Summary
Interleukin‐10 (IL‐10) is a key regulator of mucosal homeostasis. In the current study we investigated the early events after monoassociating germ‐free (GF) wild‐type (WT) mice with an Escherichia coli strain that we isolated previously from the caecal contents of a normal mouse housed under specific pathogen‐free conditions. Our results show that interferon‐γ (IFN‐γ) secreted by mesenteric lymph node (MLN) cells from both IL‐10 deficient mice and WT mice, stimulated ex vivo with E. coli lysate, was dramatically higher at day 4 after monoassociation compared with IFN‐γ secreted by cells from GF mice without E. coli colonization. Production of IFN‐γ rapidly and progressively declined after colonization of WT but not IL‐10‐deficient mice. The E. coli lysate‐stimulated WT MLN cells also produced IL‐10 that peaked at day 4 and subsequently declined, but not as precipitously as IFN‐γ. WT cells that express CD4, CD8 and NKp46 produced IFN‐γ; WT CD4‐positive cells and B cells produced IL‐10. Recombinant IL‐10 added to E. coli‐stimulated MLN cell cultures inhibited IFN‐γ secretion in a dose‐dependent fashion. MLN cells from WT mice treated in vivo with neutralizing anti‐IL‐10 receptor antibody produced more IFN‐γ compared with MLN cells from isotype control antibody‐treated mice. These findings show that a resident E. coli that induces chronic colitis in monoassociated IL‐10‐deficient mice rapidly but transiently activates the effector immune system in normal hosts, in parallel with induction of protective IL‐10 produced by B cells and CD4+ cells that subsequently suppresses this response to mediate mucosal homeostasis.
Keywords: germ‐free mice, interleukin‐10, resident bacteria
Abbreviations
- GF
germ‐free
- IFN
interferon
- IL
interleukin
- IL‐10R
interleukin‐10 receptor
- MLN
mesenteric lymph node
- SPF
specific pathogen free
- WT
wild‐type
Introduction
The gastrointestinal tract harbours a complex set of microbes, particularly in the large intestine, which is inhabited by approximately 10–100 trillion bacteria comprised of more than 1000 different species.1, 2, 3, 4, 5 Bacteria colonize the intestinal lumen after birth and interact with the host through antigens and adjuvants that stimulate either pathogenic or protective immune responses.1, 6
Interleukin‐10 (IL‐10) is a regulatory cytokine that is produced by a wide variety of cell types including T cells, B cells, macrophages, dendritic cells and epithelial cells.7, 8, 9, 10, 11, 12 This cytokine is a key inhibitor of antigen‐presenting cell function and so suppresses effector T‐cell responses and helps to maintain mucosal homeostasis.12, 13, 14 Interleukin‐10 is associated with immunological tolerance and regulatory T‐cell responses.15, 16, 17 The critical role of IL‐10 in maintaining intestinal immune regulation is demonstrated by the observation that conventionally housed IL‐10‐deficient and IL‐10‐receptor‐deficient mice spontaneously develop chronic intestinal inflammation.18, 19 In parallel, IL‐10 produced by T cells is necessary for inhibition of colitis in various murine colitis models.20 In addition to T cells, IL‐10‐secreting B cells activated by resident bacteria induce regulatory type 1 T cells, suppress effector T cells, and inhibit colitis in several murine model systems.21, 22 Soon after IL‐10‐deficient mice became available, we re‐derived them into germ‐free (GF) conditions and observed that the GF cohort, unlike the conventional IL‐10‐deficient mice, did not develop intestinal inflammation or activation of the gut‐associated immune system.23 These results demonstrate the importance of resident intestinal microorganisms in the development of intestinal inflammation in IL‐10‐deficient mice.
Numerous publications report IL‐10‐mediated inhibition of intestinal inflammation and investigators have identified pivotal molecules in the IL‐10 signalling pathway;14, 24, 25 however, most of these studies focus on chronic inflammation. Few have investigated the immune response at very early time‐points after exposure to resident intestinal bacteria. In a previous study,13 we examined the early stages of immune activation in experiments designed to determine the role of T cell‐derived IL‐10 compared to antigen‐presenting‐cell‐derived IL‐10 in regulating colitis. Our kinetic analysis of cytokine production after adoptive transfer of T cells to specific pathogen‐free (SPF) immunodeficient hosts demonstrated the rapid induction of interferon‐γ (IFN‐γ) in response to components of resident intestinal microorganisms. This response was subsequently down‐regulated in the presence but not in the absence of IL‐10‐producing antigen‐presenting cells. These results are particularly interesting because the adoptive transfer model permits evaluation of the parameters of T‐cell responses as they develop during homeostatic proliferation. El Aidy et al.26, 27 observed similarly rapid induction of cytokine gene expression after GF wild‐type (WT) mice were colonized with complex murine resident intestinal microbiota. We now report results that confirm and extend the previous studies, showing that IFN‐γ and IL‐10 were both produced rapidly after GF WT mice were monoassociated with a single intestinal resident bacteria designated Escherichia coli NC101 that we isolated from the caecum of an SPF WT mouse. Interferon‐γ subsequently declined; IL‐10 also declined, but not as precipitously as IFN‐γ. The IFN‐γ is produced by CD4‐positive, CD8‐positive and NKp46‐positive cells while CD4‐positive cells and B cells produce IL‐10. In vitro addition of recombinant murine IL‐10 suppressed E. coli ‐induced IFN‐γ production, while blocking the IL‐10 receptor (IL‐10R) in vivo enhanced IFN‐γ production, confirming that IL‐10 is involved in down‐regulating the early immune response to a resident E. coli in normal hosts.
Materials and methods
Mice
Interleukin‐10‐deficient mice on a 129S6/SvEv background, and WT 129S6/SvEv mice were maintained in GF conditions in flexible film isolators at the National Gnotobiotic Rodent Resource Center at the University of North Carolina, Chapel Hill and the Gnotobiotic Core of the Center for Gastrointestinal Biology and Disease (CGIBD) at North Carolina State University. We confirm GF status each time we remove mice from isolators for an experiment. All mice used were older than 8 weeks of age. Mice were monoassociated with E. coli NC101 by swabbing the mouth and anus with an overnight bacterial culture and were subsequently euthanized at different time‐points after initial bacterial colonization. Animal use protocols were approved by the University of North Carolina at Chapel Hill and the North Carolina State University Institutional Animal Care and Use Committees.
Bacterial strain and lysates
The murine E. coli strain designated E. coli NC101 was originally isolated from WT mice housed in SPF conditions. Lysates were prepared from E. coli cultures grown in brain–heart infusion broth as described previously.28 Protein concentrations were determined using a standard assay (Bio‐Rad Laboratories, Hercules, CA) and sterility of the lysates was confirmed by culturing on brain–heart infusion agar plates. Lysates were divided into aliquots and stored at −80°.
