Abstract
When infecting a host plant, the fungus Fusarium oxysporum secretes several effector proteins into the xylem tissue to promote virulence. However, in a host plant with an innate immune system involving analogous resistance proteins, the fungus effector proteins may trigger resistance, rather than promoting virulence. Identity of the effector genes of Fusarium oxysporum f. sp. niveum (Fon) races that affect watermelon (Citrullus lanatus) are currently unknown. In this study, the SIX6 (secreted in xylem protein 6) gene was identified in Fon races 0 and 1 but not in the more virulent Fon race 2. Disrupting the FonSIX6 gene in Fon race 1 did not affect the sporulation or growth rate of the fungus but significantly enhanced Fon virulence in watermelon, suggesting that the mutant ΔFon1SIX6 protein allowed evasion of R protein-mediated host resistance. Complementation of the wild-type race 2 (which lacks FonSIX6) with FonSIX6 reduced its virulence. These results provide evidence supporting the hypothesis that FonSIX6 is an avirulence gene. The identification of FonSix6 as an avirulence factor may be a first step in understanding the mechanisms of Fon virulence and resistance in watermelon and further elucidating the role of Six6 in Fusarium-plant interactions.
Watermelon [Citrullus lanatus (Tunb.) Matsum. & Nakai] is an important cucurbit crop accounting for 7% of the agricultural land area devoted to vegetable production worldwide. The total annual production of watermelon is approximately 90 million tons, making it among the top five most consumed fresh fruits (http://faostat.fao.org). Fusarium wilt, caused by the soil-borne fungus Fusarium oxysporum f. sp. niveum (Fon), is a major disease of watermelon throughout the world, with a large adverse impact on watermelon yield and quality1.
There are three common physiological races (0, 1 and 2) of this pathogen, classified according to their reactions with differential watermelon genotypes (Table 1)2,3,4. Race 0 is pathogenic only in watermelon cultivars with no resistance genes. Race 1 is the predominant race throughout commercial watermelon regions worldwide, and several watermelon cultivars, such as cv. Calhoun Gray, are resistant to this race. Race 2 is highly aggressive to all current commercial watermelon cultivars and hybrids. Race 3, the most virulent race of Fon described to date, was shown to cause over 90% wilt on PI296341-FR, whereas no disease was caused by a race 2 isolate5.
Table 1. Watermelon genotypes used to differentiate races of Fusarium oxysporum f. sp. niveum.
| Cultivar or genotype | Disease response to:* |
||
|---|---|---|---|
| Race 0 | Race 1 | Race 2 | |
| Sugar Baby | S | S | S |
| Charleston Gray | R | S | S |
| Calhoun Gray | R | R | S |
| PI 296341-FR | R | R | R |
*S = susceptible. R = resistant.
The co-evolution of plants and microorganisms involves complex mechanisms of attack and defence, implicating the innate immune system of plants and virulence factors of pathogens6. The first layer of plant defence, known as basal immunity, is based on the recognition of conserved microbial molecules but can be suppressed by microbial virulence factors known as “effectors”. Plants respond to this suppression by employing a second layer of defence, resistance (R) gene-based immunity, which relies on the recognition of effectors7. Finally, the pathogen evolves further and escapes detection by the R gene product by eliminating the detected virulence factor or by suppressing the defence induced by R gene products8. Effectors may be defined as pathogen proteins and small molecules that alter host-cell structure and function. These alterations either facilitate infection (virulence factors and toxins) or trigger defence responses (avirulence factors and elicitors), or both9.
The secreted effector proteins of F. oxysporum f. sp. lycopersici (Fol) infecting tomato have been identified through proteomic analysis of xylem sap from tomato plants infected with Fol. These proteins have been designated the Six (secreted in xylem) proteins and include Six1 to Six710,11. Several functions of the Six proteins have been identified thus far. Avr3 (Six1) is required for I-3-mediated resistance12, and Avr1 (Six4) is required for I-mediated resistance13. Additionally, both proteins have functions other than triggering avirulence: Avr3 is required for full virulence14, whereas Avr1 suppresses I-2- and I-3-mediated disease resistance13. Subsequently, Avr2 (Six3) shows both activities: it is required for full virulence in susceptible tomato host plants while triggering resistance in plants carrying the resistance gene I-215. Six5 is required for full virulence in susceptible plants, and knockout of this gene can breach I-2-mediated disease resistance. Avr2 and Six5 interact in yeast two-hybrid assays as well as in planta. The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato16, while Six6 is a true effector that enhances virulence and simultaneously suppresses I-2-mediated cell death17. Screening of effector proteins indicated that the three AVR gene sequences (AVR1, AVR2, and AVR3) and the SIX5 gene sequence are not present in Fon races, while the SIX6 gene homologue is present11, although its biological function has not been determined18.
