Abstract
Eosinophilic esophagitis (EoE) is a chronic allergic inflammatory disease that is triggered by food and/or environmental allergens and is characterized by a clinical and pathologic phenotype of progressive esophageal dysfunction due to tissue inflammation and fibrosis. EoE is suspected in patients with painful swallowing, among other symptoms, and is diagnosed by the presence of 15 or more eosinophils per high-power field in one or more of at least four esophageal biopsy specimens. The prevalence of EoE is increasing and has now reached rates similar to those of other chronic gastrointestinal disorders such as Crohn’s disease. In recent years, our understanding of the immunologic mechanisms underlying this condition has grown considerably. Thanks to new genetic, molecular, cellular, animal, and translational studies, we can now postulate a detailed pathway by which exposure to allergens results in a complex and coordinated type 2 inflammatory cascade that, if not intervened upon, can result in pain on swallowing, esophageal strictures, and food impaction. Here, we review the most recent research in this field to synthesize and summarize our current understanding of this complex and important disease.
Keywords: Eosinophilic esophagitis, Food allergy, Immunology, Inflammation
Introduction
Eosinophilic esophagitis (EoE) is a chronic allergic inflammatory disease of the esophagus that, if left untreated, can result in significant impairment in the quality of life due to pain on swallowing (odynophagia), food impaction, esophageal stricture formation, and in rare extreme cases, esophageal rupture [1–5]. Histopathologically, EoE is characterized by esophageal epithelial barrier defects and eosinophil infiltrates [6]. Clinically, EoE is suspected in patients with symptoms of esophageal dysfunction and/or fibrosis. In children, symptoms often mimic those of gastroesophageal reflux disease but do not respond to gastric acid suppression. Older children and adults often present with symptoms of odynophagia and food impaction caused by progressive esophageal fibrosis [7, 8]. Diagnosis is made by the presence of 15 or more eosinophils per high-power field in one or more of at least four esophageal biopsy specimens obtained via esophagogastroduodenal endoscopy, while the patient is being treated with an optimal anti-gastroesophageal reflux disease (GERD) therapeutic regimen [9, 10••].
The first case of EoE was described in 1968. Since then, the rates of EoE diagnosis in western countries have grown considerably to match those of other gastroenteropathies such as Crohn’s disease [11–16]. EoE is most common in Caucasian males and can occur sporadically or in families [11, 17–21]. The sibling risk recurrence ratio for EoE is higher than that of other allergic conditions; however, twin studies have shown the environment to be a contributor in addition to familial predisposition [22•, 23]. Both sporadic and familial EoEs seem to have similar pathologic and molecular phenotypes, suggesting that the inflammatory phase of the disease is conserved between these two groups [24]. When an EoE diagnosis is made, it is imperative that measures be taken to eliminate or control esophageal inflammation for both symptomatic relief and the prevention of fibrotic complications such as esophageal stricture and food impaction [5].
Most EoE patients have comorbid allergic conditions (allergic rhinitis, asthma, immunoglobulin E (IgE)-mediated food allergies, atopic dermatitis, etc.), suggesting that these conditions are pathophysiologically related [25]. In addition, reactions to food allergens alone are not solely responsible for the inflammation observed in EoE as some EoE patients with concurrent allergic rhinitis display seasonal exacerbations of esophageal eosinophilia [26]. Consistent with EoE falling on the allergic spectrum, the inflammation observed in EoE responds to allergen avoidance and/or topical steroid applications. Elemental diets have been shown to be highly effective in inducing histologic and clinical remission in children and adolescents with EoE, though adherence to these diets is poor [27–30]. Alternative diets have been examined such as empiric elimination diets, based on the most commonly identified etiologic foods, and allergy testing-directed diets [29, 31–34]. Subsequent examination of these two approaches has shown them to be equally effective in inducing disease remission; there is no added benefit to skin testing over empiric elimination of commonly allergic foods [28]. Orally administered topical steroids are an alternative therapy for EoE that are highly effective with resolution rates up to 80 % depending on the dose, but with potential side effects [35–43]. Together, these observations highlight the need for new and improved therapeutic approaches to prevent and treat this growing disease.
Recent discoveries in animal models and human subjects have advanced our understanding of the immunologic mechanisms underlying EoE, and have led to exciting new therapeutic prospects. We review the most up-to-date research available spanning the disciplines of basic science, translational, and clinical research. This review is organized into three sections: “Genetics, Epigenetics, and Transcriptional Analyses,” “Cytokines, Chemokines, and Other Molecules, ” and “Pathologic and Protective Cell Populations.” In addition to reviewing immunologic discoveries in patients and animal models, we touch on new immune-directed therapies for EoE that are currently under investigation. We conclude by providing a unifying theory for the pathogenesis of EoE. We hope that this review will provide a framework for clinicians and scientists interested in improving our understanding of EoE pathogenesis and developing new preventative and therapeutic strategies for this important condition.
Genetics, Epigenetics, and Transcriptional Analyses
The genetic associations and epigenetic changes observed in EoE have been nicely reviewed recently [44]. While not the focus of this review, these studies have provided important launching points for subsequent experimental approaches in mice and humans designed to elucidate the immunologic mechanisms of EoE. As such, we will touch on them briefly here. Early single-gene association studies identified polymorphisms that were more frequent in EoE patients. These include mutations in the untranslated region preceding the chemokine (C-C motif) ligand 26 (CCL26), which encodes the potent eosinophil and basophil chemotactic eotaxin-3, as well as mutations in the epidermal barrier gene filaggrin and in the promoter of the transforming growth factor beta 1 (TGF-β1) gene [35, 45–47]. More recently, genome-wide association studies have identified several loci that have a strong association with EoE. Variants in the 5q22 locus, which includes the gene which encodes thymic stromal lymphopoietin (TSLP), are associated with EoE, and TSLP is overexpressed in esophageal biopsies from individuals with EoE compared to unaffected individuals [48, 49]. Additionally, variants in a region on 2p23 which includes the gene CAPN14 (that encodes a member of the calcium-dependent, non-lysosomal cysteine protease family) are associated with EoE [50•, 51•]. CAPN14 is also up-regulated in the esophagus in active EoE, and after exposure of epithelial cells to interleukin (IL)-13 [50•]. Additional associations have been reported for variations in c11orf30 which encodes EMSY, the gene encoding signal transducer and activator of transcription (STAT)6, and ANKRD27 whose product regulates the trafficking of melanogenic enzymes to epidermal melanocytes [51•].
There is limited data about the role of epigenetic regulation in EoE pathogenesis, and most of our understanding in this regard stems from detailed studies of the CCL26 locus. Firstly, it seems that IL-13 promotes the activation of CCL26 via two STAT6 binding sites in the CCL26 promoter [52]. This signaling cascade increases acetylated histone 3 and opens the CCL26 promoter for transcription [53]. These findings may be relevant to recent observations that omeprazole blocks STAT6 binding to the CCL26 promoter, possibly explaining why patients with esophageal eosinophilia are at least in part responsive to PPI therapy (PPI-REE patients) [54, 55]. Secondly, it has been shown that portions of the CCL26 promoter are hypomethylated in esophageal epithelial cells derived from EoE patients, and that the methylation status of the CCL26 promoter dictates the propensity for STAT6 to bind to the site [56]. Together, these findings indicate that both histone acetylation and DNA methylation are mechanisms of transcriptional regulation of the CCL26 locus and that dysregulation in either system may contribute to EoE susceptibility in some patients.
MicroRNAs (miRNAs) are short, non-coding RNAs that influence the expression of genes by binding to complementary sequences in the 3' untranslated region of the target messenger RNA (mRNA) sequence and interfering with translation and/or promoting degradation of the transcript. miRNAs have been shown to play a role in multiple human inflammatory disease states including EoE [57]. Examination of miRNA expression patterns in patients with EoE revealed dysregulation of miRNAs involved in tissue remodeling and inflammation [58, 59]. Dysregulated miRNA expression is largely reversible in patients who respond to glucocorticoid treatment, suggesting that the altered miRNA expression patterns observed contribute to EoE pathogenesis [60]. Consistent with this, two of the most upregulated miRNAs observed in patients with active EoE, miR-21 and miR-223, have been shown to have functions that would support a pathologic role in EoE. miR-21 is implicated in eosinophil survival, allergic inflammation, and TGF-β-stimulated tissue fibrosis [61–63]. Similarly, miR-223 has been implicated in regulating IL-5 expression and eosinophil development [64]. Other miRNAs are downregulated in EoE. For example, miR-357, a microRNA that is suppressed by IL-13, shows significantly lower expression in patients with active EoE [59]. Together, these findings identify altered miRNA expression as a key phenotype of EoE and identify potential pathologic roles for some transcripts.
Through the utilization of PCR and microarray techniques, an EoE transcriptome has been identified with high resolution. In fact, the transcriptional fingerprint of EoE is becoming so well characterized that profiling of EoE-specific changes has been proposed as a new diagnostic modality, though it does not have additional diagnostic or predictive values compared to standard H&E analysis at the current time [65]. In total, more than 1600 transcripts have been identified that are dys-regulated in EoE including cytokines (IL1B, IL1RN, IL1F6, IL4, IL5, IL6, IL8, IL12p70, IL13, CD40L, IL1α, IL17, ABCF1), chemokines (CCL26, CXCL1, CXCL2, CXCL14, CCL1, CCL23), and receptors (IL5RA) [45, 66, 67]. Of these, the expression of CCL26, which encodes the eosinophil chemoattractant eotaxin-3, is among the most highly induced genes in the EoE transcriptome [45, 68, 69]. The role of eotaxin-3, as well as other proteins that have been identified through transcriptional analysis and implicated in the pathophysiology of EoE, will be discussed in detail in the next section of this review.
Cytokines, Chemokines, and Other Molecules
Desmoglein-1, Keratin, and Periostin
Several molecules important for epithelial integrity have been shown to be dysregulated in animal models and patients with EoE. The desmosomal cadherin desmoglein 1 (DSG1) is an intercellular adhesion molecule, belonging to the desmosomal cadherin family, that plays a critical role in maintaining suprabasal epithelial integrity [70]. There are reduced levels of DSG1 in esophageal biopsies of patients with active EoE as compared to controls, and IL-13 downregulates DSG1 expression and promotes impaired barrier function in cultured esophageal epithelial cells [71, 72•]. Additionally, a loss of DSG1 in cultured esophageal epithelial cells results in a transcriptional phenotype similar to that found in EoE, suggesting that many of the downstream mediators of inflammation in EoE may be a result of impaired epithelial integrity [72•]. Interestingly, homozygous mutations in DSG1 have recently been shown to result in a loss of cell-cell adhesion, correlated with a type 2 inflammatory response, and associated with a clinical syndrome featuring severe dermatitis, multiple allergies, and metabolic wasting [73].