Mesenteric lymph node cell cultures
Mesenteric lymph nodes (MLN) were collected from individual mice, teased gently and the cell suspension was passed through a 40‐μm filter (BD Falcon, Franklin Lakes, NJ). Unseparated MLN cells (4 × 105 cells per well) were stimulated with 1 or 10 μg/ml E. coli NC101 lysate or cultured in medium alone in 96‐well flat‐bottom plates (Costar, Corning, NY) for 72 hr at 37° in a humidified incubator with 5% CO2. Culture medium was RPMI‐1640 (Cellgro, Manassas, VA) supplemented with 5% heat inactivated fetal calf serum, 2 mm l‐glutamine, 1 mm sodium pyruvate, 0·05 mm 2‐mercaptoethanol and 50 μg/ml gentamicin. Culture supernatants were collected for cytokine analysis and frozen in aliquots at −20°.
For evaluation of cytokine‐producing cells, MLN cells were stimulated overnight with 10 μg/ml E. coli NC101 lysate. Cells were then re‐stimulated with 100 ng/ml PMA (Sigma‐Aldrich, St Louis, MO) plus 1 μg/ml ionomycin (Tocris Bioscience, Minneapolis, MN) for 5 hr. Golgi stop (BD Biosciences, San Diego, CA) was added during the last 4 hr.
Recombinant IL‐10 treatment and IL‐10R blockade
To evaluate the effects of exogenous IL‐10, recombinant murine IL‐10 (PeproTech, Rocky Hill, NJ) was added to MLN cultures at 0, 250 pg/ml, 500 pg/ml, 1000 pg/ml, or 2000 pg/ml.
To block IL‐10 signalling in vivo, 0·5 mg per mouse of anti‐IL‐10R‐specific antibody (clone 1B1.3A; BioXCell, West Lebanon, NH) or purified rat IgG1 isotype control antibody (clone HRPH; BioXCell) was injected intraperitoneally 1 day before, 2 days after and 5 days after monoassociation with E. coli NC101. Mice were euthanized on day 7 after E. coli monoassociation. The MLN cells were prepared and stimulated with E. coli NC101 lysate as described above.
Cytokine measurements
To measure the amount of IFN‐γ or IL‐10 secreted by stimulated MLN cells, ELISAs were performed by staff of the Advanced Analytics Core of the CGIBD at the University of North Carolina at Chapel Hill. The ELISA sets for murine IFN‐γ (catalogue no. 551866) and murine IL‐10 (catalogue no. 555252) were from BD Biosciences. Assays were performed according to the manufacturer's protocols. Cytokine levels were measured in triplicate culture supernatants and amounts were determined by comparison with standard curves generated by using the appropriate recombinant cytokine.
Real‐time polymerase chain reaction
Total RNA from transverse colon was extracted using an RNeasy Mini Kit (Qiagen Sciences, Valencia, CA) following the manufacturer's instructions and treated with amplification‐grade DNase Ι (Qiagen Sciences). First‐strand complementary DNA was synthesized using a Tetro cDNA Synthesis Kit (Bioline, Taunton, MA) and treated with Ribonuclease H (Invitrogen, Carlsbad, CA). Quantitative RT‐PCR was performed on a QuantStudio 6 Flex Real‐Time PCR System (Life Technologies, Carlsbad, CA) using SYBR green (Bio‐Rad) to quantify the gene expression of IFN‐γ, IL‐10, IL‐1β and IL‐6. Each sample was analysed in triplicate and the results were normalized to the housekeeping gene GAPDH.
Flow cytometry
Antibodies used to enumerate cell subsets directly after MLN harvest were peridinin chlorophyll protein‐labelled anti‐mouse CD4, phycoerythrin‐labelled anti‐mouse CD8, or FITC‐labelled anti‐mouse CD45R/B220 (BD Biosciences). Antibodies used to detect cell surface molecules in intracellular cytokine detection experiments were phycoerythrin‐labelled anti‐mouse CD4, CD8, CD19, CD11b and CD335 (NKp46). Alexa Fluor 488 labelled anti‐IFN‐γ or anti‐IL‐10 were used to enumerate cytokine‐producing cells (eBioscience, San Diego, CA). MLN cells were stimulated for intracellular cytokine detection as described above, collected and incubated with anti‐CD antibodies for 30 min at 4°. Cells were washed, then fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences) according to the manufacturer's instructions before incubating with anti‐IFN‐γ, anti‐IL‐10, or isotype control antibody for 1 hr at 4°. Cells were washed and analysed on an LSRII flow cytometer (BD Biosciences). We gated on the events for analysis based on forward and side scatter characteristics of viable cells. To determine the proportions of cytokine‐positive cells among cell subsets, we gated on the cell‐surface‐marker‐positive populations.
Data analysis
All statistics were performed using graphpad prism 5.0 software (GraphPad, San Diego, CA). We used two‐tailed Student's t‐tests for comparisons between two groups. Comparisons between three or more experimental groups were analysed using one‐way analysis of varaince with Tukey's multiple‐comparison post‐test. P‐values < 0·05 were considered significant.
Results
Kinetics of IFN‐γ production in E. coli NC101 monoassociated mice
To evaluate intestinal bacteria‐induced immune responses in the intestinal tract upon bacterial colonization, we used 129SvEv WT and IL‐10‐deficient mice that were born in GF conditions and monoassociated with a non‐pathogenic murine strain of Escherichia coli, designated E. coli NC101.28 Escherichia coli NC101 shares phylogenetic and genomic characteristics with several human and canine strains of adherent/invasive E. coli.29 However, our previous results show that this organism is not pathogenic because inflammation does not develop in WT mice.28
At different time‐points after monoassociation, we harvested MLN and stimulated the cells in vitro with bacterial lysates containing components of E. coli NC101. We then measured IFN‐γ and IL‐10 secretion in culture supernatants. As shown in Fig. 1, we found that IFN‐γ produced by unfractionated MLN cells from both IL‐10‐deficient and WT mice was dramatically higher at day 4 after colonization compared with GF mice. The IFN‐γ remained consistently high in supernatants of E. coli lysate‐stimulated IL‐10‐deficient MLN cells obtained at day 7 and 30 after monoassociation, corroborating our previously published results,28, 30 whereas amounts of IFN‐γ rapidly declined with time after colonization of WT mice (Fig. 1a,b). We tested two different concentrations of E. coli NC101 lysate, 10 and 1 μg/ml, for in vitro re‐stimulation. Results were consistent for both doses (data for cells stimulated with 1 μg/ml not shown). We also evaluated IL‐17 production in the supernatants of stimulated MLN cells and IL‐12p40 production in colon explant cultures. Interleukin‐17 secretion by MLN cells from IL‐10‐deficient mice followed the same kinetics after E. coli monoassociation as IFN‐γ; however, MLN cells from WT mice did not produce detectable IL‐17 (data not shown). Likewise, IL‐12p40 in colonic culture supernatants from IL‐10‐deficient mice increased with time after monoassociation but was undetectable in the supernatants of colon tissue from WT mice (data not shown).
Figure 1.
