Abstract
The CTX-M-type extended-spectrum β-lactamases (ESBLs) present a serious public health threat as they have become nearly ubiquitous among clinical gram-negative pathogens, particularly the enterobacteria. To aid in the understanding and eventual control of the spread of such resistance genes, we sought to determine the diversity of CTX-M ESBLs not among clinical isolates, but in the environment, where weaker and more diverse selective pressures may allow greater enzyme diversification. This was done by examining the CTX-M diversity in municipal wastewater and urban coastal wetlands in southern California, United States, by Sanger sequencing of polymerase chain reaction amplicons. Of the five known CTX-M phylogroups (1, 2, 8, 9, and 25), only genes from groups 1 and 2 were detected in both wastewater treatment plants (WWTPs), and group 1 genes were also detected in one of the two wetlands after a winter rain. The highest relative abundance of blaCTX-M group 1 genes was in the sludge of one WWTP (2.1 × 10−4 blaCTX-M copies/16S rRNA gene copy). Gene libraries revealed surprisingly high nucleotide sequence diversity, with 157 new variants not found in GenBank, representing 99 novel amino acid sequences. Our results indicate that the resistomes of WWTPs and urban wetlands contain diverse blaCTX-M ESBLs, which may constitute a mobile reservoir of clinically relevant resistance genes.
Introduction
β-lactam antibiotics are among the most commonly preferred treatments for many bacterial infections and are the most diverse and heavily prescribed antibiotics in the world today.7,26,31,54,57 The β-lactams, predominantly penicillins, cephalosporins, and carbapenems, as well as their derivatives, share a four-membered ring that irreversibly inhibits the transpeptidases involved in bacterial peptidoglycan cross-linking, leading to cell lysis. The most common mechanism of β-lactam resistance is found in the β-lactamase enzymes,21 which hydrolyze the β-lactam ring, rendering the drug inactive. The earliest β-lactamases, such as TEM-1 and SHV-1, acted primarily as narrow-spectrum penicillinases. However, we have increasingly come to depend on cephalosporins when penicillins fail, resulting in the emergence of various cephalosporinases, known as extended-spectrum β-lactamases (ESBLs), which convey an extended spectrum of activity toward the β-lactam antibiotics.14,41
Third- and fourth-generation cephalosporins are categorized as critically important by the World Health Organization, primarily because of their essential role in combatting infections caused by multidrug-resistant enterobacteria.64 The increasing global prevalence and distribution of CTX-M ESBLs among Enterobacteriaceae have led some regions to shift away from the later-generation cephalosporins toward carbapenems. In turn, this has contributed to the emergence of dangerous highly active carbapenemases such as NDM-1.39,61
CTX-M enzymes are Ambler class A serine ESBLs, known to hydrolyze aztreonam, penicillins, and oxyimino-cephalosporins, with typically greater activity against cefotaxime than ceftazidime and limited or no activity against cefoxitin and the carbapenems.6 As of September 2015, there are currently 168 distinct CTX-M enzyme variants known (www.lahey.org/Studies/), most of which were first discovered in clinical isolates demonstrating an ESBL phenotype. CTX-M enzymes are classified into five phylogenetic clusters (groups 1, 2, 8, 9, and 25), and alleles are numbered sequentially as they are discovered and validated, irrespective of which group they are in.
The first CTX-M ESBL, CTX-M-1, was described in 1990.4 Since then, blaCTX-M genes have come to dominate the ESBL gene pool.6,14 For example, CTX-M occurrence among clinical Escherichia coli isolates from one U.S. hospital increased from 25% to 89% in a 5-year period.30 The rapid global dissemination and penetration of CTX-M threaten to undermine current antimicrobial chemotherapy strategies, which rely heavily on the third-generation cephalosporins to control infections by gram-negative bacteria.44
The blaCTX-M genes appear to have mobilized from chromosomal DNA in Kluyvera spp. to plasmids by way of mobile genetic elements such as insertion sequences and integrons.6,9,46,59 For instance, the blaCTX-M group 1 genes, believed to have originated from a chromosomal bla gene in K. cryocrescens,15 are commonly associated with the insertion sequence, ISEcp1.43 The blaCTX-M group 2 genes appear to have originated in K. ascorbata,20 while groups 8, 9, and 25 likely came from K. georgiana.6
The purpose of this study was to advance our understanding of the diversity of CTX-M ESBLs by searching not among clinical isolates, but in the environment where selective pressures may be less strong and constant, allowing for greater standing genetic variation. To avoid the inherent biases of cultivation-based methods, we used polymerase chain reaction (PCR) amplification, cloning, and sequencing to investigate CTX-M diversity in municipal wastewater and urban coastal wetlands near the adjacent cities of Tijuana, Mexico, and San Diego, United States. Our results demonstrate that considerable as yet undiscovered diversity remains to be described among the CTX-M-type ESBLs.
Materials and Methods
Site descriptions and sampling
The Famosa Slough (FAM) and the Tijuana River Estuary (TRE) are coastal wetlands in urban San Diego County, California (USA). FAM receives urban runoff, while TRE receives both urban and agricultural runoff, as well as sporadic untreated sewage from Tijuana, Mexico, particularly during the rainy season (November–March). A brief overview of these sites is given here, but a more thorough description of the two wetlands, along with detailed maps of their locations and sampling sites within them, was published by us previously.11
For this study, sterile spatulas were used to collect quadruplicate surface sediment samples (∼50 g from the top 5 mm) from two locations (sites FAM-2 and TRE-1)11 during low tide events in both the rainy season (February 2011) and the dry season (June 2012). Samples were transported to the laboratory on ice. Each 50-g sample was homogenized, but replicates were kept separate.
The Point Loma Wastewater Treatment Plant (PL WWTP) treats ∼175 million gallons of sewage per day from the greater San Diego area, including residents living within the San Diego River watershed, whose estuary includes FAM. Thus, any wastewater within the watershed that escapes containment (e.g., breaks, leaks, spills) is carried to the remaining San Diego River wetlands, including FAM, before discharge into the Pacific Ocean. The South Bay International WWTP (SBI WWTP) treats 25 million gallons of sewage per day from the greater Tijuana, Mexico, area. Much of Tijuana's municipal wastewater is directed into the Tijuana River, which flows across the international border into the United States where it is diverted and treated at the SBI WWTP before release into the ocean. Winter rains overwhelm the treatment plant and the sewage-laden storm water is allowed to flow, untreated, into the estuary and ocean. Both WWTPs are considered advanced primary treatment facilities. Operators of the treatment plants provided 1 L (0.001 m3) each from primary influent and effluent, as well as ∼100 g of sludge in May 2013.