In this study, we identified and analysed the biological function of the SIX6 gene in Fon (FonSIX6) and demonstrated that FonSIX6 is an AVR gene playing a key role in the Fon-watermelon pathosystem.
Results
Cloning and analysis of FonSIX6 and flanking sequences
Using the genome sequence of Fol (http://www.broadinstitute.org/) as a reference for constructing PCR primers, we cloned a SIX6 gene of Fon. Here, we used primers that annealed immediately outside the FolSIX6 gene ORF11. The resultant gene was designated FonSIX6. Genome searches using the FonSIX6 ORF sequence as a query showed high sequence homology of SIX6 with F. oxysporum f. sp. melonis (Fom, 100%) and Fol (94.91%) (Supplementary Fig. S1). To obtain additional information about FonSIX6, the Fol genome was used as a reference for designing specific PCR primers for the flanking sequence. However, no PCR fragments were amplified, suggesting that the Fol and Fon genomic sequences are different. Finally, the 1974 bp upstream sequence (directly adjacent to the start codon, −1974 bp) and the 453 bp downstream (directly 3′ to the stop codon, +453 bp) of the FonSIX6 open reading frame were cloned via chromosome walking (Supplementary Fig. S2).
Analysis of the conserved FonSIX6 homologue sequence in a watermelon-infecting strain
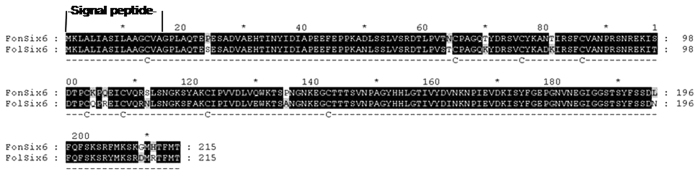
The deduced FonSix6 protein consists of 215 amino acids and contains 8 cysteine residues (Fig. 1). This intronless gene is predicted to encode a 21.85 kDa mature protein (after cleavage of the predicted N-terminal signal peptide) (http://web.expasy.org/compute_pi/). Amino acid sequence comparisons between FonSix6 and FolSix6 (NCBI GenBank:ACN69116.1) showed 90.23% identity, with minor differences. Using the SignalP 4.0 Server, the FonSix6 protein was predicted to contain a signal peptide consisting of 16 amino acids at its N-terminus (http://www.cbs.dtu.dk/services/SignalP) (Fig. 1). Among the three physiological races (0, 1 and 2) of the Fon pathogen, the FonSIX6 gene sequence exists in the genomes of Fon races 0 and 1 but not in the more aggressive race 2 (Fig. 2).
Figure 1. Amino acid alignment of putative Six6 from FonSix6 (Fusarium oxysporum f. sp. niveum) with FolSix6 (F. oxysporum f. sp. lycopersici).
The signal peptide sequence predicted by the Signal P program (http://www.cbs.dtu.dk/services/SignalP/) is depicted, and the eight cysteine residues are marked below the sequence. FonSix6: the putative amino acid sequence from Fusarium oxysporum f. sp. niveum, the nucleotide sequence cloned from wild race 1; FolSix6: Fusarium oxysporum f. sp. lycopersici (ACY39286.1).
Figure 2. The presence of SIX6 in F. oxysporum f. sp. niveum.
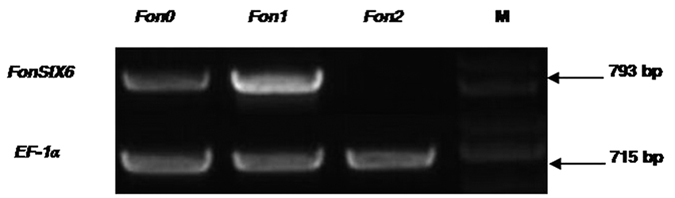
Fon 0, Fon 1, Fon 2: F. oxysporum f. sp. niveum wild race 0, race 1 and race 2. FonSIX6: the SIX6 gene from F. oxysporum f. sp. niveum; EF-1α: elongation factor of F. oxysporum f. sp. niveum. M: marker lane (DL 2000, Takara).