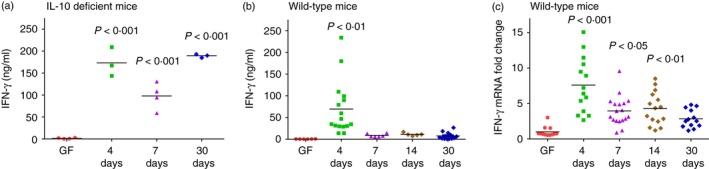
Kinetics of interferon‐γ (IFN‐γ) production in 129SvEv wild‐type (WT) and interleukin‐10 (IL‐10) ‐deficient mice monoassociated with Escherichia coli NC101. (a) IFN‐γ production by unseparated mesenteric lymph node (MLN) cells from germ‐free (GF) or E. coli NC101 monoassociated IL‐10‐deficient mice, 129SvEv background. (b) IFN‐γ production by unseparated MLN cells from GF or E. coli NC101 monoassociated 129SvEv WT mice. MLN cells were stimulated in vitro with 10 μg/ml E. coli NC101 lysate. Supernatants were collected 72 hr later and IFN‐γ was quantified by ELISA. (c) IFN‐γ mRNA expression in transverse colon of GF or E. coli NC101 monoassociated 129SvEv WT mice determined by quantitative PCR. Copy number is normalized to GAPDH and expressed as fold change relative to GF mice. Each dot represents an individual mouse analysed in either two independent experiments (GF or day 4, 7, 14, 30 WT mice or GF IL‐10‐deficient mice) or one experiment (IL‐10‐deficient mice day 4, 7 and 30). Day after monoassociation is indicated on the x‐axis. Data were analysed using one‐way analysis of variance with Tukey's multiple‐comparison post‐test. P‐values indicate statistically significant differences between IFN‐γ produced by E. coli NC 101 lysate‐stimulated MLN cells in (a) and (b) or IFN‐γ mRNA expressed in transverse colon in (b) from GF mice compared with mice at 4, 7, 14 or 30 days after monoassociation with E. coli NC101. In addition, statistically significant differences in (b) 4‐day versus 7‐day P < 0·05, 4‐day versus 14‐day P < 0·05, 4‐day versus 30‐day P < 0·001; in (c), 4‐day versus 7‐day P < 0·001, 4‐day versus 14‐day P < 0·01, 4‐day versus 30‐day P < 0·001.
We next investigated IFN‐γ mRNA expression in the transverse colon of GF and E. coli monoassociated WT mice. The IFN‐γ mRNA expressed in the transverse colon showed the same pattern as IFN‐γ production in the supernatants of bacterial lysate‐stimulated WT MLN cells, with significantly higher IFN‐γ mRNA levels 4 days after monoassociation with E. coli compared with GF and lower amounts at later time‐points (Fig. 1c). Together, these data show that IFN‐γ is produced in normal, WT mice at early time‐points in response to resident intestinal bacteria.
Kinetics of IL‐10, IL‐1β and IL‐6 production in E. coli NC101 monoassociated mice
Interleukin‐10‐mediated inhibition of IFN‐γ production has been established in numerous model systems.12, 31, 32 Therefore, we explored IL‐10 production in the same kinetic study. We measured IL‐10 secretion in supernatants of E. coli lysate‐stimulated unseparated MLN cells and IL‐10 mRNA expression in transverse colon from the same monoassociated WT mice. We found that IL‐10 secretion in the supernatants was higher at day 4 after colonization compared with GF levels and decreased thereafter (Fig. 2a). Interleukin‐10 mRNA expression in transverse colon from WT mice showed the same patterns with higher levels of IL‐10 mRNA at day 4 after monoassociation compared with GF mice, progressively decreasing at later time‐points (Fig. 2b). Together, these results show that IFN‐γ and IL‐10 are produced rapidly and concomitantly in normal hosts upon bacterial monoassociation.
Figure 2.
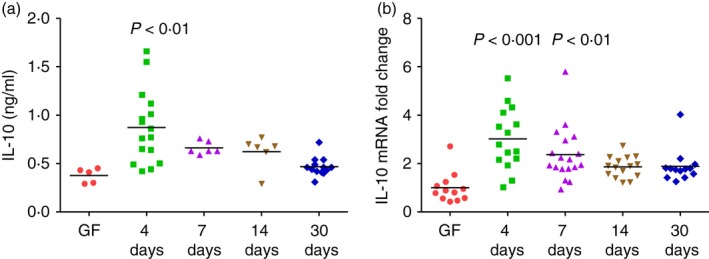
Kinetics of interleukin‐10 (IL‐10) production in 129SvEv wild‐type (WT) mice monoassociated with Escherichia coli NC101. (a) IL‐10 production by unseparated mesenteric lymph node (MLN) cells from germ‐free (GF) or E. coli NC101 monoassociated 129SvEv WT mice. Day after monoassociation is indicated on the x‐axis. MLN cells were stimulated in vitro with 10 μg/ml E. coli NC 101 lysate. Supernatants were collected 72 hr later and IL‐10 was quantified by ELISA. (b) IL‐10 mRNA expression in transverse colon of GF or E. coli NC101 monoassociated 129SvEv WT mice determined by quantitative PCR. Copy number is normalized to GAPDH and expressed as fold change relative to GF. Each dot represents an individual mouse evaluated in experiments described for Figure 1. Data were analysed using one‐way analysis of variance with Tukey's multiple‐comparison post‐test. P‐values indicate statistically significant differences between IL‐10 produced by E. coli NC 101 lysate‐stimulated MLN cells in (a) or IL‐10 mRNA expressed in transverse colon in (b) from GF mice compared with mice at 4, 7, 14 or 30 days after E. coli NC101 monoassociation. In addition, statistically significant differences in (a), 4‐day versus 7‐day not significant, 4‐day versus 14‐day not significant, 4‐day versus 30‐day P < 0·001; in (b) 4‐day versus 7‐day not significant, 4‐day versus 14‐day P < 0·01, 4‐day versus 30‐day P < 0·05.
We also quantified IL‐1β and IL‐6 mRNA expression in the transverse colon from monoassociated WT mice (Fig. 3). The IL‐1β mRNA expression was approximately fourfold higher 4 days after colonization compared with GF, then decreased at later time‐points (Fig. 3a). Interleukin‐6 mRNA expression (Fig. 3b) was similar to IL‐1β, with fivefold higher levels at day 4 compared with GF followed by a subsequent decline. These data show that like IFN‐γ and IL‐10, both IL‐1β and IL‐6 are activated rapidly after monoassociation of normal mice with a single non‐pathogenic bacterial species, then are subsequently down‐regulated.
Figure 3.
