Abstract
We discuss the tripartite pathophysiological circuit of inflammatory bowel disease (IBD), involving the intestinal microbiota, barrier function, and immune system. Dysfunction in each of these physiological components (dysbiosis, leaky gut, and inflammation) contributes in a mutually interdependent manner to IBD onset and exacerbation. Genetic and environmental risk factors lead to disruption of gut homeostasis: genetic risks predominantly affect the immune system, environmental risks predominantly affect the microbiota, and both affect barrier function. Multiple genetic and environmental ‘hits’ are likely necessary to establish and exacerbate disease. Most conventional IBD therapies currently target only one component of the pathophysiological circuit, inflammation; however, many patients with IBD do not respond to immune-modulating therapies. Hope lies in new classes of therapies that target the microbiota and barrier function.
Keywords: barrier function, Crohn’s disease, inflammatory bowel disease, leaky gut, microbiome
Introduction
Inflammatory bowel disease (IBD) is a chronic relapsing inflammatory condition of the gastrointestinal tract which includes two partially overlapping clinical entities: Crohn’s disease (CD), characterized by patchy transmural inflammation that can involve the entire gastrointestinal tract, and ulcerative colitis (UC), characterized by mucosal inflammation limited to the colon [Feldman et al. 2015]. The incidence of CD and UC is rising worldwide [Molodecky et al. 2012] and despite current medical treatments which focus primarily on immunosuppression [Talley et al. 2011], over 20% of patients with CD still require surgery and over 10% of patients with UC still require colectomy [Rungoe et al. 2014]. The pathogenesis of IBD is multifactorial with genetic and environmental contributions believed to play a role in potentiating the immune system [Zhang and Li, 2014]. Recent work has also highlighted the importance of the intestinal microbiome and mucosal barrier function in disease pathophysiology [Kostic et al. 2014; Merga et al. 2014].
The intestinal microbiome has been likened to a virtual organ composed of microorganisms exhibiting complex bidirectional crosstalk with the environment and other organ systems [O’Hara and Shanahan, 2006; Sun and Chang, 2014]. The intestinal mucosal barrier is a virtual wall of tightly connected epithelial cells, buttressed by antimicrobial factors and mucus, that limits interaction between the microbiome and immune system [Turner, 2009]. In health, homeostasis exists between the intestinal microbiome, mucosal barrier, and immune system. In IBD, this homeostasis is disrupted leading to durable alterations in the intestinal microbiome (dysbiosis), disrupted barrier function (leaky gut), and immune system activation (inflammation) (Figure 1). Both genetic and environmental factors can influence transitions between health and disease. In this review, we discuss these factors with a focus on the microbiome and barrier function. While most current therapies modulate inflammation, we highlight new microbiome and barrier function based therapies under investigation for IBD.
Figure 1.
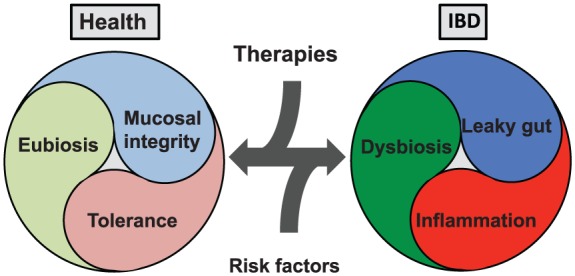
The tripartite pathophysiological circuit of inflammatory bowel disease (IBD). The microbiome, barrier function, and immune system all play critical roles in IBD pathophysiology with eubiosis, mucosal integrity and tolerance seen in health and dysbiosis, leaky gut, and inflammation seen in disease. Risk factors (environmental and genetic) push these pathophysiological components in the direction of disease. Therapeutic targets (microbiome, barrier function, and immune system based) push the components in the direction of health.
The gastrointestinal microbiome
New research tools employed by initiatives like the Human Microbiome [Human Microbiome Project Consortium, 2012; Integrative HMP (iHMP) Research Network Consortium, 2014] and metagenomics of the human intestinal tract (MetaHIT) [Arumugam et al. 2011] have led to rapid advances in our understanding of the microbes present on and within our body. These microbes are collectively referred to as the microbiota and the complement of their genomic content is termed the microbiome [Ursell et al. 2012]. Massively parallel deep sequencing of bacterial 16S ribosomal RNA and yeast 18S ribosomal RNA has allowed taxonomic categorization of the microbiota without the need to grow individual organisms, the majority of which remain uncultured [Rajilic-Stojanovic et al. 2007; Hamady and Knight, 2009; Metzker, 2010]. Metagenomics, metatranscriptomics, metaproteomics, and metabolomics have helped us understand the metabolic pathways present within the microbiome [Lepage et al. 2013]. Model systems like gnotobiotic mice [Faith et al. 2010] and ex vivo systems [Roeselers et al. 2013] are allowing us to investigate host microbe interactions and understand the contributions of isolated microbes under controlled conditions to health and disease.
Within the gastrointestinal tract, the microbiota varies lengthwise (mouth to rectum) and cross sectionally (lumen to mucosa) [Eckburg et al. 2005; Wang et al. 2005]. It contains all divisions of life: archaea, prokarya, eukarya (mostly fungi) as well as viruses (mostly bacteriophage) [Ley et al. 2006; Scanlan and Marchesi, 2008; Ianiro et al. 2014; Scarpellini et al. 2015]. The majority of studies to date have focused on the 10–100 trillion bacterial cells present throughout the gastrointestinal tract [Eckburg et al. 2005]. Ninety percent of the bacteria fall into the two phyla: Bacteroidetes and Firmicutes [Eckburg et al. 2005]. Other phyla, including Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia, are also present in lower quantities [Eckburg et al. 2005]. Diversity estimates place the total number of species between 1000 and 5000 [Zoetendal et al. 2008], and only a fraction of these, the ‘core’ microbiota, are commonly present in most individuals [Turnbaugh et al. 2009; Sekelja et al. 2011]. Individual microbiomes have been classified into different enterotypes (or faecotypes) [Arumugam et al. 2011; Jeffery et al. 2012], characterized by predominant species and metabolic pathways that correlate with long-term dietary preferences (high protein and animal fat versus high carbohydrate) [Wu et al. 2011].
The intestinal microbiome is shaped by both genetic and environmental factors [Spor et al. 2011]. Interestingly, the microbiomes of monozygotic twin pairs are more similar than mother–child pairs, which are more similar than unrelated pairs irrespective of physical separation [Turnbaugh et al. 2009]. From the time of birth, environmental factors like mode of delivery (cesarean section versus vaginal) and feeding preference (breast feeding versus formula feeding) shape the gut microbiome [Penders et al. 2006; Fallani et al. 2010]. This microbiome is highly dynamic in the first year of life with relative stabilization in the transition to an adult diet [Koenig et al. 2011]. Short- and long-term food preferences including vegetarian versus meat-based diets have significant effects on the microbiome [Wu et al. 2011; David et al. 2014]. Environmental factors such as level of hygiene, exposure to infections, antibiotics, and other drugs can also modify the microbiome [Spor et al. 2011].
In health, the microbiome plays key roles in metabolism of food and drugs, development of the gastrointestinal epithelium, development and modulation of the immune system, and protection from infections [Sekirov et al. 2010]. A healthy microbiota, established at an early age, exhibits resilience; multiple environmental inputs are likely necessary to effect a sustained and clinically relevant change [Lozupone et al. 2012]. Alterations in the microbiome have been associated with a surprising range of conditions, including neuropsychiatric diseases [Collins et al. 2012], asthma and atopic diseases [Van Nimwegen et al. 2011], obesity and metabolic syndrome [Nicholson et al. 2012], colorectal cancer [Louis et al. 2014], enteric infections [Kamada et al. 2013], irritable bowel syndrome [Dupont, 2014], and IBD [Manichanh et al. 2012].
The microbiome in IBD
The microbiome in IBD is known to be different from that of healthy individuals [Ott et al. 2004; Manichanh et al. 2006; Frank et al. 2007; Michail et al. 2012; Nagalingam and Lynch, 2012; Rajilic-Stojanovic et al. 2013; Bellaguarda and Chang, 2015; Sheehan et al. 2015] (Figure 2). The extent to which these changes are a cause or a consequence of inflammation remains a debate, but both may be accurate in that dysbiosis and inflammation are likely to be mutually reinforcing in patients with IBD. Similar to other forms of inflammatory diarrhea, there is a loss of diversity and stability of the microbiota in IBD. One of the most consistent findings is a decrease in the commensal spore-forming and butyrate-producing Clostridium clusters IV and XIVa (Firmicutes phylum) [Manichanh et al. 2006; Frank et al. 2007; Sartor, 2008]. These species are known to stimulate regulatory T cells (Tregs), leading to immune tolerance and reduction in gastrointestinal inflammation [Atarashi et al. 2011]. One member of this cluster, Faecalibacterium prausnitzii, is decreased in IBD [Fujimoto et al. 2013; Machiels et al. 2014] and predicts the recurrence of disease after ileal resection in CD [Sokol et al. 2008]. Similar decreases in Clostridium clusters IV and XIVa are seen in Clostridium difficile infection and C. difficile-negative nosocomial diarrhea and are therefore likely to be both a generic effect of and predisposing factor for inflammatory diarrhea [Antharam et al. 2013].
Figure 2.
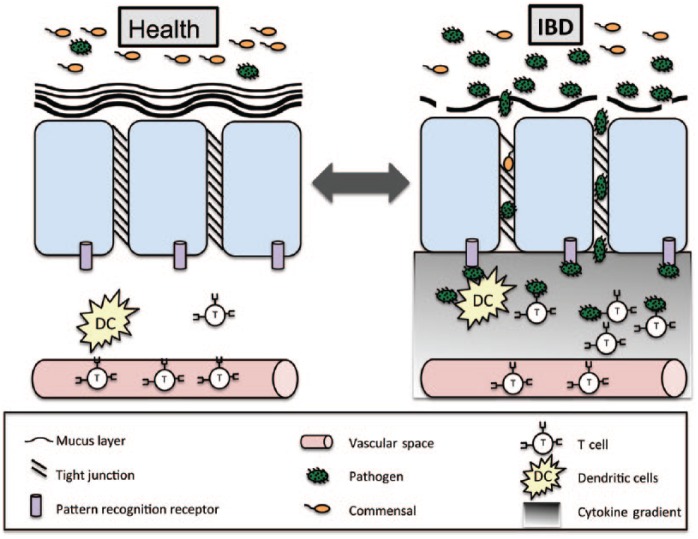
Mechanisms of inflammatory bowel disease (IBD) pathophysiology. IBD involves three pathophysiological components (dysbiosis, leaky gut, and inflammation) that are mutually dependent. In health, the mucosal barrier including two mucus layers, epithelial cells, and tight junctions separate the microbiota from the immune system. Breakdown of the mucosal barrier due to environmental and genetic factors leads to translocation of gastrointestinal organisms and activation of the innate and adaptive immune system. Genetic and environmental factors also contribute to dysbiosis and immune system activation leading to further breakdown of the mucosal barrier.