Impact of FonSIX6 disruption on fungal development
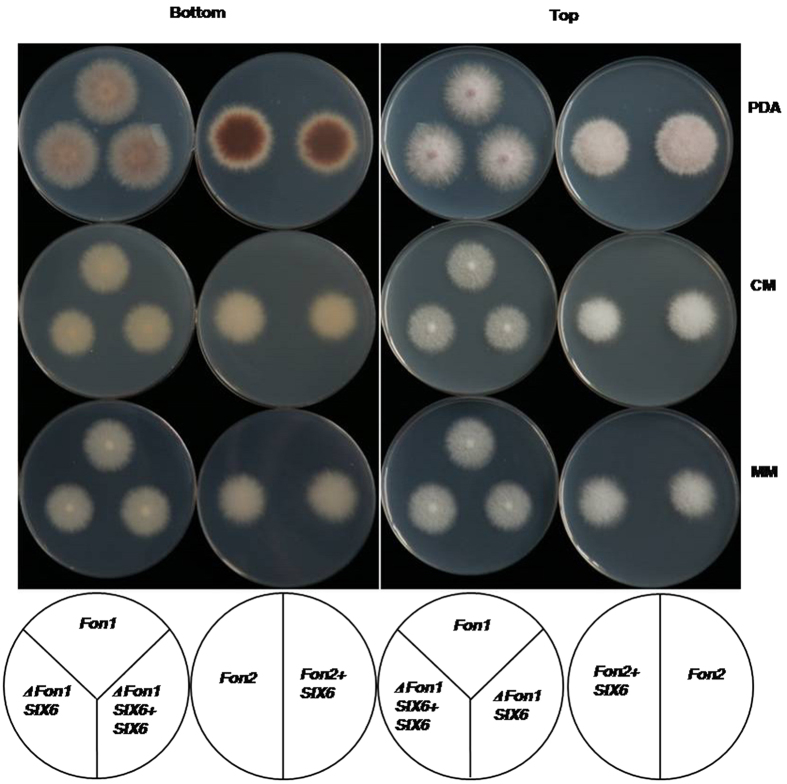
To determine whether FonSIX6 affects fungal growth and microsclerotia production in Fon, the growth patterns of the gene disruption mutant ΔFonSIX6 on potato dextrose agar (PDA), minimal medium (MM) and complete medium (CM) media were compared with those of the wild-type Fon strain and the gene-complemented ΔFon1SIX6 + SIX6 and Fon2 + SIX6 strains.
During growth on PDA, the Fon mycelium produced red-brown pigments, whereas no red-brown colour was observed on MM or CM medium. Measurement of colony diameter on PDA, MM, or CM medium during the first 4 days of culture indicated that the radial growth of the FonSIX6-disrupted and complemented strains did not differ from that of the wild-type strain (Fig. 3). The morphology and quantity of spores also did not differ substantially, as observed under a microscope. These observations indicate that the FonSIX6 gene is not essential for Fon growth and development.
Figure 3. Colony growth of deletion and complementation mutants compared with wild-type Fon on PDA, CM, and MM.
Fon1: Fusarium oxysporum f. sp. niveum wild race 1; Fon2: Fusarium oxysporum f. sp. niveum wild race 2; ΔFon1SIX6: race 1 with SIX6 disrupted by gene replacement; ΔFon1SIX6 + SIX6: ΔFon1SIX6 transformed with SIX6; Fon2 + SIX6: race 2 transformed with SIX6; PDA: potato dextrose agar medium; MM: minimal medium; CM: complete medium. The photographs were taken from the top and bottom of the plates 4 days after incubation.
FonSIX6 is expressed 3 days after Fon infection
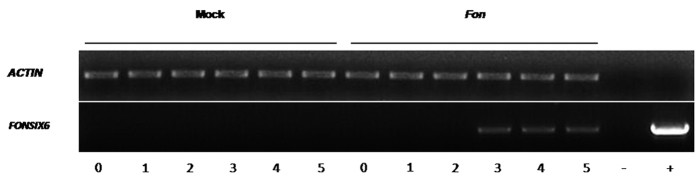
To determine whether FonSIX6 was expressed during earlier stages of infection, RNA was collected from Fon-infected watermelon roots on the 1st, 2nd, 3rd, 4th and 5th day post-inoculation (DPI). The expression of FonSIX6 was monitored using reverse-transcriptase polymerase chain reaction (RT-PCR). FonSIX6 transcripts could be detected on the 3rd through the 5th DPI in infected plants, whereas the mock-inoculated controls did not produce this transcript (Fig. 4).