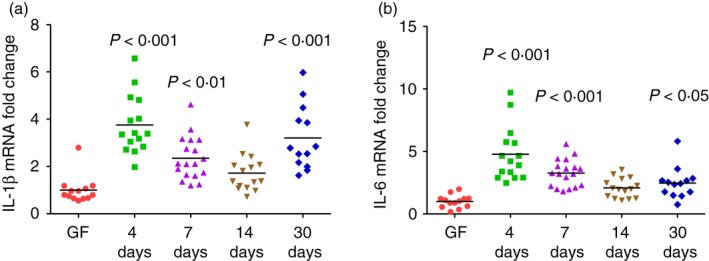
Kinetics of interleukin‐1β (IL‐1β) and IL‐6 expression in transverse colon of 129SvEv wild‐type (WT) mice monoassociated with Escherichia coli NC101. (a) IL‐1β mRNA expression in transverse colon of germ‐free (GF) or E. coli NC101 monoassociated 129SvEv WT mice determined by quantitative PCR. Day after monoassociation is indicated on the x‐axis. Copy number is normalized to GAPDH and expressed as fold change relative to GF. Each dot represents an individual mouse. Data were analysed using one‐way analysis of variance with Tukey's multiple‐comparison post‐test. P‐values indicate statistically significant differences between IL‐1β expressed in transverse colon from GF mice compared with mice at 4, 7, 14 or 30 days after monoassociation. In addition, statistically significant differences comparing 4‐day versus 7‐day P < 0·01 4‐day versus 14‐day P < 0·001 4‐day versus 30‐day not significant. (b) IL‐6 mRNA expression in transverse colon of GF or E. coli NC101 monoassociated 129SvEv WT mice determined by quantitative PCR. Copy number is normalized to GAPDH and expressed as fold change relative to GF. Each dot represents an individual mouse evaluated in experiments described for Fig. 1. Data were analysed using one‐way analysis of variance with Tukey's multiple‐comparison post‐test. P‐values indicate statistically significant differences between IL‐6 expressed in transverse colon from GF mice compared with mice at 4, 7, 14 or 30 days after monoassociation. In addition, statistically significant differences in comparing 4‐day versus 7‐day P < 0·05, 4‐day versus 14‐day P < 0·001, 4‐day versus 30‐day P < 0·001.
Cell number and phenotype
While the total number of cells obtained from MLN of E. coli monoassociated mice was significantly higher than the number harvested from MLN of GF mice (Fig. 4c), the proportions of CD4‐positive cells, CD8‐positive cells and B cells in MLN of GF and E. coli monoassociated mice were essentially the same (Fig. 4a), with the exception of a slightly higher percentage of CD4‐positive cells at 14 days after colonization with E. coli. Therefore, the increased amounts of IFN‐γ (Fig. 1b) and of IL‐10 (Fig. 2a) secreted by E. coli lysate‐stimulated MLN cells obtained at day 4 after E. coli monoassociation is not the result of changes in cell subset proportions among the fixed number of MLN cells (4 × 105 per well) from GF and E. coli monoassociated mice that were stimulated in vitro.
Figure 4.
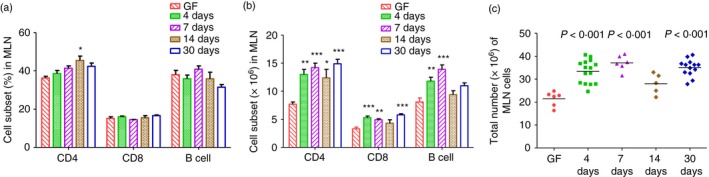
Total cell number and cell subsets in mesenteric lymph nodes (MLN) of 129SvEv wild‐type (WT) mice monoassociated with Escherichia coli NC101 or germ‐free (GF). Unseparated MLN cells from GF or E. coli NC101 monoassociated 129SvEv WT mice were collected and analysed by flow cytometry in experiments described for Fig. 1. (a) Percent CD4‐positive cells, CD8‐positive cells, or B cells in MLN, (b) Total numbers of CD4‐positive cells, CD8‐positive cells, or B cells in MLN. In (a) and (b), data are presented as mean ± SEM. Asterisks indicate statistically significant differences compared with GF in each cell type. *P < 0·05, **P < 0·01, ***P < 0·001. (c) Total MLN cell numbers. Each dot represents an individual mouse evaluated in experiments described for Fig. 1. P‐values indicate statistically significant differences in total numbers of MLN cells obtained from mice at 4, 7, 14 or 30 days after monoassociation with E. coli NC 101 compared with MLN cells obtained from GF mice. Data were analysed using one‐way analysis of variance with Tukey's multiple‐comparison post‐test.
Cell types that produce IFN‐γ and IL‐10
Next we designed experiments to determine which types of cells in MLN produce IFN‐γ and IL‐10 after normal mice are monoassociated with intestinal bacteria. We harvested MLN cells from WT mice monoassociated with E. coli NC101 for 4 days and re‐stimulated them with E. coli lysate overnight, followed by a 5‐hr stimulation with ionomycin plus PMA. For comparison, we also evaluated MLN cells from GF WT mice stimulated according to the same activation protocol. Representative intracellular cytokine analysis using MLN cells from E. coli monoassociated mice (Fig. 5a) shows that a small proportion of CD4+ cells and a higher proportion of CD8+ cells produce IFN‐γ. Cells that express the NK cell and innate lymphoid cell marker NKp4633, 34 also produce IFN‐γ. IL‐10 production can be readily detected in cells that express CD19; a smaller proportion CD4+ cells produce IL‐10. Interestingly, the potent activation signals provided by in vitro stimulation with E. coli lysate followed by ionomycin plus PMA are sufficient for detection of cytokine‐producing cells in MLN of GF mice (Fig. 5b,c). Although the amounts of IFN‐γ in supernatants of MLN cells from GF mice stimulated with E. coli lysate for 72 hr ranged from undetectable to ~0·6 ng/ml (Fig. 1b), the intracellular IFN‐γ results show that MLN cells from GF mice have the capacity to produce this cytokine. However, MLN from GF mice contain fewer IFN‐γ producing CD4‐positive and NKp46‐positive cells compared with MLN from E. coli monoassociated mice (Fig. 5b).
Figure 5.
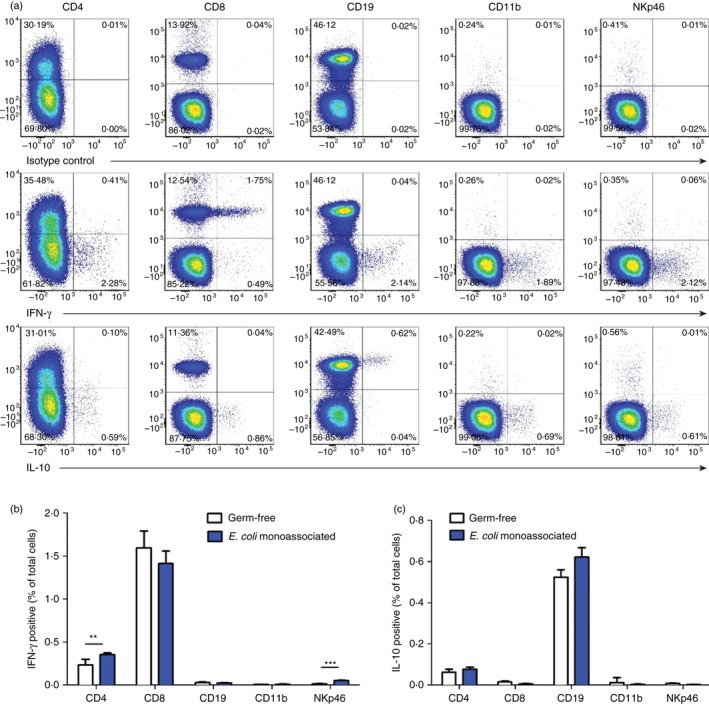
Cell types that produce interferon‐γ (IFN‐γ) and interleukin‐10 (IL‐10). Unseparated mesenteric lymph node (MLN) cells from 129SvEv wild‐type (WT) mice monoassociated with Escherichia coli NC101 for 4 days or from 129SvEv WT germ‐free (GF) mice were stimulated overnight with 10 μg/ml E. coli NC101 lysate, followed by 5 hr with ionomycin plus PMA. Golgi stop was added during the last 4 hr. (a) Representative flow cytometry plots showing IFN‐γ‐ or IL‐10‐producing CD4, CD8, CD19, CD11b and NKp46 cells in MLN from E. coli monoassociated mice, (b) mean percent (minus mean percent isotype control) ± SEM of IFN‐γ‐positive MLN cells from GF mice (n = 6) or E. coli monoassociated mice (n = 4), (c) mean percent (minus mean percent isotype control) ± SEM of IL‐10‐positive MLN cells from GF mice or E. coli monoassociated mice. All four E. coli monoassociated mice were evaluated in one experiment, representative of three separate experiments. The six GF mice were evaluated in two separate experiments, three mice in each. Data were analysed by unpaired Student's t‐test. Asterisks indicate statistically significant differences. **P < 0·01, ***P < 0·001. No significant differences were observed for IL‐10 detection in any of the cell types evaluated.
