Abstract
Macrophages play important roles in maintaining intestinal homeostasis via their ability to orchestrate responses to the normal microbiota as well as pathogens. One of the most important steps in beginning to understand the functions of these cells is the ability to effectively isolate them from the complex intestinal environment. Here, we detail methodology for the isolation and phenotypic characterization of macrophages from the mouse small and large intestine.
Keywords: Intestine, Antigen-presenting cell, Macrophage, Dendritic cell
1 Introduction
The mammalian intestine is constantly exposed to a variety of microbes and food antigens [1]. Antigen-presenting cells (APCs), comprised primarily of macrophages and dendritic cells (DCs), are central components of the mucosal immune system that foster homeostasis in the intestine [2–9]. Just beneath the intestinal epithelial layer, the lamina propria (LP) contains a large population of macrophages that are ideally positioned to sample luminal contents and perform surveillance activity [10–12]. The positioning of macrophages in the LP suggests these cells have an important role in modulating innate and adaptive immune responses toward the microbiota [10–19]; however, the mechanisms by which these cells interact with foreign antigens and orchestrate effective immune reactivity remains as area of active investigation [20–25]. Paramount to identifying the functions of intestinal macrophages is the ability to efficiently isolate these cells from a complex cellular environment [26]. While many studies have begun to define the function of intestinal macrophages, continued advancements in the identification and characterization of these cells in the steady state and during inflammatory processes in mouse and humans are desired. Here, we summarize detailed methodology for the isolation and purification of intestinal macrophages that may be employed to investigate these important cell types and the role they play in regulating mucosal immunity.
2 Materials
2.1 Equipment
MaxQ 4450 benchtop orbital shaker; any orbital shaker with sufficient capacity should suffice.
LS MACS columns and a QuadroMACS separator (Miltenyi Biotec).
LSR II benchtop flow cytometer (BD) or other analyzer.
FACSAria II benchtop cell sorter (BD) or other sorter.
2.2 Reagents and Solutions
1× PBS, Ca2+- and Mg2+-free (CMF PBS).
Hank's balanced salt solution (HBSS) with phenol red, Ca2+-and Mg2+-free (CMF HBSS); HBSS is commonly used for isolation of intestinal immune cells.
CMF HBSS with 5 % FBS (CMF HBSS/FBS) and 2 mM EDTA.
Sodium bicarbonate.
1 M HEPES in 0.85% NaCl.
Fetal bovine serum, heat-inactivated.
0.5 M EDTA (pH 8.0).
Collagenase type VIII.
DNase I; Stock solution: 100 mg/mL.
Working Collagenase VIII/DNAse I solution: 1.5 mg/mL of collagenase VIII and 40 μg/mL of DNase I in CMF HBSS/FBS.
2.3 Antibodies and Staining Reagents
Ice-cold staining buffer: CMF PBS + 5 % FBS.
LIVE/DEAD Fixable Aqua Dead Cell Stain Kit for 405 nm excitation; Use at 1:1000 in ice-cold PBS.
CD45-PerCP mAb (30 F11); Use at 1:100 dilution in ice-cold staining buffer.
CD103-PE mAb (M290); Use at 1:100 dilution in ice-cold staining buffer.
FcγRIII/II mAb (2.4G2); Use at 1:200 dilution in ice-cold staining buffer.
CD11c-APC mAb (N418); Use at 1:100 dilution in ice-cold staining buffer.
MHC-II (I-Ab)-Alexa Fluor 700 mAb (M5/114); Use at 1:100 dilution in ice-cold staining buffer.
CD11b-eFluor 450 mAb (M1/70); Use at 1:200 dilution in ice-cold staining buffer.
F4/80-PE-Cy7 mAb (BM8); Use at 1:200 dilution in ice-cold staining buffer.
CD11b microbeads.
CD11c microbeads.
2.4 Disposable Reagents
50 mL conical tubes.
Single mesh wire strainer.
Small weigh boat.
100 μm cell strainer.
40 μm cell strainer.
5 mL polystyrene round-bottom tubes.
3 Methods
3.1 Isolation of Mouse Small and Large Intestine
- Prepare the following reagents and equipment:
- Warm Ca2+/Mg2+-free PBS (CMF PBS) to room temperature.
- Warm Ca2+/Mg2+-free HBSS with 5 % FBS (CMF HBSS/FBS) and 2 mM EDTA to room temperature.