The transcript for another molecule important for epithelial integrity, keratin, is also expressed by esophageal epithelial cells and is downregulated in patients with active EoE [74]. Conversely, the matricellular protein periostin, which interacts with several extracellular matrix proteins such as type 1 collagen and Notch1 to mediate cell migration and adhesion, is highly upregulated in patients with EoE [45, 75]. Periostin expression is thought to be induced by TGF-β and IL-13 signaling and may play an important role in promoting eosinophil adhesion. Periostin can also induce expression of TSLP, a molecule which may be central to EoE pathogenesis and is discussed in detail below [76]. Finally, periostin-deficient mice are protected against allergic inflammatory responses in the esophagus [75]. Together, these findings implicate baseline impaired mucosal integrity as a risk factor for allergen exposure and development of EoE [77]. Consistent with this hypothesis, the prevalence of EoE in patients with inherited connective tissue disorders (such as Marfan and Ehlers-Danlos syndromes) is eightfold higher than the general population [78].
Eotaxin
While many genes are known to be dysregulated in patients with EoE, the eosinophil chemoattractant eotaxin-3 is one of the best characterized. A potent eosinophil chemotactic, eotaxin-3 is expressed by endothelial cells, binds to CCR-3 on the surface of eosinophils, and causes upregulation of cell adhesion molecules and production of effector cytokines such as IL-13 [79]. A single-nucleotide polymorphism in the gene encoding eotaxin-3 (CCL26) has been shown to be associated with susceptibility to EoE [45]. Additionally, CCL26 is upregulated in the inflamed esophageal mucosa of EoE patients and is downregulated upon treatment with topical steroids [68]. Murine models have shown that overexpression of eotaxin results in recruitment of eosinophils to the mucosa of the gastrointestinal tract and mice lacking CCR-3 are protected against experimental EoE [45, 80–82]. Interestingly, expression of eotaxin-3 is only mildly elevated in patients with GERD, suggesting that analysis of eotaxin-3 expression may be able to differentiate these two conditions [69].
Several lines of investigation have interrogated transcriptional regulation of CCL26 in EoE. The CCL26 promoter is hypomethylated, and CCL26 expression is highly induced, in patients with EoE [45, 56]. Additionally, PARP14, a transcriptional cofactor from the poly(ADP-ribose) polymerase (PARP) family that facilitates CCL26 transcription via STAT6, is also dysregulated in EoE [83–85]. PARP14 expression is increased in biopsies of children with EoE compared to controls, and CCL26 expression strongly correlates with PARP14 expression [85]. Cotransfection of an esophageal cell line with both PARP14 and STAT6 increases the activity of a CCL26 reporter, and a PARP inhibitor attenuates IL-4 and IL-13-mediated induction of CCL26 transcription [85]. Together, these studies have helped to identify eotaxin-3 as a molecule that is often dysregulated in EoE and that likely contributes to the exaggerated eosinophilic inflammation observed in this condition.
Histamine
The histamine receptors HR1 and HR4 have increased expression in esophageal biopsies of patients with active EoE as compared to those with inactive EoE or healthy controls. HR1 and HR4 are expressed in epithelial eosinophils from biopsies of patients with active EoE, while HR2 expression is present throughout the EoE inflamed tissue [86]. Interestingly, expression of HR2 is increased in patients with both active and inactive EoE compared to controls, suggesting that aberrant epithelial expression of HR2 may predispose individuals to the development of EoE [86]. Further investigation is needed to better elucidate the pathophysiologic implications of these observations and to determine whether antihistamines or other targeted therapies may be beneficial in the treatment of EoE.
IgE
As discussed previously, most EoE patients have comorbid allergic conditions, suggesting that these conditions share a common pathophysiologic mechanism [25]. IgE has long been known to be an antigen-specific mechanism of granulocyte degranulation, and IgE plays an important role in multiple allergic conditions including allergic rhinitis, asthma, and IgE-mediated food allergies. More recently, additional roles for IgE have been identified in immune cell development and homeostasis [87–89]. Regardless of atopic history, B cells that are class switched to the IgE isotype and IgE-bound mast cells are both increased in patients with EoE [90]. Despite these findings, eosinophilic inflammation is independent of B cells in one model of EoE and independent of IgE in another [91, 92••]. As such, the role of B cells and IgE in the pathogenesis of EoE is unclear.
Additional evidence indicates that EoE is not an IgE-mediated disease. For example, IgE-mediated food allergy and EoE seem to occur independently [93, 94]. Additionally, patients with milk-induced EoE do not outgrow their disease which is in contrast to IgE-mediated allergy where 75 % of children become tolerant by the early teenage years [7, 95, 96]. Furthermore, children who do outgrow IgE-mediated milk allergy, as well as those undergoing oral immunotherapy, are at increased risk of developing EoE once milk is reintroduced into their diet [93, 97]. Finally, food elimination diets based on IgE testing are largely unsuccessful [32–34, 98].
Omalizumab is a humanized mouse monoclonal antibody that binds to the IgE molecule rendering it inactive. Omalizumab has been shown to influence allergic cell populations and improve outcomes in asthma and chronic urticaria and is being investigated in other atopic conditions [99]. Additionally, omalizumab has been studied in multiple times in the context of EoE. While one open-label, single arm, non-blinded study showed a positive effect of omalizumab therapy in a subset of EoE patients, several other studies have shown no effect [100]. In a small study of 2 patients, omalizumab therapy had no effect on esophageal eosinophils while in another 16-week open-label study of nine subjects with allergic eosinophilic gastroenteritis omalizumab treatment resulted in reduced peripheral blood, duodenal, and antral eosinophil counts but modestly increased esophageal eosinophils [101, 102]. Finally, in a prospective, randomized, double-blind, placebo-controlled trial of adults with EoE, omalizumab did not alter symptoms of eosinophilic esophagitis or esophageal eosinophil counts compared to placebo [103•]. Together, these findings indicate that IgE is unlikely to play a formative role in the pathogenesis of EoE and that omalizumab is not an effective therapeutic strategy for EoE.
IL-4
Interleukin (IL)-4 is critical for the development of T helper type 2 (Th2) cells and contributes to many type 2 inflammatory responses [104]. Several studies in humans and mice have shown that EoE is characterized by a type 2 allergic inflammatory response. Esophageal biopsies and blood samples of patients with active EoE have increased levels of the type 2 prototypical cytokines and chemokines including IL-4, IL-5, and IL-13 [67, 105–107]. IL-4 also induces secretion of eotaxin-3 by esophageal epithelial cells in vitro [108]. However, the presence of IL-4 is not necessary for the development of esophageal inflammation in some animal models of EoE, suggesting that IL-4-independent mechanisms of allergic inflammation may be more relevant in these cases [109].
IL-5
IL-5 has long been known to promote eosinophil development, activation, survival, and recruitment to sites of inflammation [110]. Patients with EoE are more likely to have polymorphisms in the gene encoding IL-5, higher numbers of circulating IL-5+ CD4+ T cells, and higher esophageal tissue levels of IL-5 and its receptor IL-5-R compared to controls [67, 68, 107, 111, 112]. Furthermore, IL-5 expression in the esophagus of patients with EoE is downregulated with topical steroid treatment, supporting a pathologic role for this cytokine [67, 68, 107, 111]. Animal studies have shown us that non-specific overproduction of IL-5 results in eosinophil accumulation in the esophagus, a phenomenon which is potentiated by eotaxin. Furthermore, epicutaneous antigen exposure primes the immune system for subsequent eosinophilic inflammation in the esophagus upon airway antigen exposure in an IL-5-dependent manner [113]. Conversely, eosinophil accumulation and fibrosis in the esophagus in response to oral or intranasal allergen administration is attenuated in the absence of IL-5 [80, 81, 114–116]. These findings suggest that IL-5 is both sufficient and necessary for eosinophil trafficking to the murine esophagus in some experimental contexts and that IL-5 and eosinophils contribute to the esophageal fibrosis observed in these experimental systems.
As IL-5 is dysregulated in patients with EoE and contributes to some experimental models of EoE, IL-5 blockade was suggested as an attractive potential therapeutic strategy for EoE. Mepolizumab and reslizumab are humanized monoclonal IgG1 antibodies targeted against IL-5 that have been studied in multiple disease contexts from hypereosinophilic syndrome to eosinophilic granulomatosis with polyangiitis [117–119]. In two small studies in adults, mepolizumab therapy caused reductions in peripheral blood and esophageal eosinophilia and a variable degree of resolution of EoE-related symptoms with overall improvement in quality of life scores [120, 121]. One double-blind, randomized, prospective study of 59 children with EoE showed that treatment with mepolizumab reduced esophageal eosinophils, but only to a peak level of 40 eosinophils/hpf which is above the threshold for EoE diagnosis [122]. In a larger double-blind, randomized, placebo-controlled trial of reslizumab in 227 children and adolescents with EoE, there was also a partial reduction in esophageal eosinophils, but again not to normal levels [123]. Importantly, improvements in symptoms in this study were equivalent in all groups including the placebo [123]. Together, these findings suggest that IL-5 blockade may not be a useful therapeutic strategy in EoE possibly due to redundant pathways for eosinophil recruitment and activation.
IL-13
IL-13 is one of the prototypical type 2 cytokines and is integral to multiple aspects of human physiology and disease [124]. Polymorphisms in the gene encoding IL-13 have also been shown to be genetically associated with EoE in a phenome-wide association study [112]. There are several mechanisms by which IL-13 has been proposed to contribute to EoE pathogenesis including upregulation of periostin, induction of AMCase expression, and influencing genes important for epithelial integrity [47, 75, 125]. Esophageal epithelial cells express the IL-13 receptor, and IL-13 induces dysregulated gene expression in the esophageal epithelium, such as overexpression of eotaxin-3, that mimics the transcription profile observed in esophageal biopsies of EoE patients [71]. Mouse models have shown that IL-13 overexpression in the lung, but not the esophagus, results in an EoE-like disease state that is partially dependent on eotaxin-1 but not eosinophils [126]. Intratracheal IL-13 also induces features of experimental EoE in mice in a IL-5-, eotaxin-1-, and STAT6-dependent manner. Importantly, IL-13-deficient mice are protected from the development of experimental EoE in some but not all model systems of EoE and anti-IL-13 treatment in mice reduces esophageal eosinophilia [109, 113, 115, 127, 128].
