Abstract
Deep-sequencing of bacterial transcriptomes using RNA-Seq technology has made it possible to identify small non-coding RNAs, RNA molecules which regulate gene expression in response to changing environments, on a genome-wide scale in an ever-increasing range of prokaryotes. However, a simple and reliable automated method for identifying sRNA candidates in these large datasets is lacking. Here, after generating a transcriptome from an exponential phase culture of Mycobacterium tuberculosis H37Rv, we developed and validated an automated method for the genome-wide identification of sRNA candidate-containing regions within RNA-Seq datasets based on the analysis of the characteristics of reads coverage maps. We identified 192 novel candidate sRNA-encoding regions in intergenic regions and 664 RNA transcripts transcribed from regions antisense (as) to open reading frames (ORF), which bear the characteristics of asRNAs, and validated 28 of these novel sRNA-encoding regions by northern blotting. Our work has not only provided a simple automated method for genome-wide identification of candidate sRNA-encoding regions in RNA-Seq data, but has also uncovered many novel candidate sRNA-encoding regions in M. tuberculosis, reinforcing the view that the control of gene expression in bacteria is more complex than previously anticipated.
Keywords: RNA-Seq, transcriptome, non-coding RNA, Mycobacterium tuberculosis
Introduction
The causal agent of tuberculosis (TB), Mycobacterium tuberculosis, is one of the most ancient and successful pathogens and causes substantial mortality worldwide [1]. New approaches in both drug and vaccine development, urgently needed to reduce the global burden of TB, will be greatly facilitated by a greater understanding of the basic biology underlying the response of this pathogen to the harsh environments it encounters during pathogenesis. An increasing number of studies have demonstrated that bacterial small non-coding RNAs (sRNAs) play an important role in the fine-tuning of gene expression in response to environmental changes [2,3] and stress conditions [4,5] such as those encountered during host invasion, but the study of sRNAs in M. tuberculosis is still in its infancy.
sRNAs are classified according to their genomic location: antisense (as) or cis-encoded sRNAs which are oriented antisense to their target protein-encoding mRNA, and intergenic region (IGR) or trans-encoded sRNAs which are located in IGRs and act in trans on targets that can be located at some distance from the sRNA [4,6,7]. Earlier studies on sRNAs in M. tuberculosis using classical experimental and computational approaches [8–10] identified 26 sRNAs, including IGR sRNAs, asRNAs and potential regulatory untranslated regions (UTRs). Deep sequencing has subsequently been applied to identify novel sRNA candidates in M. tuberculosis [11–13], increasing the total number of sRNAs identified in M. tuberculosis to 59 (reviewed in Ref. [14]).
Although RNA-Seq has now been applied for genome-wide identification of sRNAs in a broad range of bacteria, a simple, effective, and systematic method for identifying potential sRNA candidates for further investigation within RNA-Seq datasets is not yet available. Arnvig et al. [15], for example, identified sRNA candidates in M. tuberculosis based on visual examination of reads coverage maps after mapping RNA-Seq data to the H37Rv reference genome. Cortes et al. [16] later applied the dRNA-Seq approach for global mapping of transcription start sites (TSS) to M. tuberculosis. In addition to providing information on TSS and regulatory mechanisms associated with already annotated genes or operons, dRNA-Seq also facilitates the discovery of novel regulatory elements, including sRNAs expressed from IGRs or antisense to ORFs [16–19]. Cortes et al. [16] identified 4164 TSS, and reported that 8% of IGRs had a TSS that was not associated with a previously reported ncRNA. They identified 758 annotated CDSs that were associated with antisense TSS, but did not provide additional verification of these transcripts. Pellin et al. [12], also working on M. tuberculosis, developed a more systematic method for genome-scale identification of sRNA candidates, which integrates examination of the characteristics of RNA-Seq reads coverage maps and ‘conservation maps’ (based on the conservation of nucleotides in each candidate sRNA sequence across the mycobacteria). However, only a relatively small portion (258/1373; 19%) of the candidate sRNAs identified using their method could be validated in a later study using microarrays [13].
Here, we describe and validate an alternative method for genome-wide identification of sRNA candidate-encoding regions within RNA-Seq datasets based on sequencing libraries of different lengths to capture the whole cellular transcriptome and subsequent analysis of the characteristics of reads coverage maps.
Materials and Methods
Bacterial strains and culture conditions
Mycobacterium tuberculosis H37Rv was cultured in Middlebrook 7H9 medium (Difco Laboratories, Detroit, USA) with 10% oleic acid-albumin-dextrose-catalase (OADC) enrichment. Cultures were harvested at an OD600 of about 0.6.
RNA extraction and purification
Total RNA was extracted using a FastRNA™ Pro Blue kit (MP Biomedicals, Santa Ana, USA). Frozen cell pellets were thawed on ice, resuspended in 1 ml RNApro™ solution, lysed using a FastPrep-24 (6.0 M/s, 45 s) (MP Biomedicals), and centrifuged at 12,000 g for 10 min at 4°C according to the manufacturer’s instructions. RNA was then precipitated at −80°C overnight using an equal volume of isopropanol, 1/10 volume of 3 M sodium acetate (pH 5.3), and 2 μl glycogen (5 mg/ml). RNA was quantified after every manipulation step using a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Houston, USA). Samples containing RNA were treated with RNA-free DNase I (Fermentas, Glen Burnie, USA) to remove any residual DNA and purified by TRIzol (Invitrogen, Carlsbad, USA) extraction according to the manufacturer’s instructions. Ten micrograms of total RNA was subject to further purification; 16S and 23S rRNA were depleted using a MICROBExpress Kit (Ambion, Foster City, USA) according to the manufacturer’s instructions. Purified RNA samples were finally resuspended in 6 μl of RNase-free water and stored at −80°C until required.
cDNA library construction and sequencing
Total RNA was separated into three fractions of different lengths using 6% 7 M urea polyacrylamide gel electrophoresis (PAGE). Fraction 1 contained RNAs of 18–40 nt in length, fraction 2 contained RNAs of 40–80 nt in length, and fraction 3 contained RNAs of 80–140 nt in length. The remaining rRNA-depleted RNA (fraction 4) was sheared into smaller fragments of ~140 nt at elevated temperatures using divalent cations.
