed homogeneously to further distances away from the point of administration (Figure 3A, 3B and S4). The distribution of DNA-NP as a function of rostral–caudal distance from the plane of infusion was uniformly greater for PBAE-BPN, with their sagittal distribution spanning 4.1 mm; in comparison, PBAE-CP distributed only 1.8 mm sagittally (Figure 3C). Overall, PBAE-BPN demonstrated 20-fold greater volume of distribution compared to PBAE-CP within the striatum (Figures 3D, S4); DNA-NP distribution in the white matter was not quantified. In accordance with our recently published work,11d CED of appropriately designed DNA-NP capable of diffusing through the brain ECS resulted in a synergistic improvement in their distribution in the brain. These findings suggest that the short-term ex vivo diffusion behavior is well reflected in the long-term in vivo distribution pattern, although a direct quantitative correlation cannot be drawn due to the introduction of an external convective force in the in vivo experiments.
Figure 3. In vivo distribution of DNA-NP following CED.
(A) Representative distribution of Cy5-labeled PBAE-BPN (red) and Cy3-labeled PBAE-CP (green) in rat striatum following co-infusion via CED. DAPI staining (blue) indicates cell nuclei. Scale bar = 1 mm. (B) Representative 3D-rendered image showing distribution of PBAE-BPN (red) and PBAE-CP (green) distribution in the rat striatum following CED. Scale bar = 2 mm. (C) Image-based MATLAB quantification of area of distribution of individually administered PBAE-BPN and PBAE-CP as a function of distance from the plane of infusion (N ≥ 4 rats). (D) Summated image-based MATLAB quantification of volume of distribution for PBAE-BPN and PBAE-CP (N ≥ 4 rats). *p < 0.01.
2.4 CED of PBAE-BPN leads to widespread and high overall levels of in vivo transgene expression in healthy brain
The diffuse nature of most CNS disorders, including neurodegenerative diseases and brain tumors, mandates widespread therapeutic transgene expression throughout the brain parenchyma at therapeutically relevant levels.8 We thus investigated whether the improved distribution of PBAE-BPN (Figure 3) yields broad coverage of reporter transgene expression in the striatum. Consistent with the DNA-NP distribution patterns, PBAE-BPN provided widespread transgene expression far away from the point of administration (Figure 4A), whereas the transgene expression mediated by PBAE-CP was primarily confined to the needle tract (Figure 4B). PBAE-BPN provided 11-fold higher volume of transgene expression compared to PBAE-CP on average (Figure 4C and 4D).
Figure 4. In vivo transgene expression following CED of DNA-NP.
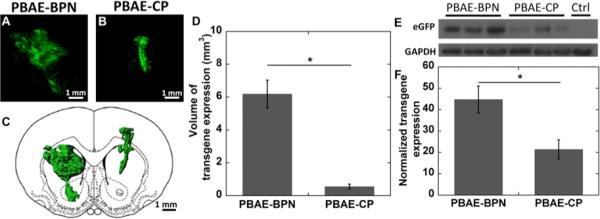
Representative stacked and aligned confocal images of enhanced green fluorescent protein (eGFP) expression (green) following CED of (A) PBAE-BPN and (B) PBAE-CP in the rat striatum. Scale bar = 1mm. (C) Representative 3D-rendered image of distribution of in vivo eGFP expression of respective PBAE-based DNA-NP. Scale bar = 1 mm (D) Image-based MATLAB quantification of the volume of eGFP expression (N = 4-6 rats). *p < 0.05. (E) Representative western blot of brain striatum samples following CED of respective DNA-NP. (F) Normalized transgene expression following CED of DNA-NP in the rat striatum. The expression level of eGFP was normalized to a non-treated control brain. Data represents the mean ± SEM (N = 8 rats).
Similar to uncoated PBAE-CP, leading virus-based and synthetic gene vectors have demonstrated dissipation of transgene expression in cells no more than 1 mm away from the point of administration following CED.11a, b, 17 We show that PBAE-BPN provide transgene expression at more than double the previously achieved distance (> 2 mm). Although CED of adeno-associated virus (AAV) has mediated transfection at even greater distances away from the point of administration, the transgene expression at farther distances was confined to the vicinity of the perivascular spaces, suggesting that the flow of AAV occurred through these fluid-filled channels rather than through the parenchymal ECS.18 This spatially confined and non-uniform transgene expression pattern has partially limited the clinical success of gene therapy of neurological diseases to date.19 Here, we observed a uniform distribution pattern of transgene expression over 70% of the striatum in the sagittal axis following CED of PBAE-BPN, without indication of preferential gene transfer surrounding the blood vessel tracts.