We have also determined cytokine‐producing cells as a proportion of each MLN cell subset. The results in Table 1 show that although only 0·44% of MLN cells from mice monoassociated with E. coli express NKp46, an average of 12% of these cells produce IFN‐γ. A similar proportion of CD8‐positive cells produce this cytokine, whereas IL‐10 is produced by much lower proportions of CD4‐positive (0·25%) or CD19‐positive (1·4%) cells.
Table 1.
Proportion of interferon‐γ‐ or IL‐10‐positive cells in mesenteric lymph node cell subsets
| IFN‐γ positive cells | ||||
|---|---|---|---|---|
| Germ‐free | E. coli monoassociated for 4 days | |||
| Percent of subset in MLNa | Percent IFN‐γ‐positive of subsetb | Percent of subset in MLN | Percent IFN‐γ‐positive of subset | |
| CD4 | 32·8 ± 1·6 | 0·74 ± 0·06 | 31·5 ± 1·9 | 1·13 ± 0·06** |
| CD8 | 14·8 ± 0·7 | 10·68 ± 0·83 | 12·3 ± 0·7 | 11·46 ± 0·55 |
| NKp46 | 0·39 ± 0·01 | 3·55 ± 0·83 | 0·44 ± 0·01 | 12·42 ± 1·23*** |
| IL‐10 positive cells | ||||
|---|---|---|---|---|
| Germ‐free | E. coli monoassociated for 4 days | |||
| Percent of subset in MLN | Percent IL‐10 positive of subsetc | Percent of subset in MLN | Percent IL‐10 positive of subset | |
| CD4 | 32·8 ± 1·6 | 0·17 ± 0·04 | 31·5 ± 1·9 | 0·25 ± 0·04 |
| CD19 | 37·5 ± 2·0 | 1·40 ± 0·05 | 43·5 ± 1·7 | 1·43 ± 0·07 |
Values represent mean ± SEM of cells expressing the indicated cell surface marker evaluated after overnight stimulation with E. coli NC101 lysate followed by 5 hr incubation with ionomycin plus PMA, with Golgi stop during the last 4 hr. MLN from E. coli NC101 monoassociated mice n = 4; MLN from GF mice n = 6.
Values represent mean ± SEM of the proportion of IFN‐γ positive cells determined after gating on each subset.
Values represent mean ± SEM of the proportion of IL‐10 positive cells determined after gating on each subset.
**P < 0·01 compared to dual CD4 positive – IFN‐γ positive cells in MLN of GF mice; ***P < 0·001 compared to dual NKp46 positive – IFN‐γ positive cells in MLN of GF mice. Otherwise differences between percent of cytokine positive cells of each subset from E. coli monoassociated and GF mice are not statistically significant.
Recombinant IL‐10 treatment in vitro
We then evaluated the effects of exogenous IL‐10 on IFN‐γ production in vitro in our model system. We harvested MLN cells from WT mice monoassociated with E. coli NC101 for 4 days and stimulated the cells with E. coli NC101 lysate in the presence of 0, 250, 500, 1000 or 2000 pg/ml recombinant IL‐10. As shown in Fig. 6, addition of recombinant IL‐10 caused a dose‐dependent decrease in IFN‐γ production by MLN cells from each mouse evaluated.
Figure 6.
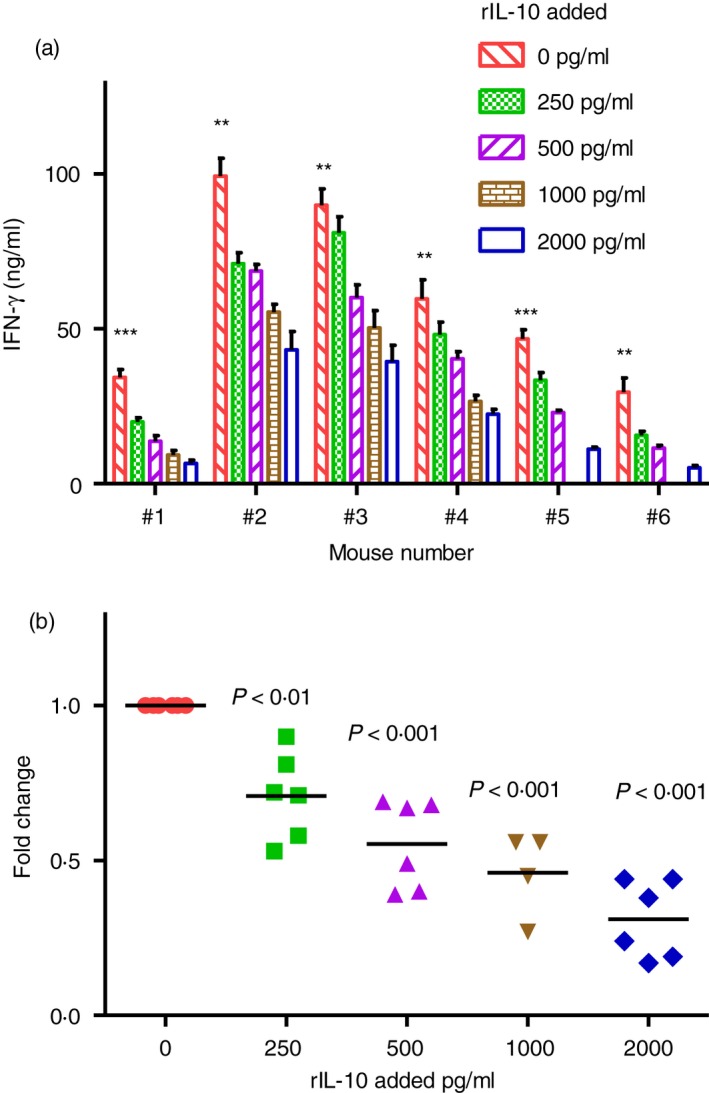
Interferon‐γ (IFN‐γ) production by mesenteric lymph nodes (MLN) cells treated with recombinant interleukin‐10 (IL‐10) in vitro. IFN‐γ production by unseparated MLN from 129SvEv wild‐type (WT) mice monoassociated with Escherichia coli NC101 for 4 days. MLN cells were stimulated in vitro with 10 μg/ml E. coli NC101 lysate. Supernatants were collected 72 hr later and IFN‐γ was quantified by ELISA. Cultures contained 0, 250, 500, 1000 or 2000 pg/ml rIL‐10 added at the time of in vitro stimulation. (a) Mean ± SEM of IFN‐γ produced by cells from individual mice. Data were analysed by unpaired Student's t‐test. Asterisks indicate statistically significant differences comparing IFN‐γ in supernatants of cultures without rIL‐10 to IFN‐γ in supernatants of cultures containing 2000 pg/ml rIL‐10. **P < 0·01, ***P < 0·001. (b) Fold change in IFN‐γ compared with amounts produced by cells cultured without rIL‐10. MLN cultures contain 0, 250, 500 or 2000 pg/ml rIL‐10 (n = 6 mice); MLN cultures contain 1000 pg/ml rIL‐10 (n = 4 mice). Each dot represents an individual mouse. Data were analysed by one‐way analysis of variance with Tukey's multiple‐comparison post‐test. P‐values indicate statistically significant differences compared with cultures without added rIL‐10.