To prepare for Illumina single-end sequencing, RNAs from fractions 1 and 2 were first isolated from total RNA by PAGE. After ligation of 5′ and 3′ RNA adapters, cDNA constructs were prepared by reverse transcription followed by low cycle polymerase chain reaction (PCR) amplification (initial denaturation at 98°C for 30 s, followed by 15 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 15 s, and then 72°C for 10 min). PCR products were collected by gel purification and sequenced on an Illumina Genome Analyzer II (Illumina, San Diego, USA) platform according to the manufacturer’s instructions (45 cycles for 18–40 nt library and 81 cycles for 40–80 nt library).
For Illumina paired-end sequencing, strand-specific libraries were constructed for RNA from fractions 3 (80–140 nt) and 4 (>140 nt) (isolated by PAGE as above) according to a previously reported dUTP method [20] with some modifications. Briefly, 200 ng of total RNA was fragmented by heating at 98°C for 40 min in 0.2 mM sodium citrate, pH 6.4 (Ambion). After concentration to 5 μl, the RNA was mixed with 3 μg random hexamers, incubated at 70°C for 10 min, and then chilled on ice. First-strand cDNA was synthesized with this RNA primer mix by adding 4 μl of 5× first-strand buffer, 2 μl of 100 mM DTT, 1 μl of 10 mM dNTPs, 4 μg of actinomycin D, 200 U of SuperScript III and 20 U of SUPERase-In, incubating at room temperature for 10 min followed by incubation at 55°C for 1 h. The first-strand cDNA was purified by PCIA extraction (twice) and ethanol precipitation with 0.1 volume 5 M ammonium acetate to remove dNTPs, and then resuspended in 104 μl H2O. Second-strand cDNA was synthesized by adding 4 μl of 5× first-strand buffer, 2 μl of 100 mM DTT, 4 μl of 10 mM dNTPs [dTTP was replaced by dUTP (Sigma, St Louis, USA)], 30 μl of 5× second-strand buffer, 40 U of Escherichia coli DNA polymerase, 10 U of E. coli DNA ligase, and 2 U of E. coli RNase H, and then incubating at 16 °C for 2 h. The paired-end library for Illumina sequencing was prepared using a TruSeq mRNA kit (Illumina) according to the instructions provided, with the following modifications: (i) five times less adapter mix was added to the cDNAs; (ii) the dsDNA product was incubated with 1 U Uracil-DNA Glycosylase (Invitrogen) at 37°C for 15 min followed by 5 min of incubation at 95°C before PCR to excise dUTP; (iii) PCR was performed with Phusion High-Fidelity DNA polymerase (New England Biolabs, Ipswich, USA) with GC buffer and 2 M betaine; and (iv) PCR primers were removed using 1.8 volumes of AMPure beads (Beckman-Coulter, Brea, USA). Sequencing of 91 bp paired-end reads was then carried out on the Illumina Genome Analyzer II following the manufacturer’s protocols, and image analysis and base calling were performed on the Genome Analyzer pipeline (Illumina) using default parameters.
Processing and analysis of sequenced reads
Trimmomatic (v 0.32) [21] was used to filter reads as follows: (i) adapter sequences were removed; (ii) reads with a mean Phred quality score <15 or a Phred quality score <15 across the whole read (4-bp sliding window) were discarded; (iii) reads <16 nt in length were discarded. Trimmed reads were then aligned to the M. tuberculosis H37Rv reference genome (NC_000962.3) using Bowtie2 [22] with default parameters, allowing up to two mismatches. Strand-specific reads coverage maps were generated for each genomic base based on Bowtie2 alignments [22] using SAMtools [23] and BEDTools [24]. Sequence coverage at each genomic position was visualized with integrated genome browser (IGB) [25].
Identification of candidate sRNA-encoding regions
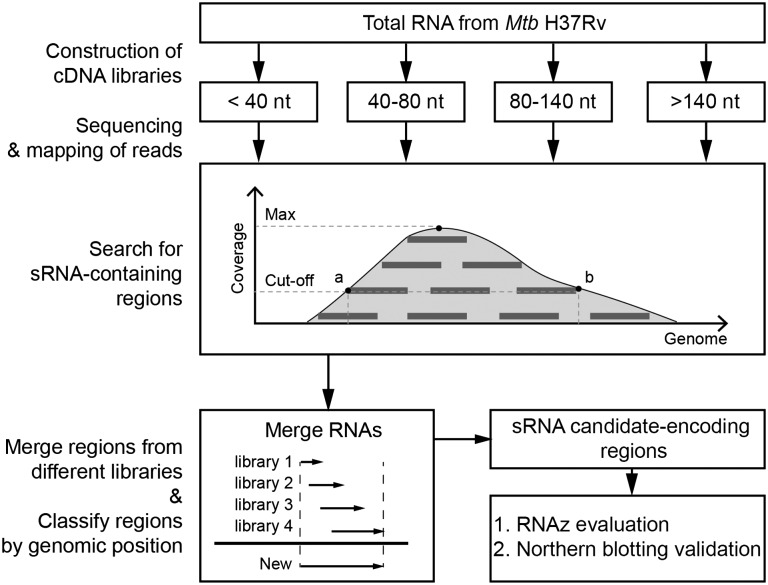
Candidate sRNA-encoding regions were identified based on coverage depth at each base. Each library was screened for putative sRNA-encoding regions with a customized Perl script (unpublished work) according to the following principles: (i) coverage depth at each base should be higher than a certain cut-off value (100); (ii) mean coverage of the selected regions should be at least twice as high as the cut-off value; (iii) 5′/3′ ends of the transcript should be at least 100/60 bp, respectively, from the nearest protein-encoding ORFs (to avoid misclassification of UTRs as sRNAs); and (iv) transcripts should be ≥40 nt and ≤500 nt in length. sRNA candidate-encoding regions from the four libraries, which had overlapping genome coordinates, were then merged and the distance between the 5′ and 3′ ends of the merged fragments and the nearest coding ORFs was reexamined. Those that met criteria (iii) were retained as candidate sRNA-encoding regions.