Figure 4. FonSIX6 is expressed during the early stages of infection.
Reverse-transcriptase polymerase chain reaction analysis of watermelon actin (ACTIN) or FonSIX6 expression using RNA isolated from the roots of watermelon seedlings, which were either mock or Fusarium oxysporum f. sp. niveum inoculated and harvested on the 1st, 2nd, 3rd, 4th and 5th day post-inoculation (DPI). Water was included as a negative control (−), while genomic DNA from Fusarium oxysporum f. sp. niveum (+) was used as a positive control.
Watermelon inoculation with wild-type and transformants
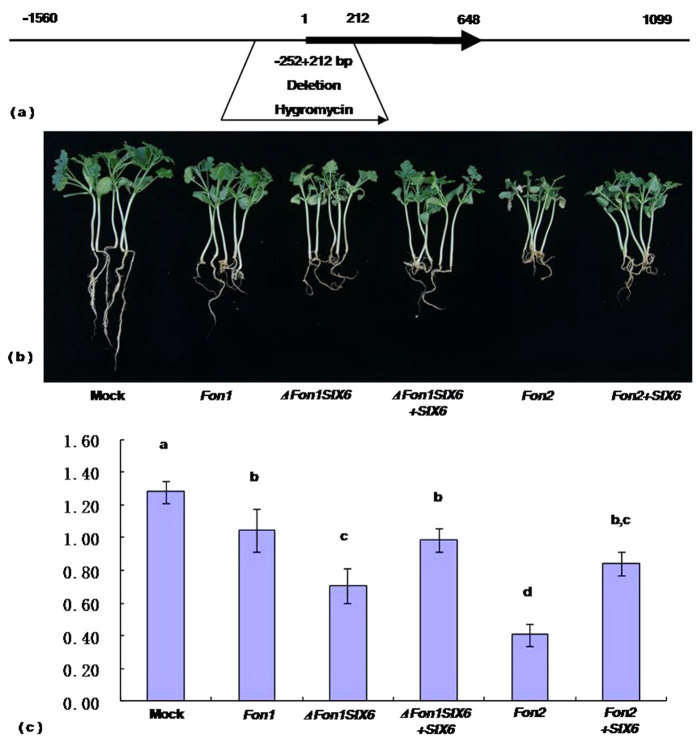
To determine the role of FonSIX6 in the infection of watermelon, FonSIX6 knockout mutants (ΔFon1SIX6) were generated in Fon race 1 by replacing FonSIX6 with a hygromycin resistance cassette (Fig. 5a). Then, the virulence of the ΔFonSIX6 strain was assessed by inoculating watermelon seedlings (cv. Calhoun Gray, resistant to Fon 1). To our surprise, the severity of disease symptoms in watermelon plants inoculated with ΔFon1SIX6 transformants were significantly enhanced (severe, Fig. 5b,c). Reintroduction of the FonSIX6 gene into ΔFon1SIX6 led to disease symptoms similar to those associated with wild-type Fon 1 (mild, Fig. 5b,c). On the other hand, when symptom expression was compared in watermelon plants inoculated with Fon2 + SIX6 transformants and the more aggressive wild-type Fon 2 (lack of SIX6), the severity of disease symptoms was significantly reduced in plants inoculated with Fon2 + SIX6 transformants compared with those inoculated with wild-type Fon 2 (Fig. 5b,c). These results indicated that the mutant ΔFonSIX6 protein allowed evasion of host resistance mediated by the R protein. Therefore, FonSIX6 is an avirulence factor. Complementation of wild-type race 2 with the FonSIX6 gene reduced race 2 virulence, further confirming that FonSIX6 is an AVR gene.
Figure 5. Watermelon plants were inoculated with Fon 1, ΔFon1SIX6, ΔFon1SIX6 + SIX6, Fon 2 and Fon 2 + SIX6 and the development of disease symptoms was assessed.
Seedlings (cv. Calhoun Gray, resistance to Fon 1) at the first true leaf stage were inoculated with a fungal spore suspension, and disease symptoms were scored after 15 days. (a) The ΔFonSIX6 mutants were generated using Agrobacterium-mediated targeted disruption of the SIX6 gene. (b) Representative plants are shown 15 days post-inoculation. (c) Quantification of disease assays by weight. The outcomes of the disease assays depicted in (b) were quantified based on the average plant weight from each inoculation. Error bars indicate standard deviation and letters indicate values that are significantly different from each other (P < 0.01, all pairs Student’s t-test). All of the assays were repeated at least three times.