DNA was extracted from sediment and sludge samples using the Fast DNA SPIN Kit for Soil (MP Biomedicals) according to the manufacturer's instructions. Bacteria from WWTP influent and effluent (750 mL) were either captured on 0.22-μm filters and collected with a sterile cotton swab or pelleted by centrifugation (16,000 g, 20 min), followed by DNA extraction with the same kit.
Conventional PCR, cloning, and sequencing
All extracted DNA samples were diluted to a final concentration of 50 ng/μL before conventional (end point) PCR. To ensure that the environmental DNA samples were of sufficient quality and that potential inhibitors were at subinhibitory concentrations for successful PCR, 16S rRNA genes were first amplified using primers, Bac331F and Bac797R.37 blaCTX-M genes from groups 1, 2, 8, 9, and 25 were amplified in triplicate from each environmental DNA using previously published PCR primers13,43 (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/mdr). The originally published CTX-M-F primer43 had an unintended G at the 5′ end (Yvonne Pfeifer, personal communication), which was removed in our study. Thirty cycles of PCR (annealing temp. 55°C for all primers) were carried out using Green GoTaq Master Mix (Promega) with 100 ng template DNA and 10 μg bovine serum albumin (Roche). Reaction specificity was confirmed by agarose gel electrophoresis and sequence analysis (see below).
Replicate CTX-M-positive PCR reactions from a given environmental site were combined and purified using the Wizard PCR Preps DNA Purification System (Promega), ligated into the pGEM-T Easy vector (Promega), and used to transform E. coli JM109 by heat shock according to the manufacturer's instructions. Approximately 125 putative transformants were randomly selected from each library and screened for full-length inserts by colony PCR amplification using primers, M13F and M13R (Promega), which flank the multiple cloning site on the vector (same cycling protocol as above). Amplicons were purified as described above and sequenced by the Sanger method at least once in each direction using the original amplification primers (Retrogen, Inc.).
Sequence data assembly and analyses
Nucleotide sequence chromatogram files were assembled de novo, trimmed, and manually curated using Geneious software (version 6; Biomatters [www.geneious.com/]).
All subsequent analyses were performed with MEGA5.56 Both nucleotide and amino acid sequences were aligned using the ClustalW algorithm. The best-fit evolutionary model for all sequences found in this study was identified as GTR + G (general time reversible with nonuniformity of evolutionary rates among sites fit with a discrete Gamma distribution). Phylogenetic trees were constructed using the maximum likelihood and neighbor-joining algorithms; tree topology was similar, thus only maximum likelihood trees are presented. Bootstrap analysis on these trees was performed using 1,000 replicates. Only validated CTX-M nucleotide and amino acid sequences from the Lahey Clinic (www.lahey.org/Studies/) were used as references.
A site-by-site analysis of the amino acid sequence diversity across each library was calculated using the Shannon entropy method.10 The Shannon entropy for each of the 291 positions in CTX-M was calculated using −Σi pi log(pi), where pi is the proportion of sequences in each library having amino acid i at a particular position.
In keeping with β-lactamase numbering conventions,3 CTX-M amino acid numbering in this report originates with S70 and counts forward and backward from there. Additionally, as CTX-M enzymes do not have an amino acid at position 239 relative to staphylococcal penicillinases (Karen Bush, personal communication), the sequences in this report skip from position 238 to 240. Thus, our sequences are numbered from −2 to 238 (including 0), followed by 240 to 289.
Real-time quantitative PCR
Real-time quantitative PCR (qPCR) targeting blaCTX-M group 1 genes was based on SYBR Green I chemistry as described previously.11 Briefly, DNA templates (5 μL each in triplicate) were first incubated at 95°C (10 min), followed by 35 cycles at 95°C (1 min), 60°C (1.5 min), and 72°C (0.5 min) using QuantiTect SYBR Green PCR Master Mix (Qiagen) in a Bio-Rad Chromo4 real-time PCR instrument. Fluorescence was read after every cycle. Primers targeting blaCTX-M group1, generating a 232-bp product, were obtained from Ellem et al.16
The number of copies of blaCTX-M genes was normalized to both total DNA mass and to the number of copies of 16S rRNA genes present. For qPCR targeting the 16S rRNA gene, primers, Bac331F and Bac797R, were applied with the same chemistry and thermocycling protocol as for blaCTX-M.
Quantitation standards were prepared from dilutions of a blaCTX-M and a 16S rRNA gene previously amplified, cloned, and sequenced from TRE sediments. Ten-fold serial dilutions of PCR products ranging in concentration from 101 to 106 copies per μL (blaCTX-M) and 105 to 109 copies per μL (16S rRNA gene) generated slopes of −3.469 (blaCTX-M) and −3.376 (16S rRNA gene) and R2 values of 0.978 (blaCTX-M) and 0.996 (16S rRNA genes). Melting curve analysis of the PCR products suggested that no primer dimers were formed and that the amplicons were uniform in size and sequence for both primer sets.
Nucleotide accession numbers
The blaCTX-M group 1 nucleotide sequences reported in this study have been deposited in the GenBank database5 (www.ncbi.nlm.nih.gov/genbank) under the accession numbers, KJ802486-KJ802720.
Results
Distribution and abundance of blaCTX-M ESBLs in wetlands and WWTPs
We determined the presence and relative abundance of genes encoding CTX-M enzymes in multiple environmental samples by both conventional PCR with group-specific primers and by qPCR (Tables 1 and 2). Only blaCTX-M genes from two of the five known groups were detected. In the TRE sediments, only group 1 genes were detected and only during the wet season. In the FAM sediments, no blaCTX-M genes were detected in either the wet or dry seasons. Both WWTPs contained blaCTX-M genes from groups 1 and 2. Genes from blaCTX-M groups 8, 9, and 25 were not detected in any of the samples. The presence/absence of blaCTX-M group 1 was confirmed by qPCR, with measurable quantities only observed in TRE (wet) sediments as well as the two WWTPs (Table 2). The TRE wet season sediments showed a relative abundance of 4.39 ± 2.08 × 10−5 (mean ± 1 standard deviation, n = 4). Among the other qPCR-positive sites, relative gene abundance as expressed in copies blaCTX-M/copy 16S rRNA gene ranged from 2.85 × 10−5 in the effluent of the SBI WWTP to 2.13 × 10−4 in the raw sludge of the same plant. Interestingly, at the PL WWTP, blaCTX-M was only detected in the effluent.