Experimental findings in mice and humans suggest that IL-13 may also be sufficient and necessary for EoE-like inflammation in some cases, leading to the hypothesis that anti-IL-13 treatment may be a useful therapeutic modality. Anti-IL-13 treatment in adult patients with EoE normalized expression of EoE disease-related transcripts, decreased esophageal eosinophil counts (though not to normal levels), but only resulted in a non-significant trend towards improved symptoms [129]. Therefore, while animal and translational studies support a role for IL-13 in the pathogenesis of EoE, further study is needed to determine whether IL-13 blockade will ultimately become a useful therapy for EoE patients.
IL-15
IL-15 is a cytokine with a structural similarity to IL-2 that binds to and signals through a complex composed of the IL-2/IL-15 receptor beta chain (CD122) and the common gamma chain (CD132). Produced predominantly by monocytes, macrophages, and dendritic cells, IL-15 plays an important role in antiviral immunity by supporting the development, survival, proliferation, and activation of multiple lymphocyte lineages including natural killer cells [130]. Additionally, IL-15 has recently been shown to support type 2 allergic inflammatory responses. For example, mast cells express a distinct receptor for IL-15 (IL-15RX) which signals through the JAK2/STAT5 pathway and murine mast cells treated with IL-15 engage the IL-2/IL-15Rγ chain causing phosphorylation of Tyk2/STAT6 and the production of IL-4 [131, 132].
Both IL-15 and IL-15Ra transcripts are elevated in esophageal biopsies of patients with EoE compared to controls [133]. IL-15 transcripts are also elevated in the esophagus of mice with experimental EoE, IL-15-mediated signaling causes CD4+ T cell proliferation and production of IL-5 and IL-13, and IL-15-deficient mice are protected against experimental EoE [133]. Finally, coculture with IL-15 causes primary esophageal epithelial cells to increase the expression of eotaxin proteins which could promote eosinophil chemotaxis [133]. Together, these findings support a tentative role for IL-15 in contributing to EoE pathogenesis and identify one potential mechanism by which IL-15 may promote esophageal eosinophilia.
IL-18
IL-18 is a cytokine with structural homology to IL-1 that is produced by both immune and non-immune cells and mediates both immunity and pathologic inflammation in various contexts [134]. While originally characterized for its role with IL-12 in stimulating Th1 cell differentiation, IL-18 is also known to influence type 2 inflammation as polymorphisms in IL-18 are protective for some atopic conditions and IL-18 promotes allergic inflammation in some animal models [135, 136]. Recently, a role for IL-18 in the pathogenesis of EoE has also been postulated. Esophageal biopsies of patients with EoE showed higher expression levels of IL-18 mRNA and a higher proportion of IL-18Rα+ cells, and IL-18 can influence IL-5 and IL-13 cytokine production by invariant natural killer T cells (iNKTs) providing one potential pathologic mechanism for this cytokine [137]. Further research may continue to elucidate a role for IL-18 in EoE pathogenesis.
TGF-β and Tumor Necrosis Factor Alpha
TGF-β1 is a member of the transforming growth factor beta superfamily of cytokines. It is a secreted protein that performs many cellular functions including the control of cell growth, proliferation, differentiation, and apoptosis [138]. TGF-β is also a potent stimulator of the synthesis of extracellular matrix proteins and plays a prominent role in the development of tissue fibrosis [139]. Tumor necrosis factor alpha (TNF-α) is an inflammatory cytokine that is produced by multiple immune cell types and has many roles including mediating the activation and survival of eosinophils [140, 141].
Both TGF-β and TNF-α are thought to play a role in EoE pathogenesis. Polymorphisms in the promoter for TGF-β1 are associated with EoE susceptibility, and TGF-β1+ cells are overrepresented in the esophagus of patients with EoE [35, 77]. Similarly, TNF-α is upregulated in EoE and is highly expressed by epithelial cells of the esophagus in patients with active disease [45, 105]. TGF-β1 is produced by eosinophils and mast cells and promotes collagen production and tissue fibrosis by signaling through the Smad3 pathway [142–144]. Consistently, Smad3-deficient mice are protected against esophageal fibrosis and angiogenesis [145]. It is thought that the inflamed epithelial cells prime esophageal fibroblasts to secrete the profibrogenic cytokines IL-1β and TNF-α, which in turn promote epithelial-to-mesenchymal transition and esophageal fibrosis [142, 146]. In one study, coculture of primary epithelial or muscle cells derived from patients with EoE with eosinophils caused increased cell line secretion of fibronectin and collagen I, a response that was inhibited by blocking TGF-β1 [147]. Coculture of eosinophils with cultured muscle cells also resulted in reduced contractility, an observation that may be the result of TGF-β1-induced phospholamban expression [147, 148].
As the aforementioned studies suggest, TGF-β and TNF-α may promote esophageal fibrosis in EoE. Based on these observations, it was hypothesized that blockade of TNF-α with infliximab (a chimeric IgG1 mAb directed against TNF-α) could be beneficial in the treatment of patients with EoE. In a small study of three adult patients, infliximab therapy did not influence esophageal inflammation or symptoms [149]. However, given the limited size of this study, it is reasonable that further investigation may support a role for TNF-α blockade in EoE therapy.
TSLP
As discussed previously, variants at chromosome 5q22 encompassing the TSLP and WDR36 genes are strongly associated with EoE [49]. TSLP is overexpressed in esophageal biopsies from individuals with EoE as compared to unaffected individuals, while WDR36 expression is unaltered, implicating the 5q22 locus (and TSLP in particular) in the pathogenesis of EoE [48]. Animal models also support a role for TSLP in EoE pathogenesis [150•]. In a murine epicutaneous sensitization model of EoE, esophageal inflammation is dependent in part of TSLP and basophils, but independent of IgE. In the same study, elevated TSLP expression and exaggerated basophil responses were observed in esophageal biopsies of patients with EoE, and a gain-of-function polymorphism in TSLP was found to be associated with increased basophil responses in EoE patients [92••]. Further investigation may continue to elucidate a role for TSLP in EoE pathogenesis.
Prostaglandin 2
Prostaglandin D2 (PGD2) is a prostanoid that is produced by mast cells and promotes allergic responses. Introduction of exogenous prostaglandins results in eosinophil infiltration into the esophagus of animal models, while prostaglandin antagonists protect against EoE-like inflammation [151]. Chemoattractant receptor expressed on Th2 cells (CRTH2) is the receptor for PGD2 and mediates chemotaxis of Th2 cells, eosinophils, and basophils [152]. One study examined the utility of a CRTH2 inhibitor in treating EoE. In this randomized, double-blind, placebo-controlled trial of 26 adult EoE patients that were dependent or resistant to corticosteroids, 8 weeks of anti-CRTH2 therapy resulted in reduced esophageal eosinophil load compared to treatment with a placebo. Assessments of disease activity also improved, which correlated with reduced extracellular deposits of eosinophil peroxidase and tenascin C (which are markers of esophageal remodeling), suggesting that anti-CRTH2 therapy may have utility in refractory EoE patients [153].
Pathologic and Protective Cell Populations
Antigen-Presenting Cells
Classically, CD4+ T helper cells are known to be activated by professional antigen-presenting cells (APCs) such as dendritic cells, macrophages, and B cells [154]. Additionally, non-professional APCs are now known to be able to process and present antigens to T cells in certain settings [155]. The roles of professional and non-professional APCs in processing and presenting antigen to T cells in EoE are only beginning to be understood. In models of EoE, epithelial cells have been shown to express MHCII and can induce T helper cell lymphocyte proliferation in vitro [156]. Additionally, eosinophils seem to express some of the molecules necessary for T cell activation though it has not been shown that eosinophils act as APCs in models of EoE [157]. Despite these advances, questions remain to be answered. For example, it is unclear at present where APCs encounter antigen to promote adaptive responses in EoE. Potential locations include the esophageal epithelium, the lamina propria, or organized lymphoid structures. Additionally, it is not clear which APCs are most critical to initiating adaptive response in EoE. Further investigation is therefore warranted.
Basophils
Basophils are a long-known innate granulocyte that circulate in the blood and were initially regarded as a redundant granulocyte population lacking unique functions. However, the past decade of research has revealed an important pathologic role for the basophil in multiple inflammatory disease states [158]. In particular, basophils have been shown to be activated by key allergic signals such as TSLP and to respond to allergen exposures by homing to lymphatics or sites of tissue inflammation where they can help to initiate and propagate allergic responses [88, 159]. As discussed previously, mouse models suggest that TSLP and basophils contribute to an epicutaneous sensitization model of EoE [92••]. Consistently, altered TSLP and basophil responses are present in patients with EoE [92••]. Together, these findings support a tentative role for basophils in EoE pathogenesis.
Eosinophils
The presence of eosinophils in the esophageal mucosa of patients is the classic observation from which EoE derives its name. Highlighting the perceived importance of eosinophils in EoE pathogenesis, the diagnosis of EoE requires the presence of 15 or more eosinophils per high-power field in one or more of at least four esophageal biopsies [9, 10••]. However, eosinophils in the esophagus is not pathognomonic for EoE as many other conditions such as GERD, proton pump inhibitor-responsive esophageal eosinophilia, drugs, infection, autoimmune conditions, and primary hypereosinophilic syndromes have a similar histopathologic appearance [160, 161]. One commonly considered differential diagnosis for EoE is GERD, and as such, it is imperative that a patient is being treated with an optimal anti-GERD therapeutic regimen prior to considering a diagnosis of EoE. However, recent findings that a subset of patients with esophageal eosinophilia have improvement or resolution of disease with PPI therapy alone has led some investigators and clinicians to examine the relationship between EoE, esophageal eosinophilia, and GERD. Specifically, proton pump inhibitor-responsive esophageal eosinophilia is an active area of research [162]. Nevertheless, it is likely that the underlying inflammatory mechanisms of GERD and EoE are distinct and that the presence of eosinophils in the esophagus represents a common inflammatory endpoint [163, 164].
Mast Cells
Mast cells are tissue-resident granulocytes that bind IgE to their surface and, when activated, release histamine and other allergic mediators [165]. Mast cell-associated genes are upregulated in adult EoE, and mast cells are present in increased numbers in esophageal biopsies from both pediatric and adult EoE patients [105, 143, 166, 167]. Implementation of a selective food elimination diet or topical corticosteroid therapy results in downregulation of mast cell-associated genes and significantly reduces epithelial mast cell numbers [143, 166–168]. Furthermore, therapy with mepolizumab, a humanized monoclonal antibody directed against human IL-5, results in decreased numbers of esophageal mast cells in pediatric but not adult EoE patients [121, 169]. Tissue mast cells are not thought to be directly responsive to IL-5 signaling, and it has therefore been hypothesized that the effects of mepolizumab on tissue mast cells are indirect and perhaps mediated through reduction in IL-9+ eosinophils [169]. The precise role of mast cells in EoE pathogenesis is yet unclear.