Discussion
The vascular pathogen F. oxysporum is an asexual fungus with a broad host range that causes wilt and root diseases in many economically important crop plants, including watermelon19. In Fol, 14 ‘Secreted in xylem’ (Six) proteins (Six1~14) have been identified from Fol-infected tomato plants10,20. Of the Six proteins, Six6 contributes to virulence and suppresses I-2-mediated cell death. Although a Six6 homologue sequence has been identified in Fon isolates, its functional has not been characterized17,18. In this report, we describe the identification and functional analysis of the SIX6 gene of Fon.
Earlier studies showed that the FonSIX6 gene was present in the forma specialis niveum in isolates 546 and 704 but not in isolates 703, 705, CBS 187.60, CBS 418.90, and CBS 419.9011,18. Three generally accepted physiological races (0, 1 and 2) of the Fon pathogen have been identified to date, according to their effects in differential watermelon genotypes2,3,4. This study indicates the possibility that the FonSIX6 gene is present in races 0 and 1 but not in race 2. Based on these results, we speculate that isolates 703, 705, CBS 187.60, CBS 418.90, and CBS 419.90 likely belong to race 2 while the other isolates are likely from races 0 or 1.
Although the pathogenicity of Fon is gradually increased in races 0, 1 and 2, there is growing speculation that the distinction between race 0 and race 1 may be more quantitative than qualitative. Consequently, races 0 and 1 might be strains of race 1 with varying aggressiveness. On the other hand, race 2 is highly aggressive to all current commercial watermelon cultivars and hybrids and is clearly a distinct race2. Here, the association of the FonSIX6 gene with the pathogenicity of different Fon races may provide a potential cultivar-specific pathogenicity marker that is useful for defining host targets and evolutionary bottlenecks that control the Fon-watermelon pathosystem.
In Fol, most SIX genes are located in the same lineage-specific (LS) genomic region- chromosome (chromosome 14), also known as the pathogenicity chromosome, and are associated with chromosomal sub-regions enriched for DNA transposons. The LS genome regions could have been acquired through horizontal transfer from another species, leading to the hypothesis that horizontal chromosome transfer in F. oxysporum can generate new pathogenic lineages21,22. FolSIX6 is located on a supernumerary chromosome 14, an LS chromosome21. Searches carried out using the FonSIX6 ORF sequence as a query showed strong sequence identity to SIX6 of Fom (100%) and Fol (94.91%) (Supplementary Fig. S1). Comparison of the FonSIX6 ORF and its flanking sequences between the Fom and Fol genomes in the −609 ~ +157 and −433 ~ +157 regions showed nucleotide sequence identities of 99.58% and 96.29%, respectively (Supplementary Fig. S2). The genome of F. oxysporum 4287 (FO2) has been sequenced and is available (http://www.broadinstitute.org/). BLAST searches of this genome sequence database using the FonSIX6 sequence segment (−1974 ~ +453) as a query showed the presence of high sequence identity (90 ~ 100%) in 5 distinct segments. Segment 1 (−1974 ~ −686) is located in supercontig 37 of Chr15 (91.47% nucleotide sequence identity). Segments 2 ~ 4 are located on supercontig 22 of Chr14 at different positions with various nucleotide sequence identities, with segment 2 (−685 ~ −654) showing 100% identity, segment 3 (−653 ~ −434) showing 95% identity and the segment 4 (−433 ~ +157) showing 96.29% identity, while segment 5 (+158 ~ +453), located on nonpositional scaffolds, exhibited 98.31% nucleotide sequence identity. These analyses suggested that the −433 ~ +157 sequence segment identified in Fon may contain the full gene sequences that are necessary to complete the function of FonSIX6.
Here, we generated FonSIX6 gene knockout mutants in race 1, evaluated the complementation of the knockout mutants and wild-type race 2 with the FonSIX6 gene, and observed that the disruption of FonSIX6 did not affect the growth rate or fungal sporulation. These results demonstrate that FonSIX6 is not absolutely necessary for Fon growth and development. Therefore, the change in the virulence of ΔFonSIX6 is not associated with fungal growth or development and is instead due to the effector’s key role in the Fon-watermelon pathosystem.