Table 1.
Detection of blaCTX-M ESBLs in Wetlands and Wastewater Treatment Plants by Conventional PCR
| Site | Group 1 | Group 2 | Groups 8/25a | Group 9 |
|---|---|---|---|---|
| TRE (wet) | + | − | − | − |
| TRE (dry) | − | − | − | − |
| FAM (wet) | − | − | − | − |
| FAM (dry) | − | − | − | − |
| SBI WWTPb | + | + | − | − |
| PL WWTPb | + | + | − | − |
Positive PCR reactions were confirmed by cloning and sequencing.
The primers used cannot distinguish group 8 from group 25.
If any fraction from the wastewater treatment process was positive, the site is reported as positive.
ESBLs, extended-spectrum β-lactamases; FAM, Famosa Slough; PCR, polymerase chain reaction; PL WWTP, Point Loma Wastewater Treatment Plant; SBI WWTP, South Bay International Wastewater Treatment Plant; TRE, Tijuana River Estuary; wet/dry, wet season or dry season wetland samples.
Table 2.
Abundance of 16S rRNA Genes and blaCTX-M Group 1 ESBLs in Wetlands and Wastewater Treatment Plants Estimated by Real-Time Quantitative PCR
| Site | blaCTX-Mgp1 | 16S rRNA genes | blaCTX-M/16S rRNA gene |
|---|---|---|---|
| TRE (wet) | 1.34 ± 0.619 | 3.06 × 104 ± 1.44 × 103 | 4.39 × 10−5 ± 2.08 × 10−5 |
| TRE (dry) | BDL | 3.29 × 103 ± 3.34 × 103 | |
| FAM (wet) | BDL | 1.58 × 104 ± 6.05 × 103 | |
| FAM (dry) | BDL | 1.27 × 104 ± 5.67 × 103 | |
| SBI-E | 0.550 | 1.93 × 104 | 2.85 × 10−5 |
| SBI-R | 0.423 | 7.71 × 103 | 5.48 × 10−5 |
| SBI-RS | 1.98 | 9.30 × 103 | 2.13 × 10−4 |
| SBI-L | 0.596 | 1.12 × 104 | 5.32 × 10−5 |
| PL-E | 14.4 | 7.80 × 104 | 1.85 × 10−4 |
| PL-R | BDL | 1.87 × 104 | |
| PL-RS | BDL | 2.81 × 104 | |
| PL-DS | BDL | 1.13 × 104 |
Abundance of blaCTX-M group 1 and 16S rRNA genes reported as copies per nanogram of environmental DNA. Values for TRE and FAM samples represent mean ± 1 standard deviation (n = 4). Values for SBI and PL samples represent the means of triplicate measurements, but not true replication (thus, no variance is reported).
BDL, below the detection limit of 0.2 copies per nanogram DNA.
DS, digested sludge; E, effluent; L, lime-treated sludge; R, raw influent; RS, raw sludge.
Diversity of blaCTX-M group 1 nucleotide sequences
To assess the diversity of blaCTX-M group 1 genes, PCR amplicons were cloned and sequenced. The PCR primers used to target blaCTX-M group 1 anneal within ISEcp1, the most common genetic environment for group 1 genes, at locations flanking the β-lactamase gene, covering the entire protein-coding region.43 Clone libraries were prepared from blaCTX-M group 1 PCR amplicons from TRE wet season sediments, SBI WWTP (all samples combined), and PL WWTP (only the effluent, sample PL-E, was PCR positive) (Table 3). A total of 237 blaCTX-M group 1 sequences were obtained from all three libraries. One hundred sixty-five of the 237 clones were nonredundant across the libraries, representing 157 novel nucleotide sequences that were not reported previously in the GenBank database. When compared with the 51 validated blaCTX-M group 1 sequences in the Lahey Clinic database, these new variants dramatically increase our knowledge of blaCTX-M group 1 diversity. Furthermore, the low frequency of redundant sequence variants in the libraries suggests that considerable blaCTX-M diversity remains to be discovered in these environmental compartments.
Table 3.
Summary of blaCTX-M Group 1 Clone Libraries from Wetlands and WWTPs
| Environment | Lib sizea | Unique (nt)b | Unique (aa)b | Novel (nt)c | Novel (aa)c |
|---|---|---|---|---|---|
| TRE (wet) | 80 | 61 | 45 | 56 | 41 |
| SBI WWTPd | 84 | 55 | 36 | 49 | 30 |
| PL WWTPe | 73 | 63 | 38 | 57 | 32 |
| TOTALf | 237 | 179 | 119 | 164 | 103 |
| Combinedg | 165 | 108 | 157 | 99 |
Total number of cloned environmental blaCTX-M group 1 DNA sequences analyzed from each library.
Nonredundant within a given library at either the nucleotide (nt) or amino acid (aa) level.
Nonredundant in the GenBank database at either the (nt) or (aa) level.
All positive PCR reactions from the SBI WWTP were pooled to create a single library.
Only the effluent of the PL WWTP was PCR positive.
Sum of sequences across all three libraries.
TOTAL less any clones that were observed in multiple libraries.
Lib, Library.
BLAST alignments of the 237 full-length nucleotide sequences confirmed their affiliation with blaCTX-M group 1, revealing the greatest similarities (0–8 mismatched nucleotides) to 11 reference sequences from the Lahey Clinic database (Fig. 1). Close variants of blaCTX-M-3 were the most common in all three libraries, comprising 37.5% (TRE), 43.8% (PL WWTP), and 76.2% (SBI WWTP) of the cloned sequences from each. Other high-frequency sequences included variants of blaCTX-M-30 and blaCTX-M-36. Variants of the globally ubiquitous blaCTX-M-1 and blaCTX-M-15 were only observed in the PL WWTP effluent where they comprised 2.7% and 15.1% of the library, respectively. Phylogenetic trees were constructed using the cloned blaCTX-M nucleotide sequences from the environment as well as group 1 representatives from GenBank (Supplementary Fig. S1). The results indicated that the environmental DNA sequences were more similar to plasmid-encoded clinical ESBLs than to genes from the chromosomes of their putative original hosts, Kluyvera spp.
FIG. 1.