Increases in certain bacteria are also observed in IBD. It remains unclear to what extent these specific increases are the driving forces of the inflammatory process (keystone pathogen) [Hajishengallis et al. 2012] versus opportunistic contributors to an already established inflammatory process (pathobiont) [Chow et al. 2011]. Some of the most consistently elevated bacterial species in IBD are members of the family Enterobacteraceae (phylum Proteobacteria). These include the iconic gut pathogens Campylobacter spp., Salmonella spp., Shigella spp., and Escherichia coli. Indeed, there is an extensive line of research linking adherent-invasive E. coli to ileal Crohn’s disease [Darfeuille-Michaud et al. 2004]. The gut pathogen C. difficile is also increased in prevalence in IBD [Clayton et al. 2009; Berg et al. 2013]. A large multicenter study of patients with new-onset patients demonstrated increases in E. coli, Fusobacterium nucleatum, Haemophilus parainfluenzae, and Veillonella parvula; this increase in combination with a decrease in other species and an overall decline in species diversity correlated strongly with inflammation [Gevers et al. 2014]. Other studies have found increases in the intracellular bacteria Mycobacterium paratuberculosis in CD [Mcnees et al. 2015] and adherent invasive bacteria, Fusobacterium, in UC [Strauss et al. 2011].
While most research has focused on bacteria, work has begun to interrogate the role of fungal and viral components of the microbiota and unlike the bacterial microbiota, the diversity of the mycobiome [Richard et al. 2015] and virome [Norman et al. 2015; Ray, 2015] appear to be increased. The pathophysiological significance of these changes is an area of active investigation. Related immune system studies are also ongoing, including evaluation of the role of C-type lectin receptor dectin 1 (CLEC7A); a polymorphism of this receptor, which appears to interact with the mycobiome, may be linked to severe UC [Iliev et al. 2012].
Intestinal mucosal barrier function in IBD
The intestinal mucosal barrier separates the microbiota, food, and other luminal contents from the innate and adaptive immune system (Figure 2). It is composed of inner and outer mucus layers impregnated with antimicrobial factors and underlying intestinal epithelial cells stitched together with connecting protein networks called tight junctions [Turner, 2009]. In a healthy gut, the microbiota does not touch epithelial cells and is sampled in a controlled manner via specialized microfold (M) cells located in Peyer’s patches along the distal small intestine [Hooper and Macpherson, 2010]. Depending on the microbe and the immune system, this can lead to either immune tolerance or activation. In IBD, this mucosal barrier is disrupted, resulting in translocation of the intestinal microbiota and potentiation of the immune system [Merga et al. 2014]. As with dysbiosis, it is debated whether changes seen in barrier function are the result or the cause of the disease.
The inner mucus layer while devoid of bacteria in healthy controls [Johansson et al. 2008], shows increased permeability in IBD allowing interaction of the microbiota with the normally inaccessible epithelial surface [Schultsz et al. 1999; Swidsinski et al. 2005; Johansson et al. 2014]. The increased permeability may be due to altered composition of the mucus components secreted by goblet cells, including decreased mucin [Moehle et al. 2006], decreased glycosylation products [Theodoratou et al. 2014], decreased trefoil factor [Aamann et al. 2014] or due to decreases in antimicrobial factors secreted into the mucus by epithelial cells (Reg3γ), Paneth cells (defensins) and plasma cells [immunoglobulin A (IgA)] [MacDermott et al. 1989; Ramasundara et al. 2009; Hooper and Macpherson, 2010]. In UC but not CD, the mucus layers are thinner or absent and the goblet cells responsible for mucus production are depleted [Johansson et al. 2014]. Certain members of the IBD-associated microbiota use mucus as an energy source and tightly regulate its production, thus there is evidence that the mucus changes may be as much the result of dysbiosis as a cause [Deplancke and Gaskins, 2001; Derrien et al. 2004; Png et al. 2010].
The network of proteins called tight junctions connecting epithelial cells also show increased permeability in IBD [Michielan and D’Inca, 2015]. Both environmental (microbes, diet) and genetic factors can influence tight junction integrity [Ulluwishewa et al. 2011]. Disruption allows microbes to translocate beyond the mucosal surface resulting in access to the immunologically active submucosa and systemic space. Endotoxemia (lipopolysaccharide) is well documented in IBD [Pastor Rojo et al. 2007] and other microbial components (flagellin, pilli, and lipoteichoic acid) are also likely responsible for stimulating the immune system [Klapproth and Sasaki, 2010].
Immune system in IBD
The immune system plays a critical role in the development of IBD and it is likely that invading microorganisms are necessary for potentiating its effects [Geremia et al. 2014] (Figure 2). Microorganisms that invade epithelial cells, the submucosa, or systemic space can stimulate various components of the immune system, including autophagy [Parkes, 2012], innate immunity [Abraham and Medzhitov, 2011], and adaptive immunity [Kato et al. 2014]. Dysfunction in these pathways plays a role in IBD pathogenesis.
Autophagy, the regulated mechanism by which cells process and destruct organelles and intracellular pathogens, is disrupted in some forms of CD. Mutations in key autophagy genes like nucleotide oligomerization domain 2 (NOD2) and ATG16L1 are associated with terminal ileal Crohn’s, and certain intracellular pathogens are able to manipulate autophagy to form autophagic vacuoles where they remain protected from immune responses [Parkes, 2012].
Microorganisms that reach the submucosa through disrupted tight junctions can interact with the basolateral surface of epithelial cells that are covered in pattern recognition receptors such as toll-like receptors that recognize various components of microbial pathogens [Man et al. 2011]. They stimulate the release of inflammatory cytokines recruiting phagocytic cells and components of the adaptive immune system. Toll-like receptors and regulating molecules have been shown to have altered expression in both active and inactive IBD [Fernandes et al. 2015].
IBD risk factors
Both genetic and environmental risk factors influence the development of IBD (Figure 3). Understanding how these factors impact disease susceptibility, onset, and exacerbation can guide future investigations aimed at identifying targets for disease treatment and prevention.
Figure 3.
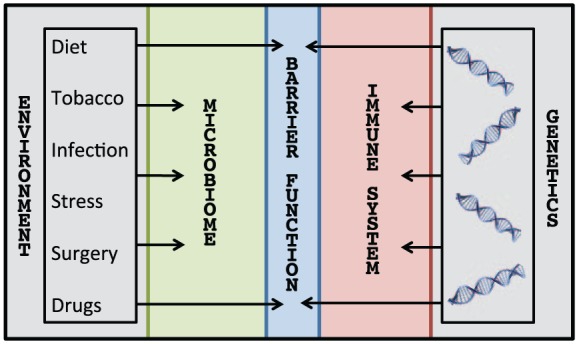
Risk factors for inflammatory bowel disease (IBD). There are multiple risk factors, both genetic and environmental, that mutually contribute to the development and exacerbation of IBD. Genetics directly affect barrier function and the immune system, whereas environmental factors (e.g. diet, tobacco use, infections, stress, surgical procedures, and medications, including antibiotic exposure) directly affect barrier function and the microbiota.
Genetic risks
Monozygotic twin concordance rates are only 15–20% in UC and less than 50% in CD [Halme et al. 2006], indicating that although genes confer increased susceptibility to IBD, they are not sufficient for the development of disease. Twins and family members of patients with IBD often demonstrate abnormal gut parameters, including leaky gut [Buhner et al. 2006; D’Inca et al. 2006] and altered microbiome without developing IBD, suggesting that genetics predispose people to gut dysfunction but other environmental factors are necessary for triggering disease onset [Willing et al. 2010; Halfvarson, 2011; Hedin et al. 2014].
Genome-wide association studies (GWAS) have identified over 200 IBD-associated loci [Ogura et al. 2001; Jostins et al. 2012; Knights et al. 2013; McGovern et al. 2015]. All gene variants carry a low penetrance with the highest odds ratio of 7 conferred by NOD2, a gene coding for an intracellular receptor that recognizes a component of bacterial cell walls. Other gene associations have odds ratios of less than 1. A mutation in the IL23 receptor has been shown to offer a two- to threefold protection against developing IBD [McGovern et al. 2015]. Genes likely influence all three components of IBD pathophysiology (microbiome, barrier function, and immunity) but primarily affect barrier function and immunity through their impact on mucus production, tight junctions, autophagy, and the innate and adaptive immune response [Jostins et al. 2012; Dalal and Chang, 2014; Van Limbergen et al. 2014; Bianco et al. 2015].
Environmental risks
Epidemiologic studies have described a rising incidence of both UC and CD as countries undergo industrialization and demographic transition [Bernstein and Shanahan, 2008]. These epidemiologic changes are brisk beyond the speed of genetic alterations at the population level, suggesting there is likely a strong environmental component [Sheehan et al. 2015]. There are multiple environmental factors that may synergistically contribute to the rising incidence of IBD, including hygiene, diet, medications, tobacco use, surgical practices, and stress [Shanahan, 2012]. These environmental factors are likely to have effects on all three components of IBD pathophysiology (microbiome, barrier function, and immunity), but may exert a disproportionate effect on the gut microbiome and barrier function.
Hygiene
The hygiene hypothesis has been discussed as a contributing factor to the rise of autoimmune diseases in the developed world [Rook, 2012]. Central to this hypothesis is the idea that developed countries see less pathogenic burden (e.g. viral infections, Helicobacter pylori, and helminths) due to improved cleanliness in comparison to developing countries. Lack of exposure to infectious antigens during early immune development in childhood may have lasting impact on the immune system by switching the predominance of T lymphocyte subtypes (T helper 1 to T helper 2) that potentiate autoimmune phenomena including IBD [Koloski et al. 2008].
Diet
Several studies describe a potential association between the rising incidence of IBD and a ‘Western’ diet composed largely of processed foods that are high in fat and protein and low in fiber (fruits and vegetables) [Chapman-Kiddell et al. 2010; Hou et al. 2011]. This may be due to direct effects of diet on the microbiota [Wu et al. 2013] and barrier function [Martinez-Medina et al. 2014]. Diets low in fiber have consistently been linked to IBD perhaps due to decreases in short chain fatty acid (SCFA) production by commensal bacteria (Clostridium clusters IV and IVXa) whose preferred energy source is fiber [Van Immerseel et al. 2010]. The SCFA butyrate is critical for colonic health as the preferred energy source for colonocytes [Van Immerseel et al. 2010]. It also contributes to tight junction integrity and is a regulator of Treg cells [Smith et al. 2013].
Associations between high fat, protein, and sugar diets have been less consistent in epidemiologic studies [Hou et al. 2011], although animal studies have shown a strong link between high fat, high simple sugar diets [Martinez-Medina et al. 2014]. It may be that particular fats versus overall fat intake is important for disease development. For example, saturated milk fat, but not polyunsaturated fat, leads to the expansion of the sulfite reducing pathobiont Bilophila wadsworthia and to the development of colitis in a mouse model of IBD [Devkota et al. 2012]. Interestingly, mode of feeding after birth (breastfeeding versus formula) influences the microbiota and may have a long-term effect on IBD incidence [Barclay et al. 2009; Guaraldi and Salvatori, 2012].