While the non-adhesive surface coating of PBAE-BPN allows for the aforementioned favorable distribution pattern, it has been widely reported that surface PEGylation can undermine cellular entry and intracellular trafficking of synthetic gene vectors, thereby leading to lower transfection in vitro.20 We found here that PBAE-BPN, regardless of their surface PEG coatings, provided 2-fold higher overall in vivo transgene expression compared to PBAE-CP (Figure 4E and 4F). This is most likely due to the improved brain distribution of PBAE-BPN that allowed for transgene delivery in greater number of cells compared to PBAE-CP.11d This finding emphasizes that in vitro screening alone is limited in nature and the effects of extracellular environments must be factored in for the development of synthetic gene vectors for in vivo and ultimately clinical applications.
3. CONCLUSION
We formulated highly compacted, biodegradable DNA-NP that exhibit outstanding stability in physiological conditions and rapid diffusivity within the brain parenchyma. These PBAE-BPN provided widespread and high levels of in vivo transgene expression following CED in the brain, which could not be achieved by conventional PBAE-CP. These findings resolve critical drawbacks of the otherwise superior PBAE-based gene delivery platform, and provide an avenue for its therapeutic use in brain.
4. EXPERIMENTAL SECTION
4.1 Materials and Reagents
The reagents for polymer synthesis were purchased from different companies as follows: 1,4-butanediol diacrylate and 4-amino-1-butanol from Alfa Aesar (Ward Hill, MA); 1,3-diaminopropane from Sigma-Aldrich (St. Louis, MO); 2-(3-aminopropylamino)ethanol from Oakwood Chemical (West Columbia, SC); methoxy-PEG-succinimidyl succinate from JenKem Technology (Plano, TX). Solvents were purchased as follows: ethyl ether anhydrous from Fischer Scientific (Pittsburgh, PA); dimethyl sulfoxide anhydrous (DMSO) and tetrahydrofuran (THF) from Sigma-Aldrich.
4.2 Polymer Synthesis
The uncapped PBAE polymers were synthesized using a two-step Michael addition reaction as previously reported 4b-i, 16 by reacting 1,4-butanediol diacrylate and 4-amino-1-butanol at molar ratios of 1.1:1 and 1.2:1 for 24 h at 90°C to yield PBAE of 6 kDa and 4 kDa, respectively (Figure S1). The polymers were precipitated in cold ether, washed, dried under vacuum and lyophilized. The molecular weight (MW) was estimated using nuclear magnetic resonance (NMR). Assuming two end acrylate groups per PBAE polymer, the number of repeating units was calculated to be 19-20 and 13-14 for 6 kDa and 4 kDa PBAE polymers, respectively (Figure S2A and S2B; peaks j, k, l, m). The MW was also characterized by gel permeation chromatography (GPC). Subsequently, 6 kDa PBAE was dissolved in THF at 100 mg ml−1 and 30 molar excess of one of the six selected capping groups was added (Figure S1), and reaction was undertaken while stirring for 4-5 h at room temperature. The end-capped PBAE polymers were retrieved by crashing out the polymers in cold ether and subsequently drying under vaccum/lyophilization.
For the synthesis of PBAE-PEG polymers, 4 kDa PBAE was first capped with capping group 1 and purified using the method described earlier. This end-capped PBAE polymer and 2.05 molar excess of 5 kDa methoxy-PEG-succinimidyl succinate were transferred to a glass viral, vacuumed and purged with nitrogen. The mixture of reactants was dissolved in anhydrous THF and reacted at room temperature overnight. The final PBAE-PEG polymers were precipitated and washed with cold ether and dried under vacuum. PEG conjugation was confirmed with NMR (Figure S2C). The final polymer products were dissolved in DMSO at 100mg ml−1 and stored in −20°C for further use.
4.3 Nuclear magnetic resonance
1H NMR spectra of the polymer products dissolved in deuterated methanol (MeOH-d4; Cambridge Iso. Lab., Inc., Andover, MA) were recorded using a Bruker spectrometer (500 MHz; Billerica, MA), the MeOH peak was used as an internal standard and 1H chemical shifts were reported in ppm (δ). Data were processed using the MestRe-C software (Mestrelab Research, Escondido, CA) (Figure S2).
4.4 Gel permeation chromatography
Polymer MW was measured by GPC using a Waters Breeze System and three Styragel Columns (7.8 × 300 mm) in series HR 1, HR 3, and HR 4 (Waters, Milford, MA). A solution of 95% THF/5% DMSO/0.1 M piperidine was used as a mobile phase and the samples were eluted at a flow rate of 1 ml min−1.