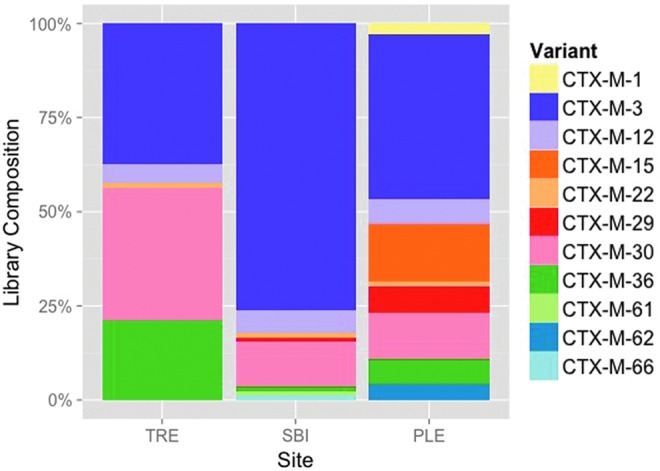
Distribution of blaCTX-M group 1 cloned gene sequences in each library grouped according to the most similar reference sequence variants. Only validated blaCTX-M sequences from www.lahey.org/Studies/ were used as references. TRE, Tijuana River Estuary; PLE, Point Loma Wastewater Treatment Plant effluent; SBI, South Bay International Wastewater Treatment Plant. Color images available online at www.liebertpub.com/mdr
Diversity of CTX-M group 1 amino acid sequences
While the nucleotide sequence diversity may reflect both selection and genetic drift, it is the amino acid sequences that can suggest the potential for functional variability. Supplementary Table S2 identifies the putative amino acid substitutions predicted for all 179 unique blaCTX-M group 1 cloned sequences from the environment. Over 80% of the sequences in the environmental libraries were most similar to one of three CTX-M reference sequences: CTX-M-3, -30, and -36 (Fig. 1 and Supplementary Table S2). The most abundant environmental sequences were 100% identical to CTX-M-3 (Supplementary Table S2). CTX-M-30 and CTX-M-36 were not only most abundant in TRE but also present in the other two libraries. Relatively few environmental sequences were similar to the globally ubiquitous CTX-M-1- and CTX-M-15-like enzymes (Fig. 1 and Supplementary Table S2).
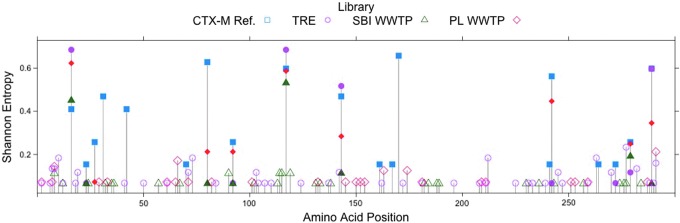
Another approach to understanding the effects of selective pressures on CTX-M enzyme diversity is to examine the distributions and frequencies of substitutions in the different libraries. Plots of the position-by-position variability for each library were constructed by calculating the Shannon Entropy at each amino acid site (Fig. 2). It should be noted that the number of unique amino acid sequences in a population appears to be independent of the number of sequences sampled (Table 3; compare Lib size to Unique [aa]). The Shannon entropy at a particular amino acid position is zero only if the position is conserved in the library. Otherwise, the heights of the bars indicate the relative frequencies of (generally) biallelic substitutions at each position (i.e., short bars indicate a rare allele and long bars indicate two common alleles). Figure 2 indicates that rare variants are seen more often in the TRE population than in the others, while the reference sequences show the greatest position-specific variability, but the least variation along the gene. Of the 19 variable amino acid positions within the CTX-M group 1 reference library, 10 are found within these samples and 6 are found in all 3 samples. Figure 3 summarizes the number of shared variable amino acid positions between libraries. The majority of the variable amino acid positions are unique within each library. These results suggest that evolution of the clinical CTX-M enzymes has been more constrained, possibly due to stronger and more persistent selective pressures, than that of the environmental proteins, where selection may be weak and infrequent.
FIG. 2.
Shannon entropy of each amino acid position calculated across each library and among all CTX-M group 1 reference sequences from the Lahey Clinic (www.lahey.org/Studies/). Bars greater than zero indicate the presence of two or more alleles, with greater height of the bar generally indicating greater frequency of the rare allele and rarely the presence of more than two alleles in the library. Solid icons indicate amino acid positions with variability shared with the CTX-M group 1 reference sequences. Figure created using R (R Foundation for Statistical Computing, Vienna, Austria [www.R-project.org/]). Amino acid numbering is absolute (i.e., not referenced to S70 and position 239 is included). Color images available online at www.liebertpub.com/mdr
FIG. 3.
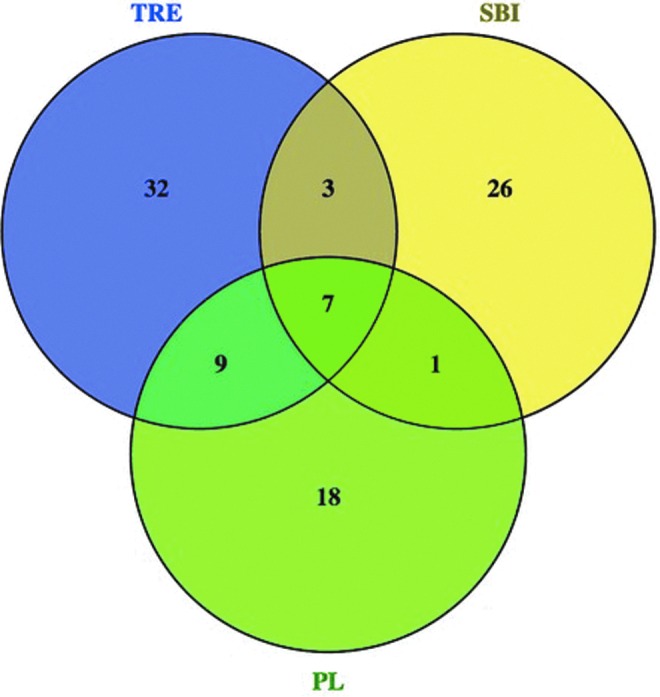
Venn diagram of variable amino acid positions shared between environmental blaCTX-M gene libraries. Figure created using Venny (Venny. An interactive tool for comparing lists with Venn diagrams [bioinfogp.cnb.csic.es/tools/venny/index.html]). Color images available online at www.liebertpub.com/mdr
Discussion
Epidemiological studies of antibiotic-resistant bacteria and their resistance genes have been focused primarily on their spread among clinical and community pathogens.29 However, the role of environmental reservoirs in their dissemination and penetration remains unclear. It has been proposed that the human gut microbiome may be the primary location for selection of antibiotic resistance traits.42,47,51 Accordingly, it is not surprising that wastewater is a rich reservoir of resistance genes and the mobile genetic elements that disseminate them.48,55 The ultimate release of wastewater, treated or untreated, into the environment can cause spread and accumulation of resistance genes in downstream receptors.11,28,60 Additionally, evidence suggests that resistance genes may evolve further after release into environmental reservoirs,32,62 possibly generating novel phenotypes. Furthermore, water, soil, and sediment containing various anthropogenic contaminants may act as mixing vessels, enabling diverse microbial genomes to participate in horizontal transfer, transposition, and recombination, potentially producing novel genetic combinations.