In comparison with the watermelon plants inoculated with wild-type Fon, the severity of the disease symptoms of the watermelon plants inoculated with the ΔFon1SIX6 transformants was significantly enhanced. These results suggest that the mutant ΔFon1SIX6 protein allowed evasion of R protein-mediated host resistance. On the other hand, complementation of wild-type Fon 2 (lacking FonSIX6) with the FonSIX6 gene reduced its virulence. Taken together, these results indicate that FonSIX6 is an AVR gene. Loss of function of an AVR gene (FonSIX6) in race 2 allowed the pathogen to avoid the induction of resistance in a watermelon cultivar. Thus, the pathogen gained pathogenicity in that cultivar, and a new pathogenic race (race 2) emerged. The three known races (1, 2 and 3) carry AVR genes in different combinations in Fol. Fol race 2 emerged from race 1 by losing AVR1 and thereby allowed evasion of host resistance mediated by I (the resistance gene corresponding to AVR1). Race 3 emerged when race 2 sustained a point mutation in AVR2, allowing it to evade I-2-mediated resistance of the host15,23. The results of the present study indicate that Fon race 2 may have emerged from race 1 owing to loss of the entire FonSIX6 gene sequence or may have resulted from a mutation that impaired the function of the FonSIX6 gene, evading mediated host resistance. Additional studies are needed to further determine the differences between Fon races 1 and 2.
Methods
Alignment
DNA sequence and protein alignments were performed using the computer programs ClustalW and DNAMAN. The genome of Fol (http://www.broadinstitute.org) was used as a reference sequence for constructing PCR primers to clone the homologous SIX6 gene sequence of Fon.
Fon races and mutant lines used in this study
The following Fon strains were used: Fon 0 (race 0), Fon 1 (race 1), and Fon 2 (race 2) (a kind gift from the National Engineering Research Center for Vegetables, Beijing, China). ΔFon1SIX6 was race 1 with SIX6 disrupted by gene replacement. ΔFon1SIX6 + SIX6 was ΔFon1SIX6 transformed with SIX6. Fon2 + SIX6 was race 2 transformed with SIX6.
FonSIX6 disruption and complementation constructs
The FonSIX6 flanking sequence was cloned using the Genome Walker Universal kit (Clontech). The FonSIX6 disruption construct was generated via PCR amplification of FonSIX6 upstream and downstream sequences (with partial FonSIX6 sequences) for homologous recombination, followed by insertion in front of and behind the hygromycin resistance gene in the vector pDHt224. An upstream fragment, from 1560 to 252 bp upstream of the start codon, was cloned into pDHt2 between the EcoR I (5′>CCGGAATTCACGCTCTGTATGCCTGCTC<3′) and Sac I (5′>CGAGCTCGTCGGTGAATGGTATGTTGTTT<3′) sites, and a downstream fragment, from 212 bp after the start codon to 1099 bp downstream of the stop codon, was cloned into pDHt2 between the Sac I (5′>CGAGCTCTGACCGCTCCGTCTGCTA<3′) and Xba I (5′>TCTCCTCTAGAATCGACGCGGCTGTAAGGAT<3′) sites (Fig. 5a). Transformants were selected on hygromycin B and confirmed by PCR (Supplementary Fig. S3).
To generate a FonSIX6 complementation construct, a fragment of 2659 bp containing the FonSIX6 open reading frame, 1560 bp of upstream sequence and 451 bp of downstream sequence was amplified via PCR using primers with EcoR I and Xba I linkers (underlined) (5′>CCGGAATTCACGCTCTGTATGCCTGCTC<3′ and 5′>TCTCCTCTAGAATCGACGCGGCTGTAAGGAT<3′). This fragment was cloned into pCOM24. Transformants were selected on geneticin and confirmed through PCR (Supplementary Figs S4 and S5). Transformation of the constructs into Fon was carried out using Agrobacterium as described previously25.
Plant material and fungal strains
The following watermelon differentials were used: cv. Sugar Baby and cv. Calhoun Gray. Fon race 0 causes wilt in cv. Sugar Baby; Fon race 1 causes wilt in cv. Sugar Baby and cv. Charleston Gray but not in cv. Calhoun Gray; and Fon race 2 causes wilt in all of the differential cultivars but not in PI296341-FR4.
Pathogenicity assay
Each Fon isolate was cultured on potato sucrose broth (PSB) for 5 days at 25 °C at 120 rpm, and conidial suspensions (1.0 × 106 conidia ml−1) were prepared.