4.5 DNA nanoparicle Formulation and Characterization
An enhanced green fluorescent protein (eGFP) expressing plasmid regulated by a cytomegalovirus promoter (pEGFP) was purchased from Clontech Laboratories Inc. (Mountainview, CA). Fluorescent labeling of plasmid DNA with a Cy3 or Cy5 fluorophores was performed using the Mirus Label IT® Tracker™ Intracellular Nucleic Acid Localization Kit (Mirus Bio, Madison, WI). Either unlabeled or fluorescently labeled plasmid DNA were used for the formulation of PBAE-based DNA-NP. Conventional non-PEGylated DNA-NP (i.e. PBAE-CP) were formulated as previously described4c, 4e, 4i, 21. The DNA-NP solely based on PEG-PBAE polymers were similarly formulated. Highly compacted, PEGylated DNA-NP capable efficiently penetrating the brain parenchyma (i.e. PBAE-BPN) were formulated by the drop-wise addition of 5 volumes of plasmid DNA (0.1 mg ml−1) to 1 volume of polymer solution, consisting of a mixture of PBAE and PBAE-PEG at a w/w ratio of 2:3 based on PBAE mass and at a PBAE to plasmid DNA ratio of 60:1 w/w. Using a 0.1 M hydrochloride solution, both the DNA and the polymer solutions were pre-adjusted to pH 6.0. DNA-NP were washed with 3 volumes of ultrapure distilled water, and re-concentrated to a concentration of 1 mg ml−1 of plasmid DNA using Amicon® Ultra Centrifugal Filters (100,000 MWCO, Millipore Corp., Billerica, MA) to remove free polymers. The final DNA concentration was measured by the Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies, Grand Island, NY). The amount of polymer in a 1 mg ml−1 solution of DNA in DNA-NP form was measured by an XSE analytical balance (Mettler Toledo, Columbus, OH), following the lyophilization of DNA-NP. Conventional polyethylenimine-based DNA-NP (i.e. PEI-CP) as well as PEGylated poly-L-lysine based DNA-NP (i.e. PLL-CP) were formulated as previously described 15, 22 and used as controls for in vitro cell viability studies.
The hydrodynamic diameter and PDI of DNA-NP were measured in ultra-pure water and ζ-potential was measured in 10 mM NaCl at pH 7.0 by dynamic light scattering (DLS) and laser Doppler anemometry, respectively, using a Nanosizer ZS90 (Malvern Instruments, Southborough, MA). PBAE-CP and PBAE-BPN stability was assessed by incubating the DNA-NP in aCSF (Harvard Apparatus, Holliston, MA) at 37°C, DLS was performed every half hour for 15 h or until PDI > 0.5. DNA-NP morphology as well as stability were also assessed by transmission electron microscopy (TEM, Hitachi H7600, Japan) in ultrapure distilled water as well as in aCSF.
4.6 Cell viability
HT22 hipocampal neuronal cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen Corp., Carlsbad, CA) supplemented with 1% penicillin/streptomycin (pen/strep, Invitrogen Corp.) and 10% heat inactivated fetal bovine serum (FBS, Invitrogen). HT22 cells were seeded onto 96-well plates at an initial density of 5 × 103 cells well−1 and incubated at 37 °C. After 24 h, cells were incubated with a wide range of plasmid doses encapsulated in PBAE-BPN and conventional DNA-NP (PBAE-CP, PEI-CP, PLL-CP) in media for 24 h at 37 °C. Cell viability was assessed using the Dojindo cell counting kit-8 (Dojindo Molecular Technologies, Inc., Rockville, MD). Absorbance at 450 nm was measured spectrophotometrically using the Synergy Mx Multi-Mode Microplate Reader (Biotek, Instruments Inc. Winooski, VT).
