Abstract
The amount, timing, and location of bone morphogenetic protein 2 (BMP2) synthesis influences the differentiation of pluripotent mesenchymal cells in embryos and adults. The BMP2 3′ untranslated region (3′UTR) contains a highly conserved AU-rich element (ARE) embedded in a sequence that commonly represses gene expression in mesenchymal cells. Computational analyses indicate that this site also may bind several microRNAs (miRNAs). Although miRNAs frequently target AU-rich regions, this ARE is unusual because the miRNAs directly span the ARE. We began to characterize the factors that may regulate Bmp2 expression via this complex site. The activating protein HuR (Hu antigen R, ELAVL1, HGNC:3312) directly binds this ARE and can activate gene expression. An miRNA was demonstrated to reverse HuR-mediated activation. Mutational and RNA-interference evidence also supports an AUF1 (AU-factor-1, HNRNPD, HGNC:5036) contribution to the observed repressive activity of the 3′UTR in mesenchymal cells. A limited number of studies describe how miRNAs interact with ARE-binding proteins that bind adjacent sites. This study is among the first to describe protein/miRNA interactions at the same site.
Keywords: GENE REGULATION, POST-TRANSCRIPTIONAL, MESENCHYMAL CELL, GROWTH FACTOR
Bone morphogenetic protein 2 (BMP2, HGNC:1069, GeneID: 650) strongly influences the differentiation of pluripotent mesenchymal cells into muscle, fat, cartilage, and bone cells. The amount, timing, and location of BMP2 synthesis influence this differentiation in embryos and in adult cells. Alterations in BMP2 levels disrupt embryogenesis and contribute to adult pathologies. Pathologies associated with BMP2 include osteoporosis, osteoarthritis, and all forms of pathological calcification in the vasculature and in cardiac valves. Altered differentiation of pluripotent mesenchymal cells occurs in all BMP2-influenced pathologies. Consequently, understanding the molecules and conditions that regulate BMP2 gene expression is crucial.
Numerous factors that positively or negatively regulate Bmp2 expression have been described (reviewed in [Rogers et al., in press]). At the transcriptional level, many essential transcription factors, for example, TCF/LEF proteins, GLI proteins, CREB, NFκB, E2Fs, MEF2A, GATA6, and HOXA13 and HOXD13 have been shown to modulate Bmp2 expression. In addition highly conserved post-transcriptional processes act via the BMP2 3′UTR to modulate synthesis. In mesenchymal cells, the 3′UTR often inhibits Bmp2 gene regulation. Which positive or negative factors regulate Bmp2 expression by binding the 3′UTR remains incompletely described.
An “ultra-conserved sequence” (UCS) within the 3′untranslated region (3′UTR) functions as a regulatory switch that facilitates BMP2 down-regulation in some cells, but promotes up-regulation in other cells [Abrams et al., 2004; Fukui et al., 2006; Devaney et al., 2009; Jiang et al., 2010; Kruithof et al., 2011a; Kruithof et al., 2011b]. Reduced levels of UCS-mediated repression may contribute to pathological BMP2 synthesis. For example, microRNAs post-transcriptionally repress protein synthesis via the 3′UTR. We determined that 106 miRNAs were down-regulated in oncogenically transformed lung cell lines [Fotinos et al., 2014]. BMP2 targeting miRNAs whose level was abnormally low in lung cancer cells, were shown to be cytotoxic in lung adenocarcinoma cells that synthesize BMP2, but not in non-transformed lung cells [Fotinos et al., 2014]. Reduced levels of these repressors in tumor cells likely contribute to the excess synthesis of the pro-oncogenic BMP2 in lung tumors. Similarly pathological loss of molecules that repress Bmp2 may contribute to other pathologies.
Particularly in mesenchymal cells, Bmp2 can be transcribed, but protein synthesis may be blocked at the post-transcriptional level [Fukui et al., 2006; Devaney et al., 2009; Jiang et al., 2010; Kruithof et al., 2011a; Kruithof et al., 2011b]. The UCS or the intact 3′UTR represses diverse reporter genes in vascular and valve cells in vitro and in vivo. Reduced Bmp2 3′UTR function may contribute to the ectopic BMP2 observed in pathologically calcified vascular and valve cells [Shao et al., 2005; Yutzey et al., 2014].
Many diverse combinations of RNA-binding proteins and miRNAs may interact with the Bmp2 3′UTR (reviewed in [Rogers et al., in press]). Nucleolin (HGNC:7667), the stabilizing protein HuR (ELAVL1, HGNC:3312), and the destabilizing protein AUF1 (HNRNPD, HGNC:5036) have been shown to bind transcripts bearing the UCS [Fritz et al., 2006; Devaney et al., 2009; Jiang et al., 2010; Wu et al., 2013; Yoon et al., 2014]. In addition, a handful of miRNAs have been experimentally validated. These include mir-140-5p, mir-106a, mir-17-5p, mir-27a, mir-370, mir-34b, mir-34c-3p, and miR-486-3p [Rogers et al., in press]. How proteins and miRNAs interact to modulate BMP2 synthesis in different cell types is unknown.
The UCS in mammalian Bmp2 genes contains 10 dispersed AU-rich elements (AREs) that modulate in vitro stability of synthetic Bmp2 transcripts [Abrams et al., 2004; Fritz et al., 2004; Fritz et al., 2006]). Many of the ARE-binding proteins that bind the human UCS also bind the classical ARE in the TNFα mRNA [Fritz et al., 2006]. Currently our understanding of how ARE-binding proteins interact with miRNAs is limited. The critical miRNA seed sequence is often located in or near AU-rich sequences. However, both transcriptome-wide surveys and studies of individual mRNAs found that miRNA seed sequences tend to be adjacent to rather than overlapping the sites that bind the HuR ARE-binding protein [Lebedeva et al., 2011; Mukherjee et al., 2011; Srikantan et al., 2012]. Indeed only 89 out of 1240 (7.1%) transcripts with both miRNA and HuR sites had sites that directly coincided [Mukherjee et al., 2011]. We observed that a classical ARE in the Bmp2 UCS is computationally predicted to also target several miRNAs. This site may be positioned for a direct interaction with ARE-binding proteins such as HuR or AUF1. The data presented here is the first to describe the proteins and miRNAs that may compete or cooperate at this unusual, conserved AU-rich sequence.