Food additives may also be linked to the development of IBD. These include common dietary emulsifiers, carboxymethylcellulose (CMC) and polysorbate 80 (P80), which induce low-grade inflammation and metabolic syndrome in wild type mice and promote a robust colitis in genetically predisposed mice. The emulsifiers altered the microbiota to have more inflammatory potential and increased the number of mucolytic bacteria causing erosion of the mucus layer [Chassaing et al. 2015]. Maltodextrin, another common food emulsifier, has shown similar effects in animal models [Nickerson et al. 2015]; however, the effect of these additives in humans is less clear. Other food components such as gliadin (a glycoprotein and major component of gluten) can disrupt tight junctions and may also be critical for IBD pathophysiology [Ulluwishewa et al. 2011; Chassaing et al. 2015; Lerner and Matthias, 2015].
Medications
Different patterns of medication use have been postulated to play a role in the rising incidence of IBD via an effect on the microbiota or barrier function. Antibiotics are known to cause shifts in microbial composition and use in infancy [Shaw et al. 2010], childhood [Kronman et al. 2012], and adult life [Shaw et al. 2011] has been associated with an increased risk of IBD [Modi et al. 2014]. Tetracycline in the treatment of acne has been associated with increased risk of CD [Margolis et al. 2010; Alikhan et al. 2011]. Nonsteroidal anti-inflammatory drugs promote intestinal barrier disruption [Ananthakrishnan, 2013], alter the microbiota composition [Rogers and Aronoff, 2015], and have been associated with increased risk of IBD development and clinical relapse [Takeuchi et al. 2006; Chan et al. 2011; Ananthakrishnan et al. 2012]. Oral contraceptives have effects on the vaginal microbiota [Achilles and Hillier, 2013], also likely affect the gastrointestinal microbiota, and are variably associated with an increased risk for developing Crohn’s disease [Timmer et al. 1998; Cosnes et al. 1999; Alic, 2000]. Other medications too are associated with microbiota changes (e.g. proton pump inhibitors) [Freedberg et al. 2015] and improvements in tight junction integrity (e.g. β blockers) [Reiberger et al. 2013] and should be evaluated for their association with increased and decreased risk of IBD.
Tobacco use
Nicotine interestingly has a beneficial effect on tight junction integrity and it is possible that different modes of action of tobacco on the microbiota versus gut barrier function may explain the differential effects of tobacco on CD and UC [McGilligan et al. 2007]. Tobacco use is associated with an increased risk for CD and reduced risk for UC and tobacco cessation reverses this effect [Cosnes, 2008; Parkes et al. 2014]. Smokers with active CD have a clinically relevant gastrointestinal dysbiosis, and smoking cessation induces profound changes in the microbiota [Benjamin et al. 2012; Biedermann et al. 2013].
Surgical practices
Appendectomies are associated with a decreased risk for UC [Kaplan et al. 2008; Radford-Smith, 2008; Cheluvappa et al. 2014] and possibly an increased risk for CD [Andersson et al. 2003; Kaplan et al. 2007]. Patients with UC, after appendectomies, experience fewer flares, fewer colectomies and a decreased need for immunosuppressive therapy [Naganuma et al. 2001; Radford-Smith et al. 2002; Radford-Smith, 2008]. Studies evaluating the microbiota of the resected inflamed appendix show increases in pathogenic organisms, including the adherent invasive bacterium Fusobacterium [Swidsinski et al. 2011; Guinane et al. 2013]. Similar organisms have increased incidence in active IBD and thus appendectomies may be protective in UC by eliminating a pathogen reservoir [Strauss et al. 2011]. Cesarean sections versus vaginal births are also associated with alterations in the microbiota and an increased risk for IBD [Dominguez-Bello et al. 2010; Bager et al. 2012] perhaps due to differences in early microbial colonization of the gastrointestinal tract.
Stress
Stress correlates with IBD relapse [Singh et al. 2009] and improvements in stress via counseling correlate with decreased IBD symptoms [Wahed et al. 2010]. Stress has also been shown to have profound effects on the microbiota [Lutgendorff et al. 2008; Bangsgaard Bendtsen et al. 2012], barrier function [Soderholm et al. 2002; Camilleri et al. 2012], and intestinal inflammation [Melgar et al. 2008; Singh et al. 2009; Matsunaga et al. 2011] and thus may contribute significantly to IBD pathophysiology.
IBD therapies
The majority of IBD therapies to date have focused on modulating inflammation via the immune system. These have enabled great leaps forward in medically treating a disease that historically only had surgical treatment options. Despite therapeutic advances, many patients with IBD still require surgery. Short-circuiting IBD pathophysiology may ultimately require a therapeutic approach that integrates all three pathophysiological components (dysbiosis, leaky gut, and inflammation). Therapies will likely be highly individualized based on future diagnostics that point toward keystone dysfunction in one or more of these pathophysiological components. To this end, in addition to immune-based therapies, therapies targeting dysbiosis and leaky gut are currently being explored (Figure 4).
Figure 4.
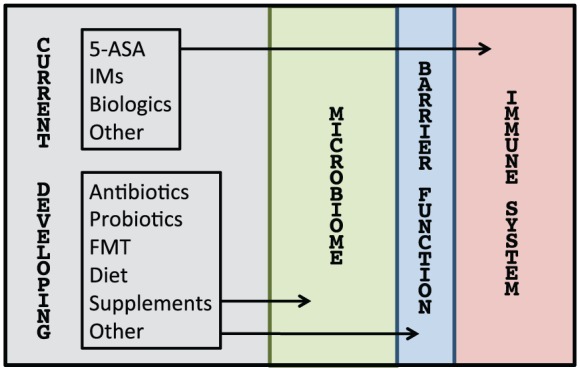
Therapies for inflammatory bowel disease (IBD). Conventional therapies like aminosalicylates (5-ASA), immunomodulators (IMs), and biologics are traditionally thought to work via the immune system but may also have direct and indirect effects on the microbiota and barrier function (not depicted). Vice versa, developing therapies like antibiotics, probiotics, fecal microbiota transplantation (FMT), diet, and supplements that are thought to target the microbiota and barrier function may have direct and indirect effects on the immune system (not depicted).
Immune-based therapies
Current IBD treatments include the use of medications that modulate the immune system, including aminosalicylates, corticosteroids, immunomodulators (e.g. methotrexate, azathioprine), antitumor necrosis factor (TNF) agents, and integrin inhibitors. Newer biologic agents that focus on other inflammatory cytokines, their receptors, and downstream pathways are also in various stages of development [Peng et al. 2014].
Immune-based therapies as their name implies have direct effects on the immune system, but some studies have also demonstrated either direct or indirect effects on the microbiota and mucosal barrier function. Mesalamine and other salicylates for example have been shown to change the intestinal microbiota and this may in part be due to direct effects on microbes [Andrews et al. 2011]. Indeed, mesalamine and other salicyaltes decrease the expression of microbial adherence factors and biofilm formation [Damman, 2013]. Unlike immunomodulators, which lead to proliferation of mucosally associated bacteria, mesalamine leads to a decrease in mucosally associated bacteria in UC [Swidsinski et al. 2007]. Studies of biologics show an ameliorating effect on the microbiome and on tight junctions [Edelblum and Turner, 2009; Busquets et al. 2015]. Indeed, TNF and other inflammatory cytokines directly disrupt tight junctions and thus a biologic’s primary mechanism of action may in large part be due to reestablishing barrier function [Li et al. 2008; Edelblum and Turner, 2009].
Microbiota-based therapies
Microbiota-based therapies include antibiotics, probiotics, fecal microbiota transplantation (FMT), and diet. These have been investigated as treatments for IBD with varying results. Therapeutic manipulation of the microbiota offers theoretical advantages over immune system and barrier function based therapies as a treatment strategy since the microbiota is more malleable than host factors under greater genetic influence.
Antibiotics
The antibiotics that have been most studied as treatments for IBD include metronidazole, rifaximin, ciprofloxacin, and antimycobacterial agents [Bejaoui et al. 2015]. Most efficacy has been demonstrated in CD, particularly in inducing remission (with less consistent data showing maintenance of remission), treatment of perianal disease, and treatment of pouchitis [Khan et al. 2011; Cammarota et al. 2015]. Antibiotics are less effective in the treatment of adult UC (Khan et al. 2011), although there may be some efficacy in the pediatric population [Turner et al. 2014]. Rifaximin and ciprofloxacin by one meta-analysis may have the greatest benefit in inducing remission in CD [Arnold et al. 2002; Prantera et al. 2006; Khan et al. 2011]. Metronidazole has particular benefit in the treatment of perianal disease [Brandt et al. 1982; Sutherland et al. 1991; Khan et al. 2011; Mowat et al. 2011].
Rifaximin’s effect on the microbiota includes decreases in certain pathogens with reciprocal increases in Bifidobacteria and F. prausnitzii in CD [Maccaferri et al. 2010; Guslandi, 2011]. Antibiotics are not without risk and while there may be some studies demonstrating benefit, there is also the potential for antibiotic-related side effects, including resistance, reduction in biodiversity, and risk for C. difficile infection.
Probiotics
Probiotics have shown some efficacy in UC and pouchitis with less efficacy in CD [Cammarota et al. 2015]. Generalizations about the efficacy of probiotics is complicated by the variability of the formulations and specific strains studied. The two probiotic formulations that have been studied most extensively in IBD are E. coli Nissle 1917 and VSL#3. E. coli Nissle 1917 has been shown to be comparable to mesalamine in maintaining remission [Kruis et al. 2004; Henker et al. 2008]. VSL#3 is a mixture of eight different bacteria (four strains of lactobacilli, three strains of Bifidobacteria, and one strain of Streptococcus) and has been shown to be effective for induction and maintenance of remission in pouchitis [Gionchetti et al. 2000; Mimura et al. 2004] in both pediatric UC [Miele et al. 2009] and adult UC [Sood et al. 2009]. VSL#3 when used in combination with conventional therapy has also demonstrated some efficacy in decreasing disease activity [Tursi et al. 2010].
Several mechanisms of action have been proposed, including promoting the growth of anti-inflammatory bacteria and inhibiting the growth of pathogenic bacteria [Dalal and Chang, 2014]. Some strains of bacteria are also able to produce SCFAs that are the preferred energy source of colonocytes. Probiotics are often formulated with prebiotics, indigestible fibers that help promote their growth. Despite some beneficial results overall in IBD, there are no current guidelines recommending the routine use of probiotics in the induction or maintenance of IBD. Additionally, one must consider the potential risk of bacterial translocation of probiotics in patients who are critically ill or immunocompromised, which can very rarely lead to sepsis and multiorgan failure [Theodorakopoulou et al. 2013].
Fecal microbiota transplantation
FMT involves the infusion of donor stool into an individual with the aim of restoring a ‘healthy’ microbiota and treating disease. It has been used most extensively and effectively (>90% cure rate) as a treatment for recurrent C. difficile infection [Gough et al. 2011; Van Nood et al. 2013] and in the wake of this success has been evaluated for other diverse indications [Borody et al. 2013].