Th2 Cells
The eosinophilic inflammation observed in EoE is thought to originate as a result of antigen-specific differentiation of T helper type 2 cells. While in some cases of EoE one or more allergenic food triggers can be identified, the analysis of human Th2 cell subsets in this condition is surprisingly limited with only one study demonstrating an increased percentage of IL-5-expressing CD4+ T cells in the peripheral blood of patients with active EoE [111]. Animal models support a role for the adaptive immune system in EoE pathogenesis. In a mouse model of EoE, B and T cell populations, as well as eosinophils, were found in increased numbers of experimental animals as compared to controls. Additionally, oral sensitization of mice with peanut protein results in EoE-like inflammation, the production of peanut-specific IgE, and the secretion of IL-4, IL-5, and IL-13 in splenocyte cultures stimulated with peanut protein [170]. Furthermore, eosinophil accumulation in the esophagus of experimental animals is absent in RAG1-or FOXN1-deficient mice that are deficient in B and T cells or T cells, respectively, but not in IgH6-deficient mice that are deficient in B cells. Moreover, CD8-deficient mice develop experimental EoE while CD4-deficient mice are partially protected from esophageal inflammation. Taken together, these studies support a role for antigen-specific adaptive immune responses, and Th2 cells in particular, in EoE pathogenesis [91].
Regulatory T Cells
Regulatory T cells (Tregs) are a subset of T lymphocytes that play an important role in the prevention and control of many autoimmune and allergic diseases [171]. Tregs are reduced in biopsies of adults with EoE as compared to controls, a finding that is irrespective of steroid therapy [172]. In contrast, Tregs seem to be increased in esophageal tissue of children with EoE [173, 174]. In animal models of EoE, epicutaneous immunotherapy (EPIT) induced Tregs in the spleen and expression of FOXP3 in the esophagus that correlated with reduced eosinophilic infiltration, while depletion of CD25 cells abrogated Tregs induction and resulted in increased esophageal inflammation. Transfer of Tregs isolated from mice who had undergone EPIT prevented peanut-induced eosinophil infiltration and eotaxin expression in the esophagus of mice [175]. Interestingly, patients with the autosomal recessive form of hyper-IgE syndrome caused by mutations in the DOCK8 gene have defective regulatory T cells and often develop EoE [176, 177]. Together, these findings show that Tregs are dysregulated in patients with EoE and that they can protect against an EoE-like disease in some cases.
iNKTs and Innate Lymphoid Cells
iNKTs are a subset of lymphocytes which can produce type 2 cytokines in response to certain stimuli and have been postulated to play a pathogenic role in some atopic diseases [178]. Recently, patients with EoE were found to have reduced peripheral blood iNKTs, and increased esophageal iNKTs, compared to controls. Interestingly, iNKTs from patients with active EoE expand more readily and were found to produce more IL-13 in response to stimulation when compared to controls [179].
Innate lymphoid cells (ILCs) are another type of innate lymphocyte population that play important roles in mouse models of infection, inflammation, and tissue repair and can be dysregulated in specific disease states [180]. Group 2 ILCs (ILC2s), a lineage-negative lymphocyte which express CRTH2, are induced by IL-33 and TSLP to produce large amounts of Th2 cytokines, and are enriched in biopsies of patients with active EoE [181]. Further research is warranted to determine whether either of these innate lymphocyte populations play a prominent role in EoE pathogenesis.
Commensal Bacteria
Commensal bacteria that colonize the intestine and other mucosal sites are now widely regarded as integral to promoting normal human physiology. Commensals are now known to influence broad aspects of the mammalian immune system and have been identified as key modifiers in multiple human disease states including allergy [182]. Despite these advances, studies of how commensals contribute to, or are modified in EoE, are comparably lacking. It is known that patients with EoE have dysregulated commensal bacterial populations compared to healthy controls [183]. Additionally, the total bacterial load is increased in patients with EoE compared to healthy subjects [184]. With regards to specific bacteria, Haemophilus is present in a higher proportion in subjects with untreated EoE [184]. Together, these findings indicate that EoE is associated with commensal bacterial dysbiosis. Whether microbial dysbiosis is a marker of, or a contributor to, the inflammation observed in EoE is yet to be determined.
Conclusions
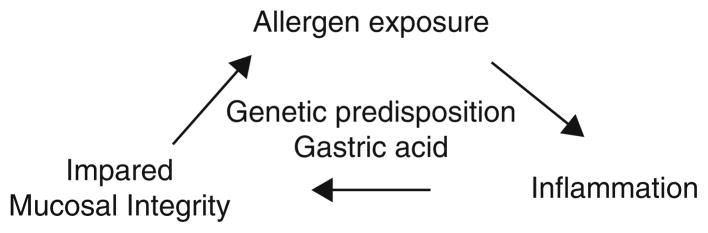
EoE is the result of a complex and coordinated inflammatory response to allergens in a patient’s environment. We have reviewed the most recent research in this field to provide an overview of what is currently known about the immunologic mechanisms underlying this condition. It is known that there are a number of genetic and epigenetic factors that predispose to the development of EoE. Additionally, gastric acid is clearly damaging to the esophageal mucosa and may predispose to inflammation and/or allergen exposure. Once the cycle of inflammation is established, it is likely that impaired mucosal integrity promotes further allergen exposure that may compound the degree of inflammation (Fig. 1). What is not well understood is where the initial antigen-APC-T cell interaction occurs (whether it be at the epithelium, within the underlying lamina propria, or in a more organized lymphoid structure) and how the inflammatory cycle is initiated. It will be important to elucidate these aspects of EoE pathogenesis as intervening upon them (for example, via intervention at the point of the APC or upon innate cytokines such as TSLP that can influence adaptive responses) may provide for new preventative or therapeutic strategies.
Fig. 1.
The inflammatory cycle of EoE
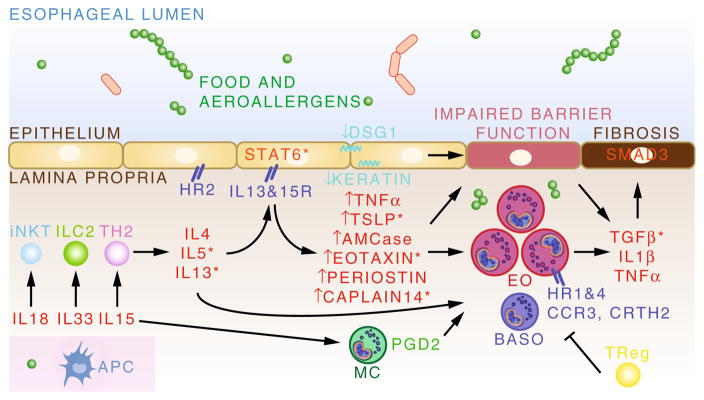
Once the inflammatory cycle is initiated, several key cell types (including Th2, iNKT, and ILC2 cells) likely provide an important source of pro-inflammatory type 2 cytokines including IL-4, IL-5, and IL-13 (Fig. 2). Interactions between these cytokines and the esophageal epithelial cell is important for subsequent upregulation of additional cytokines, chemoattractants, and other molecules which recruit inflammatory effector cell types such as eosinophils, mast cells, and basophils. This inflammatory response disrupts normal epithelial integrity through down-regulation of cell adhesion molecules such as DSG1 and keratin and upregulates profibrotic molecules such as TGF-β, IL-1β, and TNF-α. Tregs may be important in trying to control this inflammation, but prolonged exposure to allergen can lead to the development of odynophagia, fibrosis, esophageal strictures, and in rare and extreme cases, esophageal rupture [1–5]. Given that the current therapeutic approaches available to clinicians and families have significant side effects, and at times cases of refractory EoE can occur, it is important that we continue to strive to develop new and innovative preventative and therapeutic strategies for this important and growing disease.
Fig. 2.
The inflammatory mechanisms of EoE. Allergen interaction with antigen-presenting cells (APC) leads to innate and adaptive lymphocyte responses and characteristic type 2 cytokine production. Signaling in the esophageal epithelial cells broadens the inflammatory response through production of effector and chemoattractant molecules which mediate recruitment and activation of eosinophils (EO), mast cells (MC), and basophils (BASO). Downregulation of cell adhesion molecules contributes to impaired mucosal integrity. Ultimately, persistent inflammation results in the development of esophageal fibrosis
Acknowledgments
DAH is supported by a Resident Research Grant from the American Academy of Pediatrics. JMS is supported by the Stuart Starr Endowed Chair of Pediatrics, The Children’s Hospital of Philadelphia Eosinophilic Esophagitis Fund, a Food Allergy Research & Education, Inc. Clinical Network grant, and the Consortium of Eosinophilic Gastrointestinal Disease Researchers (U54 AI117804) which is part of the Rare Disease Clinical Research Network, an initiative of the National Center for Advancing Translational Sciences (NCATS) Office of Rare Disease Research, and is funded by NCATS and collaborating institute centers.
Abbreviations
- EoE
Eosinophilic esophagitis
- Th
T helper cell
- GERD
Gastroesophageal reflux disease
- Ig
Immunoglobulin
- CCL
C-C motif ligand
- TGF-β
Transforming growth factor beta
- TSLP
Thymic stromal lymphopoietin IL, Interleukin
- DSG
Desmosomal cadherin desmoglein
- miRNA
MicroRNA
- STAT
Signal transducer and activator of transcription
- CD
Cluster of differentiation
- PARP
Poly(ADP-ribose) polymerase
- TNF-α
Tumor necrosis factor alpha
- PGD
Prostaglandin
- CRTH2
Chemoattractant receptor expressed on Th2 cells
- ILCs
Innate lymphoid cells
- iNKTs
Invariant natural killer T cells
- APC
Antigen-presenting cell
- MHC
Major histocompatibility complex
- HR
Histamine receptor
Footnotes
Compliance with Ethical Standards
Conflict of Interest Dr. Spergel reports grants from NIH, DBV Technology, Aimmune Therapeutics, FARE Clinical Network, and Stanford Food Allergy Center and is a consultant for Dannone. Dr. Hill reports grants from the AAP. The authors have no financial relationships or other conflicts of interest relevant to this article to disclose.
