Abstract
Polypeptides containing the motif ((GA)mGX)n occur in silk (we refer to them as ‘silk-like’) and have a strong tendency to self-assemble. For example, polypeptides containing (GAGAGAGX)n, where X = G or H have been observed to form filaments; similar sequences but with X = Q have been used in the design of coat proteins (capsids) for artificial viruses. The structure of the (GAGAGAGX)m filaments has been proposed to be a stack of peptides in a β roll structure with the hydrophobic side chains pointing outwards (hydrophobic shell). Another possible configuration, a β roll or β solenoid structure which has its hydrophobic side chains buried inside (hydrophobic core) was, however, overlooked. We perform ground state analysis as well as atomic-level molecular dynamics simulations, both on single molecules and on two-molecule stacks of the silk-inspired sequence (GAGAGAGQ)10, to decide whether the hydrophobic core or the hydrophobic shell configuration is the most stable one. We find that a stack of two hydrophobic core molecules is energetically more favorable than a stack of two shell molecules. A shell molecule initially placed in a perfect β roll structure tends to rotate its strands, breaking in-plane hydrogen bonds and forming out-of-plane hydrogen bonds, while a core molecule stays in the β roll structure. The hydrophobic shell structure has type II’ β turns whereas the core configuration has type II β turns; only the latter secondary structure agrees well with solid-state NMR experiments on a similar sequence (GA)15. We also observe that the core stack has a higher number of intra-molecular hydrogen bonds and a higher number of hydrogen bonds between stack and water than the shell stack. Hence, we conclude that the hydrophobic core configuration is the most likely structure. In the stacked state, each peptide has more intra-molecular hydrogen bonds than a single folded molecule, which suggests that stacking provides the extra stability needed for molecules to reach the folded state.
Introduction
A growing number of proteins and other polypeptides are recognized as having the propensity to form filaments with a cross-beta structure. These include the now well-known pathogenic amyloids1, such as Aβ(1–42) and amylin which stack along an axis to form protofilaments2 containing two β sheets connected by turns3, and functional amyloids such as curli which stack to form solenoid-like structures4. Other examples of polypeptides that form filaments include α-synuclein which has a superpleated β-structure5, antifreeze protein which has a β-roll structure6 and membrane protein7 which has a β-barrel structure, etc. All of these structures have a “β-strand-turn-β-strand” motif (we call it β arch)8 in which two strands are connected by an appropriate turn and then interact via their side chains instead of their backbones. When molecules with β arch structures stack axially, they form two parallel β sheets with a hydrophobic interior. In this structure, which is found, e.g., in Aβ (1–42)2, side chain-side chain interaction and backbone-backbone hydrogen bonding find an ideal compromise.
A related class of secondary structures is beta solenoids9, such as the β-roll and the β-helix. Sequences with these structures tend to wind their chains into solenoidal structures with a fixed degree of periodicity. The repeating unit of a β solenoid is a structure containing 2 or more β strands connected by turns. At first sight, silks (from insects and spiders) may seem to be in a different class; however, these materials also feature pairs of beta sheets with a hydrophobic core. Stacks of these pairs form the crystallites that give silk its strength and toughness. An example is the bombyx Mori silk sequence involved in beta sheet formation: the hexapeptide GAGAGAGS10.
One of us recently investigated a variant of this bombyx Mori silk sequence, namely the ‘silk-like’ repeated sequence (GAGAGAGX)n, where X is a hydrophilic residue and n is the number of octapeptide repeats. This sequence, with X = glutamic acid, was introduced by Tirrell et al11, and found to be reasonably soluble in basic solutions, but to form crystalline precipitates at low pH where the glutamic moiety is largely protonated and uncharged. When we conjugated long random-coil polypeptides to both ends of (GAGAGAGX)48, with X = E or X = H, we found that these molecules, when in uncharged form, folded and slowly formed a quaternary structure: a micron sized filament with a ribbon-shaped core containing the silk-like block, and a water-rich corona containing the random coil block12. From CD spectra we deduced that the silk blocks indeed fold into a regular secondary structure, but that they only do so once they attach to the growing end of a filament12. The thickness and width of the core, as obtained from SAXS data, would seem compatible with some sort of solenoid configuration. We also studied an uncharged variant with X = Q, which also showed a strong self-assembling tendency; when conjugated with a cationic binding block and a random coil, it assembled with DNA in a manner reminiscent of rod-like viruses13.