To determine whether any highly expressed sRNA-encoding regions were misclassified as UTRs (i.e. the 5′/3′ ends were <100/60 bp, respectively, from the nearest protein-encoding ORFs), further rounds of screening were performed at progressively higher coverage depth cut-off values (1000, 10,000, and 100,000). sRNA candidate-encoding regions from further rounds of screening that met the above criteria were added to the list of candidate sRNA-encoding regions.
sRNA secondary structure conservation analysis
RNAz [26], a program for predicting structurally conserved and thermodynamically stable RNA secondary structures, was used with default parameters to detect conserved secondary structures in sRNA-encoding regions. An RNAz score of 0.5 was considered to indicate secondary structure conservation.
Calculation of the expression of sRNA-encoding regions
Raw counts for putative sRNA candidate-encoding regions were obtained for each library using featureCounts (version 1.4.6-p3) [27], with the following parameters: (i) –M: multi-mapping reads/fragments were counted N times, where N is the number of reported mapping locations; (ii) --fraction: a fractional count 1/N was generated for each multi-mapping read; and (iii) –O: multi-overlapping features were allowed. Transcripts per million (TPM) was calculated for each sRNA-encoding region as described by Wagner et al. [28].
Northern blotting
Northern blotting was performed as described by Miotto et al. [13]. Oligonucleotides used are shown in Supplementary Table S1.
Analysis of nucleotide conservation in candidate sRNA-encoding regions across Mycobacterium spp.
Conservation of candidate sRNA-encoding region nucleotide sequences was assessed across 65 complete mycobacterial genomes (Supplementary Table S2) downloaded from the NCBI ftp server (ftp://ftp.ncbi.nlm.nih.gov/genomes/archive/old_refseq/Bacteria/; August 1, 2015) using Blast-Like Alignment Tool v 36 (BLAT) [29] according to Shinhara et al. [30]. An sRNA-encoding region was considered ‘conserved’ in another organism if its sequence had: (i) an E-value of <0.01; (ii) >70% identity with the sRNA sequence in the other organism; and (iii) its length was >70% of the length of the sRNA sequence in the other organism. Conservation scores were calculated using the formula: [(nucleotide match-length) × (nucleotide identity/100)]/(nucleotide length of candidate sRNA). A given candidate sRNA-encoding region was considered to be conserved within the Mycobacterium tuberculosis complex (MTBC) if its nucleotide sequence was conserved in >90% of the MTBC strains and <10% of the non-MTBC strains included in this analysis. Hierarchical clustering of the conservation scores of candidate sRNAs was performed using the pheatmap package in R [31].
Results and Discussion
Method development and validation—detection of tRNAs and previously reported sRNAs
The success of any RNA-Seq strategy for global identification of sRNAs will depend on whether: (i) the initial cDNA library that captures the transcriptome is an accurate reflection of cellular sRNA expression and (ii) the analytical approach taken is able to accurately identify sRNAs within the dataset generated. A variety of cDNA library construction strategies for transcriptome capture are available; however, using standard library construction procedures a portion of cellular RNAs (sRNA sequencing: 18–40 nt and mRNA sequencing: >200 nt) cannot be detected in a single cDNA library due to technical limitations such as RNA ligation efficiency and the read length of Illumina sequencers. Our goal was to capture a transcriptome that reflected the sRNA composition of bacterial cells as closely as possible, bearing RNA processing and degradation in mind [32]. We thus chose to split total RNA into fractions according to RNA length and prepare cDNA libraries of different lengths (18–40 nt, 40–80 nt, 80–140 nt, and >140 nt) in order to capture the full length of all RNAs within the bacterial cell. We then designed a systematic strategy for analyzing the RNA-Seq data captured in this way from an M. tuberculosis H37Rv transcriptome (a profile of which is presented in Table 1) to automatically select putative sRNA-encoding regions (see ‘Materials and Methods’ section; Fig. 1) based on selection parameters that describe the likely length (40–500 nt), expression characteristics (coverage depth at each base of >100 and a mean coverage of >200), and genomic location (5′/3′ ends >100/60 bp, respectively, from flanking protein-encoding ORFs) of sRNAs. To take RNA processing and degradation into consideration, we merged putative sRNA-encoding regions from the four cDNA libraries according to genome coordinates in order to determine their full length, based on the fact that RNAs of different lengths derived from the same transcript were likely present within the transcriptome. Having retrieved putative sRNA-encoding regions in this automated manner, we characterized each candidate region using the RNAz algorithm [26], which identifies RNA structures based on both structural conservation and thermodynamic stability, and nucleotide conservation analysis [30] to assess its conservation among Mycobacterium spp. Selected sRNA candidate-encoding regions were subsequently validated by northern blotting.
Table 1.