Several case series [Colman and Rubin, 2014] and placebo-controlled trials [Moayyedi et al. 2015; Rossen et al. 2015] have evaluated its efficacy in IBD with mixed results [Damman et al. 2012; Hansen and Sartor, 2015]. Many of these studies have evaluated its effect in UC [Angelberger et al. 2013; Kump et al. 2013; Kunde et al. 2013; Damman et al. 2015], although some studies have also investigated the treatment for CD [Zhang et al. 2013; Cui et al. 2015; Suskind et al. 2015]. While induction of remission appears to be possible in a subset of patients with both UC and CD, this effect is neither universal, nor sustained. Several studies have measured whether engraftment of the donor stool correlates with efficacy with mixed results [Angelberger et al. 2013; Kump et al. 2013; Damman et al. 2015]. It is likely that repeated infusions are necessary for maximum efficacy and sustained effect [Damman et al. 2015]. It is possible that pretreatment with antibiotics and adjunctive treatment with diet may also augment efficacy [Damman et al. 2015].
The enthusiasm exploring FMT as a treatment for IBD has been partially tempered by observed and theoretical side effects [Rubin, 2013]. While case series have demonstrated that FMT may be safe in a diverse array of immunocompromised patients, this remains a concern in patients with IBD on immunosuppressive therapy [Kelly et al. 2014; Di Bella et al. 2015]. Fever and elevations in inflammatory markers have been observed in patients with IBD following FMT [Rubin, 2013]. Despite screening measures, there are also concerns that FMT may transmit infectious agents that may not manifest in disease for years [Bourlioux and Workgroup of the French Academy of Pharmacy, 2015]. To help mitigate this risk, several groups are evaluating the role of a rationally designed ‘artificial stool’ that contains a subset of clinically active microbes with more limited infectious risks [Petrof et al. 2013; Petrof and Khoruts, 2014].
Diet
Various diets have been proposed to prevent and treat IBD. Given the strong benefit of fiber intake, a well balanced, healthy diet with fruits and vegetables is recommended. Since a protein-rich diet with excess meat and alcohol has demonstrated increased relapse rates in UC, avoidance of these items may be beneficial [Tilg and Kaser, 2004].
Exclusive elemental nutrition (EEN), a formula-based therapy, with an efficacy rate of 85% and low side-effect profile, is recommended as first-line therapy for induction of remission [Critch et al. 2012] and helps maintain remission in pediatric CD [Wu et al. 2013]. It is equivalent to corticosteroid therapy in inducing clinical remission and superior to corticosteroids in inducing histologic remission [Gorard et al. 1993; Borrelli et al. 2006]. Efficacy of EEN in adult patients with CD appears to be less perhaps as a result of poor compliance or greater prior exposure to immunosuppressive therapies [Lee et al. 2015]. Partial enteral nutrition, a diet in which table foods are added to EEN, has also been shown to be efficacious in both adult and pediatric CD [Sigall-Boneh et al. 2014]. EEN is hypothesized to be effective by limiting antigen exposure (due to rapid transit), enhancing nutritional status, and altering the microbiome and immune response [Voitk et al. 1973; Rajendran and Kumar, 2010].
Other nonformula-based dietary interventions have also been studied. The specific carbohydrate diet (SCD), an elimination diet which removes grains, milk, and sweeteners (except honey), has been shown in small case series to be effective for inducing and maintaining remission in CD [Cohen et al. 2014; Suskind et al. 2014; Kakodkar et al. 2015] and UC [Obih et al. 2016]. The mechanism by which the SCD works may come from alteration of the microbiome or barrier function via differences in macronutritients or removal of certain dietary exposures such as emulsifiers [Martinez-Medina et al. 2014; Chassaing et al. 2015; Nickerson et al. 2015]. Interestingly, the effect of EEN and SCD on the microbiota is divergent, with microbial diversity decreasing with EEN [Gerasimidis et al. 2014; Quince et al. 2015] and increasing with SCD [Walters et al. 2014]. Other diet therapies including individually tailored exclusion [Riordan et al. 1993], IgG4 targeted exclusion [Rajendran and Kumar, 2010], lacto-ovo-vegetarian fiber rich diet [Chiba et al. 2010] and low FODMAPs (fermentable oligo-di-monosaccharides and polyols) diet [Gearry et al. 2009] have reported primarily symptomatic improvement in patients with IBD.
Barrier function based therapies
Restoring mucosal barrier integrity holds promise as a future treatment approach for IBD. It may indeed be a common denominator in all IBD therapies, including those that target the immune system or microbiota. As noted above, inhibition of TNFα and other inflammatory cytokines leads to improvement in barrier function. Remodeling the microbiota has also been shown to improve barrier function [Cani et al. 2009; Ulluwishewa et al. 2011], and exclusionary diets may improve barrier function via effects on the mucus layer [Martinez-Medina et al. 2014; Chassaing et al. 2015; Nickerson et al. 2015]. Even the mechanism of action of helminth therapies in which parasites are introduced to patients in a controlled fashion may be grounded in improving barrier function [Wolff et al. 2012].
Several drugs are known to act directly on restoring barrier function. A delayed release phosphatidylcholine, ‘LT-02’, recently completed phase II clinical trials for UC and is believed to work by helping restore the mucus layer [Stremmel et al. 2010; Karner et al. 2014]. Teduglutide is a glucagon-like 2 peptide analog that is indicated for the treatment of short gut syndrome [Jeppesen, 2012]. It has strong trophic effects on intestinal mucosa with restoration of the mucosal barrier and pilot studies have shown efficacy in the treatment of moderate to severe CD [Buchman et al. 2010; Blonski et al. 2013].
Some supplements have also been investigated for their ability to improve tight junctions [Ulluwishewa et al. 2011]. The amino acid, L-glutamine, has been studied extensively in nutrition for critically ill patients and burn victims in which tight junction disruption is known to contribute to bacteremia [Wischmeyer, 2007; Bollhalder et al. 2013], although in patients in the intensive care unit with multiorgan failure, glutamine increased mortality [Heyland et al. 2013]. In vitro cell culture studies show a direct effect on improving tight junction function [Bertrand et al. 2015]. Only limited studies have been performed in treating active IBD with negative results [Akobeng et al. 2000; Ockenga et al. 2005], but further studies are being considered to investigate the role of L-glutamine in maintenance rather than induction of remission.
Natural products often used by alternative practitioners for the treatment of IBD have shown some efficacy and may also exert their effect via modulation of tight junctions [Gilardi et al. 2014]. Curcumin (turmeric extract) and boswellia have been shown to promote improvement in tight junctions in cell culture based assays [Wang et al. 2012; Catanzaro et al. 2015]. Other dietary components (macro- and micronutrients), in addition to the microbiota effects described above, may directly impact tight junctions in IBD [Ulluwishewa et al. 2011].
Conclusion
The microbiome, barrier function, and immune system play an integrated role in the development of IBD, and all three components are likely critical for perpetuating the disease process. Genetic risk factors may be disproportionately responsible for immune system dysfunction, environmental risk factors may be disproportionately responsible for dysbiosis, and both factors likely contribute to barrier dysfunction. While currently available therapies targeting the immune system have made great strides in IBD therapy, many patients remain refractory to treatment. There is great promise in targeting the other two pathophysiological components of IBD, the microbiota and barrier function, as new primary or adjunctive therapies for IBD.
Acknowledgments
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the University of Washington.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Contributor Information
Stephen M. Vindigni, Division of Gastroenterology, Department of Medicine, University of Washington, Seattle, WA, USA
Timothy L. Zisman, Division of Gastroenterology, Department of Medicine, University of Washington, Seattle, WA, USA
David L. Suskind, Department of Pediatrics, Seattle Children’s Hospital and University of Washington, Seattle, WA, USA
Christopher J. Damman, University of Washington School of Medicine, 1959 NE Pacific Street, Box 356424, Seattle, WA 98195-6424, USA.
References
- Aamann L., Vestergaard E., Gronbaek H. (2014) Trefoil factors in inflammatory bowel disease. World J Gastroenterol 20: 3223–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham C., Medzhitov R. (2011) Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology 140: 1729–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilles S., Hillier S. (2013) The complexity of contraceptives: understanding their impact on genital immune cells and vaginal microbiota. AIDS 27(Suppl. 1): S5–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akobeng A., Miller V., Stanton J., Elbadri A., Thomas A. (2000) Double-blind randomized controlled trial of glutamine-enriched polymeric diet in the treatment of active Crohn’s disease. J Pediatr Gastroenterol Nutr 30: 78–84. [DOI] [PubMed] [Google Scholar]
- Alic M. (2000) Epidemiology supports oral contraceptives as a risk factor in Crohn’s disease. Gut 46: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhan A., Henderson G., Becker L., Sciallis G. (2011) Acne treatment and inflammatory bowel disease: what is the evidence? J Am Acad Dermatol 65: 650–654. [DOI] [PubMed] [Google Scholar]
- Ananthakrishnan A. (2013) Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y) 9: 367–374. [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan A., Higuchi L., Huang E., Khalili H., Richter J., Fuchs C., et al. (2012) Aspirin, nonsteroidal anti-inflammatory drug use, and risk for Crohn disease and ulcerative colitis: a cohort study. Ann Intern Med 156: 350–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Olaison G., Tysk C., Ekbom A. (2003) Appendectomy is followed by increased risk of Crohn’s disease. Gastroenterology 124: 40–46. [DOI] [PubMed] [Google Scholar]
- Andrews C., Griffiths T., Kaufman J., Vergnolle N., Surette M., Rioux K. (2011) Mesalazine (5-aminosalicylic acid) alters faecal bacterial profiles, but not mucosal proteolytic activity in diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther 34: 374–383. [DOI] [PubMed] [Google Scholar]
- Angelberger S., Reinisch W., Makristathis A., Lichtenberger C., Dejaco C., Papay P., et al. (2013) Temporal bacterial community dynamics vary among ulcerative colitis patients after fecal microbiota transplantation. Am J Gastroenterol 108: 1620–1630. [DOI] [PubMed] [Google Scholar]
- Antharam V., Li E., Ishmael A., Sharma A., Mai V., Rand K., et al. (2013) Intestinal dysbiosis and depletion of butyrogenic bacteria in clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51: 2884–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold G., Beaves M., Pryjdun V., Mook W. (2002) Preliminary study of ciprofloxacin in active Crohn’s disease. Inflamm Bowel Dis 8: 10–15. [DOI] [PubMed] [Google Scholar]