The detailed secondary structure of the silk block (GAGAGAGX)n in the filament still remains elusive. Schor et al.14 used Replica Exchange Molecular Dynamics simulations to investigate the structure of a silk block with sequence (GAGAGAGE)n. They predicted that the structure of the silk block is a β-roll configuration with hydrophobic side chains pointing outwards; this configuration has dimensions consistent with the ribbon dimensions as obtained from SAXS data15.
A typical β roll structure could arise by winding (GAGAGAGX) beta strands into a flat spiral (see figure 1), with the polar X residues occupying the turns and the flat parts forming hydrogen-bonded beta sheets, from which the alanine methyl groups would stick out normal to the plane of the sheet. The winding can be done in two different ways, namely with the alanine methyl groups pointing out of the spiral or into the spiral. We shall refer to these as the ‘hydrophobic shell’ and ‘hydrophobic core’ structures, respectively. The hydrophobic shell structure would have an obvious propensity to stack on account of its exposed hydrophobic side chains; this would be consistent with the formation of filaments as found experimentally. However, the hydrophobic core structure might be intrinsically more stable on its own and could possibly stack by hydrogen bonding. Hence, there is no strong a priori argument in favour of either of the two. Yet, in Schor et al.’s previous study14, the authors discarded the hydrophobic core structure on the grounds that it would probably have an unfavourable turn structure; they did not underpin this assumption with simulations. We consider this issue therefore as unsolved and revisit it here, studying both candidate structures in more detail, and comparing them. We are motivated by the fact that hydrophobic side chains tend to be buried within solenoid-like structures, rather than the opposite; many types of β solenoids indeed have a hydrophobic core, e.g., the amyloid fibrils of the HET-s (218–289) prion16.
Figure 1.
A typical representation of the β roll structure consisting of parallel β sheets (in green) both in the top and bottom layers connected by the β turns (in pink). The β roll structure has a left-handedness from N terminal to C terminal. Three consecutive strands are labeled as i, i+1 and i+2; strands i and i+2 are on top, strand i+1 is at bottom.
The central questions in the present paper are: (i) what are the stabilities of the hydrophobic core and hydrophilic shell secondary structures, and (ii) which of those structures corresponds to the global free energy minimum. The remainder of the paper is organized as follows. First we introduce the β roll structure in detail and compare the sequence and structural difference between the roll structures with hydrophobic core and hydrophobic shell. Then we carry out a ground state analysis of the potential energies of both core and shell stacks, assuming each molecule in the stack is forming a perfect β roll structure with the maximum possible number of hydrogen bonds and hydrophobic contacts. Finally, we present and analyze results from our atomistic explicit-solvent molecular dynamics simulations regarding secondary structures, hydrogen bonding behavior, and internal potential energies of both single-chain and stacked double-chain systems.