RNA sequencing profiles for an exponential phase culture of M. tuberculosis H37Rv
| Reads a which map to | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Library | Total mapped reads | rRNA | Antisense to rRNA | tRNA | Antisense to tRNA | mRNAb | Antisense to mRNAb | IGR | Known sRNAc | Novel sRNAd |
| 18–40 nt | 10.33 | 0.51 | 0.04 | 1.15 | 0.04 | 3.60 | 2.36 | 2.63 | 0.85 | 2.05 |
| 40–80 nt | 13.03 | 1.14 | 0.00 | 0.56 | 0.00 | 7.44 | 1.15 | 2.74 | 0.72 | 1.82 |
| 80–140 nt | 4.27 | 2.26 | 0.00 | 0.41 | 0.00 | 0.54 | 0.16 | 0.90 | 0.29 | 0.35 |
| >140 nt | 14.94 | 11.79 | 0.02 | 0.00 | 0.02 | 1.87 | 0.24 | 0.98 | 0.31 | 0.57 |
| % of total mapped reads | ||||||||||
| 18–40 nt | 4.93 | 0.39 | 11.10 | 0.39 | 34.85 | 22.84 | 25.50 | 8.24 | 19.83 | |
| 40–80 nt | 8.72 | 0.01 | 4.29 | 0.01 | 57.14 | 8.81 | 21.02 | 5.52 | 13.96 | |
| 80–140 nt | 52.84 | 0.01 | 9.62 | 0.01 | 12.70 | 3.82 | 20.99 | 6.76 | 8.15 | |
| >140 nt | 78.94 | 0.16 | 0.02 | 0.16 | 12.54 | 1.61 | 6.57 | 2.07 | 3.84 | |
aNumber of reads given in millions (the read count for reads that mapped to multiple sites was divided equally between the sites).
bProtein CDSs in the reference genome.
cFifty nine previously reported sRNAs (M. tuberculosis H37Rv).
dEight hundred and fifty six novel candidate sRNA-encoding regions identified using our approach.
Figure 1.
Outline of our procedure for identifying sRNA candidate-encoding regions After constructing and sequencing cDNA libraries containing RNAs (>18 nt) of different lengths, sequenced reads from each library were mapped to the H37Rv reference genome. Reads coverage maps were generated and a cut-off value (>100) was used to identify putative 5′ and 3′ ends of sRNA candidate-encoding regions in each library. sRNA candidate-encoding regions from the four libraries with overlapping genome coordinates were merged and then classified as putative antisense (as) or intergenic (IGR) sRNA candidate-encoding regions, depending on their location in the genome. Candidates were evaluated using RNAz [26]. Forty sRNA candidate-encoding regions were selected for validation by northern blotting.
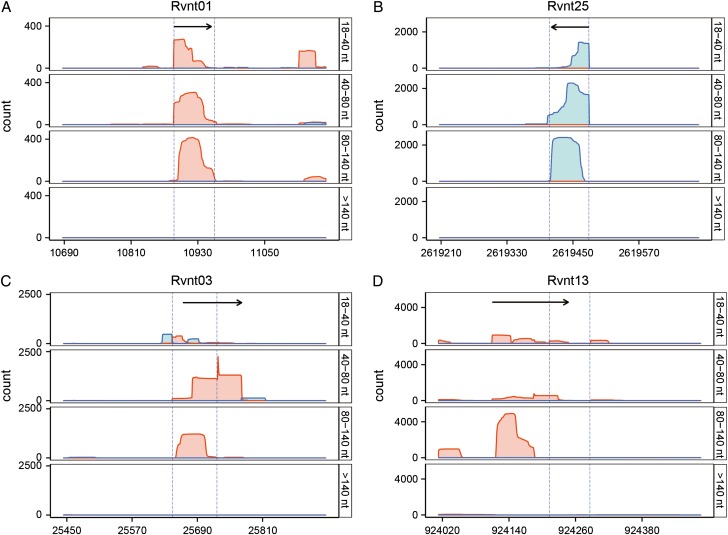
The parameters and analytical process used in our method were optimized by testing their ability to accurately retrieve M. tuberculosis tRNA coding sequences, known to be stable and conserved, and 59 previously reported M. tuberculosis sRNAs, from our RNA-Seq dataset. The expression of all 45 tRNAs was detected in our transcriptome, and the 5′ and/or 3′ ends of 35 of the 45 tRNAs were found to be identical with genome annotations (±3 bp; Fig. 2 and Supplementary Table S3). Expression of 37 of the 59 previously reported sRNAs was also detected (Fig. 3 and Supplementary Table S4). Visual examination of reads coverage maps using IGB [25] indicated that the expression of 19 of the 22 remaining previously reported sRNAs was not captured in any of the libraries, and they were thus likely not expressed in the growth phase or under the culture conditions sampled in this study (Fig. 4). Twelve of the 37 previously reported sRNAs detected here had coding regions that were too close to flanking genes (<60/100 bp) and thus did not meet our criteria for identifying sRNA-encoding regions. The eight sRNAs whose 5′ end genome coordinates were previously determined by rapid amplification of cDNA end (RACE) were detected with good accuracy (5′ to ±1 bp and 3′ to ±15 bp) (Supplementary Fig. S1). For example, MTS0997 (named as ncRv11264Ac here according to the nomenclature proposed in Ref. [33]), one of three sRNAs involved in pathogenesis identified by Arnvig et al. [15] and reported to be a 116 nt transcript, was identified by our analytical method as a 117 nt sRNA whose 5′ end was identical to that previously reported.
Figure 2.
Reads coverage maps of four representative tRNA coding regions The coverage maps from the four RNA-Seq libraries are for RVNt01 (A), Rvnt25 (B), Rvnt03 (C) and Rvnt13 (D). Red: + strand; blue: – strand. Dashed lines indicate the predicted 5′/3′ ends of the tRNAs, black arrows indicate the tRNAs identified using the approach used in this study. The 5′ or 3′ ends of 35/45 tRNAs were almost identical to their genome annotations (±3 bp, Supplementary Table S3).