Human and Animal Rights and Informed Consent This article does not contain any primary studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Schoepfer AM, Gonsalves N, Bussmann C, Conus S, Simon HU, Straumann A, et al. Esophageal dilation in eosinophilic esophagitis: effectiveness, safety, and impact on the underlying inflammation. Am J Gastroenterol. 2010;105:1062–70. doi: 10.1038/ajg.2009.657. [DOI] [PubMed] [Google Scholar]
- 2.Straumann A, Bussmann C, Zuber M, Vannini S, Simon HU, Schoepfer A. Eosinophilic esophagitis: analysis of food impaction and perforation in 251 adolescent and adult patients. Clin Gastroenterol Hepatol. 2008;6:598–600. doi: 10.1016/j.cgh.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 3.DeBrosse CW, Franciosi JP, King EC, Butz BK, Greenberg AB, Collins MH, et al. Long-term outcomes in pediatric-onset esophageal eosinophilia. J Allergy Clin Immunol. 2011;128:132–8. doi: 10.1016/j.jaci.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franciosi JP, Hommel KA, DeBrosse CW, Greenberg AB, Greenler AJ, Abonia JP, et al. Quality of life in paediatric eosinophilic oesophagitis: what is important to patients? Child Care Health Dev. 2012;38:477–83. doi: 10.1111/j.1365-2214.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoepfer AM, Safroneeva E, Bussmann C, Kuchen T, Portmann S, Simon HU, et al. Delay in diagnosis of eosinophilic esophagitis increases risk for stricture formation in a time-dependent manner. Gastroenterology. 2013;145:1230.e1-2–6.e1-2. doi: 10.1053/j.gastro.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Simon D, Radonjic-Hosli S, Straumann A, Yousefi S, Simon HU. Active eosinophilic esophagitis is characterized by epithelial barrier defects and eosinophil extracellular trap formation. Allergy. 2015;70:443–52. doi: 10.1111/all.12570. [DOI] [PubMed] [Google Scholar]
- 7.Spergel JM, Brown-Whitehorn TF, Beausoleil JL, Franciosi J, Shuker M, Verma R, et al. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48:30–6. doi: 10.1097/MPG.0b013e3181788282. [DOI] [PubMed] [Google Scholar]
- 8.Orenstein SR, Shalaby TM, Di Lorenzo C, Putnam PE, Sigurdsson L, Mousa H, et al. The spectrum of pediatric eosinophilic esophagitis beyond infancy: a clinical series of 30 children. Am J Gastroenterol. 2000;95:1422–30. doi: 10.1111/j.1572-0241.2000.02073.x. [DOI] [PubMed] [Google Scholar]
- 9.Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, et al. First International Gastrointestinal Eosinophil Research Symposium (FIGERS) subcommittees: eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342–63. doi: 10.1053/j.gastro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 10••.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20.e6. doi: 10.1016/j.jaci.2011.02.040. quiz 21–2. The most recent consensus guidelines for the treatment of eosinophlic esophagitis. [DOI] [PubMed] [Google Scholar]
- 11.Straumann A, Simon HU. Eosinophilic esophagitis: escalating epidemiology? J Allergy Clin Immunol. 2005;115:418–9. doi: 10.1016/j.jaci.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Hruz P, Straumann A, Bussmann C, Heer P, Simon HU, Zwahlen M, et al. Swiss EoE study group: escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J Allergy Clin Immunol. 2011;128:1349–50.e5. doi: 10.1016/j.jaci.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Giriens B, Yan P, Safroneeva E, Zwahlen M, Reinhard A, Nydegger A, et al. Escalating incidence of eosinophilic esophagitis in Canton of Vaud, Switzerland, 1993–2013: a population-based study. Allergy. 2015 doi: 10.1111/all.12733. [DOI] [PubMed] [Google Scholar]
- 14.van Rhijn BD, Verheij J, Smout AJ, Bredenoord AJ. Rapidly increasing incidence of eosinophilic esophagitis in a large cohort. Neurogastroenterol Motil. 2013;25:47–52.e5. doi: 10.1111/nmo.12009. [DOI] [PubMed] [Google Scholar]
- 15.Cherian S, Smith NM, Forbes DA. Rapidly increasing prevalence of eosinophilic oesophagitis in Western Australia. Arch Dis Child. 2006;91:1000–4. doi: 10.1136/adc.2006.100974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasad GA, Alexander JA, Schleck CD, Zinsmeister AR, Smyrk TC, Elias RM, et al. Epidemiology of eosinophilic esophagitis over three decades in Olmsted County, Minnesota. Clin Gastroenterol Hepatol. 2009;7:1055–61. doi: 10.1016/j.cgh.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351:940–1. doi: 10.1056/NEJM200408263510924. [DOI] [PubMed] [Google Scholar]
- 18.Patel SM, Falchuk KR. Three brothers with dysphagia caused by eosinophilic esophagitis. Gastrointest Endosc. 2005;61:165–7. doi: 10.1016/s0016-5107(04)02459-9. [DOI] [PubMed] [Google Scholar]
- 19.Meyer GW. Eosinophilic esophagitis in a father and a daughter. Gastrointest Endosc. 2005;61:932. doi: 10.1016/s0016-5107(05)00508-0. [DOI] [PubMed] [Google Scholar]
- 20.Zink DA, Amin M, Gebara S, Desai TK. Familial dysphagia and eosinophilia. Gastrointest Endosc. 2007;65:330–4. doi: 10.1016/j.gie.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 21.Franciosi JP, Tam V, Liacouras CA, Spergel JM. A case-control study of sociodemographic and geographic characteristics of 335 children with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2009;7:415–9. doi: 10.1016/j.cgh.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 22•.Alexander ES, Martin LJ, Collins MH, Kottyan LC, Sucharew H, He H, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1084–92.e1. doi: 10.1016/j.jaci.2014.07.021. One of the first studies to explore the role of the environment and genetics in EoE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118:1054–9. doi: 10.1016/j.jaci.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 24.Collins MH, Blanchard C, Abonia JP, Kirby C, Akers R, Wang N, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6:621–9. doi: 10.1016/j.cgh.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon D, Marti H, Heer P, Simon HU, Braathen LR, Straumann A. Eosinophilic esophagitis is frequently associated with IgE-mediated allergic airway diseases. J Allergy Clin Immunol. 2005;115:1090–2. doi: 10.1016/j.jaci.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Ram G, Lee J, Ott M, Brown-Whitehorn TF, Cianferoni A, Shuker M, et al. Seasonal exacerbation of esophageal eosinophilia in children with eosinophilic esophagitis and allergic rhinitis. Ann Allergy Asthma Immunol. 2015 doi: 10.1016/j.anai.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz JE, Spergel JM, Ruchelli E, Liacouras CA. Elemental diet is an effective treatment for eosinophilic esophagitis in children and adolescents. Am J Gastroenterol. 2003;98:777–82. doi: 10.1111/j.1572-0241.2003.07390.x. [DOI] [PubMed] [Google Scholar]
- 28.Henderson CJ, Abonia JP, King EC, Putnam PE, Collins MH, Franciosi JP, et al. Comparative dietary therapy effectiveness in remission of pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2012;129:1570–8. doi: 10.1016/j.jaci.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lucendo AJ. Meta-analysis-based guidance for dietary management in eosinophilic esophagitis. Curr Gastroenterol Rep. 2015;17 doi: 10.1007/s11894-015-0464-y. 464-015-0464-y. [DOI] [PubMed] [Google Scholar]
- 30.Peterson KA, Byrne KR, Vinson LA, Ying J, Boynton KK, Fang JC, et al. Elemental diet induces histologic response in adult eosinophilic esophagitis. Am J Gastroenterol. 2013;108:759–66. doi: 10.1038/ajg.2012.468. [DOI] [PubMed] [Google Scholar]
- 31.Spergel JM, Brown-Whitehorn TF, Cianferoni A, Shuker M, Wang ML, Verma R, et al. Identification of causative foods in children with eosinophilic esophagitis treated with an elimination diet. J Allergy Clin Immunol. 2012;130:461–7.e5. doi: 10.1016/j.jaci.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 32.Spergel JM, Brown-Whitehorn T, Beausoleil JL, Shuker M, Liacouras CA. Predictive values for skin prick test and atopy patch test for eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119:509–11. doi: 10.1016/j.jaci.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Spergel JM, Beausoleil JL, Mascarenhas M, Liacouras CA. The use of skin prick tests and patch tests to identify causative foods in eosinophilic esophagitis. J Allergy Clin Immunol. 2002;109:363–8. doi: 10.1067/mai.2002.121458. [DOI] [PubMed] [Google Scholar]
- 34.Spergel JM, Andrews T, Brown-Whitehorn TF, Beausoleil JL, Liacouras CA. Treatment of eosinophilic esophagitis with specific food elimination diet directed by a combination of skin prick and patch tests. Ann Allergy Asthma Immunol. 2005;95:336–43. doi: 10.1016/S1081-1206(10)61151-9. [DOI] [PubMed] [Google Scholar]
- 35.Aceves SS, Newbury RO, Chen D, Mueller J, Dohil R, Hoffman H, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65:109–16. doi: 10.1111/j.1398-9995.2009.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajan J, Newbury RO, Anilkumar A, Dohil R, Broide DH, Aceves SS. Long-term assessment of esophageal remodeling in patients with pediatric eosinophilic esophagitis treated with topical corticosteroids. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konikoff MR, Noel RJ, Blanchard C, Kirby C, Jameson SC, Buckmeier BK, et al. A randomized, double-blind, placebo-controlled trial of fluticasone propionate for pediatric eosinophilic esophagitis. Gastroenterology. 2006;131:1381–91. doi: 10.1053/j.gastro.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 38.Noel RJ, Putnam PE, Collins MH, Assa’ad AH, Guajardo JR, Jameson SC, et al. Clinical and immunopathologic effects of swallowed fluticasone for eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2004;2:568–75. doi: 10.1016/s1542-3565(04)00240-x. [DOI] [PubMed] [Google Scholar]
- 39.Butz BK, Wen T, Gleich GJ, Furuta GT, Spergel J, King E, et al. Efficacy, dose reduction, and resistance to high-dose fluticasone in patients with eosinophilic esophagitis. Gastroenterology. 2014;147:324–33.e5. doi: 10.1053/j.gastro.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straumann A, Conus S, Degen L, Felder S, Kummer M, Engel H, et al. Budesonide is effective in adolescent and adult patients with active eosinophilic esophagitis. Gastroenterology. 2010;139:1526–37.e1. doi: 10.1053/j.gastro.2010.07.048. [DOI] [PubMed] [Google Scholar]
- 41.Straumann A, Conus S, Degen L, Frei C, Bussmann C, Beglinger C, et al. Long-term budesonide maintenance treatment is partially effective for patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2011;9:400–9.e1. doi: 10.1016/j.cgh.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 42.Dohil R, Newbury R, Fox L, Bastian J, Aceves S. Oral viscous budesonide is effective in children with eosinophilic esophagitis in a randomized, placebo-controlled trial. Gastroenterology. 2010;139:418–29. doi: 10.1053/j.gastro.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Aceves SS, Bastian JF, Newbury RO, Dohil R. Oral viscous budesonide: a potential new therapy for eosinophilic esophagitis in children. Am J Gastroenterol. 2007;102:2271–9. doi: 10.1111/j.1572-0241.2007.01379.x. quiz 2280. [DOI] [PubMed] [Google Scholar]