Our understanding of the diversity of blaCTX-M genes and the ESBL enzymes they encode has come largely from those identified in third-generation cephalosporin-resistant clinical strains isolated after therapeutic failure. In this study, we chose to broadly survey and describe diverse blaCTX-M genes that have been released into the environment using PCR amplification of environmental DNA samples.
Group 1 blaCTX-M genes were detected in sediments of the TRE after a winter storm and in every fraction of the wastewater treatment process at the SBI WWTP upstream of the wetlands (Table 2). They were notably absent, however, from FAM sediments as well as all treatment fractions, except the effluent of the PL WWTP. Others have reported a similar increase in the fraction of resistant bacteria or resistance genes in WWTP effluent,12,27 including bacteria resistant to cephalosporin antibiotics,25 suggesting that current wastewater treatment methods may select for an increase in some resistance factors.
The most striking result of this study was the unexpectedly high diversity of group 1 genes and putative proteins in wastewater and coastal sediment. Phylogenetic analysis indicated closer affiliation with plasmid-encoded blaCTX-M sequences than chromosomal sequences found in Kluyvera spp. (Supplementary Fig. S1), suggesting dispersion of clinically relevant plasmids into the environment. Thus, WWTPs and associated environmental receptors can serve as reservoirs of diverse, undescribed, and uncharacterized blaCTX-M genes that represent a potential public health risk.
The prevalent enzyme variants in the environment differed from those most commonly reported in clinical settings. From clinical studies, it is believed that CTX-M-14 and CTX-M-15 are the two most broadly dispersed alleles.9,63 Other nearly ubiquitous alleles include CTX-M-1, CTX-M-3, and CTX-M-9. In this study of environmental CTX-M, however, only CTX-M-3 was common in our samples (Fig. 1). CTX-M-1 variants were rare and CTX-M-15 variants were only seen at low frequency in the samples from one of the WWTPs. Group 9 alleles, including CTX-M-9 and CTX-M-14, were not detected at all. CTX-M-3-like enzymes represented over 50% of the combined libraries and more than 75% of the cloned sequences in samples from the SBI WWTP treating Mexican sewage.
The first of the CTX-M-3-like enzymes was described in 1988 in an E. coli strain isolated from the feces of a presumably healthy dog.33 Originally named FEC-1, the β-lactamase was later recognized as a variant of CTX-M-3,6 which was first reported clinically in Citrobacter freundii isolates in Poland in 1998.19 Like CTX-M-3, FEC-1 provided resistance to various oxyimino-cephalosporins and was susceptible to β-lactamase inhibitors. CTX-M-3 was one of the first variants to truly globalize in the 1990s and 2000s9 and appears, from our study, to have gained a firm foothold in the southern California and northern Mexico populations.
The scarcity of CTX-M-1 and CTX-M-15 in our samples was unexpected given their broad global distribution. CTX-M-1, first identified in 1990 and originally named MEN-1, was the first non-TEM, non-SHV ESBL capable of hydrolyzing oxyimino-cephalosporins.4 In this study, only two variants of CTX-M-1 were identified and only in the PL WWTP (Fig. 1 and Supplementary Table S2), suggesting that they may not be as prevalent in this region as elsewhere in the world. Like the CTX-M-1 variants, CTX-M-15 was only found in the PL WWTP, but not in the SBI WWTP or the TRE sediments (Fig. 1). Originally identified in India in the late 1990s,22 CTX-M-15 is believed to be the most widespread and penetrating variant of CTX-M enzymes in the world today.9 It is particularly surprising that it was only identified in the WWTP treating wastewater from the United States and not the WWTP treating Mexican wastewater or the nearby wetlands. Similarly, the small percentage of the clones related to CTX-M-15 was not expected. While no studies of CTX-M diversity in the cities of San Diego, California, and Tijuana, Mexico, are available for comparison, CTX-M-15 appears to be the predominant CTX-M variant in the state of California2,40 and across Mexico.18,35,36,49,50
The low representation of the globally predominant variant of CTX-M in the WWTPs and wetlands may reflect regional variation in its distribution or it may be due to a unique genetic context that impeded detection by our primers. Recall that the PCR primers used in this study anneal to sequences within the ISEcp1 element, thus only allowing for the amplification of those blaCTX-M genes located within this genetic context. If blaCTX-M-15 was associated with another element, such as an integron, or if the ISEcp1 site was interrupted by another insertion sequence such as IS26, it would have gone undetected. Thus, this method may have underestimated the diversity present in the environmental samples under investigation.
Relatively little is known about the global distribution of the remaining CTX-M group 1 variants reported in Fig. 1 and Supplementary Table S2. Most reports of these alleles have come from Asia,24,52,58,65,66 with a few isolated accounts from Africa,23 Europe,8,34 Australia,67 South America,17 Russia,45 and Canada.1 Thus, it would appear that this study represents the first report of many CTX-M group 1 alleles in the United States.
Of the 108 predicted CTX-M amino acid sequences in the environmental libraries, 99 have not been reported previously in the GenBank database (Table 3). Of particular interest are three sequences from TRE sediments that appear to have substitutions at the catalytically conserved S70 residue (Supplementary Table S2). β-lactamases form an acyl–enzyme intermediate with the antibiotic at S70 with the help of K73 and E166.53 While all 109 proteins deduced from the cloned blaCTX-M nucleotide sequences have conserved catalytic residues at K73 and E166, three enzymes from the TRE, sequences TRE-1.09, TRE-1.69, and TRE-1.79, have predicted substitutions at the catalytically critical S70 position (Supplementary Table S2). In addition to a limited number of substitutions away from the active site, clone TRE-1.09 has an S70N substitution and both TRE-1.69 and TRE-1.79 have an S70I substitution. Until enzyme activity studies have been performed, however, it is impossible to speculate on the functional effects of these variants. They may have novel activities, or they may simply have been subjected to the genetic drift that appears to characterize the TRE wetland genes.