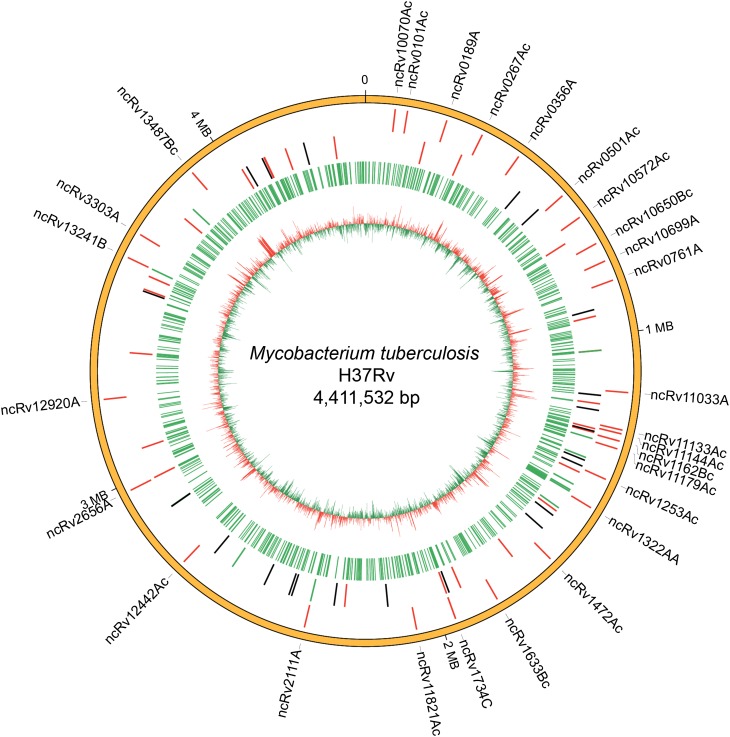
Figure 3.
Distribution of sRNA-encoding regions identified in this study on the M. tuberculosis H37Rv chromosome Novel sRNA-encoding regions validated by northern blotting are indicated on the outermost ring. Ring 2 shows the genomic positions of previously reported sRNAs (H37Rv); red: 25 sRNAs validated using our approach; green: 12 sRNAs that were expressed but did not meet the criteria here to be classified as sRNAs; black: sRNAs not identified here due to very low abundance of RNA-Seq reads. Ring 3 shows all the sRNA-encoding regions identified using our analytical approach. The innermost ring shows the distribution of high (red) and low (green) GC content across the genome.
Figure 4.
Box-plot showing the expression of previously reported sRNAs in H37Rv More than two-thirds (37/59) of the previously reported highly expressed sRNAs were detected using the methodology developed here. (A) The 25 previously reported sRNAs detected here had an average TPM of 1574.1. (B) The 12 other previously reported sRNAs, which did not meet the criteria here for classification as sRNAs, had an average TPM of 127.7. (C) The 20 previously reported sRNAs not detected here had an average TPM of 0.94. Low expression of these 20 sRNAs was the main reason that they were not detected using our approach.
The value of our strategy of using RNA libraries of different lengths and then merging putative sRNA-encoding regions from these libraries can be seen using MTS2823 (named ncRv13661B here) as an example (Supplementary Fig. S1). Its 5′ end was mapped to genome position 4,100,669 by Arnvig et al. [15] and reported to be 300 nt in length. Here, visual examination of RNA-Seq reads from <140 nt libraries showed that they mapped to both ends of the ncRv13661B encoding region and formed multiple short peaks (from 4,100,684 to 4,100,979) that could easily be mistakenly called as sRNAs if only one library was considered. Reads from the >140 nt library, however, were distributed in the middle of the ncRv13661B encoding region and formed one peak (from 4,100,692 to 4,100,950), connecting the peaks observed in the other libraries. Merging the sRNA-encoding regions from all four cDNA libraries yielded an sRNA-encoding region whose full length was 296 nt (consistent with the band observed by northern blotting), and its 5′ end mapped to 4,100,684. RNA-Seq reads for the ncRv11264Ac-encoding region mentioned above showed a similar distribution (Supplementary Fig. S1). Generally speaking, information derived from the <140 nt libraries can be used to determine the 5′/3′ borders of longer (approximately >100 nt) sRNA-encoding regions, and that from the >140 nt library to determine their full length (i.e. to determine whether adjacent peaks in reads maps are derived from the same or different sRNA-encoding regions).
During method optimization, we noted that some previously reported sRNAs that did not meet our sRNA genomic location criteria were highly expressed and well-validated sRNAs. As UTR length varies with gene, and cut-off values are therefore somewhat arbitrary, we added further rounds of selection at progressively higher reads coverage cut-off values to detect such highly expressed sRNAs more accurately in the RNA-Seq dataset. For example, a 919 nt candidate sRNA-containing region that overlapped with Rv1535 was predicted using a cut-off value of 100, but was not classified as an sRNA-encoding region due to this overlap. When the cut-off value was raised to 10,000, we identified a 50 nt sRNA-encoding region, named here as ncRv11534A, transcribed from the Rv1534-Rv1535 intergenic region. This sRNA was previously reported by Miotto et al. [13] and validated by northern blotting (Supplementary Table S4). Further rounds of selection thus improved the accuracy of our approach and prevented misclassification of sRNAs.
Multiple isoforms of the one sRNA were evident in some cases. For example, two isoforms were identified in the B11 encoding region (ncRv13660Ac: 237 nt; ncRv13660Bc: 83 nt; Supplementary Fig. S1 and Table S4), previously reported to produce an sRNA of 93 nt in length [8]. Interestingly, the expression of the ~80 nt sRNA-encoding regions (ncRv13660Bc) was 140 folds more than that of ncRv13660Ac (237 nt) (Supplementary Table S5), implying differences in the regulation or processing of these transcripts. Multiple bands (~80, ~100, ~240) were also detected by northern blotting (Supplementary Fig. S1). These results imply that transcription of multiple sRNAs from the same region can be detected using our method. Further in-depth analysis is required to unravel the functions of different isoforms and the mechanisms underlying this complex pattern of regulation.