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D., et al. (2011) Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., et al. (2011) Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager P., Simonsen J., Nielsen N., Frisch M. (2012) Cesarean section and offspring’s risk of inflammatory bowel disease: a national cohort study. Inflamm Bowel Dis 18: 857–862. [DOI] [PubMed] [Google Scholar]
- Bangsgaard Bendtsen K., Krych L., Sorensen D., Pang W., Nielsen D., Josefsen K., et al. (2012) Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One 7: e46231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay A., Russell R., Wilson M., Gilmour W., Satsangi J., Wilson D. (2009) Systematic review: the role of breastfeeding in the development of pediatric inflammatory bowel disease. J Pediatr 155: 421–426. [DOI] [PubMed] [Google Scholar]
- Bejaoui M., Sokol H., Marteau P. (2015) Targeting the microbiome in inflammatory bowel disease: critical evaluation of current concepts and moving to new horizons. Dig Dis 33(Suppl. 1): 105–112. [DOI] [PubMed] [Google Scholar]
- Bellaguarda E., Chang E. (2015) IBD and the gut microbiota – from bench to personalized medicine. Curr Gastroenterol Rep 17: 15. [DOI] [PubMed] [Google Scholar]
- Benjamin J., Hedin C., Koutsoumpas A., Ng S., McCarthy N., Prescott N., et al. (2012) Smokers with active Crohn’s disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm Bowel Dis 18: 1092–1100. [DOI] [PubMed] [Google Scholar]
- Berg A., Kelly C., Farraye F. (2013) Clostridium difficile infection in the inflammatory bowel disease patient. Inflamm Bowel Dis 19: 194–204. [DOI] [PubMed] [Google Scholar]
- Bernstein C., Shanahan F. (2008) Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut 57: 1185–1191. [DOI] [PubMed] [Google Scholar]
- Bertrand J., Ghouzali I., Guerin C., Bole-Feysot C., Gouteux M., Dechelotte P., et al. (2015) Glutamine restores tight junction protein claudin-1 expression in colonic mucosa of patients with diarrhea-predominant irritable bowel syndrome. JPEN J Parenter Enteral Nutr, 13 May 2015 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Bianco A., Girardelli M., Tommasini A. (2015) Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J Gastroenterol 21: 12296–12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann L., Zeitz J., Mwinyi J., Sutter-Minder E., Rehman A., Ott S., et al. (2013) Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One 8: e59260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonski W., Buchner A., Aberra F., Lichtenstein G. (2013) Teduglutide in Crohn’s disease. Expert Opin Biol Ther 13: 1207–1214. [DOI] [PubMed] [Google Scholar]
- Bollhalder L., Pfeil A., Tomonaga Y., Schwenkglenks M. (2013) A systematic literature review and meta-analysis of randomized clinical trials of parenteral glutamine supplementation. Clin Nutr 32: 213–223. [DOI] [PubMed] [Google Scholar]
- Borody T., Paramsothy S., Agrawal G. (2013) Fecal microbiota transplantation: indications, methods, evidence, and future directions. Curr Gastroenterol Rep 15: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli O., Cordischi L., Cirulli M., Paganelli M., Labalestra V., Uccini S., et al. (2006) Polymeric diet alone versus corticosteroids in the treatment of active pediatric Crohn’s disease: a randomized controlled open-label trial. Clin Gastroenterol Hepatol 4: 744–753. [DOI] [PubMed] [Google Scholar]
- Bourlioux P. and Workgroup of the French Academy of Pharmacy (2015) Faecal microbiota transplantation: key points to consider. Ann Pharm Fr 73: 163–168. [DOI] [PubMed] [Google Scholar]
- Brandt L., Bernstein L., Boley S., Frank M. (1982) Metronidazole therapy for perineal Crohn’s disease: a follow-up study. Gastroenterology 83: 383–387. [PubMed] [Google Scholar]
- Buchman A., Katz S., Fang J., Bernstein C., Abou-Assi S. Teduglutide Study Group (2010) Teduglutide, a novel mucosally active analog of glucagon-like peptide-2 (GLP-2) for the treatment of moderate to severe Crohn’s disease. Inflamm Bowel Dis 16: 962–973. [DOI] [PubMed] [Google Scholar]
- Buhner S., Buning C., Genschel J., Kling K., Herrmann D., Dignass A., et al. (2006) Genetic basis for increased intestinal permeability in families with Crohn’s disease: role of CARD15 3020insC mutation? Gut 55: 342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busquets D., Mas-De-Xaxars T., Lopez-Siles M., Martinez-Medina M., Bahi A., Sabat M., et al. (2015) Anti-tumour necrosis factor treatment with adalimumab induces changes in the microbiota of Crohn’s disease. J Crohns Colitis 9: 899–906. [DOI] [PubMed] [Google Scholar]
- Camilleri M., Madsen K., Spiller R., Greenwood-Van Meerveld B., Verne G. (2012) Intestinal barrier function in health and gastrointestinal disease. Neurogastroenterol Motil 24: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarota G., Ianiro G., Cianci R., Bibbò S., Gasbarrini A., Currò D. (2015) The involvement of gut microbiota in inflammatory bowel disease pathogenesis: potential for therapy. Pharmacol Ther 149: 191–212. [DOI] [PubMed] [Google Scholar]
- Cani P., Possemiers S., Van De, Wiele T., Guiot Y., Everard A., Rottier O., et al. (2009) Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58: 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro D., Rancan S., Orso G., Dall’acqua S., Brun P., Giron M., et al. (2015) Boswellia serrata preserves intestinal epithelial barrier from oxidative and inflammatory damage. PLoS One 10: e0125375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S., Luben R., Bergmann M., Boeing H., Olsen A., Tjonneland A., et al. (2011) Aspirin in the aetiology of Crohn’s disease and ulcerative colitis: a European prospective cohort study. Aliment Pharmacol Ther 34: 649–655. [DOI] [PubMed] [Google Scholar]
- Chapman-Kiddell C., Davies P., Gillen L., Radford-Smith G. (2010) Role of diet in the development of inflammatory bowel disease. Inflamm Bowel Dis 16: 137–151. [DOI] [PubMed] [Google Scholar]
- Chassaing B., Koren O., Goodrich J., Poole A., Srinivasan S., Ley R., et al. (2015) Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheluvappa R., Luo A.S., Grimm M.C. (2014) Autophagy Suppression by Appendicitis and Appendectomy Protects against Colitis. Inflamm Bowel Dis 20: 847–855. [DOI] [PubMed] [Google Scholar]
- Chiba M., Abe T., Tsuda H., Sugawara T., Tsuda S., Tozawa H., et al. (2010) Lifestyle-related disease in Crohn’s disease: relapse prevention by a semi-vegetarian diet. World J Gastroenterol 16: 2484–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J., Tang H., Mazmanian S. (2011) Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol 23: 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E., Rea M., Shanahan F., Quigley E., Kiely B., Hill C., et al. (2009) The vexed relationship between Clostridium difficile and inflammatory bowel disease: an assessment of carriage in an outpatient setting among patients in remission. Am J Gastroenterol 104: 1162–1169. [DOI] [PubMed] [Google Scholar]
- Cohen S., Gold B., Oliva S., Lewis J., Stallworth A., Koch B., et al. (2014) Clinical and mucosal improvement with specific carbohydrate diet in pediatric Crohn disease. J Pediatr Gastroenterol Nutr 59: 516–521. [DOI] [PubMed] [Google Scholar]
- Collins S., Surette M., Bercik P. (2012) The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 10: 735–742. [DOI] [PubMed] [Google Scholar]
- Colman R., Rubin D. (2014) Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 8: 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosnes J. (2008) What is the link between the use of tobacco and IBD? Inflamm Bowel Dis 14(Suppl. 2): S14–S15. [DOI] [PubMed] [Google Scholar]
- Cosnes J., Carbonnel F., Carrat F., Beaugerie L., Gendre J. (1999) Oral contraceptive use and the clinical course of Crohn’s disease: a prospective cohort study. Gut 45: 218–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critch J., Day A., Otley A., King-Moore C., Teitelbaum J., Shashidhar H., et al. (2012) Use of enteral nutrition for the control of intestinal inflammation in pediatric Crohn disease. J Pediatr Gastroenterol Nutr 54: 298–305. [DOI] [PubMed] [Google Scholar]
- Cui B., Feng Q., Wang H., Wang M., Peng Z., Li P., et al. (2015) Fecal microbiota transplantation through mid-gut for refractory crohn’s disease: safety, feasibility, and efficacy trial results. J Gastroenterol Hepatol 30: 51–58. [DOI] [PubMed] [Google Scholar]
- Dalal S., Chang E. (2014) The microbial basis of inflammatory bowel diseases. J Clin Invest 124: 4190–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damman C. (2013) Salicylates and the microbiota: a new mechanistic understanding of an ancient drug’s role in dermatological and gastrointestinal disease. Drug Development Research 74: 344–352. [Google Scholar]
- Damman C., Brittnacher M., Westerhoff M., Hayden H., Radey M., Hager K., et al. (2015) Low level engraftment and improvement following a single colonoscopic administration of fecal microbiota to patients with ulcerative colitis. PLoS One 10: e0133925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damman C., Miller S., Surawicz C., Zisman T. (2012) The microbiome and inflammatory bowel disease: is there a therapeutic role for fecal microbiota transplantation? Am J Gastroenterol 107: 1452–1459. [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A., Barnich N., et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127: 412–421. [DOI] [PubMed] [Google Scholar]
- David L., Maurice C., Carmody R., Gootenberg D., Button J., Wolfe B., et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Inca R., Annese V., Di Leo V., Latiano A., Quaino V., Abazia C., et al. (2006) Increased intestinal permeability and NOD2 variants in familial and sporadic Crohn’s disease. Aliment Pharmacol Ther 23: 1455–1461. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Gaskins H. (2001) Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr 73: 1131S-1141S. [DOI] [PubMed] [Google Scholar]
- Derrien M., Vaughan E., Plugge C., De Vos W. (2004) Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54: 1469–1476. [DOI] [PubMed] [Google Scholar]
- Devkota S., Wang Y., Musch M., Leone V., Fehlner-Peach H., Nadimpalli A., et al. (2012) Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in IL10-/- mice. Nature 487: 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bella S., Gouliouris T., Petrosillo N. (2015) Fecal microbiota transplantation (FMT) for Clostridium difficile infection: focus on immunocompromised patients. J Infect Chemother 21: 230–237. [DOI] [PubMed] [Google Scholar]
- Dominguez-Bello M., Costello E., Contreras M., Magris M., Hidalgo G., Fierer N., et al. (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107: 11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont H. (2014) Review article: evidence for the role of gut microbiota in irritable bowel syndrome and its potential influence on therapeutic targets. Aliment Pharmacol Ther 39: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Eckburg P., Bik E., Bernstein C., Purdom E., Dethlefsen L., Sargent M., et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelblum K., Turner J. (2009) The tight junction in inflammatory disease: communication breakdown. Curr Opin Pharmacol 9: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J., Rey F., O’Donnell D., Karlsson M., McNulty N., Kallstrom G., et al. (2010) Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J 4: 1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M., Young D., Scott J., Norin E., Amarri S., Adam R., et al. (2010) Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr 51: 77–84. [DOI] [PubMed] [Google Scholar]