Seeds of each cultivar were sown in vermiculite in plastic pots (6 by 6 by 5 cm, 32 cells tray−1) and grown in a greenhouse set at 24–30 °C on top of a heat pad (30 °C). The standard root dip method was used to inoculate watermelon seedlings. At the first true leaf stage, the seedlings were dipped in a conidial suspension (1.0 × 106 conidia ml−1) for 5 min and replanted to a vermiculite tray. Disease was scored at 15 days post-inoculation. The disease assay results were quantified based on the average plant weight and the typical disease symptoms of yellowing, stunting and wilting. Because the inoculation methods involving direct dipping or root cutting yielded a similar disease incidence and symptom severity, the data obtained using the two methods were combined for analysis. All of the tests were repeated at least three times.
RNA isolation and RT-PCR
For FonSIX6 gene expression analysis, RT-PCR experiments were performed using tissue harvested from Fon1-infected roots of watermelon cv. Sugar baby. The root samples were ground in liquid nitrogen. Then, total RNA was extracted with the RNAiso plus reagent (Takara), and DNA was removed with recombinant DNase I (Takara). cDNA was subsequently synthesized using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara). The primer combinations w-actinF/w-actinR (W-actinF: 5′>AATGTGCCTGCTATGTATGTCG<3′; W-actinR: 5′>GATGGAGTTGTAGGTAGTTTCG<3′) and FonSIX6F/FonSIX6R (FonSIX6F:5′>CGCTCTTATCGCATCAATCT<3′; FonSIX6R:5′>GGGTTGACTGAGGTCGTGGT<3′) were used to amplify the watermelon actin gene and FonSIX6.
Vegetative growth, conidiation and microsclerotia formation assays
For each sample, a 0.5 μL drop of a conidial suspension (1.0 × 106 conidia ml−1) was inoculated onto the centre of a 90-mm Petri dish containing potato dextrose agar (PDA), minimal medium (MM), and complete medium (CM)26 and cultured at 25 °C. The colony diameter and morphology of the vegetative mycelia were examined at 4 days after inoculation. To estimate conidial production, discs of 7 mm in diameter obtained from the edge of a 10 day-old fungal colony on PDA medium were suspended in 1 mL sterilized water, then subjected to shaking at 150 rpm for 10 min. A 100 mL drop of the conidial suspension was subsequently placed onto a haemocytometer, and the spores were counted under a microscope. All of the tests were repeated at least three times.
Additional Information
Accession codes: Sequence data for FonSIX6 with flanking sequences from this paper have been deposited with the EMBL/DDBJ/GenBank data libraries under accession no. LT160066.1.
How to cite this article: Niu, X. et al. The FonSIX6 gene acts as an avirulence effector in the Fusarium oxysporum f. sp. niveum - watermelon pathosystem. Sci. Rep. 6, 28146; doi: 10.1038/srep28146 (2016).
Supplementary Material
Acknowledgments
This research was supported by the National Natural Science Foundation of China (31301787) and (31272188), the Natural Science Foundation of Zhejiang Province (LQ13C150004), and the Major Science and Technology Project of Zhejiang Province (2012C12903).
Footnotes
Author Contributions X.N. and M.F. conceived the experiments. X.N., X.Z. and Y.S. conducted the experiments. X.N., K.-S.L. and A.L. analysed the results and wrote the manuscript. All authors reviewed the manuscript.