The above results suggest that our method can accurately detect the expression of sRNA-encoding regions, giving a good indication of the likely 5′ and 3′ ends, and can also detect the presence of sRNAs of different lengths transcribed from the same location. Accurate determination of 5′ and 3′ ends, however, requires further validation by 5′ and 3′ RACE. Taken together, these validation steps demonstrate that while our method will not detect all sRNAs due to the stringent criteria used, it is useful and reliable for detecting a high proportion of the sRNA-encoding regions in RNA-Seq data.
Genome-wide prediction of sRNA-encoding regions in M. tuberculosis H37Rv
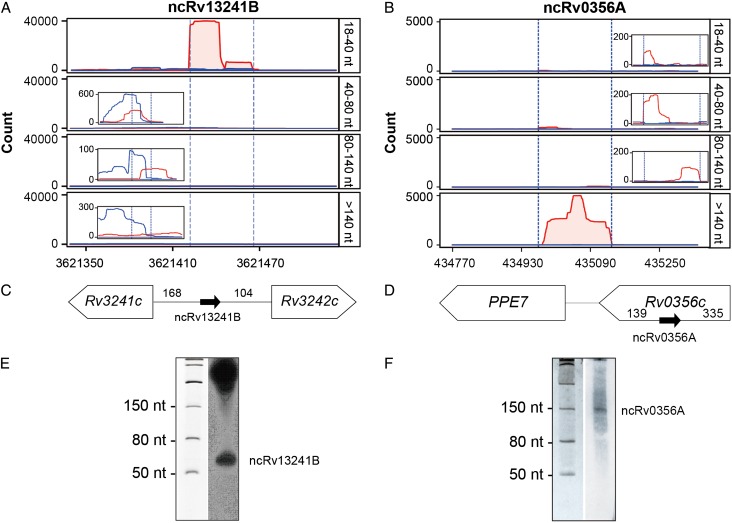
We applied the above approach to detect sRNA-encoding regions in an M. tuberculosis H37Rv transcriptome, discovering 883 putative sRNA-encoding regions (including 27, which correspond to the 25 previously reported sRNAs discussed above; Figs. 3 and 5, and Supplementary Table S5). Of the remaining 856 novel candidate sRNA-encoding regions (length: 40–449 nt; mean length: 75 nt; median length: 57 nt), 192 (22.5%) were classified as candidate IGR sRNA-encoding regions and 664 (77.5%) as candidate asRNA-encoding regions. The 10 longest sRNA-encoding regions were asRNA-encoding regions, and 86% (734 of 856) of all sRNA-encoding regions were 40–100 nt in length. Of the 20 most highly expressed sRNA-encoding regions in this transcriptome (Supplementary Table S5), 14, including ncRv2656A, the most highly expressed sRNA, were novel sRNA-encoding regions identified using our approach.
Figure 5.
Two novel sRNA-encoding regions identified in this study (A,B) Reads coverage maps from the 18–40 nt, 40–80 nt, 80–140 nt, and >140 nt RNA-Seq libraries for ncRv13241B (A) and ncRv0356A (B). Red: + strand; blue: – strand. Dashed lines indicate the predicted 5′/3′ ends of the sRNAs. (C,D) Genomic locations of the two sRNA-encoding regions. (E,F) Validation of the two sRNA candidates by northern blotting.
The high number of candidate asRNA-encoding regions detected here deserves further discussion. In recent years, pervasive antisense transcription has been widely reported in bacterial species as diverse as E. coli, Helicobacter pylori, Synechocystis sp. PCC6803, and Staphylococcus aureus [17,34–37], and it has been confirmed by independent studies using a range of techniques in some species. For example, pervasive transcription has been detected in E. coli using DNA microarray-based transcriptomics [38], promoter-reporter fusion assays [39], and RNA-Seq [34]. Although the functional significance of pervasive transcription is still a source of debate, a growing body of evidence (reviewed in Ref. [32]) suggests that RNAs resulting from pervasive transcription are more than ‘transcriptional noise’ and have important functions in gene regulation and genome evolution. Many examples of asRNAs that regulate the expression of an overlapping gene have been reported [40], some of which exert their effects even though unstable. The high proportion of reads in our dataset that map to the antisense strand (9%–23%) likely points to an abundance of ‘real’ antisense transcripts that have a regulatory function in addition to those which may be derived from transcriptional noise or RNA degradation. In the absence of further widely accepted criteria that can discriminate between functional antisense transcripts and those representing ‘noise’, it is not possible to predict exactly how many of the candidate sRNA-encoding regions identified here play a genuine functional role. While structural stability (RNAz score) and sequence conservation are not definitive criteria as such (not all known sRNAs have high RNAz scores, only 20 of the 27 regions corresponding to previously reported M. tuberculosis sRNAs identified here had RNAz scores >0.5) or are highly conserved, asRNA-encoding regions that meet these criteria are perhaps more likely to represent genuine functional asRNAs. Here, 318/856 candidate sRNA-encoding regions identified (246 asRNAs and 72 IGR sRNAs) had an RNAz score >0.5, indicating that they have highly stable secondary structures. That more than one-third of asRNA-encoding regions identified here have high RNAz scores adds further confidence to their designation as sRNAs. To evaluate nucleotide conservation, we assessed the candidate sRNA-encoding regions against 65 complete Mycobacterium spp. genomes (NCBI, 2015-08-01) using BLAT. The vast majority of the 59 previously reported M. tuberculosis H37Rv sRNAs (48, 81.4%) and 560 candidate asRNA-encoding regions, including 206 that had highly stable secondary structures, was highly conserved in the 34 MTB complex strains, but not in other Mycobacterium spp. strains (Supplementary Table S5), further demonstrating that they likely are genuine asRNA-encoding regions. In addition, comparison of our results with the M. tuberculosis TSS data reported by Cortes et al. [16] indicates that 56 candidate asRNA-encoding regions (24 of which had an RNAz score >0.5) had at least one TSS that mapped within ±1 bp of their 5′ ends (Supplementary Table S5), further indicating that they are likely to be genuine asRNAs.