- Feldman M., Friedman L., Brandt L. (2015) Sleisenger and Fordtran’s Gastrointestinal and Liver Disease. 10th ed. Philadelphia, PA: Saunders. [Google Scholar]
- Fernandes P., Macsharry J., Darby T., Fanning A., Shanahan F., Houston A., et al. (2015) Differential expression of key regulators of toll-like receptors in ulcerative colitis and Crohn’s disease: a role for Tollip and PPARgamma? Clin Exp Immunol 183: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., St Amand A., Feldman R., Boedeker E., Harpaz N., Pace N. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104: 13780–13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg D., Toussaint N., Chen S., Ratner A., Whittier S., Wang T., et al. (2015) Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology 149: 883–885 e889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T., Imaeda H., Takahashi K., Kasumi E., Bamba S., Fujiyama Y., et al. (2013) Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J Gastroenterol Hepatol 28: 613–619. [DOI] [PubMed] [Google Scholar]
- Gearry R., Irving P., Barrett J., Nathan D., Shepherd S., Gibson P. (2009) Reduction of dietary poorly absorbed short-chain carbohydrates (FODMAPs) improves abdominal symptoms in patients with inflammatory bowel disease – a pilot study. J Crohns Colitis 3: 8–14. [DOI] [PubMed] [Google Scholar]
- Gerasimidis K., Bertz M., Hanske L., Junick J., Biskou O., Aguilera M., et al. (2014) Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm Bowel Dis 20: 861–871. [DOI] [PubMed] [Google Scholar]
- Geremia A., Biancheri P., Allan P., Corazza G., Di Sabatino A. (2014) Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev 13: 3–10. [DOI] [PubMed] [Google Scholar]
- Gevers D., Kugathasan S., Denson L., Vazquez-Baeza Y., Van Treuren W., Ren B., et al. (2014) The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi D., Fiorino G., Genua M., Allocca M., Danese S. (2014) Complementary and alternative medicine in inflammatory bowel diseases: what is the future in the field of herbal medicine? Expert Rev Gastroenterol Hepatol 8: 835–846. [DOI] [PubMed] [Google Scholar]
- Gionchetti P., Rizzello F., Venturi A., Brigidi P., Matteuzzi D., Bazzocchi G., et al. (2000) Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 119: 305–309. [DOI] [PubMed] [Google Scholar]
- Gorard D., Hunt J., Payne-James J., Palmer K., Rees R., Clark M., et al. (1993) Initial response and subsequent course of Crohn’s disease treated with elemental diet or prednisolone. Gut 34: 1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough E., Shaikh H., Manges A. (2011) Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53: 994–1002. [DOI] [PubMed] [Google Scholar]
- Guaraldi F., Salvatori G. (2012) Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinane C., Tadrous A., Fouhy F., Ryan C., Dempsey E., Murphy B., et al. (2013) Microbial composition of human appendices from patients following appendectomy. MBio 4: e00366–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guslandi M. (2011) Rifaximin in the treatment of inflammatory bowel disease. World J Gastroenterol 17: 4643–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G., Darveau R., Curtis M. (2012) The Keystone-Pathogen hypothesis. Nat Rev Microbiol 10: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfvarson J. (2011) Genetics in twins with Crohn’s disease: less pronounced than previously believed? Inflamm Bowel Dis 17: 6–12. [DOI] [PubMed] [Google Scholar]
- Halme L., Paavola-Sakki P., Turunen U., Lappalainen M., Farkkila M., Kontula K. (2006) Family and twin studies in inflammatory bowel disease. World J Gastroenterol 12: 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M., Knight R. (2009) Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19: 1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Sartor R. (2015) Therapeutic manipulation of the microbiome in IBD: current results and future approaches. Curr Treat Options Gastroenterol 13: 105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedin C., McCarthy N., Louis P., Farquharson F., McCartney S., Taylor K., et al. (2014) Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn’s disease and their unaffected siblings. Gut 63: 1578–1586. [DOI] [PubMed] [Google Scholar]
- Henker J., Müller S., Laass M., Schreiner A., Schulze J. (2008) Probiotic Escherichia coli Nissle 1917 (ECN) for successful remission maintenance of ulcerative colitis in children and adolescents: an open-label pilot study. Z Gastroenterol 46: 874–875. [DOI] [PubMed] [Google Scholar]
- Heyland D., Muscedere J., Wischmeyer P., Cook D., Jones G., Albert M., et al. (2013) A randomized trial of glutamine and antioxidants in critically ill patients. N Engl J Med 368: 1489–1497. [DOI] [PubMed] [Google Scholar]
- Hooper L., Macpherson A. (2010) Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol 10: 159–169. [DOI] [PubMed] [Google Scholar]
- Hou J., Abraham B., El-Serag H. (2011) Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol 106: 563–573. [DOI] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiro G., Bruno G., Lopetuso L., Beghella F., Laterza L., D’Aversa F., et al. (2014) Role of yeasts in healthy and impaired gut microbiota: the gut mycome. Curr Pharm Des 20: 4565–4569. [DOI] [PubMed] [Google Scholar]
- Iliev I., Funari V., Taylor K., Nguyen Q., Reyes C., Strom S., et al. (2012) Interactions between commensal fungi and the C-type lectin receptor dectin-1 influence colitis. Science 336: 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Integrative HMP (iHMP) Research Network Consortium (2014) The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell Host Microbe 16: 276–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery I., Claesson M., O’Toole P., Shanahan F. (2012) Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Microbiol 10: 591–592. [DOI] [PubMed] [Google Scholar]
- Jeppesen P. (2012) Teduglutide, a novel glucagon-like peptide 2 analog, in the treatment of patients with short bowel syndrome. Therap Adv Gastroenterol 5: 159–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Gustafsson J., Holmen-Larsson J., Jabbar K., Xia L., Xu H., et al. (2014) Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut 63: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. (2008) The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105: 15064–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L., Ripke S., Weersma R., Duerr R., McGovern D., Hui K., et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakodkar S., Farooqui A., Mikolaitis S., Mutlu E. (2015) The specific carbohydrate diet for inflammatory bowel disease: a case series. J Acad Nutr Diet 115: 1226–1232. [DOI] [PubMed] [Google Scholar]
- Kamada N., Chen G., Inohara N., Nunez G. (2013) Control of pathogens and pathobionts by the gut microbiota. Nat Immunol 14: 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G., Jackson T., Sands B., Frisch M., Andersson R., Korzenik J. (2008) The risk of developing Crohn’s disease after an appendectomy: a meta-analysis. Am J Gastroenterol 103: 2925–2931. [DOI] [PubMed] [Google Scholar]
- Kaplan G., Pedersen B., Andersson R., Sands B., Korzenik J., Frisch M. (2007) The risk of developing Crohn’s disease after an appendectomy: a population-based cohort study in Sweden and Denmark. Gut 56: 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner M., Kocjan A., Stein J., Schreiber S., Von Boyen G., Uebel P., et al. (2014) First multicenter study of modified release phosphatidylcholine ‘LT-02’ in ulcerative colitis: a randomized, placebo-controlled trial in mesalazine-refractory courses. Am J Gastroenterol 109: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato L., Kawamoto S., Maruya M., Fagarasan S. (2014) The role of the adaptive immune system in regulation of gut microbiota. Immunol Rev 260: 67–75. [DOI] [PubMed] [Google Scholar]
- Kelly C., Ihunnah C., Fischer M., Khoruts A., Surawicz C., Afzali A., et al. (2014) Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol 109: 1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Ullman T., Ford A., Abreu M., Abadir A., Marshall J., et al. (2011) Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol 106: 661–673. [DOI] [PubMed] [Google Scholar]
- Klapproth J., Sasaki M. (2010) Bacterial induction of proinflammatory cytokines in inflammatory bowel disease. Inflamm Bowel Dis 16: 2173–2179. [DOI] [PubMed] [Google Scholar]
- Knights D., Lassen K., Xavier R. (2013) Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut 62: 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig J., Spor A., Scalfone N., Fricker A., Stombaugh J., Knight R., et al. (2011) Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl. 1): 4578–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koloski N., Bret L., Radford-Smith G. (2008) Hygiene hypothesis in inflammatory bowel disease: a critical review of the literature. World J Gastroenterol 14: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A., Xavier R., Gevers D. (2014) The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146: 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronman M., Zaoutis T., Haynes K., Feng R., Coffin S. (2012) Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics 130: e794–e803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruis W., Fric P., Pokrotnieks J., Lukás M., Fixa B., Kascák M., et al. (2004) Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump P., Gröchenig H., Lackner S., Trajanoski S., Reicht G., Hoffmann K., et al. (2013) Alteration of intestinal dysbiosis by fecal microbiota transplantation does not induce remission in patients with chronic active ulcerative colitis. Inflamm Bowel Dis 19: 2155–2165. [DOI] [PubMed] [Google Scholar]
- Kunde S., Pham A., Bonczyk S., Crumb T., Duba M., Conrad H., Jr, et al. (2013) Safety, tolerability, and clinical response after fecal transplantation in children and young adults with ulcerative colitis. J Pediatr Gastroenterol Nutr 56: 597–601. [DOI] [PubMed] [Google Scholar]
- Lee D., Albenberg L., Compher C., Baldassano R., Piccoli D., Lewis J., et al. (2015) Diet in the pathogenesis and treatment of inflammatory bowel diseases. Gastroenterology 148: 1087–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage P., Leclerc M., Joossens M., Mondot S., Blottiere H., Raes J., et al. (2013) A metagenomic insight into our gut’s microbiome. Gut 62: 146–158. [DOI] [PubMed] [Google Scholar]
- Lerner A., Matthias T. (2015) Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun Rev 14: 479–489. [DOI] [PubMed] [Google Scholar]
- Ley R., Peterson D., Gordon J. (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124: 837–848. [DOI] [PubMed] [Google Scholar]
- Li Q., Zhang Q., Wang M., Zhao S., Ma J., Luo N., et al. (2008) Interferon-gamma and tumor necrosis factor-alpha disrupt epithelial barrier function by altering lipid composition in membrane microdomains of tight junction. Clin Immunol 126: 67–80. [DOI] [PubMed] [Google Scholar]
- Louis P., Hold G., Flint H. (2014) The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12: 661–672. [DOI] [PubMed] [Google Scholar]