- Warm Orbital shaker to 37 °C.
Euthanize mice and spray 70 % ethanol onto the abdomen (see Note 1).
Make a horizontal incision in abdomen with a scissor and peel back the skin and cut open the peritoneum.
Cut the intestine at the pyloric sphincter to separate the stomach from the upper small intestine (see Note 2).
Carefully remove the mesentery using forceps and cut at the ileo- cecal valve to separate the small intestine from the large intestine.
Continue to tease apart the mesentery from the large intestine and make a cut at the anal verge. Place the large intestine in CMF PBS on ice while first attending to the small intestine.
Place the entire small intestine on paper towels pre-wet with CMF PBS. Remove the Peyer's patches along the anti-mesenteric side of the small intestine and cut open longitudinally using scissors and forceps (see Note 3).
Place the intestine in a Petri dish containing room temperature CMF PBS and rapidly move the intestine around using forceps until the PBS becomes cloudy with luminal contents. Move the intestine into a new Petri dish with fresh CMF PBS and repeat this process until the PBS no longer becomes cloudy (usually 3–5 Petri dishes).
Cut the small intestine into approximately 1.5 cm pieces and place into a 50 mL conical tube containing 30 mL of pre-warmed CMF HBSS/FBS and 2 mM EDTA (see Note 4).
Cut open the colon longitudinally using scissors and wash off feces and mucus in CMF PBS at room temperature as per steps 7 and 8 of Subheading 3.1 (see Note 5).
3.2 Removal of Epithelial Layer
Place each 50 mL conical tube horizontally into an orbital shaker and stabilize using tape. Shake at 250 rpm for 20 min at 37 °C in order to begin removing epithelial cells and intraepithelial lymphocytes.
After the shaking is done, pour the contents of each 50 mL conical tube through a wire mesh strainer to recover the 1.5 cm pieces of intestine while allowing the epithelial cells and intraepithelial lymphocytes pass through. If analyses of epithelial cells and/or intraepithelial lymphocytes are desirable place a collection tube under the wire mesh strainer during this step.
-
Repeat steps 1 and 2 of Subheading 3.2 one additional time for a total of two shaking cycles.
At this stage, the intestine is stripped of most of the epithelial cell layer and is ready for tissue digestion.
3.3 Digestion of Intestinal Tissue
- Prepare the following reagents and equipment:
- Pre-warmed working Collagenase VIII/DNAse I solution (see Note 6).
- Pre-warmed CMF HBSS/FBS and 2 mM EDTA.
- Pre-warmed orbital shaker at 37 °C.
- Ice-cold CMF HBSS/FBS.
Transfer the 1.5 cm pieces of intestine to a small plastic weigh boat after dabbing away excess media using a paper towel (see Note 7).
Rapidly mince the 1.5 cm pieces of intestine using sharp dissection scissors directly in the weigh boat for approximately 10–20 s and then add minced intestine to 20 mL of pre-warmed collagenase solution (see Note 8).
Horizontally place each 50 mL conical tube into an orbital shaker and digest at 200 rpm for 10–20 min at 37 °C (see Notes 9 and 10).
Vortex remaining intestinal tissue for 5-10 s to ensure thorough dissociation and then filter through a 100 μm cell strainer directly into a 50 mL conical tube (see Note 11).
Add CMF HBSS/FBS to top off each 50 mL conical tube and then centrifuge at 1500 rpm for 5 min at 4 °C (see Note 12). Repeat this wash step once more.
After pouring off the supernatant, resuspend the cell pellets in ice- cold CMF HBSS/FBS and place samples on ice (see Notes 13 and 14).
3.4 Antibody Staining for Multi-Color Flow Cytometric Analyses
Transfer cells into a 5 mL polystyrene round-bottom (FACS) tube or if staining multiple samples use a 96-well V-bottom plate.
Wash cells twice using ice-cold CMF PBS.
Incubate samples with LIVE/DEAD Fixable Aqua Dead Cell Stain for 15 min on ice in the dark (see Note 15).
Wash cells twice in 200 μL ice-cold CMF PBS.
Block non-specific binding of antibodies to cells by using 2.4G2 anti-FcγRIII/II in ice-cold staining buffer for 10 min on ice.
Wash cells in ice-cold staining buffer.
Incubate samples with specific antibody staining cocktail for 20 min on ice in the dark.