4.7 Multiple Particle Tracking
MPT was performed to measure the MSD of fluorescently labeled DNA-NP in ex vivo rodent brain slices as previously published.13b, 23 Briefly, brain was harvested from healthy adult Fisher rats and incubated in aCSF for 10 min. Coronal slices (1.5 mm) of rat brains were prepared by Zivic brain matrix slicer (Zivic Instruments, Pittsburgh, PA) and placed on custom made slides. Half a microliter of fluorescently labeled DNA-NP was injected in the cerebral cortex at a depth of 1 mm using a 50 ul Hamilton Neuro Syringe (Hamilton, Reno, NV) mounted on a stereotaxic frame. Tissues were covered by a 22 mm × 22 mm coverslip to reduce tissue movement. Particle trajectories were recorded over 20 s at an exposure time of 66.7 ms (15 frames s−1) by an Evolve 512 EMCCD camera (Photometrics, Tucson, AZ) mounted on an inverted epifluorescence microscope (Axio Observer D1; Carl Zeiss, Hertfordshire, UK) equipped with a 100x/1.46 NA oil-immersion objective. Movies were analyzed with a custom-made MATLAB code to extract x, y-coordinates of DNA-NP centroids over time and calculate the MSD of each nanoparticle as a function of time.13b, 24
4.8 Animal Studies
Female Fischer 344 rats purchased from Harlan Laboratories (Frederick, MD) were used. Animals were treated in accordance with the policies and guidelines of the Johns Hopkins University Animal Care and Use Committee (Protocol number: RA12M440). Rats were anesthetized with a mixture of ketamine (75 mg kg−1) and xylazine (7.5 mg kg−1), as previously described.25 A midline scalp incision was made to expose the coronal and sagittal sutures and a burr whole was drilled 3 mm lateral to the sagittal suture and 0.5 mm posterior to the bregma. A 33 gauge 50 μl Hamilton Neuro Syringe mounted onto a stereotaxic frame was lowered to a depth of 3.5 mm and a 20 μl solution of DNA-NP in normal saline was administered using a Chemyx Inc. Nanojet Stereotaxic syringe pump at the infusion rate of 0.33 μl min−1, (Chemyx, Stafford, TX). To qualitatively assess the DNA-NP distribution following CED, we co-administered Cy3-labeled PBAE-CP and Cy5-labeled PBAE-BPN at a plasmid concentration of 500 μg ml−1 for each DNA-NP type (N = 4 per condition). To quantify the area of distirbution for each slice as well as the total volume of distribution, we similarly administered Cy-3 labeled DNA-NP, either PBAE-CP or PBAE-BPN, at a plasmid concentration of 1 mg ml−1 (N ≥ 4 per group). The animals were sacrificed 5 h following the injection. For the evalutation of in vivo transgene expression mediated by DNA-NP following CED, we administered either PBAE-CP or PBAE-BPN carrying pEGFP at a plasmid concentration of 1 mg ml−1. The distribution and the overall level of transgene expression were quantified by an image-based analysis (N = 4-6 per group) and western blot (N = 8 per group), respectively, 48 h after the CED administration.
4.9 Imaging Analysis
Harvested brains were fixed in 4% paraformaldehyde overnight, followed by incubation in a gradient of sucrose solutions (10%, 20% and 30%) prior to cryosection. Tissues were sectioned coronally into 50 μm slices using a Leica CM 1905 cryostat (Leica Biosystems, Buffalo Grove, IL). Slices were stained with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes, Eugene, OR) to visualize the cell nuclei, and imaged for the fluorescence originated from DAPI, Cy3 and Cy5 or eGFP using confocal LSM 710 microscope (Carl Zeiss) under 5x and 10x magnification. Laser power, pinhole, gain, offset and digital gain were optimized so as to avoid background fluorescence based on non-injected control rat brains.
We quantified the volume of DNA-NP distribution and transgene expression expression following CED, using a custom MATLAB script that subtracted the background fluorescence and thresholded the fluorescent intensities at 10% of the maximum intensity. DNA-NP fluorescence in the corpus callosum due to backflow was excluded from the quantification. Every other 50 μm slice within 3 mm of the injection plane was imaged. The area of distribution on each slice was summated to calculate the total volume of DNA-NP distribution. The same process was employed to analyze the distribution of reporter transgene expression in the rodent striatum. To reconstruct three dimensional images of DNA-NP distribution and transgene expression within the rodent striatum, we stacked and aligned the images of individual coronal brain sections using the Metamorph® Microscopy Automation & Image Analysis Software (Molecular Devices, Sunnyvale, CA), and used Imaris® Software (Bitplane, South Windsor, CT) to create 3D isosurfaces of the reconstructed images.
4.10 Antibodies and Western blotting
To quantify the overall level of in vivo transgene expression following the CED of DNA-NP carrying pEGFP, antibodies, anti-GFP (B-2): sc-9996 and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (6C5): sc-32233 (Santa Cruz Biotechnology, Santa Cruz, CA), were utilized for the detection of transgene expression (eGFP) and housekeeping protein (GAPDH), respectively. Whole brain tissues were lysed using 1 ml of Tissue Protein Extraction reagent (T-PER; Thermo Scientific, Pittsburgh, PA). Sampling buffer (10% glycerol, 2% SDS, 62.5 mM Tris–HCl, 2% β-mercaptoethanol, pH 6.8) was added and samples were boiled at 95°C for 10 min. Samples were resolved by SDS–polyacrylamide gel electrophoresis (PAGE) and proteins on the gels were transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA) using a wet electroblotting system (Bio-Rad). The membrane was blocked with 3% bovine serum albumin (BSA) in a mixture of Tris-buffered saline and Tween 20 (TBST; 10 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% Tween-20) and incubated overnight with primary antibodies at 4 °C. Immunoblots were visualized by enhanced chemiluminescence method. Quantification of western blot results was performed using the gels analysis tool of the ImageJ Software.26
4.11 Statistical Analysis
Statistical significance was analyzed with a two-tailed Student's t-test assuming unequal variances. Multiple comparisons were performed using one-way analysis of variance (ANOVA), followed by post hoc test using SPSS 18.0 software (SPSS Inc., Chicago, IL).