- 44.Sherrill JD, Rothenberg ME. Genetic and epigenetic underpinnings of eosinophilic esophagitis. Gastroenterol Clin N Am. 2014;43:269–80. doi: 10.1016/j.gtc.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 47.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–41. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:160–5.e3. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. Identify new region for EoE-CAPN14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51•.Sleiman PM, Wang ML, Cianferoni A, Aceves S, Gonsalves N, Nadeau K, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nat Commun. 2014;5:5593. doi: 10.1038/ncomms6593. Identify four new regions for EoE-CAPN14, STAT-6, EMSY, and ANKR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanchard C, Durual S, Estienne M, Emami S, Vasseur S, Cuber JC. Eotaxin-3/CCL26 gene expression in intestinal epithelial cells is up-regulated by interleukin-4 and interleukin-13 via the signal transducer and activator of transcription 6. Int J Biochem Cell Biol. 2005;37:2559–73. doi: 10.1016/j.biocel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CREB-binding protein-mediated histone 3 acetylation. J Biol Chem. 2011;286:13193–204. doi: 10.1074/jbc.M110.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Cheng E, Huo X, Yu C, Zhang Q, Pham TH, et al. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS ONE. 2012;7:e50037. doi: 10.1371/journal.pone.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dohil R, Newbury RO, Aceves S. Transient PPI responsive esophageal eosinophilia may be a clinical sub-phenotype of pediatric eosinophilic esophagitis. Dig Dis Sci. 2012;57:1413–9. doi: 10.1007/s10620-011-1991-5. [DOI] [PubMed] [Google Scholar]
- 56.Lim E, Rothenberg ME. Demethylation of the human eotaxin-3 gene promoter leads to the elevated expression of eotaxin-3. J Immunol. 2014;192:466–74. doi: 10.4049/jimmunol.1302454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Connell RM, Rao DS. Baltimore D: microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 58.Lu S, Mukkada VA, Mangray S, Cleveland K, Shillingford N, Schorl C, et al. MicroRNA profiling in mucosal biopsies of eosinophilic esophagitis patients pre and post treatment with steroids and relationship with mRNA targets. PLoS ONE. 2012;7:e40676. doi: 10.1371/journal.pone.0040676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu TX, Lim EJ, Wen T, Plassard AJ, Hogan SP, Martin LJ, et al. MiR-375 is downregulated in epithelial cells after IL-13 stimulation and regulates an IL-13-induced epithelial transcriptome. Mucosal Immunol. 2012;5:388–96. doi: 10.1038/mi.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129:1064–75.e9. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wong CK, Lau KM, Chan IH, Hu S, Lam YY, Choi AO, et al. MicroRNA-21* regulates the prosurvival effect of GM-CSF on human eosinophils. Immunobiology. 2013;218:255–62. doi: 10.1016/j.imbio.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 63.Liu X, Hong Q, Wang Z, Yu Y, Zou X, Xu L. Transforming growth factor-beta-sphingosine kinase 1/S1P signaling upregulates microRNA-21 to promote fibrosis in renal tubular epithelial cells. Exp Biol Med (Maywood) 2015 doi: 10.1177/1535370215605586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu TX, Lim EJ, Besse JA, Itskovich S, Plassard AJ, Fulkerson PC, et al. MiR-223 deficiency increases eosinophil progenitor proliferation. J Immunol. 2013;190:1576–82. doi: 10.4049/jimmunol.1202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145:1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sherrill JD, Kiran KC, Blanchard C, Stucke EM, Kemme KA, Collins MH, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15:361–9. doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–17. e1–7. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lucendo AJ, De Rezende L, Comas C, Caballero T, Bellon T. Treatment with topical steroids downregulates IL-5, eotaxin-1/ CCL11, and eotaxin-3/CCL26 gene expression in eosinophilic esophagitis. Am J Gastroenterol. 2008;103:2184–93. doi: 10.1111/j.1572-0241.2008.01937.x. [DOI] [PubMed] [Google Scholar]
- 69.Bhattacharya B, Carlsten J, Sabo E, Kethu S, Meitner P, Tavares R, et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol. 2007;38:1744–53. doi: 10.1016/j.humpath.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 70.Getsios S, Simpson CL, Kojima S, Harmon R, Sheu LJ, Dusek RL, et al. Desmoglein 1-dependent suppression of EGFR signaling promotes epidermal differentiation and morphogenesis. J Cell Biol. 2009;185:1243–58. doi: 10.1083/jcb.200809044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 72•.Sherrill JD, Kc K, Wu D, Djukic Z, Caldwell JM, Stucke EM, et al. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7:718–29. doi: 10.1038/mi.2013.90. This manuscript explores the role of epithelial function in EoE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samuelov L, Sarig O, Harmon RM, Rapaport D, Ishida-Yamamoto A, Isakov O, et al. Desmoglein 1 deficiency results in severe dermatitis, multiple allergies and metabolic wasting. Nat Genet. 2013;45:1244–8. doi: 10.1038/ng.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kc K, Rothenberg ME, Sherrill JD. In vitro model for studying esophageal epithelial differentiation and allergic inflammatory responses identifies keratin involvement in eosinophilic esophagitis. PLoS ONE. 2015;10:e0127755. doi: 10.1371/journal.pone.0127755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–96. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Masuoka M, Shiraishi H, Ohta S, Suzuki S, Arima K, Aoki S, et al. Periostin promotes chronic allergic inflammation in response to Th2 cytokines. J Clin Invest. 2012;122:2590–600. doi: 10.1172/JCI58978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frischmeyer-Guerrerio PA, Guerrerio AL, Oswald G, Chichester K, Myers L, Halushka MK, et al. TGFbeta receptor mutations impose a strong predisposition for human allergic disease. Sci Transl Med. 2013;5:195ra94. doi: 10.1126/scitranslmed.3006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abonia JP, Wen T, Stucke EM, Grotjan T, Griffith MS, Kemme KA, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013;132:378–86. doi: 10.1016/j.jaci.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lampinen M, Carlson M, Hakansson LD, Venge P. Cytokine-regulated accumulation of eosinophils in inflammatory disease. Allergy. 2004;59:793–805. doi: 10.1111/j.1398-9995.2004.00469.x. [DOI] [PubMed] [Google Scholar]
- 80.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hogan SP, Mishra A, Brandt EB, Royalty MP, Pope SM, Zimmermann N, et al. A pathological function for eotaxin and eosinophils in eosinophilic gastrointestinal inflammation. Nat Immunol. 2001;2:353–60. doi: 10.1038/86365. [DOI] [PubMed] [Google Scholar]
- 82.Mishra A, Hogan SP, Brandt EB, Wagner N, Crossman MW, Foster PS, et al. Enterocyte expression of the eotaxin and interleukin-5 transgenes induces compartmentalized dysregulation of eosinophil trafficking. J Biol Chem. 2002;277:4406–12. doi: 10.1074/jbc.M110424200. [DOI] [PubMed] [Google Scholar]
- 83.Mehrotra P, Hollenbeck A, Riley JP, Li F, Patel RJ, Akhtar N, et al. Poly (ADP-ribose) polymerase 14 and its enzyme activity regulates T(H)2 differentiation and allergic airway disease. J Allergy Clin Immunol. 2013;131:521–31. e1–12. doi: 10.1016/j.jaci.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goenka S, Kaplan MH. Transcriptional regulation by STAT6. Immunol Res. 2011;50:87–96. doi: 10.1007/s12026-011-8205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krishnamurthy P, Sherrill JD, Parashette K, Goenka S, Rothenberg ME, Gupta S, et al. Correlation of increased PARP14 and CCL26 expression in biopsies from children with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;133:577–80. doi: 10.1016/j.jaci.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Merves J, Chandramouleeswaran PM, Benitez AJ, Muir AB, Lee AJ, Lim DM, et al. Altered esophageal histamine receptor expression in eosinophilic esophagitis (EoE): implications on disease pathogenesis. PLoS ONE. 2015;10:e0114831. doi: 10.1371/journal.pone.0114831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burton OT, Oettgen HC. Beyond immediate hypersensitivity: evolving roles for IgE antibodies in immune homeostasis and allergic diseases. Immunol Rev. 2011;242:128–43. doi: 10.1111/j.1600-065X.2011.01024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–46. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kitaura J, Song J, Tsai M, Asai K, Maeda-Yamamoto M, Mocsai A, et al. Evidence that IgE molecules mediate a spectrum of effects on mast cell survival and activation via aggregation of the FcepsilonRI. Proc Natl Acad Sci U S A. 2003;100:12911–6. doi: 10.1073/pnas.1735525100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vicario M, Blanchard C, Stringer KF, Collins MH, Mingler MK, Ahrens A, et al. Local B cells and IgE production in the oesophageal mucosa in eosinophilic oesophagitis. Gut. 2010;59:12–20. doi: 10.1136/gut.2009.178020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mishra A, Schlotman J, Wang M, Rothenberg ME. Critical role for adaptive T cell immunity in experimental eosinophilic esophagitis in mice. J Leukoc Biol. 2007;81:916–24. doi: 10.1189/jlb.1106653. [DOI] [PubMed] [Google Scholar]
- 92••.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013;19:1005–13. doi: 10.1038/nm.3281. This manuscript identified the role of TSLP and basophils in EoE and showed that the pathway is not IgE mediated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maggadottir SM, Hill DA, Ruymann K, Brown-Whitehorn TF, Cianferoni A, Shuker M, et al. Resolution of acute IgE-mediated allergy with development of eosinophilic esophagitis triggered by the same food. J Allergy Clin Immunol. 2014;133:1487–89.e1. doi: 10.1016/j.jaci.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 94.Hill DA, Shuker M, Cianferoni A, Wong T, Ruchelli E, Spergel JM, et al. The development of IgE-mediated immediate hypersensitivity after the diagnosis of eosinophilic esophagitis to the same food. J Allergy Clin Immunol Pract. 2015;3:123–4. doi: 10.1016/j.jaip.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Skripak JM, Matsui EC, Mudd K, Wood RA. The natural history of IgE-mediated cow’s milk allergy. J Allergy Clin Immunol. 2007;120:1172–7. doi: 10.1016/j.jaci.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 96.Spergel JM. Natural history of cow’s milk allergy. J Allergy Clin Immunol. 2013;131:813–4. doi: 10.1016/j.jaci.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 97.Sanchez-Garcia S, Rodriguez Del Rio P, Escudero C, Martinez-Gomez MJ, Ibanez MD. Possible eosinophilic esophagitis induced by milk oral immunotherapy. J Allergy Clin Immunol. 2012;129:1155–7. doi: 10.1016/j.jaci.2011.11.042. [DOI] [PubMed] [Google Scholar]