Wash cells with ice-cold staining buffer twice and resuspend samples in 400 μL of ice-cold staining buffer and pass through 40 μm filter cap on FACS tubes.
Acquire samples on LSR II cytometer (BD) or other appropriate FACS analyzer and analyze as defined by gating strategy in Subheading 3.5 and Fig. 1.
Fig. 1.
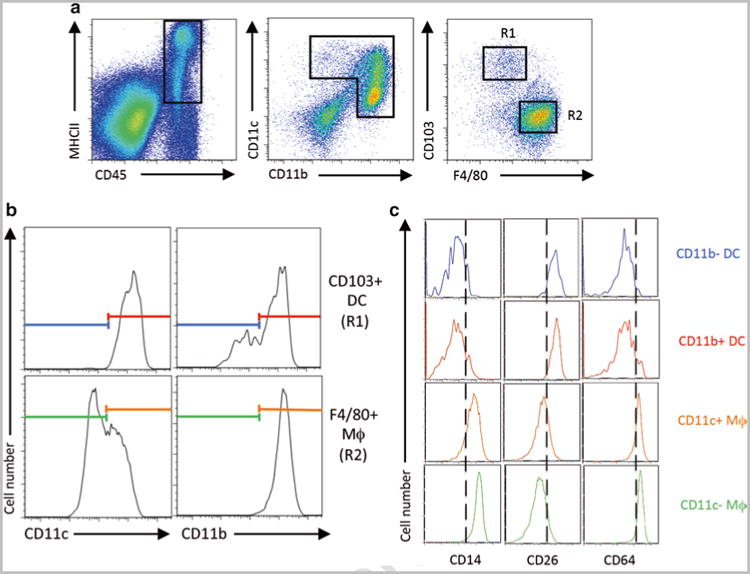
Representative analysis for macrophages and DCs in the intestine by multi-color flow cytometry. (a) Small intestinal lamina propria living CD45 + MHCII+ cells were analyzed for CD11c and CD11b expression and consequently the resulting populations were separated for CD103+ DCs (R1) and F4/80+ macrophages (R2). (b) CD103+ DCs (R1) and F4/80+ macrophages (R2) were divided by CD11c and CD11b expression. (c) Expression of CD14, CD26, and CD64 was analyzed for CD11b- DC, CD11b + DC, CD11c + macrophages, and CD11c- macrophages with colors corresponding to the populations defined in panel b
3.5 Antibody Staining for Multi-Color Flow Cytometric Analyses
After gating out dead cells, create another dot plot and further gate on CD45+ and I-Ab+ cells, which encompasses both macrophages and DCs (Fig. 1a).
Next, using a separate dot plot, gate cells that express either CD11b and/or CD11c. When this population is further analyzed for CD103 and F4/80 expression, two major subsets of cells (R1 and R2; Fig. 1a) will be evident. Cells in the R1 gate are CD103+ F4/80dull/− cells, which are mostly DCs, while cells in region R2 are CD103−F4/80+ cells, which are mostly macrophages.
Analyze regions R1 and R2 further for CD11c and CD11b expression to further differentiate macrophage and DC subsets, respectively [27]. CD103+F4/80dull/− cells in R1 universally express high levels of CD11c and are either CD11b + or CD11bdull/-. Alternatively, CD103-F4/80+ cells in R2 universally express high levels of CD11b and are either CD11chi or CD11cint (Fig. 1b).
Recently several additional markers including CD14, CD26, and CD64 have been used to discriminate intestinal macrophages and DCs [28, 29]. As shown in Fig. 1c, these markers can be used to further confirm the DC and macrophage subsets (see Note 16).
3.6 Magnetic Bead-Based Enrichment of Intestinal Macrophages
(Optional—only if more highly-purified cells are desired for functional studies).
Prepare the following reagents and equipment:
-
Ice-cold staining buffer (CMF PBS + 5 % FBS).
Incubate the cell suspension obtained from step 7 of Subheading 3.3 with CD11b and/or CD11c MACS beads according to the manufacturer's instructions (see Note 17).
Wash cells with ice-cold staining buffer followed by centrifugation.
Discard supernatant and resuspend the cell pellet in 1 mL ice-cold staining buffer and pass through a 100 μm cell strainer followed by a 40 μm cell strainer (see Note 18).
Enrich for magnetic bead-attached cells by positive selection using MACS LS magnetic column.
Repeat step 4.2 and discard supernatant.