Supplementary Material
Acknowledgements
The funding was provided by the National Institutes of Health (R01CA164789, R01EB020147 and R01CA197111). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We also thank B. Schuster for his help in high-throughput MPT analysis.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Panagiotis Mastorakos, Center for Nanomedicine, at the Wilmer Eye Institute, Johns Hopkins School of Medicine, 400 N. Broadway, Baltimore, MD 21231, USA Department of Ophthalmology, The Wilmer Eye Institute, Johns Hopkins School of Medicine, 600 N. Wolfe Street, Baltimore, MD 21297, USA.
Eric Song, Center for Nanomedicine, at the Wilmer Eye Institute, Johns Hopkins School of Medicine, 400 N. Broadway, Baltimore, MD 21231, USA; Center for Biotechnology Education, Krieger School of Arts and Sciences, Johns Hopkins University, 3400 N. Charles Street, Baltimore, MD 21218, USA.
Clark Zhang, Center for Nanomedicine, at the Wilmer Eye Institute, Johns Hopkins School of Medicine, 400 N. Broadway, Baltimore, MD 21231, USA; Department of Biomedical Engineering, Johns Hopkins University School of Medicine, 720 Rutland Av., Baltimore, MD 21205, USA.
Sneha Berry, Center for Nanomedicine, at the Wilmer Eye Institute, Johns Hopkins School of Medicine, 400 N. Broadway, Baltimore, MD 21231, USA; Center for Biotechnology Education, Krieger School of Arts and Sciences, Johns Hopkins University, 3400 N. Charles Street, Baltimore, MD 21218, USA.
Hee Won Park, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, 3400 N. Charles Street, Baltimore, MD 21218, USA.
Young Eun Kim, Department of Chemical and Biomolecular Engineering, Johns Hopkins University, 3400 N. Charles Street, Baltimore, MD 21218, USA.
Dr. Jong Sung Park, Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University, 601 N. Caroline St, Baltimore, MD 21287, USA
Prof. Seulki Lee, Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University, 601 N. Caroline St, Baltimore, MD 21287, USA
Dr. Jung Soo Suk, Center for Nanomedicine, at the Wilmer Eye Institute, Johns Hopkins School of Medicine, 400 N. Broadway, Baltimore, MD 21231, USA; Department of Ophthalmology, The Wilmer Eye Institute, Johns Hopkins School of Medicine, 600 N. Wolfe Street, Baltimore, MD 21297, USA.
Prof. Justin Hanes, Center for Nanomedicine, at the Wilmer Eye Institute, Johns Hopkins School of Medicine, 400 N. Broadway, Baltimore, MD 21231, USA; Department of Ophthalmology, The Wilmer Eye Institute, Johns Hopkins School of Medicine, 600 N. Wolfe Street, Baltimore, MD 21297, USA; Departments of Neurosurgery, Oncology, and Pharmacology & Molecular Sciences, Johns Hopkins School of Medicine, 600 N. Wolfe Street, Baltimore, MD 21287, USA..