MATERIALS AND METHODS
CELL CULTURE
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, D5796 Sigma Aldrich, St. Louis, MO) with 5% fetal bovine serum without antibiotics. F9 embryonal carcinoma cells were grown in DMEM supplemented with 10% heat-inactivated newborn calf serum on dishes pre-coated with 1% gelatin in 10% CO2. F9 cells were induced to differentiate into parietal endoderm by adding 1 μM all-trans retinoic acid, 250 μM dibutyryl cAMP, and 500 μM theophylline (RACT). MC3T3-E1 cells were grown in non-ascorbic acid-containing α-MEM (Invitrogen #01-0083D) with 10% fetal bovine serum. C3H10T½ and rat vascular smooth muscle cells (VSMCs) from the aortic media of male Sprague-Dawley rats were cultured in DMEM supplemented with 10% fetal bovine serum. Normal Human Umbilical Artery Smooth Muscle Cells (UASMC) were obtained from Lonza Walkersville, Inc., Walkersville, MD and grown according to the manufacturer’s instructions. All media was supplemented with 2 mM glutamine. Except for F9 cells, all cells were grown in 5–7% CO2 at 37°C.
LUCIFERASE PLASMIDS
Bmp2Luc (mouse nt −1,237 to 471 relative to the distal promoter, aka pGL1.7XX) and Bmp2Luc-mUCS (mouse nt −1,237 to 471 and mouse 9,574 nt to 9,938 nt relative to the distal promoter or +83 to 446 relative to the stop codon, aka pGLB2-5′ mouseCNS) were described previously [Fritz et al., 2004].
RPL10-Luc-3′UTR (1254 nt of full-length BMP2 3′UTR including both polyadenylation signals) was described in [Kruithof et al., 2011a]. RPL10-Luc-Stop-855 was generated by digesting with the restriction enzymes Sbf1 and Xho1 and deleting a 470 nt dropout fragment bearing the distal 3′UTR with the two endogenous polyA sites. RPL10-Luc-855-1254 was generated by digesting with NheI and Sbf1and deleting an 855 nt dropout fragment bearing the ultra-conserved sequence (UCS). The mutated plasmids in Figure 4A were generated by first digesting RPL10-Luc-3′UTR with EcoRv and XhoI and deleting a 377 nt. dropout fragment. The QuikChange Site Directed Mutagenesis kit II (Agilent, Santa Clara, CA) was used to introduce the indicated mutations. All new plasmids were sequenced by the NJ Medical School Molecular Resource Facility.
Fig. 4.
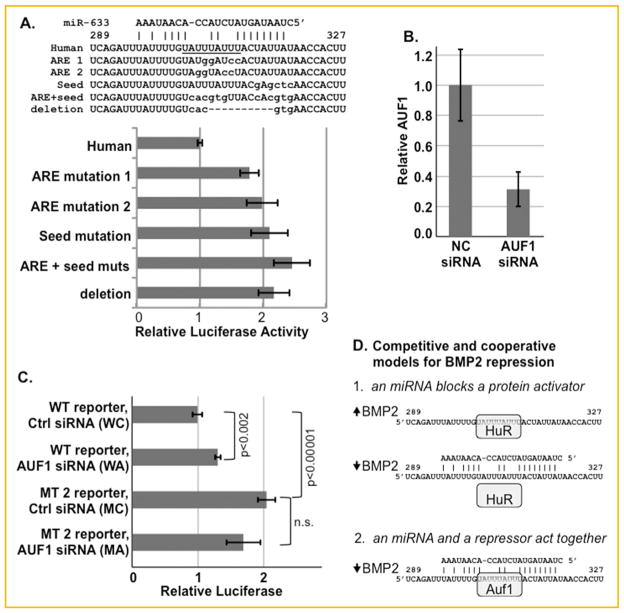
AUF1 mediated repression. A: Reporter genes were constructed with two different mutations restricted to the ARE, a mutation restricted to the seed motif of the miR-633 target site, mutations within both the ARE and the seed, or a deletion of both motifs within RPL10Luc-Stop-855 (Fig. 2B). The mutated nucleotides are shown lower case. All mutated plasmids were expressed significantly more than the wild-type human plasmid in MC3T3E1 cells (P ≤0.0002). The differences between each mutation were not significant. B, C: 3 × 104 MC3T3E1 cells were transfected with 100 pmol AUF1 siRNA (n =4) or negative control siRNA (n =3) each day for 3 days. On the third day, the cells also were transfected with either wild-type (WT) RPL10Luc-Stop-855 or this reporter with ARE mutation 2 (MT 2). B. shows the level of AUF1 protein in C3H10T½ cells as determined by Western blotting. C. shows the relative luciferase activity of each reporter in the AUF1 knockdown cells relative to control cells. Reduced AUF1 levels activated the wild-type vector (P <0.002). As shown in A, the ARE mutation induced reporter gene expression (P <0.00001). However, AUF1 knockdown did not activate the mutated vector D. Two models for hypothesized mechanisms by which factors may repress Bmp2 gene expression via the overlapping ARE and miRNA binding sites. HuR, Auf1, and miR-633 are shown as potential regulatory factors; however, the precise combination and identity of factors may be cell-specific.