We selected 40 putative sRNA candidate-encoding regions that represented the diversity within our dataset for verification by northern blotting, selecting randomly within categories to include a range of sizes (short, 23; longer, 17), types (IGR, 14; asRNA, 26), expression levels (low, 7; medium, 18; high, 15), and RNAz scores (RNAz >0.5: 21). Of these, bands corresponding to 28 (i.e. 70%) of the candidates (13 IGR sRNAs and 15 asRNAs; RNAz >0.5: 16) were detected (Supplementary Fig. S2), 22 of which corresponded closely to the predicted length of the sRNA-encoding region. For example, ncRv13241B was predicted to be 45 nt in length (Supplementary Table S5) and a ~50 nt band was detected on northern blots (Fig. 5), and ncRv0356A was predicted to be an asRNA-containing region of 171 nt in length and a band of ~150 nt was detected (Fig. 5). Discrepancies in band size for the remaining sRNAs, with bands observed generally being significantly larger than expected, raise the possibility that some relatively short sRNA candidate-encoding regions identified here may be derived from longer transcripts that have undergone processing or even degradation during RNA sample preparation. The high percentage of putative sRNA candidate-encoding regions yielding positive northern blotting results further confirms the feasibility and accuracy of the analytical approach developed here for identifying sRNA candidate-encoding regions in RNA-Seq data.
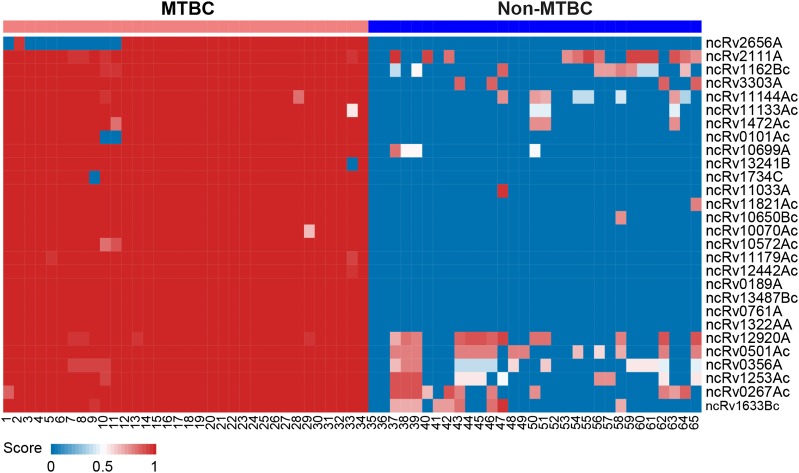
As a preliminary investigation of the putative functions of the 28 validated sRNAs, we compared their expression in H37Rv with expression data obtained in a separate RNA-Seq study on the virulent strain H37Ra (cultured under the same conditions; unpublished data). Six of the 12 sRNAs present in both the H37Rv and H37Ra transcriptomes showed significantly higher expression in H37Rv relative to H37Ra (Supplementary Table S6). For example, highly expressed sRNA ncRv2656A, transcribed from the antisense strand of Rv2656c, a gene that probably encodes a PhiRv2 phage protein, was significantly down-regulated in H37Ra to only 7.68% of its level in H37Rv (P = 2e−16). In addition, all 28 novel sRNA-encoding regions were highly conserved in the 34 MTB complex strains, but not in other Mycobacterium spp. strains (Fig. 6 and Supplementary Table S7). The possible roles of these differentially expressed sRNAs in M. tuberculosis virulence are worthy of further investigation.
Figure 6.
Sequence conservation of the 28 novel sRNA-encoding regions identified in this study Sequence conservation of the 28 novel sRNA-encoding regions across the 65 mycobacterial strains that have complete genome sequences in NCBI. Red indicates high nucleotide conservation and blue indicates no conservation. The name of each sRNA-encoding region is shown on the right and the order of the genomes is shown at the bottom (see Supplementary Tables S1 and S7).
Comparison of our approach with previous work
A previous report by Pellin et al. [12] described a bioinformatic pipeline that was used to identify sRNAs in an M. tuberculosis RNA-Seq dataset based on analysis of expression and conservation maps. Only a relatively low percentage of the sRNAs predicted (19%), however, could be validated in a later study using microarrays [13]. Here, we were able to validate 28 of 40 candidate sRNA-encoding regions (70%) examined by northern blotting. While both methods are based on the evaluation of RNA-Seq reads coverage maps, our approach, by using multiple cDNA libraries that cover the full range of RNA lengths (>18 nt) in cellular RNA and improved selection parameters, may have facilitated more accurate determination of the full length of sRNA-encoding regions. For example, 277 of the sRNA-encoding regions identified here had similar genome coordinates to 304 [267 Type A (high reads coverage), 16 Type B (medium reads coverage and conservation), and 21 Type C (highly conserved)] of the 1948 sRNA candidates identified by Pellin et al. In a number of cases, sRNA-encoding regions identified here to encode single sRNAs corresponded to several adjacent sRNA candidate regions identified by Pellin et al. For example, sRNA candidates 561, 562, and 654 were all designated here as having originated from the MTS2823 coding region: they cover a 294 nt region of the genome (4,100,684–4,100,977) almost identical to the genomic region covered by ncRv13661B (4,100,684–4,100,979) identified here (Supplementary Fig. S1, and Supplementary Table S4). In our method, the step of merging and reexamining sRNA-encoding regions with overlapping genome coordinates from the four libraries to determine the borders of sRNA-encoding regions helps to avoid the miscalling of multiple sRNAs arising from the same region.