Interleukin‐10 receptor blockade in vivo
To determine the role of endogenous IL‐10 in the immune response that develops in vivo, we investigated whether blockade of IL‐10 signalling will regulate IFN‐γ secretion. Either anti‐IL‐10R‐specific antibody or isotype control antibody was given 1 day before, 2 days after and 5 days after colonization of GF mice with E. coli NC101 and the mice were evaluated 7 days after monoassociation. Interferon‐γ measured in supernatants of E. coli NC101 lysate‐activated MLN cells from anti‐IL‐10R‐treated mice was significantly higher than in supernatants of MLN cells from mice given isotype control antibody (Fig. 7a). The IFN‐γ mRNA expression in the transverse colon from anti‐IL‐10R antibody‐treated mice was also significantly higher compared with isotype‐control‐treated mice (Fig. 7b). In addition, we detected higher expression of both IL‐1β and IL‐6 mRNA in intestinal tissue of anti‐IL10R‐treated mice compared with isotype‐control‐treated mice (Fig. 7c,d). These data show that blockade of IL‐10 signalling in vivo by anti‐IL‐10 receptor antibody increased IFN‐γ production and also mRNA expression of several cytokines, and indicate that endogenous IL‐10, produced at early time‐points after exposure to commensal bacteria, plays a role in suppressing these immune responses.
Figure 7.
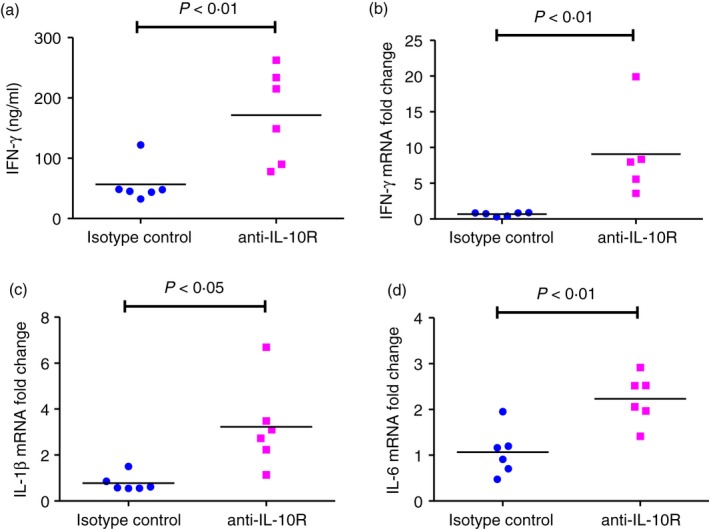
Interferon‐γ (IFN‐γ) production in 129SvEv wild‐type (WT) mice treated in vivo with anti‐interleukin‐10 (IL‐10) receptor specific antibody before and during Escherichia coli NC 101 monoassociation. Germ‐free (GF) 129SvEv WT mice were injected intraperitoneally with antibody that blocks the IL‐10 receptor (anti‐IL‐10R) or with isotype control antibody 1 day before, 2 days and 5 days after colonization with E. coli NC101. Mice were euthanized 7 days after monoassociation. (a) IFN‐γ production by unseparated mesenteric lymph node (MLN) cells stimulated in vitro with 10 μg/ml E. coli NC 101 lysate. Supernatants were collected 72 hr later and IFN‐γ was quantified by ELISA. (b) IFN‐γ mRNA expression, (c) IL‐1β mRNA expression, and (d) IL‐6 mRNA expression in transverse colon were determined by quantitative PCR. Copy number is normalized to GAPDH and expressed as fold change relative to isotype control. Each dot represents an individual mouse. Data were analysed by unpaired Student's t‐test. P‐values indicate statistically significant differences between IFN‐γ for mice treated with isotype control antibody compared with mice treated with anti‐IL‐10R.
Discussion
The intestine of normal adult hosts contains a fully competent immune system and the lumen is occupied by enormous numbers of microorganisms that produce numerous components with the potential to activate innate and acquired immune responses. However, the normal intestinal tract is in a state of “physiologic inflammation” devoid of overt aggressive responses to luminal microbes and their products. Here we report the results of our studies that were designed to elucidate the immune response in the intestinal tract after initial exposure of normal lamina propria host cells and tissues to resident microorganisms. We demonstrate that a single strain of non‐pathogenic E. coli rapidly induces a transient innate and adaptive effector immune response in previously GF mice, and that IL‐10 produced by B cells and CD4+ cells is a key mediator of the subsequent suppression of this response.
We initially embarked on the studies described in this report because we observed an early and then rapidly down‐regulated immune response in the WT mice that served as controls for our experiments investigating intestinal inflammation in IL‐10‐deficient mice colonized with normal resident bacteria. Subsequently, in a separate study, we demonstrated a similar phenomenon at the earliest time‐point evaluated (1 week) after adoptive transfer of IL‐10‐deficient CD4+ T cells into Rag2−/− immunodeficient SPF recipients.13 The Rag2−/− recipients were crossed to normal mice or to IL‐10‐deficient mice providing either Rag2−/− × IL‐10+/+ or Rag2−/− × IL‐10−/− recipients. These mice were born and raised under SPF conditions and therefore harbour a full complement of resident microorganisms, but they lack lymphoid cells before adoptive transfer and so provide an environment for homeostatic proliferation of donor cells. Relatively high and also very similar amounts of IFN‐γ were produced by MLN cells obtained from both recipient types 1 week after T‐cell transfer. By 2 weeks, the MLN cells obtained from IL‐10‐replete but not IL‐10‐deficient recipients no longer produced IFN‐γ. These results indicate that the transferred T cells that undergo homeostatic proliferation in SPF Rag2−/− recipients are rapidly activated to produce IFN‐γ in vivo by components of the intestinal microbiota, and this early IFN‐γ response is inhibited by recipient‐derived IL‐10 produced by non‐lymphoid cells.