REPORTER GENE ASSAYS
Cells were plated and transfected with plasmid using FuGene6 Transfection Reagent (Roche, Indianapolis, IN). Cells were lysed with 1×Passive Lysis Buffer (Promega, Madison, WI) and luciferase activities were measured using the Luciferase Assay System (Promega). The luciferase activity of a co-transfected, constitutively expressed Renilla luciferase reporter plasmid was used to control for transfection efficiency. Protein concentration was used to control for cell density.
IN VITRO TRANSCRIPTION PLASMIDS
pGBmp2-PvuIIPst (9,574–10,202) was linearized with Acc I to make sense probe spanning nt 85–449 relative to the stop codon [Fritz et al., 2004]. pGBmp2-PvuIIRsa (nt 85–246) and pGBmp2-RsaAcc (nt 246–449) were linearized with Hind III. The QuikChange Site Directed Mutagenesis kit II (Agilent, Santa Clara, CA) was used to introduce the mutation indicated in Figure 1A into pGBmp2-RsaAcc. pGemB2-KA (nt 9,455–9,938) and plasmids containing the homologous regions from human, chick, and zebrafish [Fritz et al., 2004] were linearized with BamHI. Wild-type or mutated TNFα plasmids [Mukherjee et al., 2002] were linearized with Hind III.
Fig. 1.
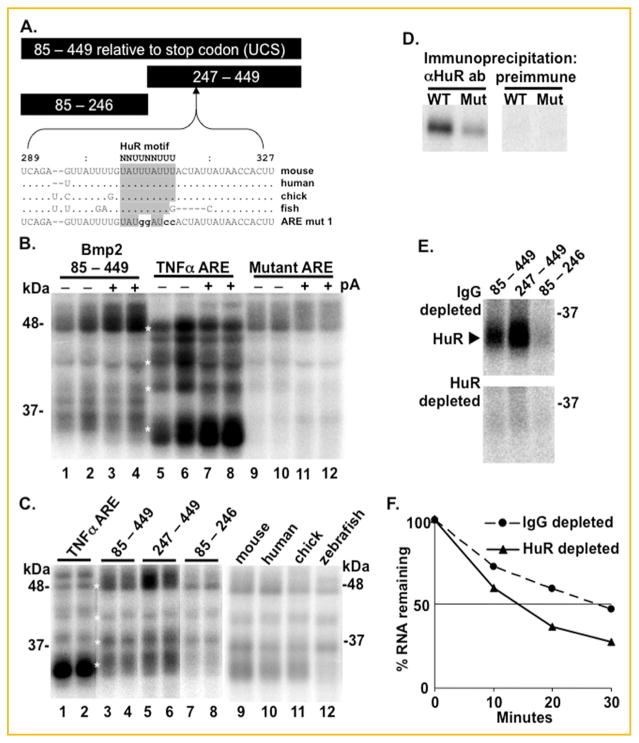
Conservation of an AU-rich HuR binding site. A: Bars show the relative lengths and positions of BMP2 RNAs relative to the termination codon used for UV-crosslinking. The mouse, human, and chicken sequences uniquely conform to a putative HuR binding site as defined by [Lopez de Silanes et al., 2004; Meisner et al., 2004]. The zebrafish sequence has a G substitution and a deletion that alters the HuR motif. The sequence of the mutated RNA used in panel D is shown. B: In vitro synthesized, radiolabeled transcripts containing mouse Bmp2 conserved sequence (nt. 85 to 449 relative to the termination codon) or the wild-type (AAUUAUUUAUUAUUUAUUUAUUAUUUAUUUAUUUAA) or mutated (AAUGAUGUACUACUUGUUCAUGAUGUUCUUCUUGAA) ARE from TNFα were UV cross-linked with HeLa cell S100 extracts without (− pA) or with (+ pA) excess nonradioactive poly(A) competitor. Duplicate lanes show bound proteins from two independently prepared HeLa cell extracts. The wild-type TNFα ARE was shown previously to bind HuR [Ford et al., 1999]. White asterisks mark co-migrating bands. C: Transcripts containing the TNFα ARE or three transcripts containing the three indicated Bmp2 RNAs or the homologous regions from mouse, human, chick, and zebrafish were incubated in HeLa extracts with excess poly(A) competitor. ARE binding proteins were most efficiently labeled by the two RNAs containing the HuR consensus site. The mammalian, avian, and fish RNAs UV cross-linked several ARE binding proteins. However the zebrafish RNA which lacks a canonical HuR site failed to label a protein migrate at 32 kDa. D: HeLa extracts were incubated with radiolabeled wild-type mouse (WT) RNA (nt. 85−449) or an RNA with its ARE mutated (MT) as shown in panel A. After UV crosslinking, the extracts were immunoprecipitated with preimmune IgG or with anti-HuR antibody (Santa Cruz, sc5261). E: Extracts from retinoid-treated F9 cells, which express elevated BMP2 levels, were immunodepleted with anti-HuR antibody or preimmune IgG, then UV cross-linked to BMP2 RNA, and finally immunoprecipitated with the HuR antibody. This experiment shows that the anti-HuR antibody fully immunodepleted HuR in the extracts used for panel F. F: Radiolabeled wild-type mouse RNA (nt. 85−449) was incubated in the immunodepleted extracts described in E. Intact RNA was plotted as a % of RNA in unincubated extracts at time 0.
IN VITRO STABILITY MEASUREMENTS
After linearization, plasmids were transcribed with SP6 RNA polymerase with 7meGpppG and α32P-UTP. Capped and labeled RNAs were incubated in S18 cytoplasmic extracts. RNAs and degradation products were visualized and quantified on an 8 M urea-containing 5% polyacrylamide (37.5:1, acrylamide:bis-acryl-amide) gel using a Molecular Dynamics PhosphorImager and ImageQuant software. Detailed methods are referenced in [Fritz et al., 2006].