REFERENCES
- 1.a O'Mahony AM, Godinho BM, Cryan JF, O'Driscoll CM. Non-viral nanosystems for gene and small interfering RNA delivery to the central nervous system: formulating the solution. J Pharm Sci. 2013;102(10):3469–84. doi: 10.1002/jps.23672. [DOI] [PubMed] [Google Scholar]; b Lentz TB, Gray SJ, Samulski RJ. Viral vectors for gene delivery to the central nervous system. Neurobiol Dis. 2012;48(2):179–88. doi: 10.1016/j.nbd.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a Ramamoorth M, Narvekar A. Non viral vectors in gene therapy- an overview. Journal of clinical and diagnostic research : JCDR. 2015;9(1):GE01–6. doi: 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Olsen NJ, Stein CM. New drugs for rheumatoid arthritis. N Engl J Med. 2004;350(21):2167–79. doi: 10.1056/NEJMra032906. [DOI] [PubMed] [Google Scholar]
- 3.Bhise NS, Shmueli RB, Gonzalez J, Green JJ. A novel assay for quantifying the number of plasmids encapsulated by polymer nanoparticles. Small. 2012;8(3):367–73. doi: 10.1002/smll.201101718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a Green JJ, Zugates GT, Langer R, Anderson DG. Poly(beta-amino esters): procedures for synthesis and gene delivery. Methods Mol Biol. 2009;480:53–63. doi: 10.1007/978-1-59745-429-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zugates GT, Peng W, Zumbuehl A, Jhunjhunwala S, Huang YH, Langer R, Sawicki JA, Anderson DG. Rapid optimization of gene delivery by parallel end-modification of poly(beta-amino ester)s. Mol Ther. 2007;15(7):1306–12. doi: 10.1038/mt.sj.6300132. [DOI] [PubMed] [Google Scholar]; c Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(beta-amino ester)s optimized for highly effective gene delivery. Bioconjugate chemistry. 2003;14(5):979–88. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]; d Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(beta-amino esters). Mol Ther. 2005;11(3):426–34. doi: 10.1016/j.ymthe.2004.11.015. [DOI] [PubMed] [Google Scholar]; e Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjug Chem. 2006;17(5):1162–9. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]; f Greenland JR, Liu H, Berry D, Anderson DG, Kim WK, Irvine DJ, Langer R, Letvin NL. Beta-amino ester polymers facilitate in vivo DNA transfection and adjuvant plasmid DNA immunization. Molecular therapy : the journal of the American Society of Gene Therapy. 2005;12(1):164–70. doi: 10.1016/j.ymthe.2005.01.021. [DOI] [PubMed] [Google Scholar]; g Guerrero-Cazares H, Tzeng SY, Young NP, Abutaleb AO, Quinones-Hinojosa A, Green JJ. Biodegradable polymeric nanoparticles show high efficacy and specificity at DNA delivery to human glioblastoma in vitro and in vivo. ACS Nano. 2014;8(5):5141–53. doi: 10.1021/nn501197v. [DOI] [PMC free article] [PubMed] [Google Scholar]; h Keeney M, Ong SG, Padilla A, Yao Z, Goodman S, Wu JC, Yang F. Development of poly(beta-amino ester)-based biodegradable nanoparticles for nonviral delivery of minicircle DNA. ACS nano. 2013;7(8):7241–50. doi: 10.1021/nn402657d. [DOI] [PMC free article] [PubMed] [Google Scholar]; i Zugates GT, Tedford NC, Zumbuehl A, Jhunjhunwala S, Kang CS, Griffith LG, Lauffenburger DA, Langer R, Anderson DG. Gene delivery properties of end-modified poly(beta-amino ester)s. Bioconjugate chemistry. 2007;18(6):1887–96. doi: 10.1021/bc7002082. [DOI] [PubMed] [Google Scholar]; j Mintzer MA, Simanek EE. Nonviral vectors for gene delivery. Chem Rev. 2009;109(2):259–302. doi: 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]; k Jones CH, Chen M, Ravikrishnan A, Reddinger R, Zhang G, Hakansson AP, Pfeifer BA. Mannosylated poly(beta-amino esters) for targeted antigen presenting cell immune modulation. Biomaterials. 2015;37:333–44. doi: 10.1016/j.biomaterials.2014.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cells and tissue. Adv Drug Deliv Rev. 2003;55(3):329–47. doi: 10.1016/s0169-409x(02)00228-4. [DOI] [PubMed] [Google Scholar]; b Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine) and its role in gene delivery. J Control Release. 1999;60(2-3):149–60. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]; c Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22(5):471–80. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 6.Mangraviti A, Tzeng SY, Kozielski KL, Wang Y, Jin Y, Gullotti D, Pedone M, Buaron N, Liu A, Wilson DR, Hansen SK, Rodriguez FJ, Gao GD, DiMeco F, Brem H, Olivi A, Tyler B, Green JJ. Polymeric nanoparticles for nonviral gene therapy extend brain tumor survival in vivo. ACS Nano. 2015;9(2):1236–49. doi: 10.1021/nn504905q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allard E, Passirani C, Benoit JP. Convection-enhanced delivery of nanocarriers for the treatment of brain tumors. Biomaterials. 