- Lozupone C., Stombaugh J., Gordon J., Jansson J., Knight R. (2012) Diversity, stability and resilience of the human gut microbiota. Nature 489: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgendorff F., Akkermans L., Soderholm J. (2008) The role of microbiota and probiotics in stress-induced gastro-intestinal damage. Curr Mol Med 8: 282–298. [DOI] [PubMed] [Google Scholar]
- Maccaferri S., Vitali B., Klinder A., Kolida S., Ndagijimana M., Laghi L., et al. (2010) Rifaximin modulates the colonic microbiota of patients with Crohn’s disease: an in vitro approach using a continuous culture colonic model system. J Antimicrob Chemother 65: 2556–2565. [DOI] [PubMed] [Google Scholar]
- MacDermott R., Nash G., Nahm M. (1989) Antibody secretion by human intestinal mononuclear cells from normal controls and inflammatory bowel disease patients. Immunol Invest 18: 449–457. [DOI] [PubMed] [Google Scholar]
- Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., et al. (2014) A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63: 1275–1283. [DOI] [PubMed] [Google Scholar]
- Man S., Kaakoush N., Mitchell H. (2011) The role of bacteria and pattern-recognition receptors in Crohn’s disease. Nat Reva Gastroenterol Hepatol 8: 152–168. [DOI] [PubMed] [Google Scholar]
- Manichanh C., Borruel N., Casellas F., Guarner F. (2012) The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 9: 599–608. [DOI] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., et al. (2006) Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis D., Fanelli M., Hoffstad O., Lewis J. (2010) Potential association between the oral tetracycline class of antimicrobials used to treat acne and inflammatory bowel disease. Am J Gastroenterol 105: 2610–2616. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M., Denizot J., Dreux N., Robin F., Billard E., Bonnet R., et al. (2014) Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut 63: 116–124. [DOI] [PubMed] [Google Scholar]
- Matsunaga H., Hokari R., Ueda T., Kurihara C., Hozumi H., Higashiyama M., et al. (2011) Physiological stress exacerbates murine colitis by enhancing proinflammatory cytokine expression that is dependent on IL-18. Am J Physiol Gastrointest Liver Physiol 301: G555–G564. [DOI] [PubMed] [Google Scholar]
- McGilligan V., Wallace J., Heavey P., Ridley D., Rowland I. (2007) The effect of nicotine in vitro on the integrity of tight junctions in Caco-2 cell monolayers. Food Chem Toxicol 45: 1593–1598. [DOI] [PubMed] [Google Scholar]
- McGovern D., Kugathasan S., Cho J. (2015) Genetics of inflammatory bowel diseases. Gastroenterology 149: 1163–1176.e1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNees A.L., Markesich D., Zayyani N.R., Graham D.Y. (2015) Mycobacterium paratuberculosis as a cause of Crohn’s disease. Expert Rev Gastroenterol Hepatol 9: 1523–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melgar S., Engstrom K., Jagervall A., Martinez V. (2008) Psychological stress reactivates dextran sulfate sodium-induced chronic colitis in mice. Stress 11: 348–362. [DOI] [PubMed] [Google Scholar]
- Merga Y., Campbell B., Rhodes J. (2014) Mucosal barrier, bacteria and inflammatory bowel disease: possibilities for therapy. Dig Dis 32: 475–483. [DOI] [PubMed] [Google Scholar]
- Metzker M. (2010) Sequencing technologies – the next generation. Nat Rev Genet 11: 31–46. [DOI] [PubMed] [Google Scholar]
- Michail S., Durbin M., Turner D., Griffiths A., Mack D., Hyams J., et al. (2012) Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm Bowel Dis 18: 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielan A., D’Inca R. (2015) Intestinal permeability in inflammatory bowel disease: pathogenesis, clinical evaluation, and therapy of leaky gut. Mediators Inflamm 2015: 628157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele E., Pascarella F., Giannetti E., Quaglietta L., Baldassano R., Staiano A. (2009) Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol 104: 437–443. [DOI] [PubMed] [Google Scholar]
- Mimura T., Rizzello F., Helwig U., Poggioli G., Schreiber S., Talbot I., et al. (2004) Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut 53: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moayyedi P., Surette M., Kim P., Libertucci J., Wolfe M., Onischi C., et al. (2015) Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149: 102–109 e106. [DOI] [PubMed] [Google Scholar]
- Modi S., Collins J., Relman D. (2014) Antibiotics and the gut microbiota. J Clin Invest 124: 4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C., Ackermann N., Langmann T., Aslanidis C., Kel A., Kel-Margoulis O., et al. (2006) Aberrant intestinal expression and allelic variants of mucin genes associated with inflammatory bowel disease. J Mol Med (Berl) 84: 1055–1066. [DOI] [PubMed] [Google Scholar]
- Molodecky N., Soon I., Rabi D., Ghali W., Ferris M., Chernoff G., et al. (2012) Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142: 46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- Mowat C., Cole A., Windsor A., Ahmad T., Arnott I., Driscoll R., et al. (2011) Guidelines for the management of inflammatory bowel disease in adults. Gut 60: 571–607. [DOI] [PubMed] [Google Scholar]
- Nagalingam N., Lynch S. (2012) Role of the microbiota in inflammatory bowel diseases. Inflamm Bowel Dis 18: 968–984. [DOI] [PubMed] [Google Scholar]
- Naganuma M., Iizuka B., Torii A., Ogihara T., Kawamura Y., Ichinose M., et al. (2001) Appendectomy protects against the development of ulcerative colitis and reduces its recurrence: results of a multicenter case-controlled study in Japan. Am J Gastroenterol 96: 1123–1126. [DOI] [PubMed] [Google Scholar]
- Nicholson J., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., et al. (2012) Host-gut microbiota metabolic interactions. Science 336: 1262–1267. [DOI] [PubMed] [Google Scholar]
- Nickerson K., Chanin R., McDonald C. (2015) Deregulation of intestinal anti-microbial defense by the dietary additive, maltodextrin. Gut Microbes 6: 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman J., Handley S., Baldridge M., Droit L., Liu C., Keller B., et al. (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obih C., Wahbeh G., Lee D., Braly K., Giefer M., Shaffer M., et al. (2016) Specific carbohydrate diet for pediatric inflammatory bowel disease in clinical practice within an academic IBD center. Nutrition 32: 418–425. [DOI] [PubMed] [Google Scholar]
- Ockenga J., Borchert K., Stuber E., Lochs H., Manns M., Bischoff S. (2005) Glutamine-enriched total parenteral nutrition in patients with inflammatory bowel disease. Eur J Clin Nutr 59: 1302–1309. [DOI] [PubMed] [Google Scholar]
- Ogura Y., Bonen D., Inohara N., Nicolae D., Chen F., Ramos R., et al. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature 411: 603–606. [DOI] [PubMed] [Google Scholar]
- O’Hara A., Shanahan F. (2006) The gut flora as a forgotten organ. EMBO Rep 7: 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott S., Musfeldt M., Wenderoth D., Hampe J., Brant O., Fölsch U., et al. (2004) Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes G., Whelan K., Lindsay J. (2014) Smoking in inflammatory bowel disease: impact on disease course and insights into the aetiology of its effect. J Crohns Colitis 8: 717–725. [DOI] [PubMed] [Google Scholar]
- Parkes M. (2012) Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis 30: 330–333. [DOI] [PubMed] [Google Scholar]
- Pastor Rojo O., Lopez San Roman A., Albeniz Arbizu E., De La Hera Martinez A., Ripoll Sevillano E., Albillos Martinez A. (2007) Serum lipopolysaccharide-binding protein in endotoxemic patients with inflammatory bowel disease. Inflamm Bowel Dis 13: 269–277. [DOI] [PubMed] [Google Scholar]
- Penders J., Thijs C., Vink C., Stelma F., Snijders B., Kummeling I., et al. (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118: 511–521. [DOI] [PubMed] [Google Scholar]
- Peng J., Shen J., Ran Z. (2014) Novel agents in the future: therapy beyond anti-TNF agents in inflammatory bowel disease. J Dig Dis 15: 585–590. [DOI] [PubMed] [Google Scholar]
- Petrof E., Gloor G., Vanner S., Weese S., Carter D., Daigneault M., et al. (2013) Stool substitute transplant therapy for the eradication of clostridium difficile infection: ‘repoopulating’ the gut. Microbiome 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof E., Khoruts A. (2014) From stool transplants to next-generation microbiota therapeutics. Gastroenterology 146: 1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png C., Linden S., Gilshenan K., Zoetendal E., McSweeney C., Sly L., et al. (2010) Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105: 2420–2428. [DOI] [PubMed] [Google Scholar]
- Prantera C., Lochs H., Campieri M., Scribano M., Sturniolo G., Castiglione F., et al. (2006) Antibiotic treatment of Crohn’s disease: results of a multicentre, double blind, randomized, placebo-controlled trial with rifaximin. Aliment Pharmacol Ther 23: 1117–1125. [DOI] [PubMed] [Google Scholar]
- Quince C., Ijaz U.Z., Loman N., Eren A.M., Saulnier D., Russell J., et al. (2015) Extensive Modulation of the Fecal Metagenome in Children with Crohn’s Disease During Exclusive Enteral Nutrition. Am J Gastroenterol 110: 1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford-Smith G. (2008) What is the importance of appendectomy in the natural history of IBD? Inflamm Bowel Dis 14(Suppl. 2): S72–S74. [DOI] [PubMed] [Google Scholar]
- Radford-Smith G., Edwards J., Purdie D., Pandeya N., Watson M., Martin N., et al. (2002) Protective role of appendicectomy on onset and severity of ulcerative colitis and Crohn’s disease. Gut 51: 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran N., Kumar D. (2010) Role of diet in the management of inflammatory bowel disease. World J Gastroenterol 16: 1442–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Shanahan F., Guarner F., De Vos W. (2013) Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis 19: 481–488. [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M., Smidt H., De Vos W. (2007) Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9: 2125–2136. [DOI] [PubMed] [Google Scholar]
- Ramasundara M., Leach S., Lemberg D., Day A. (2009) Defensins and inflammation: the role of defensins in inflammatory bowel disease. J Gastroenterol Hepatol 24: 202–208. [DOI] [PubMed] [Google Scholar]
- Ray K. (2015) IBD. Gut microbiota in IBD goes viral. Nat Rev Gastroenterol Hepatol 12: 122. [DOI] [PubMed] [Google Scholar]
- Reiberger T., Ferlitsch A., Payer B., Mandorfer M., Heinisch B., Hayden H., et al. (2013) Non-selective betablocker therapy decreases intestinal permeability and serum levels of LBP and IL-6 in patients with cirrhosis. J Hepatol 58: 911–921. [DOI] [PubMed] [Google Scholar]
- Richard M., Lamas B., Liguori G., Hoffmann T., Sokol H. (2015) Gut fungal microbiota: the yin and yang of inflammatory bowel disease. Inflamm Bowel Dis 21: 656–665. [DOI] [PubMed] [Google Scholar]
- Riordan A., Hunter J., Cowan R., Crampton J., Davidson A., Dickinson R., et al. (1993) Treatment of active Crohn’s disease by exclusion diet: East Anglian multicentre controlled trial. Lancet 342: 1131–1134. [DOI] [PubMed] [Google Scholar]
- Roeselers G., Ponomarenko M., Lukovac S., Wortelboer H. (2013) Ex vivo systems to study host-microbiota interactions in the gastrointestinal tract. Best Pract Res Clin Gastroenterol 27: 101–113. [DOI] [PubMed] [Google Scholar]
- Rogers M., Aronoff D. (2015) The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook G. (2012) Hygiene hypothesis and autoimmune diseases. Clin Rev Allergy Immunol 42: 5–15. [DOI] [PubMed] [Google Scholar]