UV CROSS-LINKING REACTIONS
Briefly, 20–50 fmol of radiolabeled RNA was incubated in the cell extract under in vitro decay assay conditions for 10 min at 37°C. EDTA was added to a concentration of 1 mM to prevent RNA decay while proteins bound to the RNA. The reaction mixtures were irradiated with ultraviolet light for 10 min using a 15W germicidal lamp at 2 μJ/s at room temperature. 100 ng RNase A was added and incubated for 15 min at 37°C to degrade the labeled RNA. Labeled proteins were solubilized by heating at 95°C for 5 min in 8% glycerol, 1.0% SDS, 75 mM dithiothreitol, 0.05% Bromophenol Blue, 62.5 mM Tris, pH 6.8, analyzed on a 10% polyacrylamide gel (37.5:1, acrylamide:bis-acrylamide) in running buffer (192 mM glycine, 25 mM Tris-HCl, 0.1% SDS), and quantified as above. Detailed methods are referenced in [Fritz et al., 2006].
IMMUNODEPLETION
F9 cytoplasmic extracts were incubated with an anti-HuR antibody (SC5261, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) or control (pre-immune) IgG at 4°C for 1 h. Protein A/G-agarose beads (Amersham Pharmacia Biotech, Piscataway, NJ) were added and incubated overnight at 4°C with rotation. Beads were pelleted and the extracts were analyzed by UV cross-linking or decay assays as described above.
AUF1 KNOCKDOWN
Cells were plated and transfected with control or anti-AUF1 siRNA using XtremeGene Transfection Reagent (Roche) according to the manufacturer’s directions and as described in [Gummadi et al., 2012]. To silence all four isoforms of AUF1, two separate siRNA oligos were used in combination to target exon 3, which is common to the 4 isoforms (CACUCUGAAGUUAGAUCCUAUCACA and UUUAGGAUCAAUCACCUUCCCAUUC). Reporter plasmids were transfected after 2 days. Knockdown was assessed by standard immunoblotting using an AUF1 antibody provided by Dr. Gary Brewer [Wu et al., 2013].
RESULTS
A CONSERVED HuR MOTIF
We extended our analyses of the ARE binding proteins that bind the Bmp2 UCS with the transcripts shown in Figure 1A. Figure 1B shows that 32P-labeled RNAs bearing the entire mouse Bmp2 UCS can UV cross-link several proteins that co-migrate with proteins labeled by the classical tumor necrosis factor (TNF) α ARE. RNAs bearing the TNF α ARE mutated with guanine and cytosine substitutions failed to label proteins below 48 kDa. The protein-RNA interactions were not notably perturbed by the presence or absence of excess poly(A) competitor. AREs are distributed throughout the Bmp2 UCS. Figure 1C compares the proteins labeled by the entire UCS or two shorter transcripts (see Fig. 1A). The band patterns suggest that proteins of similar weight were labeled by all three transcripts. This suggests that a common motif is present throughout the transcripts. The multiple dispersed AUUUA or AUUUUA motifs (AREs) are likely candidates. However, the decreased labeling efficiency of the nt. 85–246 transcript relative to the nt. 247–449 transcript suggests that this latter RNA has motifs with an increased affinity.
Indeed, one AU-rich element conforms more closely to an HuR binding motif than any other Bmp2 ARE (Fig. 1A [Lopez de Silanes et al., 2004; Meisner et al., 2004]). This ARE is perfectly conserved in the mouse, human, and chick RNAs. Although the sequence conservation is evident in the zebrafish RNA, the HuR motif is marred by a guanosine substitution and a 5 nucleotide deletion. We side-by-side compared 32P-labeled RNAs bearing the full-length mouse, human, chick, and zebrafish Bmp2 UCS (Fig. 1C). The mammalian and avian RNAs labeled several HeLa cell proteins that co-migrate with proteins labeled by the classical TNF α ARE. The fish RNA labeled most of these proteins except a 32 kDa protein. In HeLa cells, a protein of this molecular weight was shown to be HuR [Ford et al., 1999]. We previously confirmed that long human RNAs bearing the putative HuR motif were capable of UV-crosslinking HuR [Devaney et al., 2009]. Because Bmp2 RNAs contain several dispersed AREs in addition to the HuR motif shown in Figure 1A, we used a mutated mouse transcript to test if this specific motif accounts for the HuR-Bmp2 RNA interaction. An RNA with guanine and cytosine substitutions in the HuR motif labeled the protein precipitated by the HuR antibody much less efficiently (Fig. 1D, Supplemental Fig. S1A).
HuR can stabilize ARE-containing transcripts [Fan and Steitz, 1998]. We previously showed that Bmp2 transcript abundance is induced in retinoic acid (RA)-treated F9 embryonal carcinoma cells [Rogers et al., 1992; Glozak and Rogers, 1996]. In addition, Bmp2 RNAs were more stable in extracts from RA-treated cells relative to extracts from untreated cells that don’t express Bmp2 [Fritz et al., 2004]. We tested the hypothesis that HuR may stabilize the Bmp2 mRNA in these cells. Extracts from RA-treated cells were immuno-depleted with HuR antibody (Supplemental Fig. S1B). Figure 1E and Supplemental Figure S1C show that the control IgG-treated extracts retained HuR that was efficiently labeled with the two Bmp2 RNAs bearing the HuR motif (nt. 85–449 and 247–449), but not the RNA without the HuR motif (nt. 85–246). In contrast, little HuR, if any, remained in the cell lysates extracted by the HuR antibody. This experiment shows that that these extracts were fully immunodepleted. Figure 1F shows that a synthetic RNA bearing the Bmp2 UCS decayed more rapidly (t½ =14 min) in the HuR-depleted extract relative to the control extract (t½ =27 min). Together, these experiments demonstrated that HuR specifically binds a Bmp2 ARE located between 302 and 310 nt relative to the termination codon and is able to stabilize an RNA bearing this sequence in vitro.