References
- Wechter W. P., Kousik C. S., McMillan M. L. & Levi A. Identification of resistance to Fusarium oxysporum f. sp. niveum race 2 in Citrullus lanatus var. citroides plant introductions. HortScience 47, 334–338 (2012). [Google Scholar]
- Egel D. S. & Martyn R. D. Fusarium wilt of watermelon and other cucurbits. The Plant Health Instructor, 10.1094/PHI-I-2007-0122-01 (2007). [DOI] [Google Scholar]
- Martyn R. D. & Bruton B. D. An initial survey of the United State for races of Fusarium oxysporum f. sp. niveum. HortScience 26, 696–698 (1989). [Google Scholar]
- Martyn R. D. & Netzer D. Resistance to races 0, 1 and 2 of Fusarium wilt of watermelon in Citrullus sp. PI-296341-FR. Hortscience 26, 429–432 (1991). [Google Scholar]
- Zhou X. G., Everts K. L. & Bruton B. D. Race 3, a New and Highly Virulent Race of Fusarium oxysporum f. sp. niveum Causing Fusarium Wilt in Watermelon. Plant Disease 94, 92–98, 10.1094/pdis-94-1-0092 (2010). [DOI] [PubMed] [Google Scholar]
- Stahl E. A. & Bishop J. G. Plant-pathogen arms races at the molecular level. Curr Opin Plant Biol 3, 299–304 (2000). [DOI] [PubMed] [Google Scholar]
- Jones J. D. & Dangl J. L. The plant immune system. Nature 444, 323–329, 10.1038/nature05286 (2006). [DOI] [PubMed] [Google Scholar]
- Bent A. F. & Mackey D. Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45, 399–436, 10.1146/annurev.phyto.45.062806.094427 (2007). [DOI] [PubMed] [Google Scholar]
- Hogenhout S. A., Van der Hoorn R. A., Terauchi R. & Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol Plant Microbe Interact 22, 115–122, 10.1094/MPMI-22-2-0115 (2009). [DOI] [PubMed] [Google Scholar]
- Houterman P. M. et al. The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Molecular plant pathology 8, 215–221, 10.1111/j.1364-3703.2007.00384.x (2007). [DOI] [PubMed] [Google Scholar]
- Lievens B., Houterman P. M. & Rep M. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS microbiology letters 300, 201–215, 10.1111/j.1574-6968.2009.01783.x (2009). [DOI] [PubMed] [Google Scholar]
- Rep M. et al. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Molecular microbiology 53, 1373–1383, 10.1111/j.1365-2958.2004.04177.x (2004). [DOI] [PubMed] [Google Scholar]
- Houterman P. M., Cornelissen B. J. & Rep M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS pathogens 4, e1000061, 10.1371/journal.ppat.1000061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M., Meijer M., Houterman P. M., van der Does H. C. & Cornelissen B. J. Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Molecular plant-microbe interactions: MPMI 18, 15–23, 10.1094/MPMI-18-0015 (2005). [DOI] [PubMed] [Google Scholar]
- Houterman P. M. et al. The effector protein Avr2 of the xylem-colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. The Plant journal: for cell and molecular biology 58, 970–978, 10.1111/j.1365-313X.2009.03838.x (2009). [DOI] [PubMed] [Google Scholar]
- Ma L. et al. The AVR2-SIX5 gene pair is required to activate I-2-mediated immunity in tomato. The New phytologist 208, 507–518, 10.1111/nph.13455 (2015). [DOI] [PubMed] [Google Scholar]
- Gawehns F. et al. The Fusarium oxysporum effector SIX6 contributes to virulence and suppresses I-2-mediated cell death. Molecular plant-microbe interactions: MPMI 27, 336–348, 10.1094/MPMI-11-13-0330-R (2014). [DOI] [PubMed] [Google Scholar]
- Chakrabarti A. et al. Variation in potential effector genes distinguishing Australian and non-Australian isolates of the cotton wilt pathogen Fusarium oxysporum f. sp. vasinfectum. Plant Pathol 60, 232–243 (2010). [Google Scholar]
- Michielse C. B. & Rep M. Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol 10, 311–324, 10.1111/j.1364-3703.2009.00538.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S. M. et al. MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC genomics 14, 119, 10.1186/1471-2164-14-119 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. J. et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464, 367–373, 10.1038/nature08850 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M. & Kistler H. C. The genomic organization of plant pathogenicity in Fusarium species. Curr Opin Plant Biol 13, 420–426, 10.1016/j.pbi.2010.04.004 (2010). [DOI] [PubMed] [Google Scholar]
- Inami K. et al. A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: inactivation of avirulence gene AVR1 by transposon insertion. PLoS One 7, e44101, 10.1371/journal.pone.0044101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Zhao J., Guo W. & Zhang T. Functional analysis of autophagy genes via Agrobacterium-mediated transformation in the vascular Wilt fungus Verticillium dahliae. J Genet Genomics 40, 421–431, 10.1016/j.jgg.2013.04.006 (2013). [DOI] [PubMed] [Google Scholar]
- Mullins E. D. et al. Agrobacterium-Mediated Transformation of Fusarium oxysporum: An Efficient Tool for Insertional Mutagenesis and Gene Transfer. Phytopathology 91, 173–180, 10.1094/PHYTO.2001.91.2.173 (2001). [DOI] [PubMed] [Google Scholar]
- Leslie J. F. & Summerell B. A. The Fusarium Laboratory Manual 11 (Blackwell Professional; Ames, IA, USA 2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.