A larger fraction of the blaCTX-M nucleotide sequences in the TRE wet season library translated into distinct amino acid sequences (0.74) relative to the libraries constructed from the WWTPs (0.65 and 0.60 in the SBI and PL WWTPs, respectively) (unique amino acids/unique nucleotides; Supplementary Table S2). The large number of variable amino acid positions in the Shannon entropy analysis also supports this assertion (Figs. 2 and 3). Of the 51 variable amino acid positions in TRE, only 10 are shared with SBI, while 16 are shared with PL, which is unexpected because SBI spills into TRE during the rainy season, but there is no direct connection between TRE and PL. Furthermore, ∼63% of the variable positions within TRE are unique to TRE, 70% of those in SBI are unique, and 51% of those in PL. Thus, the majority of the variability within each library is unique to the library. Since we were unable to detect blaCTX-M genes in the wetlands during the dry season, it is unclear where this diversification is taking place. It is possible that many of the blaCTX-M genes washing into the wetlands during the rainy season are originating somewhere other than the SBI WWTP. It is also possible that limitations of the PCR methods (e.g., inhibitors, primer specificity) masked the presence of blaCTX-M genes during the dry season.
In the current study, we estimated the density of the blaCTX-M group 1 genes in TRE rainy season sediments to be 4.39 × 10−5 copies/copy 16S rRNA gene, which was nearly 500-fold less abundant than our previously reported estimates of the quinolone resistance gene qnrA at the same location.11 Although the qnrA genes were apparently more abundant than the blaCTX-M group 1 genes, the opposite was true of their diversity. In this study, 165 nonredundant blaCTX-M group 1 DNA sequences were seen of 237 cloned genes (70%), whereas only 10 nonredundant qnrA DNA sequences were seen of 89 clones (11%). While the previous study may have uncovered the majority of the qnrA diversity present in the sediments, the current results suggest that considerable as yet unidentified blaCTX-M diversity remains to be discovered.
Conclusion
The results of this study indicate that the diversity of CTX-M enzymes known from clinical isolates vastly underestimates the diversity that exists in nature. A better understanding of this diversity is a key component of any comprehensive plan to mitigate the further spread of CTX-M ESBLs among clinically relevant bacteria.
Supplementary Material
Acknowledgments
This work was supported by a Public Health Service grant 1R15GM102995-01A1 from the National Institute of General Medical Sciences and grant MRI-0619057 from the National Science Foundation. C.J.B. was also supported by a Public Health Service grant P20 GM103408. The authors also wish to thank the PLNU Alumni Association, the Wesleyan Center for 21st Century Studies, and the PLNU Office of the Provost for financial support.
The authors are grateful to Steve Smullen (South Bay International WWTP) and Tom Burger (Point Loma WWTP) for providing wastewater and sludge samples, and Lori Charett (City of San Diego) and Brian Collins (U.S. Fish and Wildlife Service) for access to the wetlands. DNA sequencing was performed by Retrogen, Inc.
Disclosure Statement
No competing financial interests exist.
References
- 1.Abdalhamid B., Pitout J.D.D., Moland E.S., and Hanson N.D. 2004. Community-onset disease caused by Citrobacter freundii producing a novel CTX-M β-lactamase, CTX-M-30, in Canada. Antimicrob. Agents Chemother. 48:4435–4437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams-Sapper S., Sergeevna-Selezneva J., Tartof S., Raphael E., Diep B.A., Perdreau-Remington F., and Riley L.W. 2012. Globally dispersed mobile drug-resistance genes in Gram-negative bacterial isolates from patients with bloodstream infections in a US urban general hospital. J. Med. Microbiol. 61(Pt7):968–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambler R. P., Coulson A.F.W., Frère J.-M., Ghuysen J.-M., Joris B., Forsman M., Levesque R.C., Tiraby G., and Waley S.G. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauernfeind A., Grimm H., and Schweighart S. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294–298 [DOI] [PubMed] [Google Scholar]
- 5.Benson D.A., Clark K., Karsch-Mizrachi I., Lipman D.J., Ostell J., and Sayers E.W. 2014. GenBank. Nucl. Acids Res. 42:D32–D37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonnet R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botica M.V., Botica I., Stamenić V., Andrasević A.T., Kern J., and Spehar S.S. 2013. Antibiotic prescription rate for upper respiratory tract infections and risks for unnecessary prescription in Croatia. Coll. Antropol. 37:449–54 [PubMed] [Google Scholar]
- 8.Brasme L., Nordmann P., Fidel F., Lartigue M.F., Bajolet O., Poirel L., et al. 2007. Incidence of class A extended-spectrum beta-lactamases in Champagne-Ardenne (France): a 1 year prospective study. J. Antimicrob. Chemother. 60:956–964 [DOI] [PubMed] [Google Scholar]
- 9.Cantón R., González-Alba J.M., and Galán J.C. 2012. CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cover T., and Thomas J. 1991. Elements of Information Theory. John Wiley and Sons, New York [Google Scholar]
- 11.Cummings D.E., Archer K.F., Arriola D.J., Baker P.A., Faucett K.G., Laroya J.B., Pfeil K.L., Ryan C.R., Ryan K.R.U., and Zuill D.E. 2011. Broad-dissemination of plasmid-mediated quinolone resistance genes in sediments of two urban coastal wetlands. Environ. Sci. Technol. 45:447–454 [DOI] [PubMed] [Google Scholar]
- 12.Czekalski N., Berthold T., Caucci S., Egli A., and Bürgmann H. 2012. Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into Lake Geneva, Switzerland. Front. Microbiol. 3:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallenne C., Da Costa A., Decré D., Favier C., and Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 14.D'Andrea M.M., Arena F., Pallecchi L., and Rossolini G.M. 2013. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int. J. Med. Microbiol. 303:305–317 [DOI] [PubMed] [Google Scholar]