- 98.Gonsalves N, Yang GY, Doerfler B, Ritz S, Ditto AM, Hirano I. Elimination diet effectively treats eosinophilic esophagitis in adults; food reintroduction identifies causative factors. Gastroenterology. 2012;142:1451–59.e1. doi: 10.1053/j.gastro.2012.03.001. quiz e14-5. [DOI] [PubMed] [Google Scholar]
- 99.Hill DA, Siracusa MC, Ruymann KR, Tait Wojno ED, Artis D, Spergel JM. Omalizumab therapy is associated with reduced circulating basophil populations in asthmatic children. Allergy. 2014;69:674–7. doi: 10.1111/all.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Loizou D, Enav B, Komlodi-Pasztor E, Hider P, Kim-Chang J, Noonan L, et al. A pilot study of omalizumab in eosinophilic esophagitis. PLoS ONE. 2015;10:e0113483. doi: 10.1371/journal.pone.0113483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Foroughi S, Foster B, Kim N, Bernardino LB, Scott LM, Hamilton RG, et al. Anti-IgE treatment of eosinophil-associated gastrointestinal disorders. J Allergy Clin Immunol. 2007;120:594–601. doi: 10.1016/j.jaci.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rocha R, Vitor AB, Trindade E, Lima R, Tavares M, Lopes J, et al. Omalizumab in the treatment of eosinophilic esophagitis and food allergy. Eur J Pediatr. 2011;170:1471–4. doi: 10.1007/s00431-011-1540-4. [DOI] [PubMed] [Google Scholar]
- 103•.Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology. 2014;147:602–9. doi: 10.1053/j.gastro.2014.05.036. This paper is the first paper to explore the possible role of IgG4 in EoE. [DOI] [PubMed] [Google Scholar]
- 104.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 105.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–61. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 106.Straumann A, Kristl J, Conus S, Vassina E, Spichtin HP, Beglinger C, et al. Cytokine expression in healthy and inflamed mucosa: probing the role of eosinophils in the digestive tract. Inflamm Bowel Dis. 2005;11:720–6. doi: 10.1097/01.mib.0000172557.39767.53. [DOI] [PubMed] [Google Scholar]
- 107.Kinoshita Y, Furuta K, Ishimura N, Ishihara S. Elevated plasma cytokines in Japanese patients with eosinophilic esophagitis and gastroenteritis. Digestion. 2012;86:238–43. doi: 10.1159/000341421. [DOI] [PubMed] [Google Scholar]
- 108.Cheng E, Zhang X, Huo X, Yu C, Zhang Q, Wang DH, et al. Omeprazole blocks eotaxin-3 expression by oesophageal squamous cells from patients with eosinophilic oesophagitis and GORD. Gut. 2013;62:824–32. doi: 10.1136/gutjnl-2012-302250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Niranjan R, Rayapudi M, Mishra A, Dutt P, Dynda S, Mishra A. Pathogenesis of allergen-induced eosinophilic esophagitis is independent of interleukin (IL)-13. Immunol Cell Biol. 2013;91:408–15. doi: 10.1038/icb.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kouro T, Takatsu K. IL-5- and eosinophil-mediated inflammation: from discovery to therapy. Int Immunol. 2009;21:1303–9. doi: 10.1093/intimm/dxp102. [DOI] [PubMed] [Google Scholar]
- 111.Bullock JZ, Villanueva JM, Blanchard C, Filipovich AH, Putnam PE, Collins MH, et al. Interplay of adaptive Th2 immunity with eotaxin-3/c-C chemokine receptor 3 in eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45:22–31. doi: 10.1097/MPG.0b013e318043c097. [DOI] [PubMed] [Google Scholar]
- 112.Namjou B, Marsolo K, Caroll RJ, Denny JC, Ritchie MD, Verma SS, et al. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to eosinophilic esophagitis. Front Genet. 2014;5:401. doi: 10.3389/fgene.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Akei HS, Mishra A, Blanchard C, Rothenberg ME. Epicutaneous antigen exposure primes for experimental eosinophilic esophagitis in mice. Gastroenterology. 2005;129:985–94. doi: 10.1053/j.gastro.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 114.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168:2464–9. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 115.Akei HS, Brandt EB, Mishra A, Strait RT, Finkelman FD, Warrier MR, et al. Epicutaneous aeroallergen exposure induces systemic TH2 immunity that predisposes to allergic nasal responses. J Allergy Clin Immunol. 2006;118:62–9. doi: 10.1016/j.jaci.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 116.Mishra A, Wang M, Pemmaraju VR, Collins MH, Fulkerson PC, Abonia JP, et al. Esophageal remodeling develops as a consequence of tissue specific IL-5-induced eosinophilia. Gastroenterology. 2008;134:204–14. doi: 10.1053/j.gastro.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rothenberg ME, Klion AD, Roufosse FE, Kahn JE, Weller PF, Simon HU, et al. Treatment of patients with the hypereosinophilic syndrome with mepolizumab. N Engl J Med. 2008;358:1215–28. doi: 10.1056/NEJMoa070812. [DOI] [PubMed] [Google Scholar]
- 118.Mepolizumab: 240563, anti-IL-5 monoclonal antibody—GlaxoSmithKline, anti-interleukin-5 monoclonal antibody—GlaxoSmithKline, SB 240563. Drugs R&D. 2008;9:125–130. doi: 10.2165/00126839-200809020-00006. [DOI] [PubMed] [Google Scholar]
- 119.Kim S, Marigowda G, Oren E, Israel E, Wechsler ME. Mepolizumab as a steroid-sparing treatment option in patients with Churg-Strauss syndrome. J Allergy Clin Immunol. 2010;125:1336–43. doi: 10.1016/j.jaci.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 120.Stein ML, Collins MH, Villanueva JM, Kushner JP, Putnam PE, Buckmeier BK, et al. Anti-IL-5 (mepolizumab) therapy for eosinophilic esophagitis. J Allergy Clin Immunol. 2006;118:1312–9. doi: 10.1016/j.jaci.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 121.Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut. 2010;59:21–30. doi: 10.1136/gut.2009.178558. [DOI] [PubMed] [Google Scholar]
- 122.Assa’ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA, et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology. 2011;141:1593–604. doi: 10.1053/j.gastro.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 123.Spergel JM, Rothenberg ME, Collins MH, Furuta GT, Markowitz JE, Fuchs G, 3rd, et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2012;129:456–63. e1–3. doi: 10.1016/j.jaci.2011.11.044. [DOI] [PubMed] [Google Scholar]
- 124.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–56. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 125.Cho JY, Rosenthal P, Miller M, Pham A, Aceves S, Sakuda S, et al. Targeting AMCase reduces esophageal eosinophilic inflammation and remodeling in a mouse model of egg induced eosinophilic esophagitis. Int Immunopharmacol. 2014;18:35–42. doi: 10.1016/j.intimp.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zuo L, Fulkerson PC, Finkelman FD, Mingler M, Fischetti CA, Blanchard C, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol. 2010;185:660–9. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125:1419–27. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 128.Blanchard C, Mishra A, Saito-Akei H, Monk P, Anderson I, Rothenberg ME. Inhibition of human interleukin-13-induced respiratory and oesophageal inflammation by anti-human-interleukin-13 antibody (CAT-354) Clin Exp Allergy. 2005;35:1096–103. doi: 10.1111/j.1365-2222.2005.02299.x. [DOI] [PubMed] [Google Scholar]
- 129.Rothenberg ME, Wen T, Greenberg A, Alpan O, Enav B, Hirano I, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135:500–7. doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 130.Becknell B, Caligiuri MA. Interleukin-2, interleukin-15, and their roles in human natural killer cells. Adv Immunol. 2005;86:209–39. doi: 10.1016/S0065-2776(04)86006-1. [DOI] [PubMed] [Google Scholar]
- 131.Tagaya Y, Burton JD, Miyamoto Y, Waldmann TA. Identification of a novel receptor/signal transduction pathway for IL-15/T in mast cells. EMBO J. 1996;15:4928–39. [PMC free article] [PubMed] [Google Scholar]
- 132.Masuda A, Matsuguchi T, Yamaki K, Hayakawa T, Kubo M, LaRochelle WJ, et al. Interleukin-15 induces rapid tyrosine phosphorylation of STAT6 and the expression of interleukin-4 in mouse mast cells. J Biol Chem. 2000;275:29331–7. doi: 10.1074/jbc.M910290199. [DOI] [PubMed] [Google Scholar]
- 133.Zhu X, Wang M, Mavi P, Rayapudi M, Pandey AK, Kaul A, et al. Interleukin-15 expression is increased in human eosinophilic esophagitis and mediates pathogenesis in mice. Gastroenterology. 2010;139:182–93.e7. doi: 10.1053/j.gastro.2010.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakanishi K, Yoshimoto T, Tsutsui H, Okamura H. Interleukin-18 regulates both Th1 and Th2 responses. Annu Rev Immunol. 2001;19:423–74. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 135.Cheng D, Hao Y, Zhou W, Ma Y. The relationship between interleukin-18 polymorphisms and allergic disease: a meta-analysis. Biomed Res Int. 2014;2014:290687. doi: 10.1155/2014/290687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsutsui H, Yoshimoto T, Hayashi N, Mizutani H, Nakanishi K. Induction of allergic inflammation by interleukin-18 in experimental animal models. Immunol Rev. 2004;202:115–38. doi: 10.1111/j.0105-2896.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- 137.Niranjan R, Rajavelu P, Ventateshaiah SU, Shukla JS, Zaidi A, Mariswamy SJ, et al. Involvement of interleukin-18 in the pathogenesis of human eosinophilic esophagitis. Clin Immunol. 2015;157:103–13. doi: 10.1016/j.clim.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–30. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Urban ML, Manenti L, Vaglio A. Fibrosis—a common pathway to organ injury and failure. N Engl J Med. 2015;373:95–6. doi: 10.1056/NEJMc1504848. [DOI] [PubMed] [Google Scholar]
- 140.Zeck-Kapp G, Czech W, Kapp A. TNF alpha-induced activation of eosinophil oxidative metabolism and morphology—comparison with IL-5. Exp Dermatol. 1994;3:176–88. doi: 10.1111/j.1600-0625.1994.tb00275.x. [DOI] [PubMed] [Google Scholar]