2009;30(12):2302–18. doi: 10.1016/j.biomaterials.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 8.a Voges J, Reszka R, Gossmann A, Dittmar C, Richter R, Garlip G, Kracht L, Coenen HH, Sturm V, Wienhard K, Heiss WD, Jacobs AH. Imaging-guided convection-enhanced delivery and gene therapy of glioblastoma. Ann Neurol. 2003;54(4):479–87. doi: 10.1002/ana.10688. [DOI] [PubMed] [Google Scholar]; b MacKay JA, Li W, Huang Z, Dy EE, Huynh G, Tihan T, Collins R, Deen DF, Szoka FC. HIV TAT peptide modifies the distribution of DNA nanolipoparticles following convection- enhanced delivery. Molecular Therapy. 2008;16(5):893–900. doi: 10.1038/mt.2008.36. [DOI] [PubMed] [Google Scholar]; c Stiles DK, Zhang Z, Ge P, Nelson B, Grondin R, Ai Y, Hardy P, Nelson PT, Guzaev AP, Butt MT, Charisse K, Kosovrasti V, Tchangov L, Meys M, Maier M, Nechev L, Manoharan M, Kaemmerer WF, Gwost D, Stewart GR, Gash DM, Sah DW. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp Neurol. 2012;233(1):463–71. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 9.Sykova E, Nicholson C. Diffusion in brain extracellular space. Physiol Rev. 2008;88(4):1277–340. doi: 10.1152/physrev.00027.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a Costa PM, Cardoso AL, Custodia C, Cunha P, Pereira de Almeida L, Pedroso de Lima MC. MiRNA-21 silencing mediated by tumor-targeted nanoparticles combined with sunitinib: A new multimodal gene therapy approach for glioblastoma. J Control Release. 2015;207:31–39. doi: 10.1016/j.jconrel.2015.04.002. [DOI] [PubMed] [Google Scholar]; b Iwami K, Natsume A, Wakabayashi T. Gene therapy for high-grade glioma. Neurol Med Chir (Tokyo) 2010;50(9):727–36. doi: 10.2176/nmc.50.727. [DOI] [PubMed] [Google Scholar]; c Lang FF, Bruner JM, Fuller GN, Aldape K, Prados MD, Chang S, Berger MS, McDermott MW, Kunwar SM, Junck LR, Chandler W, Zwiebel JA, Kaplan RS, Yung WK. Phase I trial of adenovirus-mediated p53 gene therapy for recurrent glioma: biological and clinical results. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2003;21(13):2508–18. doi: 10.1200/JCO.2003.21.13.2508. [DOI] [PubMed] [Google Scholar]; d Walther W, Stein U. Viral vectors for gene transfer: a review of their use in the treatment of human diseases. Drugs. 2000;60(2):249–71. doi: 10.2165/00003495-200060020-00002. [DOI] [PubMed] [Google Scholar]; e Smitt PS, Driesse M, Wolbers J, Kros M, Avezaat C. Treatment of relapsed malignant glioma with an adenoviral vector containing the herpes simplex thymidine kinase gene followed by ganciclovir. Molecular therapy : the journal of the American Society of Gene Therapy. 2003;7(6):851–8. doi: 10.1016/s1525-0016(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 11.a MacKay JA, Deen DF, Szoka FC., Jr. Distribution in brain of liposomes after convection enhanced delivery; modulation by particle charge, particle diameter, and presence of steric coating. Brain research. 2005;1035(2):139–53. doi: 10.1016/j.brainres.2004.12.007. [DOI] [PubMed] [Google Scholar]; b Kenny GD, Bienemann AS, Tagalakis AD, Pugh JA, Welser K, Campbell F, Tabor AB, Hailes HC, Gill SS, Lythgoe MF, McLeod CW, White EA, Hart SL. Multifunctional receptor-targeted nanocomplexes for the delivery of therapeutic nucleic acids to the Brain. Biomaterials. 2013;34(36):9190–200. doi: 10.1016/j.biomaterials.2013.07.081. [DOI] [PubMed] [Google Scholar]; c Writer MJ, Kyrtatos PG, Bienemann AS, Pugh JA, Lowe AS, Villegas-Llerena C, Kenny GD, White EA, Gill SS, McLeod CW, Lythgoe MF, Hart SL. Lipid peptide nanocomplexes for gene delivery and magnetic resonance imaging in the brain. J Control Release. 2012;162(2):340–8. doi: 10.1016/j.jconrel.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Mastorakos P, Zhang C, Berry S, Oh Y, Lee S, Eberhart CG, Woodworth GF, Suk JS, Hanes J. Highly PEGylated DNA nanoparticles provide uniform and widespread gene transfer in the brain. Advanced healthcare materials. 2015 doi: 10.1002/adhm.201400800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mastorakos P, Silva A. L. d., Chisholm J, Song E, Choi WK, Boyle MP, Morales MM, Hanes J, Suk JS. Highly Compacted Biodegradable DNA Nanoparticles Capable of Overcoming the Mucus Barrier for Inhaled Lung Gene Therapy. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1502281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a Nance E, Zhang C, Shih TY, Xu Q, Schuster BS, Hanes J. Brain-penetrating nanoparticles improve paclitaxel efficacy in malignant glioma following local administration. ACS Nano. 2014;8(10):10655–64. doi: 10.1021/nn504210g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Nance EA, Woodworth GF, Sailor KA, Shih TY, Xu Q, Swaminathan G, Xiang D, Eberhart C, Hanes J. A dense poly(ethylene glycol) coating improves penetration of large polymeric nanoparticles within brain tissue. Science translational medicine. 2012;4(149):149ra119. doi: 10.1126/scitranslmed.3003594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krauze MT, Saito R, Noble C, Bringas J, Forsayeth J, McKnight TR, Park J, Bankiewicz KS. Effects of the perivascular space on convection-enhanced delivery of liposomes in primate putamen. Experimental neurology. 2005;196(1):104–11. doi: 10.1016/j.expneurol.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Suk JS, Kim AJ, Trehan K, Schneider CS, Cebotaru L, Woodward OM, Boylan NJ, Boyle MP, Lai SK, Guggino WB, Hanes J. Lung gene therapy with highly compacted DNA nanoparticles that overcome the mucus barrier. Journal of controlled release : official journal of the Controlled Release Society. 2014;178C:8–17. doi: 10.1016/j.jconrel.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Accounts of chemical research. 2008;41(6):749–59. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh S, Pluhar GE, McNeil EA, Kroeger KM, Liu C, Castro MG, Lowenstein PR, Freese A, Ohlfest JR. Efficacy of nonviral gene transfer in the canine brain. Journal of neurosurgery. 2007;107(1):136–44. doi: 10.3171/JNS-07/07/0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salegio EA, Streeter H, Dube N, Hadaczek P, Samaranch L, Kells AP, San Sebastian W, Zhai Y, Bringas J, Xu T, Forsayeth J, Bankiewicz KS. Distribution of nanoparticles throughout the cerebral cortex of rodents and non-human primates: Implications for gene and drug therapy. Front Neuroanat. 2014;8:9. doi: 10.3389/fnana.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a Dent P, Yacoub A, Park M, Sarkar D, Shah K, Curiel DT, Grant S, Fisher PB. Searching for a cure: gene therapy for glioblastoma. Cancer Biol Ther. 2008;7(9):1335–40. doi: 10.4161/cbt.7.9.6408. [DOI] [PubMed] [Google Scholar]; b Lim ST, Airavaara M, Harvey BK. Viral vectors for neurotrophic factor delivery: a gene therapy approach for neurodegenerative diseases of the CNS. Pharmacological research : the official journal of the Italian Pharmacological Society. 2010;61(1):14–26. doi: 10.1016/j.phrs.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. European journal of cell biology. 2004;83(3):97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 21.a Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32(23):5402–10. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Anderson DG, Lynn DM, Langer R. Semi-automated synthesis and screening of a large library of degradable cationic polymers for gene delivery. Angewandte Chemie. 2003;42(27):3153–8. doi: 10.1002/anie.200351244. [DOI] [PubMed] [Google Scholar]
- 22.a Mastorakos P, Zhang C, Berry S, Oh Y, Lee S, Eberhart CG, Woodworth Graeme F., Suk JS, Hanes J. Highly PEGylated DNA Nanoparticles Provide Uniform and Widespread Gene Transfer in the Brain. Advanced Healthcare Materials. 2015 doi: 10.1002/adhm.201400800. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Konstan MW, Davis PB, Wagener JS, Hilliard KA, Stern RC, Milgram LJ, Kowalczyk TH, Hyatt SL, Fink TL, Gedeon CR, Oette SM, Payne JM, Muhammad O, Ziady AG, Moen RC, Cooper MJ. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum Gene Ther. 2004;15(12):1255–69. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- 23.Mastorakos P, Zhang C, Berry S, Oh Y, Lee S, Eberhart CG, Kim AJ, Woodworth GF, Suk JS, Hanes J. Brain Penetrating DNA Nanoparticles for Efficient Gene Transfer to the Central Nervous System. 2014 In Press. [Google Scholar]
- 24.Schuster BS, Suk JS, Woodworth GF, Hanes J. Nanoparticle diffusion in respiratory mucus from humans without lung disease. Biomaterials. 2013;34(13):3439–46. doi: 10.1016/j.biomaterials.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Recinos VR, Tyler BM, Bekelis K, Sunshine SB, Vellimana A, Li KW, Brem H. Combination of intracranial temozolomide with intracranial carmustine improves survival when compared with either treatment alone in a rodent glioma model. Neurosurgery. 2010;66(3):530–7. doi: 10.1227/01.NEU.0000365263.14725.39. discussion 537. [DOI] [PubMed] [Google Scholar]
- 26.Elliott SL, Cullen CF, Wrobel N, Kernan MJ, Ohkura H. EB1 is essential during Drosophila development and plays a crucial role in the integrity of chordotonal mechanosensory organs. Mol Biol Cell. 2005;16(2):891–901. doi: 10.1091/mbc.E04-07-0633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.