- 15.Decousser J.W., Poirel L., and Nordmann P. 2001. Characterization of a chromosomally encoded extended-spectrum class A β-lactamase from Kluyvera cryocrescens. Antimicrob. Agents Chemother. 45:3595–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellem J., Partridge S.R., and Iredell J.R. 2011. Efficient direct extended-spectrum β-lactamase detection by multiplex real-time PCR: accurate assignment of phenotypes by use of a limited set of genetic markers. J. Clin. Micorbiol. 49:3074–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Espinal P., Garza-Ramos U., Reyna F., Rojas-Moreno T., Sanchez-Perez A., Carrillo B., Martinez P., Mattar S., and Silva-Sanchez J. 2010. Identification of SHV-type and CTX-M-12 extended-spectrum beta-lactamases (ESBLs) in multiresistant Enterobacteriaceae from Colombian Caribbean hospitals. J. Chemother. 22:160–164 [DOI] [PubMed] [Google Scholar]
- 18.Garza-González E., Mendoza-Ibarra S. I., Llaca-Díaz J. M., and Gonzalez G. M. 2011. Molecular characterization and antimicrobial susceptibility of extended-spectrum β-lactamase-producing Enterobacteriaceae isolates at a tertiary-care entre in Monterrey, Mexico. J. Med. Microbiol. 60(Pt1):84–90 [DOI] [PubMed] [Google Scholar]
- 19.Gniadkowski M., Schneider I., Paułucha A., Jungwirth R., Mikiewicz B., and Bauernfeind A. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing β-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humeniuk C., Arlet G., Gautier V., Grimont P., Labia R., and Philipon A. 2002. β-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacoby G.A., and Munoz-Price L.S. 2005. The new β-lactamases. N. Engl. J. Med. 352:380–91 [DOI] [PubMed] [Google Scholar]
- 22.Karim A., Poirel L., Nagarajan S., and Nordmann P. 2001. Plasmid-mediate extended-spectrum β-lactamase (CTX-M-3-like) from India and gene association with insertion sequence ISEcp1. FEMS Microbiol. Lett. 201:237–241 [DOI] [PubMed] [Google Scholar]
- 23.Kariuki S., Corkill J.E., Revathi G., Musoke R., and Hart C.A. 2001. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob. Agents Chemother. 45:2141–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J.S., Kim J., Kim J.S., Jeon S.E., Oh K.H., Cho S.H., Kang Y.H., Han S.Y., and Chung G.T. 2014. Characterization of CTX-M-type extended-spectrum β-lactamase-producing diarrheagenic Escherichia coli isolates in the Republic of Korea during 2008–2011. J. Microbiol. Biotechnol. 24:421–426 [DOI] [PubMed] [Google Scholar]
- 25.Korzeniewska E., and Harnisz M. 2013. Extended-spectrum beta-lactamase (ESBL)-positive Enterobacteriaceae in municipal sewage and their emission to the environment. J. Environ. Management 128:904–911 [DOI] [PubMed] [Google Scholar]
- 26.Kumar R., Indira K., Rizvi A., Rizvi T., and Jeyaseelan L. 2008. Antibiotic prescribing practices in primary and secondary health care facilities in Uttar Pradesh, India. J. Clin. Pharm. Ther. 33:625–34 [DOI] [PubMed] [Google Scholar]
- 27.Laht M., Karkman A., Voolaid V., Ritz C., Tenson T., Virta M., and Kisand V. 2014. Abundances of tetracycline, sulphonamide and beta-lactam antibiotic resistance genes in conventional wastewater treatment plants (WWTPs) with different waste load. PLoS One 9:e103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaPara T.M., Burch T.R., McNamara P.J., Tan D.T., Yan M., and Eichmiller J.J. 2011. Tertiary-treated municipal wastewater is a significant point source of antibiotic resistance genes into Duluth-Superior Harbor. Environ. Sci. Technol. 45:9543–9549 [DOI] [PubMed] [Google Scholar]
- 29.Lautenbach E., and Perencevich E.N. 2014. Addressing the emergence and impact of multidrug-resistant Gram-negative organisms: a critical focus for the next decade. Infect. Control Hosp. Epidemiol. 35:333–335 [DOI] [PubMed] [Google Scholar]
- 30.Lewis J.S., Herrera M., Wickes B., Patterson J.E., and Jorgensen J.H. 2007. First report of the emergence of CTX-M-type extended-spectrum β-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Anitmicrob. Agents Chemother. 51:4015–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marra F., Monnet D.L., Patrick D.M., Chong M., Brandt C.T., Winters M., Kaltoft M.S., Tyrrell G.J., Lovgren M., and Bowie W.R. 2007. A comparison of antibiotic use in children between Canada and Denmark. Ann. Pharmacother. 41:659–66 [DOI] [PubMed] [Google Scholar]
- 32.Martinez J.L. 2009. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. Biol. Sci. 276:2521–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto Y., Ikeda F., Kamimura T., Yokota Y., and Mine Y. 1988. Novel plasmid-mediated β-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 32:1243–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendonça N., Ferreira E., Louro D., ARParticipants SIP, and Caniça M. 2009. Molecular epidemiology and antimicrobial susceptibility of extended- and broad-spectrum beta-lactamase-producing Klebsiella pneumoniae isolated in Portugal. Int. J. Antimicrob. Agents. 34:29–37 [DOI] [PubMed] [Google Scholar]
- 35.Morfín-Otero R., Mendoza-Olazarán S., Silva-Sánchez J., Rodríguez-Noriega E., Llaca-Díaz J., Tinoco-Carillo P., Petersen L., López P., Reyna-Flores F., Alcantar-Curiel D., Garza-Ramos U., and Garza-González E. 2013. Characterization of Enterobacteriaceae isolates obtained from a tertiary care hospital in Mexico, which produces extended-spectrum β-lactamase. Microb. Drug Resist. 19:378–383 [DOI] [PubMed] [Google Scholar]
- 36.Muro S., Garza-González E., Camacho-Ortiz A., González G. M., Llaca- Díaz J. M., Bosques F., and Rositas F. 2012. Risk factors associated with extended-spectrum β-lactamase-producing Enterobacteriaceae nosocomial bloodstream infections in a tertiary care hospital: a clinical and molecular analysis. Chemother. 58:217–224 [DOI] [PubMed] [Google Scholar]
- 37.Nadkarni M.A., Martin F.E., Jacques N.A., and Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiol. 148:257–266 [DOI] [PubMed] [Google Scholar]
- 38.Nei M., and Kumar S. 2000. Molecular Evolution and Phylogenetics. Oxford University Press, New York [Google Scholar]
- 39.Nordmann P., Poirel L., Toleman M.A., and Walsh T.R. 2011. Does broad-spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria?. J. Antimicrob. Chemother. 66:689–692 [DOI] [PubMed] [Google Scholar]