In summary, our study has shown that systematic evaluation of reads coverage maps alone provides a simple and reliable method for identifying sRNA candidate-encoding genomic regions in M. tuberculosis and can accurately detect their expression and give a good indication of the likely 5′ and 3′ ends of candidate sRNA-encoding regions. This method should greatly simplify the task of analyzing RNA-Seq data for the presence of sRNAs. In addition, functional investigation of novel sRNA candidates identified here will help to unravel the complexity of sRNA-mediated regulation of gene expression in M. tuberculosis.
NCBI short read archive accession number
Raw sequencing data in *.bedgraph format is available under Accession Number SPR015765.
Supplementary Data
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31170132), the Chinese Academy of Sciences (Nos XDA09030308 and KSZD-EW-Z-006), the National Basic Research Program of China (Nos 2011CB910302 and 2012CB518703), and the Key Project Specialized for Infectious Diseases from the Chinese Ministry of Health (Nos 2012ZX10003002-011 and 2013ZX10003006).
Supplementary Material
References
- 1.World Health Organisation Global Tuberculosis Report. 2015.
- 2.Papenfort K, Vanderpool CK.. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 2015, 39: 362–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopf M, Hess WR.. Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol Rev 2015, 39: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgardt K, Šmídová K, Rahn H, Lochnit G, Robledo M, Evguenieva-Hackenberg E.. The stress-related, rhizobial small RNA RcsR1 destabilizes the autoinducer synthase encoding mRNA sinI in Sinorhizobium meliloti. RNA Biol 2015. 10.1080/15476286.2015.1110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billenkamp F, Peng T, Berghoff BA, Klug G.. A cluster of four homologous small RNAs modulates C1 metabolism and the pyruvate dehydrogenase complex in Rhodobacter sphaeroides under various stress conditions. J Bacteriol 2015, 197: 1839–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogel J, Wagner EG.. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol 2007, 10: 262–270. [DOI] [PubMed] [Google Scholar]
- 7.Barquist L, Vogel J.. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu Rev Genet 2015, 49: 367–394. [DOI] [PubMed] [Google Scholar]
- 8.Arnvig KB, Young DB.. Identification of small RNAs in Mycobacterium tuberculosis. Mol Microbiol 2009, 73: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiChiara JM, Contreras-Martinez LM, Livny J, Smith D, McDonough KA, Belfort M.. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res 2010, 38: 4067–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelly S, Bishai WR, Lamichhane G.. A screen for non-coding RNA in Mycobacterium tuberculosis reveals a cAMP-responsive RNA that is expressed during infection. Gene 2012, 500: 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog 2011, 7: e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellin D, Miotto P, Ambrosi A, Cirillo DM, Di Serio C.. A genome-wide identification analysis of small regulatory RNAs in Mycobacterium tuberculosis by RNA-Seq and conservation analysis. PLoS One 2012, 7: e32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miotto P, Forti F, Ambrosi A, Pellin D, Veiga DF, Balazsi G, Gennaro ML, et al. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS One 2012, 7: e51950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haning K, Cho SH, Contreras LM.. Small RNAs in mycobacteria: an unfolding story. Front Cell Infect Microbiol 2014, 4: 96 10.3389/fcimb.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog 2011, 7: e1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB.. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Reports 2013, 5: 1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma CM. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010, 464: 250–255. [DOI] [PubMed] [Google Scholar]
- 18.Bischler T, Tan HS, Nieselt K, Sharma CM.. Differential RNA-seq (dRNA-seq) for annotation of transcriptional start sites and small RNAs in Helicobacter pylori. Methods 2015, 86: 89–101. [DOI] [PubMed] [Google Scholar]
- 19.Thomason MK, Bischler T, Eisenbart SK, Förstner KU, Zhang A, Herbig A, Nieselt K, et al. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J Bacteriol 2015, 197: 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 2009, 37: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012, 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinlan AR, Hall IM.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicol JW, Helt GA, Blanchard SG Jr, Raja A, Loraine AE.. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 2009, 25: 2730–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruber AR, Neubock R, Hofacker IL, Washietl S.. The RNAz web server: prediction of thermodynamically stable and evolutionarily conserved RNA structures. Nucleic Acids Res 2007, 35: W335–W338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao Y, Smyth GK, Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 28.Wagner GP, Kin K, Lynch VJ.. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 2012, 131: 281–285. [DOI] [PubMed] [Google Scholar]
- 29.Kent WJ. BLAT─the BLAST-like alignment tool. Genome Res 2002, 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinhara A, Matsui M, Hiraoka K, Nomura W, Hirano R, Nakahigashi K, Tomita M, et al. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC Genom 2011, 12: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolde R. pheatmap: Pretty Heatmaps. R package. 2012, version 107.
- 32.Wade JT, Grainger DC.. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 2014, 12: 647–653. [DOI] [PubMed] [Google Scholar]
- 33.Lamichhane G, Arnvig KB, McDonough KA.. Definition and annotation of (myco)bacterial non-codin. RNA. Tuberculosis 2013, 93: 26–29. [DOI] [PubMed] [Google Scholar]
- 34.Dornenburg JE, Devita AM, Palumbo MJ, Wade JT.. Widespread antisense transcription in Escherichia coli. mBio 2010, 1: e00024-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitschke J. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc Natl Acad Sci USA 2011, 108: 2124–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raghavan R, Sloan DB, Ochman H.. Antisense transcription is pervasive but rarely conserved in enteric bacteria. mBio 2012, 3: e00156-00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA 2011, 108: 20172–20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selinger DW. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol 2000, 18: 1262–1268. [DOI] [PubMed] [Google Scholar]
- 39.Kawano M, Storz G, Rao BS, Rosner JL, Martin RG.. Detection of low-level promoter activity within open reading frame sequences of Escherichia coli. Nucleic Acids Res 2005, 33: 6268–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Georg J, Hess WR.. cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 2011, 75: 286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.