- Rossen N., Fuentes S., Van Der Spek M., Tijssen J., Hartman J., Duflou A., et al. (2015) Findings from a randomized controlled trial of fecal transplantation for patients with ulcerative colitis. Gastroenterology 149: 110–118 e114. [DOI] [PubMed] [Google Scholar]
- Rubin D. (2013) Curbing our enthusiasm for fecal transplantation in ulcerative colitis. Am J Gastroenterol 108: 1631–1633. [DOI] [PubMed] [Google Scholar]
- Rungoe C., Langholz E., Andersson M., Basit S., Nielsen N., Wohlfahrt J., et al. (2014) Changes in medical treatment and surgery rates in inflammatory bowel disease: a nationwide cohort study 1979–2011. Gut 63: 1607–1616. [DOI] [PubMed] [Google Scholar]
- Sartor R. (2008) Microbial influences in inflammatory bowel diseases. Gastroenterology 134: 577–594. [DOI] [PubMed] [Google Scholar]
- Scanlan P., Marchesi J. (2008) Micro-eukaryotic diversity of the human distal gut microbiota: qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J 2: 1183–1193. [DOI] [PubMed] [Google Scholar]
- Scarpellini E., Ianiro G., Attili F., Bassanelli C., De Santis A., Gasbarrini A. (2015) The human gut microbiota and virome: potential therapeutic implications. Dig Liver Dis 47: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultsz C., Van Den Berg F., Ten Kate F., Tytgat G., Dankert J. (1999) The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 117: 1089–1097. [DOI] [PubMed] [Google Scholar]
- Sekelja M., Berget I., Naes T., Rudi K. (2011) Unveiling an abundant core microbiota in the human adult colon by a phylogroup-independent searching approach. ISME J 5: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I., Russell S., Antunes L., Finlay B. (2010) Gut microbiota in health and disease. Physiol Rev 90: 859–904. [DOI] [PubMed] [Google Scholar]
- Shanahan F. (2012) The gut microbiota – a clinical perspective on lessons learned. Nat Rev Gastroenterol Hepatol 9: 609–614. [DOI] [PubMed] [Google Scholar]
- Shaw S., Blanchard J., Bernstein C. (2010) Association between the use of antibiotics in the first year of life and pediatric inflammatory bowel disease. Am J Gastroenterol 105: 2687–2692. [DOI] [PubMed] [Google Scholar]
- Shaw S., Blanchard J., Bernstein C. (2011) Association between the use of antibiotics and new diagnoses of Crohn’s disease and ulcerative colitis. Am J Gastroenterol 106: 2133–2142. [DOI] [PubMed] [Google Scholar]
- Sheehan D., Moran C., Shanahan F. (2015) The microbiota in inflammatory bowel disease. J Gastroenterol 50: 495–507. [DOI] [PubMed] [Google Scholar]
- Sigall-Boneh R., Pfeffer-Gik T., Segal I., Zangen T., Boaz M., Levine A. (2014) Partial enteral nutrition with a Crohn’s disease exclusion diet is effective for induction of remission in children and young adults with Crohn’s disease. Inflamm Bowel Dis 20: 1353–1360. [DOI] [PubMed] [Google Scholar]
- Singh S., Graff L., Bernstein C. (2009) Do NSAIDs, antibiotics, infections, or stress trigger flares in IBD? Am J Gastroenterol 104: 1298–1313; quiz 1314. [DOI] [PubMed] [Google Scholar]
- Smith P., Howitt M., Panikov N., Michaud M., Gallini C., Bohlooly Y., et al. (2013) The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderholm J., Yates D., Gareau M., Yang P., Macqueen G., Perdue M. (2002) Neonatal maternal separation predisposes adult rats to colonic barrier dysfunction in response to mild stress. Am J Physiol Gastrointest Liver Physiol 283: G1257–G1263. [DOI] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L., Gratadoux J., et al. (2008) Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood A., Midha V., Makharia G., Ahuja V., Singal D., Goswami P., et al. (2009) The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol 7: 1202–1209, 1209.e1201. [DOI] [PubMed] [Google Scholar]
- Spor A., Koren O., Ley R. (2011) Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9: 279–290. [DOI] [PubMed] [Google Scholar]
- Strauss J., Kaplan G., Beck P., Rioux K., Panaccione R., Devinney R., et al. (2011) Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis 17: 1971–1978. [DOI] [PubMed] [Google Scholar]
- Stremmel W., Hanemann A., Braun A., Stoffels S., Karner M., Fazeli S., et al. (2010) Delayed release phosphatidylcholine as new therapeutic drug for ulcerative colitis – a review of three clinical trials. Expert Opin Investig Drugs 19: 1623–1630. [DOI] [PubMed] [Google Scholar]
- Sun J., Chang E. (2014) Exploring gut microbes in human health and disease: pushing the envelope. Genes Dis 1: 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suskind D., Brittnacher M., Wahbeh G., Shaffer M., Hayden H., Qin X., et al. (2015) Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn’s disease. Inflamm Bowel Dis 21: 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suskind D., Wahbeh G., Gregory N., Vendettuoli H., Christie D. (2014) Nutritional therapy in pediatric Crohn disease: the specific carbohydrate diet. J Pediatr Gastroenterol Nutr 58: 87–91. [DOI] [PubMed] [Google Scholar]
- Sutherland L., Singleton J., Sessions J., Hanauer S., Krawitt E., Rankin G., et al. (1991) Double blind, placebo controlled trial of metronidazole in Crohn’s disease. Gut 32: 1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Dorffel Y., Loening-Baucke V., Theissig F., Ruckert J., Ismail M., et al. (2011) Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60: 34–40. [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Bengmark S., Lochs H., Dorffel Y. (2007) Azathioprine and mesalazine-induced effects on the mucosal flora in patients with IBD colitis. Inflamm Bowel Dis 13: 51–56. [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Weber J., Loening-Baucke V., Hale L., Lochs H. (2005) Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol 43: 3380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Smale S., Premchand P., Maiden L., Sherwood R., Thjodleifsson B., et al. (2006) Prevalence and mechanism of nonsteroidal anti-inflammatory drug-induced clinical relapse in patients with inflammatory bowel disease. Clin Gastroenterol Hepatol 4: 196–202. [DOI] [PubMed] [Google Scholar]
- Talley N., Abreu M., Achkar J., Bernstein C., Dubinsky M., Hanauer S., et al. (2011) An evidence-based systematic review on medical therapies for inflammatory bowel disease. Am J Gastroenterol 106(Suppl. 1): S2–25; quiz S26. [DOI] [PubMed] [Google Scholar]
- Theodorakopoulou M., Perros E., Giamarellos-Bourboulis E., Dimopoulos G. (2013) Controversies in the management of the critically ill: the role of probiotics. Int J Antimicrob Agents 42(Suppl.): S41–S44. [DOI] [PubMed] [Google Scholar]
- Theodoratou E., Campbell H., Ventham N., Kolarich D., Pucic-Bakovic M., Zoldos V., et al. (2014) The role of glycosylation in IBD. Nat Rev Gastroenterol Hepatol 11: 588–600. [DOI] [PubMed] [Google Scholar]
- Tilg H., Kaser A. (2004) Diet and relapsing ulcerative colitis: take off the meat? Gut 53: 1399–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer A., Sutherland L., Martin F. (1998) Oral contraceptive use and smoking are risk factors for relapse in Crohn’s disease. The Canadian Mesalamine for Remission of Crohn’s Disease Study Group. Gastroenterology 114: 1143–1150. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P., Hamady M., Yatsunenko T., Cantarel B., Duncan A., Ley R., et al. (2009) A core gut microbiome in obese and lean twins. Nature 457: 480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D., Levine A., Kolho K., Shaoul R., Ledder O. (2014) Combination of oral antibiotics may be effective in severe pediatric ulcerative colitis: a preliminary report. J Crohns Colitis 8: 1464–1470. [DOI] [PubMed] [Google Scholar]
- Turner J. (2009) Intestinal mucosal barrier function in health and disease. Nat Rev Immunol 9: 799–809. [DOI] [PubMed] [Google Scholar]
- Tursi A., Brandimarte G., Papa A., Giglio A., Elisei W., Giorgetti G., et al. (2010) Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol 105: 2218–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulluwishewa D., Anderson R., McNabb W., Moughan P., Wells J., Roy N. (2011) Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr 141: 769–776. [DOI] [PubMed] [Google Scholar]
- Ursell L., Metcalf J., Parfrey L., Knight R. (2012) Defining the human microbiome. Nutr Rev 70(Suppl. 1): S38–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Immerseel F., Ducatelle R., De Vos M., Boon N., Van De Wiele T., Verbeke K., et al. (2010) Butyric acid-producing anaerobic bacteria as a novel probiotic treatment approach for inflammatory bowel disease. J Med Microbiol 59: 141–143. [DOI] [PubMed] [Google Scholar]
- Van Limbergen J., Radford-Smith G., Satsangi J. (2014) Advances in IBD genetics. Nat Rev Gastroenterol Hepatol 11: 372–385. [DOI] [PubMed] [Google Scholar]
- Van Nimwegen F., Penders J., Stobberingh E., Postma D., Koppelman G., Kerkhof M., et al. (2011) Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol 128: 948–955 e941–943. [DOI] [PubMed] [Google Scholar]
- Van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E., De Vos W., et al. (2013) Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368: 407–415. [DOI] [PubMed] [Google Scholar]
- Voitk A., Echave V., Feller J., Brown R., Gurd F. (1973) Experience with elemental diet in the treatment of inflammatory bowel disease. is this primary therapy? Arch Surg 107: 329–333. [DOI] [PubMed] [Google Scholar]
- Wahed M., Corser M., Goodhand J., Rampton D. (2010) Does psychological counseling alter the natural history of inflammatory bowel disease? Inflamm Bowel Dis 16: 664–669. [DOI] [PubMed] [Google Scholar]
- Walters, S., Quiros, A., Rolston, M., Grishina, I., Li, J., Fenton, A., et al. (2014) Analysis of gut microbiome and diet modification in patients with Crohn’s disease. SOJ Microbiol Infect Dis 2: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Ahrne S., Jeppsson B., Molin G. (2005) Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol 54: 219–231. [DOI] [PubMed] [Google Scholar]
- Wang N., Wang G., Hao J., Ma J., Wang Y., Jiang X., et al. (2012) Curcumin ameliorates hydrogen peroxide-induced epithelial barrier disruption by upregulating heme oxygenase-1 expression in human intestinal epithelial cells. Dig Dis Sci 57: 1792–1801. [DOI] [PubMed] [Google Scholar]
- Willing B., Dicksved J., Halfvarson J., Andersson A., Lucio M., Zheng Z., et al. (2010) A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139: 1844–1854.e1841. [DOI] [PubMed] [Google Scholar]
- Wischmeyer P. (2007) Glutamine: mode of action in critical illness. Crit Care Med 35: S541–544. [DOI] [PubMed] [Google Scholar]
- Wolff M., Broadhurst M., Loke P. (2012) Helminthic therapy: improving mucosal barrier function. Trends Parasitol 28: 187–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Bushmanc F., Lewis J. (2013) Diet, the human gut microbiota, and IBD. Anaerobe 24: 117–120. [DOI] [PubMed] [Google Scholar]
- Wu G., Chen J., Hoffmann C., Bittinger K., Chen Y., Keilbaugh S., et al. (2011) Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wang H., Wang M., Cui B., Fan Z., Ji G. (2013) Fecal microbiota transplantation for severe enterocolonic fistulizing Crohn’s disease. World J Gastroenterol 19: 7213–7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li Y. (2014) Inflammatory bowel disease: pathogenesis. World J Gastroenterol 20: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E., Rajilic-Stojanovic M., De Vos W. (2008) High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57: 1605–1615. [DOI] [PubMed] [Google Scholar]