Results and Discussion
Shell and core structures both have β roll backbone but different alanine side chain directions
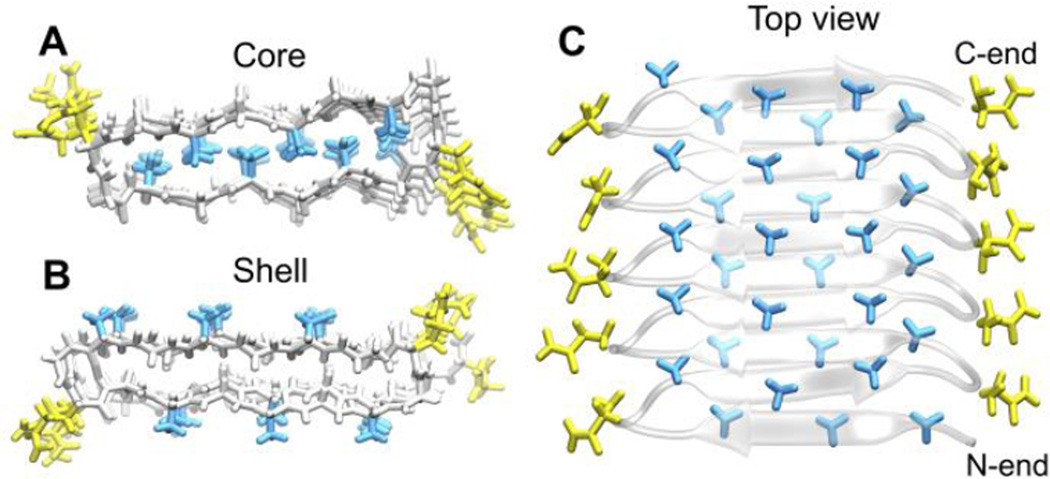
For the sequence (GAGAGAGQ)10 we constructed the hydrophobic core structure from the PDB structure for the silk block (2SLK)17 containing (GAGAGA). We obtained the hydrophobic shell structure from Schor et al. and used it as the initial configuration for our simulations14. Both the hydrophobic shell and the hydrophobic core structures have the β roll backbone shown in Figure 1. A typical β roll structure has two flat surfaces containing parallel β sheets that are connected by β turns; it is left-handed starting from the N terminus. Since both the core and shell structures have a β-roll backbone, the difference between the two structures resides in the directionalities of the alanine side chains as shown on the left panel of Figure 2. For the hydrophobic core structure, all of the hydrophobic side chains lie inside the roll, while for the shell structure, they lie on the surface of the roll.
Figure 2.
Front view of both hydrophobic core (A) and hydrophobic shell (B) structures. All the backbone atoms are in white. Side chains of alanine are in blue, glycine in white, and glutamines in yellow. In the top view of hydrophobic core structure (C), hydrophobic side chains in bright blue are pointing towards us, side chains in shaded blue are pointing away from us into the paper.
Two things are worth noticing in Figure 2 for both core and shell structures. First, the alanine (methyl group) side chains on strands in a surface align perpendicular to the direction of the β sheets (Figs 2A and 2B), and second, the alanine side chains in the core structure emanating from the top surface interdigitate with those emanating from the bottom surface as shown in Figure 2A and 2C. The side view of the shell structure (Fig 2B) shows that if a molecule in that structure has another molecule stacked on top of it, the hydrophobic side chains between them will interdigitate in the same way as they do in the single molecule core structure. This interdigitation in both the core and shell structures of the β roll also occurs for the (GAGAGA) blocks in silk, as is seen in the crystal structure (2SLK)17. Figure 1 indicates that the shell and core structures have very similar dimensions to each other. These are consistent with the dimensions observed in experiment (the distance between the turns on the left and the right sides of the roll is around 2.8 nm in Figure 2A and Figure 2B)15. To determine which structure is the more probable, we next look in detail at the dihedral angles for both structures.
Core structure has type II β turns that match NMR observations
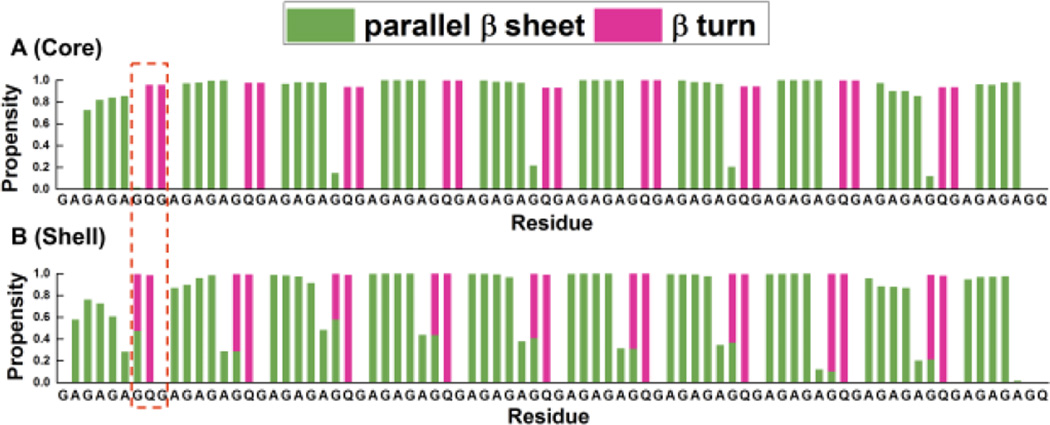
Since the directions of the alanine side chains in the core and shell structures are opposite, the dihedral angles of the residues, especially the ones on the turns, have to be different, because they govern the orientation of the alanine side chains on the strand. After the construction of turns to obtain β rolls, as described above, we simulated both a single shell and a single core molecule in explicit solvent to relax the structures, and then investigated the secondary structure propensities of the residues in these two structures. Simulation details can be found in the methods section. The ‘secondary structure propensity’ (defined as the percentage of the time during which a residue belongs to a particular secondary structure) of all the amino residues in both structures is shown in Figure 3.