- 141.Kankaanranta H, Ilmarinen P, Zhang X, Adcock IM, Lahti A, Barnes PJ, et al. Tumour necrosis factor-alpha regulates human eosinophil apoptosis via ligation of TNF-receptor 1 and balance between NF-kappaB and AP-1. PLoS ONE. 2014;9:e90298. doi: 10.1371/journal.pone.0090298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119:206–12. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 143.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–204.e4. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 144.Le AV, Cho JY, Miller M, McElwain S, Golgotiu K, Broide DH. Inhibition of allergen-induced airway remodeling in Smad 3-deficient mice. J Immunol. 2007;178:7310–6. doi: 10.4049/jimmunol.178.11.7310. [DOI] [PubMed] [Google Scholar]
- 145.Cho JY, Doshi A, Rosenthal P, Beppu A, Miller M, Aceves S, et al. Smad3-deficient mice have reduced esophageal fibrosis and an-giogenesis in a model of egg-induced eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2014;59:10–6. doi: 10.1097/MPG.0000000000000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Muir AB, Lim DM, Benitez AJ, Modayur Chandramouleeswaran P, Lee AJ, Ruchelli ED, et al. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Exp Cell Res. 2013;319:850–9. doi: 10.1016/j.yexcr.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rieder F, Nonevski I, Ma J, Ouyang Z, West G, Protheroe C, et al. T-helper 2 cytokines, transforming growth factor beta1, and eosinophil products induce fibrogenesis and alter muscle motility in patients with eosinophilic esophagitis. Gastroenterology. 2014;146:1266–77. e1–9. doi: 10.1053/j.gastro.2014.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Beppu LY, Anilkumar AA, Newbury RO, Dohil R, Broide DH, Aceves SS. TGF-beta1-induced phospholamban expression alters esophageal smooth muscle cell contraction in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1100–7.e4. doi: 10.1016/j.jaci.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Straumann A, Bussmann C, Conus S, Beglinger C, Simon HU. Anti-TNF-alpha (infliximab) therapy for severe adult eosinophilic esophagitis. J Allergy Clin Immunol. 2008;122:425–7. doi: 10.1016/j.jaci.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 150•.Collison AM, Sokulsky LA, Sherrill JD, Nightingale S, Hatchwell L, Talley NJ, et al. TNF-related apoptosis-inducing ligand (TRAIL) regulates midline-1, thymic stromal lymphopoietin, inflammation, and remodeling in experimental eosinophilic esophagitis. J Allergy Clin Immunol. 2015 doi: 10.1016/j.jaci.2015.03.031. This paper explores the various mechanisms of fibrosis in EoE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang S, Wu X, Yu S. Prostaglandin D2 receptor D-type prostanoid receptor 2 mediates eosinophil trafficking into the esophagus. Dis Esophagus. 2014;27:601–6. doi: 10.1111/dote.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pettipher R, Hansel TT, Armer R. Antagonism of the prostaglandin D2 receptors DP1 and CRTH2 as an approach to treat allergic diseases. Nat Rev Drug Discov. 2007;6:313–25. doi: 10.1038/nrd2266. [DOI] [PubMed] [Google Scholar]
- 153.Straumann A, Hoesli S, Bussmann C, Stuck M, Perkins M, Collins LP, et al. Anti-eosinophil activity and clinical efficacy of the CRTH2 antagonist OC000459 in eosinophilic esophagitis. Allergy. 2013;68:375–85. doi: 10.1111/all.12096. [DOI] [PubMed] [Google Scholar]
- 154.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kambayashi T, Laufer TM. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat Rev Immunol. 2014;14:719–30. doi: 10.1038/nri3754. [DOI] [PubMed] [Google Scholar]
- 156.Mulder DJ, Pooni A, Mak N, Hurlbut DJ, Basta S, Justinich CJ. Antigen presentation and MHC class II expression by human esophageal epithelial cells: role in eosinophilic esophagitis. Am J Pathol. 2011;178:744–53. doi: 10.1016/j.ajpath.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Le-Carlson M, Seki S, Abarbanel D, Quiros A, Cox K, Nadeau KC. Markers of antigen presentation and activation on eosinophils and T cells in the esophageal tissue of patients with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013;56:257–62. doi: 10.1097/MPG.0b013e3182758d49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Siracusa MC, Comeau MR, Artis D. New insights into basophil biology: initiators, regulators, and effectors of type 2 inflammation. Ann N Y Acad Sci. 2011;1217:166–77. doi: 10.1111/j.1749-6632.2010.05918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Siracusa MC, Saenz SA, Hill DA, Kim BS, Headley MB, Doering TA, et al. TSLP promotes interleukin-3-independent basophil haematopoiesis and type 2 inflammation. Nature. 2011;477:229–33. doi: 10.1038/nature10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sridhara S, Ravi K, Smyrk TC, Kita H, Kephart GM, Weiler CR, et al. Increased numbers of eosinophils, rather than only etiology, predict histologic changes in patients with esophageal eosinophilia. Clin Gastroenterol Hepatol. 2012;10:735–41. doi: 10.1016/j.cgh.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 161.Rodrigo S, Abboud G, Oh D, DeMeester SR, Hagen J, Lipham J, et al. High intraepithelial eosinophil counts in esophageal squamous epithelium are not specific for eosinophilic esophagitis in adults. Am J Gastroenterol. 2008;103:435–42. doi: 10.1111/j.1572-0241.2007.01594.x. [DOI] [PubMed] [Google Scholar]
- 162.Kia L, Hirano I. Distinguishing GERD from eosinophilic oesophagitis: concepts and controversies. Nat Rev Gastroenterol Hepatol. 2015;12:379–86. doi: 10.1038/nrgastro.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zukerberg L, Mahadevan K, Selig M, Deshpande V. Esophageal intrasquamous IgG4 deposits: an adjunctive marker to distinguish eosinophilic esophagitis from reflux esophagitis. Histopathology. 2015 doi: 10.1111/his.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Moawad FJ, Wells JM, Johnson RL, Reinhardt BJ, Maydonovitch CL, Baker TP. Comparison of eotaxin-3 biomarker in patients with eosinophilic oesophagitis, proton pump inhibitor-responsive oesophageal eosinophilia and gastro-oesophageal reflux disease. Aliment Pharmacol Ther. 2015;42:231–8. doi: 10.1111/apt.13258. [DOI] [PubMed] [Google Scholar]
- 165.Bischoff SC. Role of mast cells in allergic and non-allergic immune responses: comparison of human and murine data. Nat Rev Immunol. 2007;7:93–104. doi: 10.1038/nri2018. [DOI] [PubMed] [Google Scholar]
- 166.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:140–9. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hsu Blatman KS, Gonsalves N, Hirano I, Bryce PJ. Expression of mast cell-associated genes is upregulated in adult eosinophilic esophagitis and responds to steroid or dietary therapy. J Allergy Clin Immunol. 2011;127:1307–8.e3. doi: 10.1016/j.jaci.2010.12.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Arias A, Lucendo AJ, Martinez-Fernandez P, Gonzalez-Castro AM, Fortea M, Gonzalez-Cervera J, et al. Dietary treatment modulates mast cell phenotype, density, and activity in adult eosinophilic esophagitis. Clin Exp Allergy. 2015 doi: 10.1111/cea.12504. [DOI] [PubMed] [Google Scholar]
- 169.Otani IM, Anilkumar AA, Newbury RO, Bhagat M, Beppu LY, Dohil R, et al. Anti-IL-5 therapy reduces mast cell and IL-9 cell numbers in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2013;131:1576–82. doi: 10.1016/j.jaci.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mondoulet L, Dioszeghy V, Larcher T, Ligouis M, Dhelft V, Puteaux E, et al. Epicutaneous immunotherapy (EPIT) blocks the allergic esophago-gastro-enteropathy induced by sustained oral exposure to peanuts in sensitized mice. PLoS ONE. 2012;7:e31967. doi: 10.1371/journal.pone.0031967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: differentiation, specification, subphenotypes. Nat Immunol. 2009;10:689–95. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 172.Stuck MC, Straumann A, Simon HU. Relative lack of T regulatory cells in adult eosinophilic esophagitis—no normalization after corticosteroid therapy. Allergy. 2011;66:705–7. doi: 10.1111/j.1398-9995.2010.02525.x. [DOI] [PubMed] [Google Scholar]
- 173.Fuentebella J, Patel A, Nguyen T, Sanjanwala B, Berquist W, Kerner JA, et al. Increased number of regulatory Tcells in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2010;51:283–9. doi: 10.1097/MPG.0b013e3181e0817b. [DOI] [PubMed] [Google Scholar]
- 174.Tantibhaedhyangkul U, Tatevian N, Gilger MA, Major AM, Davis CM. Increased esophageal regulatory T cells and eosinophil characteristics in children with eosinophilic esophagitis and gastro-esophageal reflux disease. Ann Clin Lab Sci. 2009;39:99–107. [PubMed] [Google Scholar]
- 175.Dioszeghy V, Mondoulet L, Dhelft V, Ligouis M, Puteaux E, Dupont C, et al. The regulatory T cells induction by epicutaneous immunotherapy is sustained and mediates long-term protection from eosinophilic disorders in peanut-sensitized mice. Clin Exp Allergy. 2014;44:867–81. doi: 10.1111/cea.12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hsu AP, Davis J, Puck JM, Holland SM, Freeman AF. Autosomal dominant hyper IgE syndrome. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, et al., editors. GeneReviews(R) Seattle: University of Washington; 1993. [PubMed] [Google Scholar]
- 177.Janssen E, Morbach H, Ullas S, Bannock JM, Massad C, Menard L, et al. Dedicator of cytokinesis 8-deficient patients have a breakdown in peripheral B-cell tolerance and defective regulatory T cells. J Allergy Clin Immunol. 2014;134:1365–74. doi: 10.1016/j.jaci.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jyonouchi S, Abraham V, Orange JS, Spergel JM, Gober L, Dudek E, et al. Invariant natural killer T cells from children with versus without food allergy exhibit differential responsiveness to milk-derived sphingomyelin. J Allergy Clin Immunol. 2011;128:102–9.e13. doi: 10.1016/j.jaci.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jyonouchi S, Smith CL, Saretta F, Abraham V, Ruymann KR, Modayur-Chandramouleeswaran P, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy. 2014;44:58–68. doi: 10.1111/cea.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sonnenberg GF, Artis D. Innate lymphoid cells in the initiation, regulation and resolution of inflammation. Nat Med. 2015;21:698–708. doi: 10.1038/nm.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Doherty TA, Baum R, Newbury RO, Yang T, Dohil R, Aquino M, et al. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J Allergy Clin Immunol. 2015;136:792–4.e3. doi: 10.1016/j.jaci.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hill DA, Artis D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–67. doi: 10.1146/annurev-immunol-030409-101330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome. 2015;3 doi: 10.1186/s40168-015-0085-6. 23-015-0085-6. eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS ONE. 2015;10:e0128346. doi: 10.1371/journal.pone.0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]