In the experiments described in the current report, all of the components of the immune system are simultaneously exposed to luminal bacteria, whereas in the adoptive transfer model we were able to observe the adaptive immune response of the transferred CD4+ T cells upon their activation in vivo by resident microbiota. The conclusion is similar in both model systems, namely that resident bacteria rapidly activate lymphoid cells (primarily CD8‐positive cells but also cells that express CD4 or NKp46) to produce IFN‐γ and that IL‐10 plays a key role in subsequent suppression of this response. In the adoptive transfer model, we concluded that non‐lymphoid cells of the Rag2−/− recipients produce immunoregulatory IL‐10 because the transferred CD4+ T cells were obtained from IL‐10‐deficient donors.13 In the model using E. coli monoassociated WT mice described in the current report, we identified MLN CD4+ cells and B cells that produce IL‐10 after in vitro re‐stimulation with E. coli lysates. It is likely that other cell types, including myeloid cells and possibly also NK cells and innate lymphoid cells are capable of producing IL‐10; however, we did not detect IL‐10 production by CD11b‐positive MLN cells or NKp46‐positive MLN cells from E. coli monoassociated mice.
Numerous studies have identified a critical role for IL‐10 in maintaining intestinal homeostasis.14, 35 Both IL‐10‐deficient mice18 and IL‐10R‐deficient mice19 spontaneously develop colitis that can be ameliorated by administration of exogenous IL‐10 immunoglobulin fusion protein that mimics IL‐10 activity.36 Our focus on IL‐10 in the present study stems from previous investigations using SPF and gnotobiotic IL‐10‐deficient mice as rodent models of chronic intestinal inflammation.13, 23, 28 Therefore, in the current experiments, we either provided exogenous recombinant IL‐10 in vitro or injected anti‐IL‐10R‐specific antibody in vivo to directly evaluate the role of IL‐10 during the initial period after monoassociation of GF mice with E. coli NC101. The results of these studies indicate that IL‐10 signalling indeed inhibits IFN‐γ production and are therefore consistent with the conclusion that IL‐10 plays a key role in maintaining a quiescent immune response in the intestinal tract. This conclusion is supported by the observations of Berg et al.37 that systemic administration of recombinant IL‐10 in vivo can attenuate the onset of colitis in IL‐10‐deficient mice and prevent progression of inflammation, but not reverse established colitis.
Our results confirm and extend the comprehensive analyses published by El Aidy et al.26, 27 showing immune activation at early time‐points after colonizing GF mice with complex faecal microbiota obtained from conventionally housed normal mice. Both our study and El Aidy's results show that (i) the mucosal immune system recognizes and reacts to the components of bacteria that normally reside in the intestinal tract, and (ii) that GF mice have a fully functional immune system, with the capacity to respond rapidly, demonstrated by activation of both innate and adaptive immune responses following bacterial colonization. The maturation from a naive to an activated state proceeds very rapidly upon microbial exposure of GF mice, even when the mice are monoassociated with a single species of non‐pathogenic intestinal bacteria, namely E. coli NC101. Of note, E. coli NC101 is an example of an adherent/invasive E. coli strain29 that has the capacity to induce bacterial antigen‐specific chronic T‐cell‐mediated colitis in IL‐10‐deficient mice.28 As we show in the current report, the same organism can also activate regulatory responses, including production of IL‐10 in IL‐10 replete WT mice.
Our study expands on that of El Aidy et al. in several key areas. We investigated the kinetics of immune system activation of normal GF mice elicited by exposure to a single well‐studied strain of non‐pathogenic E. coli rather than to the multitude of normal resident microbes. In addition, our in vitro and in vivo results identify the key role of IL‐10 in suppressing the initial activation of the immune system of normal germ‐free mice after bacterial colonization, and we identified B cells and CD4+ cells in MLN as the sources of regulatory IL‐10. It is also important to note that in our study, GF inbred 129S6/SvEv mice are the hosts. El Aidy et al. used GF C57BL/6 mice. There are many genetic and functional differences between these two strains. For example, SPF IL‐10‐deficient mice backcrossed to C57BL/6 mice are relatively resistant to the development of intestinal inflammation compared with SPF IL‐10‐deficient mice on the 129S6/SvEv background.37 As another example of the difference between these two inbred strains, the ileum of SPF and GF C57BL/6 mice contains a higher proportion of lysozyme‐containing Paneth cells and higher levels of mRNA expression of several isoforms of α‐defensin compared with the same tissue obtained from SPF and GF 129SvEv mice.38 Therefore, demonstrating consistent results using a different inbred mouse strain strengthens the overall conclusion of our study and that of El Aidy et al. that non‐pathogenic bacteria first activate and then induce immune regulation in the intestinal tract of normal hosts.
Taken together, our results demonstrate that the gut‐associated immune system of normal GF mice recognizes and rapidly responds to a non‐pathogenic strain of E. coli, with an initial response that includes production of pro‐inflammatory cytokines. This response rapidly progresses to a regulated homeostatic immune profile with low IFN‐γ, IL‐1β and IL‐6 levels in the presence of sustained bacterial activation of immunoregulatory IL‐10. Hence intestinal IL‐10 signalling is a key factor in suppressing the host's mucosal effector immune response to resident bacterial components.
Author contributions
CW, SLT and RBS conceived and designed the experiments; CW and SLT performed the experiments, analysed the data, and wrote the manuscript; RBS provided intellectual input, financial support and edited the manuscript; KH provided financial support and reviewed the manuscript.
Disclosures
The authors declare no financial conflicts of interest.
Acknowledgements
The authors thank Maureen Bower of the Gnotobiotic Core of the CGIBD and National Gnotobiotic Rodent Resource Center, University of North Carolina at Chapel Hill and Maria Stone, Gnotobiotic Core of the CGIBD at North Carolina State University for GF and E. coli NC101 monoassociated mice. We also thank Anna Yedinak for expert technical assistance and Sarah Schuett, Manager of the Flow Cytometry and Cell Sorting Core, College of Veterinary Medicine, North Carolina State University for flow cytometry assistance.
This work was supported by the National Institutes of Health grants P01 DK094779 and P40 OD010995 (to RBS), by the Center for Gastrointestinal Biology and Disease grant P30 DK034987, the Crohn's and Colitis Foundation of America, and by the China Scholarship Council 2011685032 (to CW).