Figure 3.
Secondary structure propensity versus residue of (A) core and (B) shell structures. Parallel β sheets are in olive and β turns are in pink. Different positions of turns are squared with dashed red line.
For these data, we used the last 20 ns of the entire 50 ns simulation. We consider only two possible types of secondary structures here: parallel β sheet and β turn. The β turns always contain four amino acid residues labeled i, i+1, i+2 and i+3. The different types of β turns are distinguished by the (ϕ, ψ) dihedral angles for residues i+1 and i+2. Therefore, when calculating the propensity for β turns, we only consider residues i+1 and i+2.
As we can see from Figure 3, the core structure (Figure 3A) has its turn (pink) on the adjacent Q and G, but the shell structure (Figure 3B) has its turn on the adjacent G and Q. We also observe that the turn in the core structure is more stable than the turn in the shell structure because the propensity for a turn at Q-G is close to 1.0 in the core structure, but the propensity for a turn at the G in G-Q is only around 0.6 in the shell structure. Moreover, we find that the G in the G-Q turn of the shell structure has a propensity to form partly parallel β sheets and partly β turns. Not only does the β turn have different locations in the sequence, the locations for the parallel β strands on the core and shell structures are also different. Strands with parallel β sheet structure in the core structure lie at residues G-A-G-A and the turns lie at residues G-Q-G-A, while in the shell structure the strands with parallel β sheet structure lie at residues A-G-A-G and the turns lie at residues A-G-Q-G.
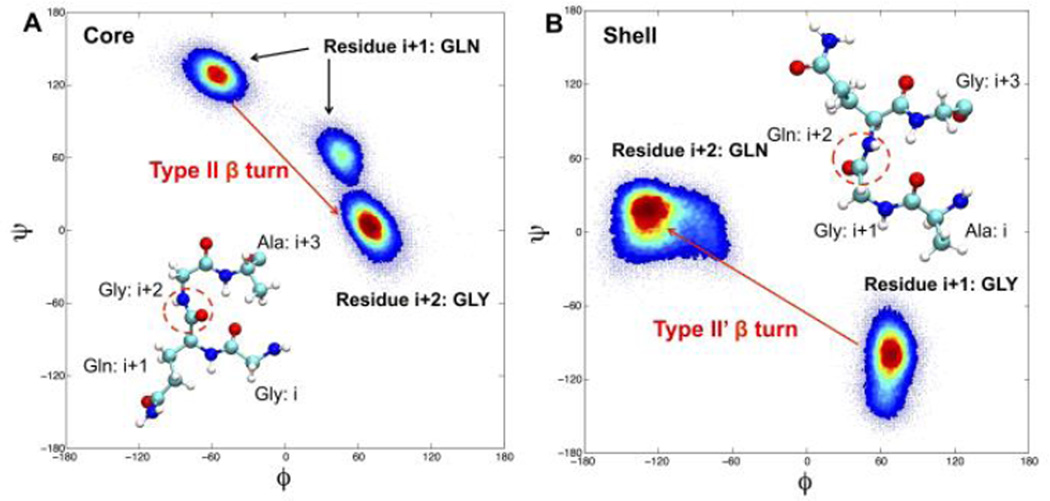
The secondary structure propensity in Figure 3 tells us the locations of the turns, but not their types. Identifying the type of β turn is useful because different types of β turns possess very different (ϕ, ψ) dihedral angles at residues i+1 and i+2, and these can be determined in experiments through solid-state NMR experiments such as those conducted by Gullion et al18. Therefore, we specifically calculated the (ϕ, ψ) angles of the residues in the Q-G turn of the core structure and in the G-Q turn of the shell structure; we show these in the Ramachandran plots in Figure 4. We can see from the left panel in Figure 4, that for core structures the turn residue Q (i+1) has its (ϕ, ψ) angle at ~ (−60, 133), and the turn residue G (i+2) has its (ϕ, ψ) angle at ~ (70,10). This is a typical type II β turn as described by Rose et al.19. This finding is consistent with a solid state NMR study conducted by Gullion et al.18 on the silk mimic sequence (GA)15, where it was found that the turn occurs right in the middle of the sequence and is a type II β turn consisting of residues G (14th), A(15th), G(16th) and A(17th). The A (15th) residue on their sequence has a (ϕ, ψ) angle of around (−63, 140) which closely matches our (ϕ, ψ) angle of Q (−60, 133). Theoretical studies on the neighbor-dependent Ramachadran probability distributions of a series of amino acids20 also reported that a Q followed by a G has a higher probability to be in the region around (−60, 130) as occurs in the Ramachadran plot of the core structure than in the region around (−130,30) as occurs in the Ramachadran plot of the shell structure.