POST-TRANSCRIPTIONAL REPRESSION IN ADDITIONAL CELL TYPES
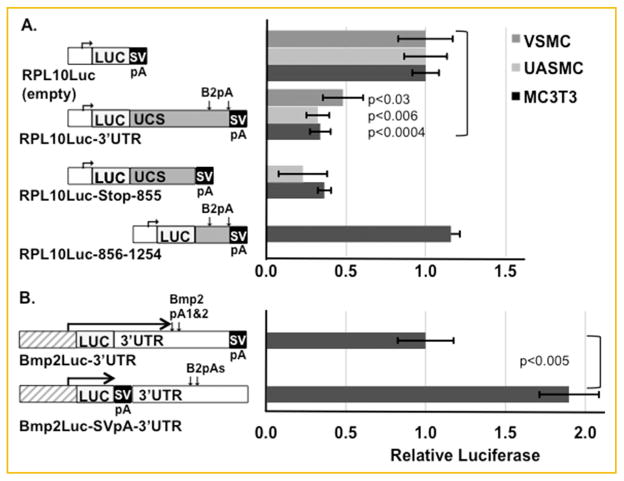
Because preventing elevated BMP2 synthesis in vascular cells is clinically relevant, we began to assess the function of the UCS and this ARE in mesenchymal cells. The Bmp2 UCS and 3′UTR can either activate [Abrams et al., 2004; Fritz et al., 2004; Fritz et al., 2006; Jiang et al., 2010] or repress [Jiang et al., 2010; Kruithof et al., 2011a, b] gene expression. Which effect depends on cell type. Therefore, we tested different regions of the 3′UTR in two primary vascular cell lines, vascular smooth muscle cells (VSMC) from rat aorta and human umbilical artery smooth muscle cells (UASMC), and in MC3T3-E1 mouse pre-osteoblast cells. Both the entire 3′UTR and the first 855 nt. bearing the UCS repressed a luciferase reporter driven by the constitutively expressed RPL10 (ribosomal protein L10) promoter (Fig. 2A, plasmids RPL10Luc-3′UTR and RPL10Luc-Stop-855). The fragment bearing only the two polyadenylation sites (plasmid RPL10Luc-856-1254 [Liu et al., 2008]) did not significantly alter expression relative to the promoter alone.
Fig. 2.
The Bmp2 3′UTR represses gene expression in immortalized and primary mesenchymal cells. A: The luciferase activities of transfected reporter genes driven by the heterologous, constitutive promoter RPL10 were compared in MC3T3-E1 immortalized preosteoblast cells, human umbilical artery smooth muscle cells (UASMC), or vascular smooth muscle cells (VSMC) from rat aorta. The indicated plasmid regions are to approximate scale. Relative to the promoter only plasmid, plasmids with the full-length human 3′UTR (1,227 nt to polyadenylation (pA) signal 2, 1,323 nt total) inserted downstream of LUC or a shorter fragment bearing the UCS (855 nt) were expressed less. The fragment bearing the two polyadenylation signals did not repress in MC3T3-E1 cells (UASMC and VSMC not done). Relative reporter activity is shown ±SEM. MC3T3-E1 cells, n =4, P <0.0004; UASMC, n =4−6, P <0.006; VSMC, n =6, P <0.03. B: Bmp2Luc-3′UTR and Bmp2Luc-SVpA-3′UTR both contain the distal mouse Bmp2 promoter (−1,237−471 relative to the distal transcription start site), the entire mouse 3′UTR (870 or 1,185 nt depending on which pA signal is used), and downstream sequence (2,212 nt total). These plasmids are identical except that the SV40 pA signal was positioned downstream or upstream of the Bmp2 3′UTR as indicated. In MC3T3-E1 cells (UASMC and VSMC not done), Bmp2Luc-SVpA-3′UTR, whose transcript excludes the 3′UTR, was nearly twice as active as Bmp2Luc-3′UTR whose transcript includes the 3′UTR. n =6, P <0.005.
In theory, the Bmp2 3′UTR sequence could contain transcriptional elements within the DNA of the plasmid or post-transcriptional elements within the transcript. To distinguish between these effects, we compared reporter plasmids whose sequences were identical except that a strong SV40 site was positioned either upstream or downstream of the 3′UTR. The SV40 site efficiently truncates the mRNA leading to luciferase transcripts with or without the 3′UTR [Jiang et al., 2010; Kruithof et al., 2011a]. Figure 2B shows that the transcript without a 3′UTR (Bmp2Luc-SVpA-3′UTR) expressed more luciferase than the transcript with the 3′UTR (Bmp2Luc-3′UTR). Thus, as previously shown in non-transformed lung cells [Jiang et al., 2010], the repression observed in many mesenchymal cells [Kruithof et al., 2011a,b]) has a post-transcriptional component.