References
- 1. Kaiko GE, Stappenbeck TS. Host–microbe interactions shaping the gastrointestinal environment. Trends Immunol 2014; 35:538–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature 2012; 489:220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006; 124:837–48. [DOI] [PubMed] [Google Scholar]
- 4. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M et al Diversity of the human intestinal microbial flora. Science 2005; 308:1635–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 1977; 31:107–33. [DOI] [PubMed] [Google Scholar]
- 6. Sartor RB. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol 2006; 3:390–407. [DOI] [PubMed] [Google Scholar]
- 7. Rivollier A, He J, Kole A, Valatas V, Kelsall BL. Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon. J Exp Med 2012; 209:139–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saraiva M, O'Garra A. The regulation of IL‐10 production by immune cells. Nat Rev Immunol 2010; 10:170–81. [DOI] [PubMed] [Google Scholar]
- 9. Jarry A, Bossard C, Bou‐Hanna C, Masson D, Espaze E, Denis MG et al Mucosal IL‐10 and TGF‐β play crucial roles in preventing LPS‐driven, IFN‐γ‐mediated epithelial damage in human colon explants. J Clin Invest 2008; 118:1132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jankovic D, Kullberg MC, Feng CG, Goldszmid RS, Collazo CM, Wilson M et al Conventional T‐bet+Foxp3− Th1 cells are the major source of host‐protective regulatory IL‐10 during intracellular protozoan infection. J Exp Med 2007; 204:273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chirdo FG, Millington OR, Beacock‐Sharp H, Mowat AM. Immunomodulatory dendritic cells in intestinal lamina propria. Eur J Immunol 2005; 35:1831–40. [DOI] [PubMed] [Google Scholar]
- 12. Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med 1989; 170:2081–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu B, Tonkonogy SL, Sartor RB. Antigen‐presenting cell production of IL‐10 inhibits T‐helper 1 and 17 cell responses and suppresses colitis in mice. Gastroenterology 2011; 141:653–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shouval DS, Ouahed J, Biswas A, Goettel JA, Horwitz BH, Klein C et al Interleukin 10 receptor signaling: master regulator of intestinal mucosal homeostasis in mice and humans. Adv Immunol 2014; 122:177–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin‐10 and the interleukin‐10 receptor. Annu Rev Immunol 2001; 19:683–765. [DOI] [PubMed] [Google Scholar]
- 16. O'Garra A, Vieira P. Regulatory T cells and mechanisms of immune system control. Nat Med 2004; 10:801–5. [DOI] [PubMed] [Google Scholar]
- 17. Roncarolo MG, Gregori S, Battaglia M, Bacchetta R, Fleischhauer K, Levings MK. Interleukin‐10‐secreting type 1 regulatory T cells in rodents and humans. Immunol Rev 2006; 212:28–50. [DOI] [PubMed] [Google Scholar]
- 18. Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin‐10‐deficient mice develop chronic enterocolitis. Cell 1993; 75:263–74. [DOI] [PubMed] [Google Scholar]
- 19. Spencer SD, Di Marco F, Hooley J, Pitts‐Meek S, Bauer M, Ryan AM et al The orphan receptor CRF2‐4 is an essential subunit of the interleukin 10 receptor. J Exp Med 1998; 187:571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Asseman C, Mauze S, Leach MW, Coffman RL, Powrie F. An essential role for interleukin 10 in the function of regulatory T cells that inhibit intestinal inflammation. J Exp Med 1999; 190:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mishima Y, Liu B, Hansen JJ, Sartor RB. Resident bacteria‐stimulated interleukin‐10‐secreting B cells ameliorate T‐cell‐mediated colitis by inducing T‐regulatory‐1 cells that require interleukin‐27 signaling. Cell Mol Gastroenterol Hepatol 2015; 1:295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mizoguchi A, Mizoguchi E, Takedatsu H, Blumberg RS, Bhan AK. Chronic intestinal inflammatory condition generates IL‐10‐producing regulatory B cell subset characterized by CD1d upregulation. Immunity 2002; 16:219–30. [DOI] [PubMed] [Google Scholar]
- 23. Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E et al Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin‐10‐deficient mice. Infect Immun 1998; 66:5224–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mittal SK, Roche PA. Suppression of antigen presentation by IL‐10. Curr Opin Immunol 2015; 34:22–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabat R, Grutz G, Warszawska K, Kirsch S, Witte E, Wolk K et al Biology of interleukin‐10. Cytokine Growth Factor Rev 2010; 21:331–44. [DOI] [PubMed] [Google Scholar]
- 26. El Aidy S, van Baarlen P, Derrien M, Lindenbergh‐Kortleve DJ, Hooiveld G, Levenez F et al Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol 2012; 5:567–79. [DOI] [PubMed] [Google Scholar]
- 27. El Aidy S, Derrien M, Aardema R, Hooiveld G, Richards SE, Dane A et al Transient inflammatory‐like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ‐free mice. Benef Microbes 2014; 5:67–77. [DOI] [PubMed] [Google Scholar]
- 28. Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J et al Variable phenotypes of enterocolitis in interleukin 10‐deficient mice monoassociated with two different commensal bacteria. Gastroenterology 2005; 128:891–906. [DOI] [PubMed] [Google Scholar]
- 29. Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL et al Inflammation‐associated adherent‐invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M‐cell translocation. Inflamm Bowel Dis 2014; 20:1919–32. [DOI] [PubMed] [Google Scholar]
- 30. Patwa LG, Fan TJ, Tchaptchet S, Liu Y, Lussier YA, Sartor RB et al Chronic intestinal inflammation induces stress‐response genes in commensal Escherichia coli . Gastroenterology 2011; 141:1842–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolfe DN, Karanikas AT, Hester SE, Kennett MJ, Harvill ET. IL‐10 induction by Bordetella parapertussis limits a protective IFN‐γ response. J Immunol 2010; 184:1392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. D'Andrea A, Aste‐Amezaga M, Valiante NM, Ma X, Kubin M, Trinchieri G. Interleukin 10 (IL‐10) inhibits human lymphocyte interferon γ‐production by suppressing natural killer cell stimulatory factor/IL‐12 synthesis in accessory cells. J Exp Med 1993; 178:1041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G et al Innate lymphoid cells – a proposal for uniform nomenclature. Nat Rev Immunol 2013; 13:145–9. [DOI] [PubMed] [Google Scholar]
- 34. Narni‐Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A et al Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci USA 2011; 108:18324–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kole A, Maloy KJ. Control of intestinal inflammation by interleukin‐10. Curr Top Microbiol Immunol 2014; 380:19–38. [DOI] [PubMed] [Google Scholar]
- 36. Nguyen DD, Wurbel MA, Goettel JA, Eston MA, Ahmed OS, Marin R et al Wiskott–Aldrich syndrome protein deficiency in innate immune cells leads to mucosal immune dysregulation and colitis in mice. Gastroenterology 2012; 143:719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berg DJ, Davidson N, Kuhn R, Muller W, Menon S, Holland G et al Enterocolitis and colon cancer in interleukin‐10‐deficient mice are associated with aberrant cytokine production and CD4+ TH1‐like responses. J Clin Invest 1996; 98:1010–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gulati AS, Shanahan MT, Arthur JC, Grossniklaus E, von Furstenberg RJ, Kreuk L et al Mouse background strain profoundly influences Paneth cell function and intestinal microbial composition. PLoS ONE 2012; 7:e32403. [DOI] [PMC free article] [PubMed] [Google Scholar]