Figure 4.
Ramachandran plot for the Q-G turn in core structure (A) and G-Q turn in shell structure (B). Different types of β turns are labeled as type II β turns for core structure and type II’ β turns for shell structure. Insets are the molecular structures of both turns, carbon atoms are in cyan, nitrogen in blue, oxygen in red and hydrogen in white. The different between the atomic arrangements of the two turns are circled in red.
In our shell structure (Fig 4B), we see that the residue i+1 has a high intensity region at ~(60, −120) and the residue i+2 has a high intensity region at ~(−110, 0). This is close to a type II’ β turn21, which is different from the type II β turn reported by Gullion et al.18 for the poplypeptide with sequence (GA)15.
Ground state analysis shows that the core stack has a lower potential energy than the shell stack
We use ground state analysis to compare approximate potential energies between a stack of N molecules in the core structure and a stack of N molecules in the shell structure. To get an estimate of the potential energy, we simply take the two configurations as constructed, and calculate the number of hydrogen bonds and the number of sidechain-sidechain hydrophobic interaction contacts. Each molecule in the stack has n repeating octamers GAGAGAGQ.
First we calculate the hydrogen-bonding energy. Contributions to the number of hydrogen bonds come from the main chain, partly from beta sheets and partly from turns. Since glutamines are always aligned on the turn of the β roll structure for both shell and core structures, we neglect the interactions between glutamine side chains. Each GAGA strand contributes 4 hydrogen bonds, except when it has neighboring strands on only one side. Each turn contributes one internal hydrogen bond, and one hydrogen bond with the next molecule in the stack. Correcting for residues at the ends, the turn and sheet contributions sum up to 6n − 12 + (N − 1)(n − 2) hydrogen bonds, for both the core and the shell configuration (see Appendix for a more extensive account). Hence, at this level of approximation, there is no difference between the number of hydrogen bonds in the core stack and in the shell configuration.
There is a finite difference between the hydrophobic interaction energy between in the core and shell stacks. If we count the number of neighbors that alanine side chains have on different strands or molecules, (hence, we ignore neighbors on the same chain) we find zero for a single molecule in the shell configuration, and 5n – 5 for the hydrophobic core. However, when we stack N molecules, each interface between two shell-type molecules contributes the same number of hydrophobic interactions; there are N – 1 such interfaces in the stack. Hence the stack of the hydrophobic core structure has 5n – 5 hydrophobic contacts more than that of the hydrophilic core structure, and is therefore, at this level of approximation, the more favorable one. This is not surprising as it lacks the two hydrophobic faces at both ends of the stack, which have a higher free energy because of hydrophobic hydration.