HuR AND MicroRNA COMPETITION
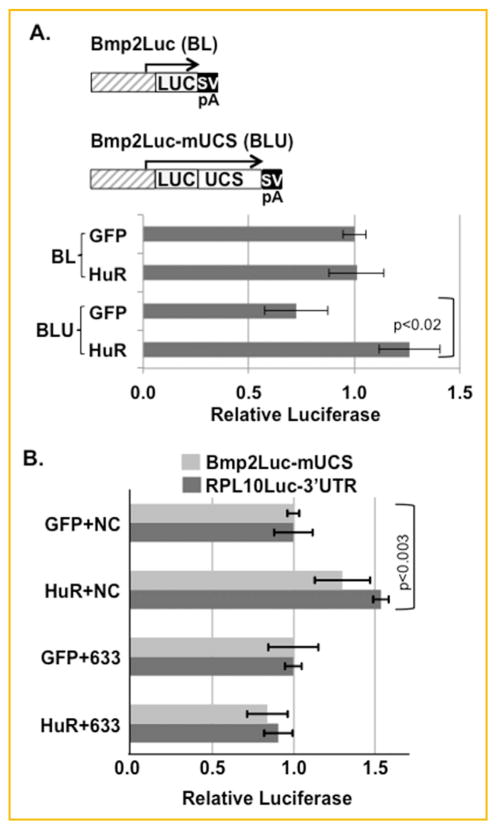
One mechanism that may explain how the UCS represses in some cells is that an activating factor such as HuR may be displaced by neutral or repressive factors. The Bmp2 3′UTR can repress various expression plasmids in C3H10T½ cells [Devaney et al., 2009; Kruithof et al., 2011a]. Figure 3A confirms that the UCS reduces the expression of a Bmp2 promoter-driven reporter plasmid when co-transfected with a control GFP-expression vector (compare Bmp2Luc with Bmp2Luc-mUCS with GFP). HuR overexpression selectively induced only the reporter plasmid with the UCS, but not the plasmid lacking the UCS. Thus, the HuR/UCS interaction up-regulates gene expression.
Fig. 3.
HuR and miR-633 competition. A: Mouse C3H10T1/2 mesenchymal cells were co-transfected with a reporter gene driven by the mouse BMP2 promoter (nt. −1,237 to 471 relative to the distal promoter, Bmp2Luc) alone or with nt. 85−449 from the mouse 3′UTR inserted downstream of luciferase (Bmp2Luc-mUCS). These cells were co-transfected with either a GFP-expressing control plasmid or a plasmid expressing HuR [Fan and Steitz, 1998]. The average luciferase reporter activity is shown ±SEM, n =12, Bmp2LucUCS with GFP vs. with HuR P <0.02. B: C3H10T1/2 cells were co-transfected with either Bmp2Luc-mUCS or RPL10Luc-3′UTR with the GFP or HuR expression vectors and with the Negative Control (NC) or miR633 (633) miRNA precursors as indicated (n =3). Although, HuR activated both the Bmp2 driven and the RPL10-driven vectors bearing the UCS or the 3′UTR respectively (P <0.003, n =3 each), HuR failed to activate in the presence of miR-633.
Several microRNAs, for example miR-633 (mirSVR score: −1.3074; PhastCons score: 0.7607, Fig. 4A), are computationally predicted to span the HuR-binding ARE (http://www.microrna.org/microrna/getMrna.do?gene=650&utr=21822&organism=9606). Functional miRNA sites are preferentially located in AU-rich regions. Indeed, three quarters of 3′UTRs with Argonaute binding sites also contained AU-rich HuR sites [Mukherjee et al., 2011]. However, in approximately 9 out of 10 messages, the Argonaute and HuR sites fail to overlap [Lebedeva et al., 2011; Mukherjee et al., 2011]. Thus the Bmp2 HuR binding site belongs in a relatively small group of sites where miRNAs are positioned for direct competition with HuR binding.
We tested if miR-633 and HuR can directly compete (Fig. 3B). HuR can induce the Bmp2-driven plasmid with only the UCS in the presence of the negative control miRNA (compare GFP+NC vs. HuR+NC). HuR also induced the constitutively-expressed RPL10Luc-3′UTR reporter gene bearing the entire human BMP2 3′UTR. This confirms that the HuR activates exclusively via the 3′UTR. In contrast, the activating effect of HuR on both plasmids was abolished by the miR-633 mimic (compare GFP+633 vs. HuR+633). These data support the principle that a miRNA can displace the activating HuR on the Bmp2 transcript.
AUF1-MEDIATED REPRESSION
Another mechanism by which the UCS might repress is to recruit proteins that function as repressors. To begin to dissect the potentially complex interactions at this ARE, we created a set of mutated reporter genes. All mutated sequences were compared to known miRNA sequences to avoid creating a new site that may bind other miRNAs. Mutations 1 and 2 disrupted the ARE with guanosine and cytosine insertions (Fig. 4A). Figure 1D demonstrated that HuR was unable to bind Mutation 1. Mutation 2 was designed to destroy the ARE without reducing the potential base pairing with miR-633. Mutation 3 disrupted the complementarity between the Bmp2 mRNA and the seed sequence of miR-633 and any other miRNAs that were predicted to bind this site. Mutation 4 and 5 mutated or deleted both the ARE and the seed respectively. These two mutations should prevent both protein and miRNA interactions. As shown in Figure 4A, all mutations significantly induced reporter gene expression. The lack of a statistically significant difference in the activities of each mutated reporter indicates that both the ARE and the seed sequence mediate repression.
If an activating protein such as HuR acts independently via this ARE, then the ARE mutations (1 and 2) might reduce reporter gene activity. However, these two mutations activated the reporter. This suggests that a repressive factor binds the ARE. The four isoforms of the ARE binding protein AUF1 (hnRNPD) promote mRNA decay [White et al., 2012]. Proteins with apparent molecular weights similar to the p37, p40, p42, and p45 AUF1 proteins also were labeled by the Bmp2 RNAs (Fig. 1B and C). We tested if AUF1 could repress the UCS-bearing reporter by knocking down AUF1 levels with an AUF1 siRNA (Fig. 4B). Indeed, AUF1 knockdown significantly activated the reporter gene (Fig. 4C, compare WC vs. WA, P <0.002). As shown in Figure 4A, a reporter with the ARE disrupted by mutation 2 was induced relative to the wild-type sequence in cells transfected with the control siRNA (compare WC vs. MC). If AUF1 repressed via this ARE and not AREs elsewhere in the sequence, then AUF1 knockdown should not activate the mutated reporter gene. Indeed, reduced levels of AUF1 failed to induce the mutated reporter (compare MC vs. MA). This confirms that AUF1 contributes to the repressive effect of the UCS in mesenchymal cells.