Incubate cells with surface marker antibodies as described in Subheadings 3.4 and 3.5.
Wash magnetic bead-enriched cells twice with ice-cold staining buffer. Resuspend cell pellets in 500 μL ice-cold staining buffer without sodium azide, and pass through 40 μm cell strainer into a FACS tube (see Note 19).
Proceed to FACS-sorting on the BD ARIA II Cell Sorter to sort intestinal macrophage (or DC) subsets of interest.
4 Notes
Steps 1–9 of Subheading 3.1 must be performed as quickly as possible to minimize the extent of cell death and to achieve maximum cell yield.
Be careful when removing the mesentery from the gut wall. Hold the intestine with one pair of forceps while gently pulling the mesentery with another pair.
To easily visualize Peyer's patches for removal, begin removing them from the ileum first and push darker luminal contents toward the jejunum and duodenum in order to contrast the light Peyer's patches from the lumen.
Each small intestine after being cut into 1.5 cm pieces is placed into a single 50 mL conical tube. When isolating more than 1 small intestine, it is important to not place more than one small intestine per single 50 mL conical tube. This prevents increased cell death.
When isolating more than one colon, up to three colons can be combined per single 50 mL conical tube.
We have had success with both type VIII and type IV collagenase from Sigma-Aldrich. While optimized lots of type VIII and type IV collagenase can both provide excellent digestion of the mouse small intestine, in our experience, collagenase type IV provides superior digestion of the mouse large intestine.
Make sure that as much excess media is dabbed away to ensure the most uniform cutting of tissue.
It is important to use sharp scissor that efficiently and thoroughly mince intestinal tissue. Also, it is important that the minced intestine is added into the collagenase solution immediately before placing in the orbital shaker. If more than one intestine is being processed, minced intestines can be left in the plastic weigh boats until they are all ready to be placed into their respective collagenase tubes.
Optimizing the concentration of collagenase and the duration of tissue digestion is important to obtain optimal cell yield without compromising viability and surface antigen expression. Under- digestion yields a low total cell number while over-digestion dramatically increases the number of dead cells and the quality of the FACS staining is compromised. Overall cell yield, viability, and surface antigen expression are affected by several factors including the source, type, concentration and enzymatic activity of collagenase, the duration of tissue digestion, the degree of mincing, the temperature of media, and the status of inflammation in the intestine [26]. Based on our experience, the concentration of collagenase is usually in the range of 1–1.5 mg/mL, and optimization of the different factors mentioned above for tissue digestion is important to ensure the highest quality and reproducibility of data.
One of the most important parameters for consideration in this protocol is the manufacturer, type, and specific lot of collagenase. Extreme variability in collagenase activity exists between different manufacturers, types of collagenase, and production lots, thus the potency of digestion may vary greatly and requires optimization.
If intestinal tissue is already dissociated after this step there is no need to perform vortexing.
If a solid pellet is not observed for colon samples after centrifugation, invert the sample several times and centrifuge again for 5 min.
Pouring off the supernatant is an important checkpoint for the quality of the cell digestion. The goal is a tightly packed cell pellet with a small ring of RBCs.
In our experience, use of a ∼45/70 % Percoll gradient to further enrich for macrophages and dendritic cells leads to reduced cell yield, likely as a result of a fraction of these cells residing on top of the upper 45 % layer.
Make sure to perform live/dead staining in PBS without serum. For a positive control for dead cell staining, we place a separate aliquot of cells into a 100 °C heat block for 1 min and then place these “dead” cells on ice and add in an equal amount of cells that were not heat-killed to ensure nice positive and negative peaks for live/dead staining. For all FACS staining, unstained intestinal cells may be utilized as a negative control to assist in the proper placement of the gates to separate positive and negative populations.
DC subsets express high levels of CD26, while macrophage subsets express high levels of CD14 and CD64. Other markers such as CD68, CX3CR1, and CD272 may also be used to identify DCs and macrophages [3, 4, 30].
If simultaneous enrichment of macrophages and DCs is desired, magnetic beads targeting CD11b and CD11c can be added at the same time. We have had success using 50 μL of beads + 450 μL buffer per each intestine.
In order to prevent clogging of the MACS LS columns, it is incredibly helpful to pass cells first through a 100 μm cell strainer followed by a 40 μm cell strainer before adding cells onto the column.
For functional studies where healthy, live cells are required, it is imperative to use ice-cold staining buffer without sodium azide.