- 40.Pannaraj P.S., Bard J.D., Cerini C., and Weissman S.J. 2015. Pediatric carbapenem-resistant Enterobacteriaceae in Los Angeles, California, a high-prevalence region in the United States. Pediatr. Infect. Dis. J. 34:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson D.L., and Bonomo R.A. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Penders J., Stobberingh E.E., Savelkoul P.H.M., and Wolffs P.F.G. 2013. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeifer Y., Matten J., and Rabsch W. 2009. Salmonella enterica serovar Typhi with CTX-M β-lactamase, Germany. Emerg. Infect. Dis. 15:1533–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitout J.D., and Laupland K.B. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8:159–66 [DOI] [PubMed] [Google Scholar]
- 45.Priamchuk S.D., Fursova N.K., Abaev I.V., Iu.Kovalev N., Shishkova N.A., Pecherskikh E.I., Korobova O.V., Astashkin E.I., Pachkunov D.M., Kruglov A.N., Ivanov D.V., Sidorenko S.V., Svetoch E.E., and Diatlov I.A. 2010. Genetic determinants of antibacterial resistance among nosocomial Escherichia coli, Klebsiella spp., and Enteorbacter spp. isolates collected in Russia within 2003–2007. Antibiot. Khimioter. 55:3–10 [PubMed] [Google Scholar]
- 46.Rossolini G.M., D'Andrea M.M., and Mugnaioli C. 2007. The spread of CTX-M-type extended-spectrum β-lactamases. Clin. Microbiol. Infect. 14(suppl. 1):33–41 [DOI] [PubMed] [Google Scholar]
- 47.Salyers A.A., Gupta A., and Wang Y. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. TRENDS Microbiol. 12:412–416 [DOI] [PubMed] [Google Scholar]
- 48.Schlüter A., Szczepanowski R., Pühler A., and Top E.M. 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31:449–477 [DOI] [PubMed] [Google Scholar]
- 49.Silva-Sánchez J., Cruz-Trujillo E., Barrios H., Reyna-Flores F., Sánchez-Pérez A.; Bacterial Resistance Consortium, and Garza-Ramos U. 2013. Characterization of plasmid-mediated quinolone resistance (PMQR) genes in extended-spectrum β-lactamase-producing Enterobacteriaceae pediatric clinical isolates in Mexico. PLoS One. 8:e77968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva-Sánchez J., Barrios H., Reyna-Flores F., Bello-Diaz M., Sánchez-Pérez A., Rojas T.; Bacterial Resistance Consortium, and Garza-Ramos U. 2011. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae isolates in Mexico. Microb. Drug Resist. 17:497–505 [DOI] [PubMed] [Google Scholar]
- 51.Sommer M.O.A., Dantas G., and Church G.M. 2009. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325:1128–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song W., Lee H., Lee K., Jeong S.H., Bae I.K., Kim J.S., and Kwak H.S. 2009. CTX-M-14 and CTX-M-15 enzymes are the dominant type of extended-spectrum beta-lactamase in clinical isolates of Escherichia coli from Korea. J. Med. Microbiol. 58:261–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strynadka N.C.J., Adachi H., Jensen S.E., Johns K., Sielecki A., Betzel C., Sutoh K., and James M.N.G. 1992. Molecular structure of the acyl-enzyme intermediate in β-lactam hydrolysis at 1.7 Å resolution. Nature 359:700–705 [DOI] [PubMed] [Google Scholar]
- 54.Suda K.J., Hicks L.A., Roberts R.M., Hunkler J.R., and Taylor T.H. 2014. Trends and seasonal variation in outpatient antibiotic prescription rates in the United States, 2006 to 2010. Antimicrob. Agents Chemother. 58:2763–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szczepanowski R., Linke B., Krahn I., Gartemann K.-H., Götzkow T., Eichler W., Pühler A., and Schlüter A. 2009. Detection of 140 clinically relevant antibiotic resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiol. 155:2306–2319 [DOI] [PubMed] [Google Scholar]
- 56.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., and Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thu T.A., Rahman M., Coffin S., Harun-Or-Rashid M., Sakamoto J., and Hung N.V. 2012. Antibiotic use in Vietnamese hospitals: a multicenter point-prevalence study. Am. J. Infect. Control. 40:840–844 [DOI] [PubMed] [Google Scholar]
- 58.Tian G.B., Wang H.N., Zhang A.Y., Fan W.Q., Xu C.W., Zeng B., Guan Z.B., and Zou L.K. 2012. Detection of clinically important β-lactamases in commensal Escherichia coli of human and swine origin in western China. J. Med. Microbiol. 61:233–238 [DOI] [PubMed] [Google Scholar]
- 59.Tzouvelekis L.S., Tzelepi E., Tassios P.T., and Legakis N.J. 2000. CTX-M-type β-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents. 14:137–142 [DOI] [PubMed] [Google Scholar]
- 60.Uyaguari M.I., Fichot E.B., Scott G.I., and Norman R.S. 2011. Characterization and quantitation of a novel β-lactamase gene found in a waste water treatment facility and the surrounding coastal ecosystem. Appl. Environ. Microbiol. 77:8226–8233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walsh T.R., Weeks J., Livermore D.M., and Toleman M.A. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11:355–362 [DOI] [PubMed] [Google Scholar]
- 62.Wellington E.M.H., Boxall A.B.A., Cross P., Feil E.J., Gaze W.H., Hawkey P.M., Johnson-Rollings A.S., Jones D.L., Lee N.M., Otten W., Thomas C.M., and Williams A.P. 2013. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13:155–165 [DOI] [PubMed] [Google Scholar]
- 63.Woerther P.L., Burdet C., Chachaty E., and Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin. Microbiol. Rev. 26:744–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.World Health Organization. 2011. Critically Important Antimicrobials for Human Medicine. 3rd revision. WHO Press, Geneva, Switzerland [Google Scholar]
- 65.Wu J.-J., Chen H.-M., Ko W.-C., Wu H.M., Tsai S.-H., and Yan J.-J. 2008. Prevalence of extended-spectrum β-lactamases in Proteus mirabilis in a Taiwanese university hospital, 1999–2005: identification of a novel CTX-M enzyme (CTX-M-66). Diagn. Microbiol. Infect. Dis. 60:169–175 [DOI] [PubMed] [Google Scholar]
- 66.Yu Y., Ji S., Chen Y., Zhou W., Wei Z., Li L., and Ma Y. 2007. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J. Infect. 54:53–57 [DOI] [PubMed] [Google Scholar]
- 67.Zong A., Partridge S.R., Thomas L., and Iredell J.R. 2008. Dominance of blaCTX-M within an Australian extended-spectrum β-lactamase gene pool. Antimicrob. Agents Chemother. 52:4198–4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.