DISCUSSION
We showed that a specific conserved ARE within the Bmp2 UCS was directly bound by the activating protein HuR. We also showed for the first time that a miRNA could reverse HuR-mediated activation in mesenchymal cells where the UCS is normally repressive. Finally, we provide evidence that AUF1 contributes to the repressive function of the UCS in mesenchymal cells.
HuR, a ubiquitous and exceedingly well-characterized ARE-binding protein, has a multifaceted relationship with miRNAs. HuR has been demonstrated to both enhance and hinder the ability of miRNAs to repress protein synthesis (reviewed in [Srikantan et al., 2012]). HuR sites can function at a distance from the targets bound by miRNAs. For example, HuR was shown to block miR-122 mediated repression at a distance of up to 50 nucleotides on the cationic amino acid transporter 1 (CAT1) mRNA [Kundu et al., 2012]. Even more remote HuR binding at 100 nucleotides enhanced the inhibition mediated by the let-7 miRNA on the c-Myc RNA [Kim et al., 2009]. The direct overlap between the Bmp2 HuR binding site and the miR-633 miRNA target is highly unusual [Lebedeva et al., 2011; Mukherjee et al., 2011]. We showed that HuR clearly binds this specific ARE and can induce gene expression via the Bmp2 3′UTR. Furthermore, exogenous miR-633 can abrogate this up-regulation. One simple model explaining the repressive effect of the Bmp2 UCS in mesenchymal cells is that endogenous miRNAs hamper the ability of HuR to interact with the Bmp2 transcript (Fig. 4D, model 1). The profiles of miRNAs present in different mesenchymal cells that support UCS-mediated repression are currently being surveyed.
However, many other proteins may bind this ARE. Indeed, AUF1, another widely expressed and well-characterized ARE-binding protein, was shown to pull-down the Bmp2 mRNA [Wu et al., 2013; Yoon et al., 2014]. AUF1 and HuR bind many of the same RNAs [Lal et al., 2004]. AUF1 is a destabilizing protein that exerts an effect opposite to HuR. The interplay between AUF1 and miRNAs has been studied on fewer mRNAs. However, like HuR, both cooperative and competitive effects have been described [Wu et al., 2013]. How AUF1 and the associated miRNAs affected transcript translation and stability was highly specific to different mRNAs. We showed that reducing the level of AUF1 stimulated the expression of a reporter gene bearing the Bmp2 UCS. Mutation of the ARE abolished this response proving that AUF1 functions at this site. However, AUF1 may not function alone, because single or combined mutations of the ARE and the miR-633 seed site activated the Bmp2 reporter gene to an equal extent. One hypothesis that explains both the effect of AUF1 knockdown and the five mutations is that AUF1 and the miRNA induced silencing complex (RISC) cooperate to promote mRNA degradation (Fig. 4D, model 2).
Where, when, and how much BMP2 is synthesized and activated must be tightly regulated in diverse cell types. The mechanisms that control Bmp2 transcription are complex [Rogers et al., in press]. Further tuning of BMP2 synthesis is accomplished by an intricate network of RNA binding proteins and non-coding RNAs. Nucleolin, the stabilizing protein HuR, and the destabilizing protein AUF1 are among the dozens of proteins that can bind the UCS (this work and [Fritz et al., 2006; Devaney et al., 2009; Jiang et al., 2010]). In addition, many miRNAs modulate BMP2 synthesis via the 3′UTR [Rogers et al., in press]. Some of these miRNAs also may coordinately regulate effectors of BMP2 signaling, such as the BMP receptor BMPR1A (miR-27a [Gong et al., 2014]), the signaling intermediary SMAD1 (miR-486 [Lin et al., 2009]), and the osteogenic transcription factor RUNX2 (mir-34c [Zhang et al., 2011]). Given the necessary complexity of Bmp2 gene regulatory mechanisms, many different combinations of ARE-binding proteins and miRNAs are likely to regulate Bmp2 in different cell types. Deciphering the protein and miRNA code that dictates the precise level and timing of BMP2 synthesis in the many BMP-responsive tissues has only just begun.
Supplementary Material
Acknowledgments
Grant sponsor: National Heart, Lung, and Blood Institute (NHLBI); Grant number: 1R01HL114751.
This study was supported by a National Heart, Lung, and Blood Institute (NHLBI) 1R01HL114751 award to MBR. Authors have no financial or commercial conflicts of interest regarding the material presented.