Acknowledgments
This work was supported by National Institutes of Health Grants 1R01DK097256 (to T.L.D.) and 1F30DK097904-03 (to D.G.).
Abbreviations
- APC
Antigen-presenting cells
- DC
Dendritic cell
- LI
Large intestine
- LP
Lamina propria
- SI
Small intestine
References
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Bain CC, Mowat AM. Intestinal macrophages—specialised adaptation to a unique environment. Eur J Immunol. 2011;41:2494–2498. doi: 10.1002/eji.201141714. [DOI] [PubMed] [Google Scholar]
- 3.Bain CC, Mowat AM. Macrophages in intestinal homeostasis and inflammation. Immunol Rev. 2014;260:102–117. doi: 10.1111/imr.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mowat AM, Bain CC. Mucosal macrophages in intestinal homeostasis and inflammation. J Innate Immun. 2011;3:550–564. doi: 10.1159/000329099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Platt AM, Mowat AM. Mucosal macrophages and the regulation of immune responses in the intestine. Immunol Lett. 2008;119:22–31. doi: 10.1016/j.imlet.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat Rev Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- 8.Bogunovic MF, Ginhoux J, Helft L, Shang D, et al. Origin of the lamina propria dendritic cell network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varol C, Vallon-Eberhard A, Elinav E, Aychek T, Shapira Y, et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 2009;31:502–512. doi: 10.1016/j.immuni.2009.06.025. [DOI] [PubMed] [Google Scholar]
- 10.Viney JL, Mowat AM, O'Malley JM, Williamson E, Fanger NA. Expanding dendritic cells in vivo enhances the induction of oral tolerance. J Immunol. 1998;160:5815–5825. [PubMed] [Google Scholar]
- 11.Hume DA, Robinson AP, MacPherson GG, Gordon S. The mononuclear phagocyte system of the mouse defined by immunohistochemical localization of antigen F4/80. Relationship between macrophages, Langerhans cells, reticular cells, and dendritic cells in lymphoid and hematopoietic organs. J Exp Med. 1983;158:1522–1536. doi: 10.1084/jem.158.5.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu LM, MacPherson GG. Antigen acquisition by dendritic cells: intestinal dendritic cells acquire antigen administered orally and can prime naive T cells in vivo. J Exp Med. 1993;177:1299–1307. doi: 10.1084/jem.177.5.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 14.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 17.Macpherson AJ, Lamarre A. BLySsful interactions between DCs and B cells. Nat Immunol. 2002;3:798–800. doi: 10.1038/ni0902-798. [DOI] [PubMed] [Google Scholar]
- 18.Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 19.Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115:153–162. doi: 10.1111/j.1365-2567.2005.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johansson-Lindbom B, Svensson M, Pabst O, Palmqvist C, Marquez G, Forster R, Agace WW. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J Exp Med. 2005;202:1063–1073. doi: 10.1084/jem.20051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pabst O, Bernhardt G. The puzzle of intestinal lamina propria dendritic cells and macrophages. Eur J Immunol. 2010;40:2107–2111. doi: 10.1002/eji.201040557. [DOI] [PubMed] [Google Scholar]
- 22.Schulz O, Jaensson E, Persson EK, Liu X, Worbs T, Agace WW, Pabst O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson EK, Uronen-Hansson H, Semmrich M, Rvollier A, et al. IRF4 transcription-factor- dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Atarashi K, Tanoue T, Shima T, Imaoka A. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov II, Atarashi K, Manel N, Brodie EL, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geem D, Medina-Contreras O, Kim W, Huang CS, Denning TL. Isolation and characterization of dendritic cells and macrophages from the mouse intestine. J Vis Exp. 2012:e4040. doi: 10.3791/4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, et al. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T Cell/APC ratio, source of mouse strain, and regional localization. J Immunol. 2011;187:733–742. doi: 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamoutounour S, Henri S, Lelouard H, de Bovis B, et al. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1- inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 29.Scott CL, Bain CC, Wright PB, Sichien D, et al. CCR2CD103 intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 2015;8:327–339. doi: 10.1038/mi.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerovic V, Bain CC, Mowat AM, Milling SW. Intestinal macrophages and dendritic cells: what's the difference? Trends Immunol. 2014;35:270–277. doi: 10.1016/j.it.2014.04.003. [DOI] [PubMed] [Google Scholar]