Footnotes
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Abrams KL, Xu J, Nativelle-Serpentini C, Dabirshahsahebi S, Rogers MB. An evolutionary and molecular analysis of Bmp2 expression. J Biol Chem. 2004;279:15916–15928. doi: 10.1074/jbc.M313531200. [DOI] [PubMed] [Google Scholar]
- Devaney JM, Tosi LL, Fritz DT, Gordish-Dressman HA, Jiang S, Orkunoglu-Suer FE, Gordon AH, Harmon BT, Thompson PD, Clarkson PM, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Brandoli C, Hoffman EP, Rogers MB. Differences in fat and muscle mass associated with a functional human polymorphism in a post-transcriptional BMP2 gene regulatory element. J Cell Biochem. 2009;107:1073–1082. doi: 10.1002/jcb.22209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. Embo J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotinos A, Nagarajan N, Martins AS, Fritz DT, Garsetti D, Lee AT, Hong CC, Rogers MB. Bone morphogenetic protein-focused strategies to induce cytotoxicity in lung cancer cells. Anticancer Res. 2014;34:2095–2104. [PMC free article] [PubMed] [Google Scholar]
- Fritz DT, Liu D, Xu J, Jiang S, Rogers MB. Conservation of Bmp2 post-transcriptional regulatory mechanisms. J Biol Chem. 2004;279:48950–48958. doi: 10.1074/jbc.M409620200. [DOI] [PubMed] [Google Scholar]
- Fritz DT, Jiang S, Xu J, Rogers MB. A polymorphism in a conserved posttranscriptional regulatory motif alters bone morphogenetic protein 2 (BMP2) RNA:protein interactions. Mol Endocrinol. 2006;20:1574–1586. doi: 10.1210/me.2005-0469. [DOI] [PubMed] [Google Scholar]
- Fukui N, Ikeda Y, Ohnuki T, Hikita A, Tanaka S, Yamane S, Suzuki R, Sandell LJ, Ochi T. Pro-inflammatory cytokine tumor necrosis factor-alpha induces bone morphogenetic protein-2 in chondrocytes via mRNA stabilization and transcriptional up-regulation. J Biol Chem. 2006;281:27229–27241. doi: 10.1074/jbc.M603385200. [DOI] [PubMed] [Google Scholar]
- Glozak MA, Rogers MB. Specific induction of apoptosis in P19 embryonal carcinoma cells by retinoic acid and BMP2 or BMP4. Devl Biol. 1996;179:458–470. doi: 10.1006/dbio.1996.0275. [DOI] [PubMed] [Google Scholar]
- Gong Y, Xu F, Zhang L, Qian Y, Chen J, Huang H, Yu Y. MicroRNA expression signature for Satb2-induced osteogenic differentiation in bone marrow stromal cells. Mol Cell Biochem. 2014;387:227–239. doi: 10.1007/s11010-013-1888-z. [DOI] [PubMed] [Google Scholar]
- Gummadi L, Taylor L, Curthoys NP. Concurrent binding and modifications of AUF1 and HuR mediate the pH-responsive stabilization of phosphoenolpyruvate carboxykinase mRNA in kidney cells. Am J Physiol Renal Physiol. 2012;303:F1545–F1554. doi: 10.1152/ajprenal.00400.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Fritz DT, Rogers MB. A conserved post-transcriptional BMP2 switch in lung cells. J Cell Biochem. 2010;110:509–521. doi: 10.1002/jcb.22567. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruithof BP, Fritz DT, Liu Y, Garsetti DE, Frank DB, Pregizer SK, Gaussin V, Mortlock DP, Rogers MB. An autonomous BMP2 regulatory element in mesenchymal cells. J Cell Biochem. 2011a;112:666–674. doi: 10.1002/jcb.22975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruithof BP, Xu J, Fritz DT, Cabral CS, Gaussin V, Rogers MB. An in vivo map of bone morphogenetic protein 2 post-transcriptional repression in the heart. Genesis. 2011b;49:841–850. doi: 10.1002/dvg.20757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. Embo J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–352. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. MiR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Fritz DT, Rogers MB, Shatkin AJ. Species-specific cis-regulatory elements in the 3′UTR direct alternative polyadenylation of bone morphogenetic protein 2 mRNA. J Biol Chem. 2008;283:28010–28019. doi: 10.1074/jbc.M804895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M, Auer M. MRNA openers and closers: Modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5:1432–1447. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. Embo J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers MB, Rosen V, Wozney JM, Gudas LJ. Bone morphogenetic proteins-2 and 4 are involved in the retinoic acid-induced differentiation of embryonal carcinoma cells. Molec Biol Cell. 1992;3:189–196. doi: 10.1091/mbc.3.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M, Shah T, Shaikh N. Turning bone morphogenetic protein 2 (BMP2) on and off in mesenchymal cells. J Cell Biochem. 116(10) doi: 10.1002/jcb.25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao JS, Cheng SL, Pingsterhaus JM, Charlton-Kachigian N, Loewy AP, Towler DA. Msx2 promotes cardiovascular calcification by activating paracrine Wnt signals. J Clin Invest. 2005;115:1210–1220. doi: 10.1172/JCI24140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: Mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2012;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chesoni S, Rondeau G, Tempesta C, Patel R, Charles S, Daginawala N, Zucconi BE, Kishor A, Xu G, Shi Y, Li ML, Irizarry-Barreto P, Welsh J, Wilson GM, Brewer G. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, Yang X, Kang MJ, Wood WH, 3rd, Noren Hooten N, Evans MK, Becker KG, Tripathi V, Prasanth KV, Wilson GM, Tuschl T, Ingolia NT, Hafner M, Gorospe M. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun. 2014;5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutzey KE, Demer LL, Body SC, Huggins GS, Towler DA, Giachelli CM, Hofmann-Bowman MA, Mortlock DP, Rogers MB, Sadeghi MM, Aikawa E. Calcific aortic valve disease: a consensus summary from the Alliance of Investigators on Calcific Aortic Valve Disease. Arterioscler Thromb Vasc Biol. 2014;34:2387–2393. doi: 10.1161/ATVBAHA.114.302523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Edwards JR, Ko SY, Dong S, Liu H, Oyajobi BO, Papasian C, Deng HW, Zhao M. Transcriptional regulation of BMP2 expression by the PTH-CREB signaling pathway in osteoblasts. PLoS ONE. 2011;6:e20780. doi: 10.1371/journal.pone.0020780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.