Other properties also show that the core structure is more energetically favorable than the shell structure
Based on the ground states analysis, we conclude that from a crude and purely energetic viewpoint (i.e., neglecting thermal motion) we should expect the core structure to be the more stable one. In the present section, we challenge that expectation by means of atomistic MD simulations, using explicit solvent. We first consider single molecules, starting from nearly perfect shell and core structures, respectively.
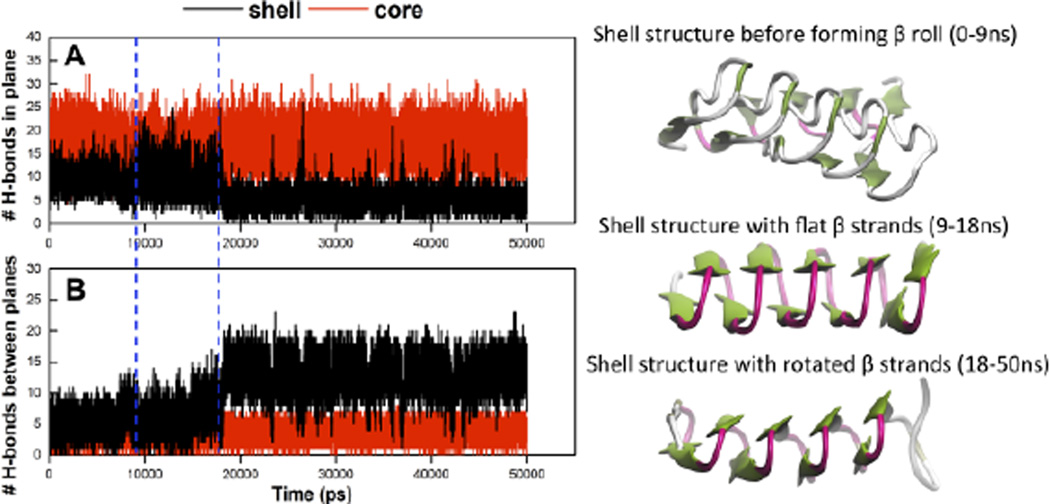
We begin by examining the number of hydrogen bonds as a function of simulation time. Hydrogen bonds were defined to occur when residues satisfied certain distance and angle criteria: the distance had to be within a cut-off of 3.5 Å, and the angle had to be lower than a cut-off of 30°. In Figure 5A, we plot the time-dependent number of hydrogen bonds formed in the top and bottom planes and in 5B the number of hydrogen bonds formed between the top and bottom planes for both core (red) and shell (black) structures.
Figure 5.
(A) Number of hydrogen bonds in top and bottom planes versus time and (B) number of hydrogen bonds between top and bottom planes (B) versus time. Number of hydrogen bonds for core structure is in red, for shell structure is in black. Shell structure in different forms are shown on the right, it does not form a perfect β roll before 9ns, from 9ns to 18ns, shell structure is in a β roll with flat β strands, after 18ns, β strands start to rotate, blue dashed lines indicate 9 ns and 18 ns separately. Turn structure is in pink, β bridge is in tan and β sheet is in green.
For the core structure, the number of in-plane H-bonds is stable over the entire simulation (Figure 5A). In contrast, the number of in-plane H-bonds for the shell structure increases somewhat after ~ 9 ns, stays constant for a short time and then abruptly decreases at 18ns, reaching a plateau which continues until the end of the simulation. We also notice that the number of in-plane H-bonds for the core structure is always higher than that for the shell structure. This indicates that the core structure conformation is better able to maintain the initially-constructed β roll structure than the shell structure.
As shown in Figure 5B, the number of out-of-plane H-bonds (between the top and bottom planes) for the core structure remains stable and lower than that for the shell structure over the entire simulation. The number of out-of-plane H-bonds for the shell structure increases until ~ 18 ns (Figure 5B), where the in-plane H-bonds break (Figure 5A). The right side of Figure 6 shows three representative shell structures associated with the three distinct time periods 0–9 ns, 9–18 ns and 18–50 ns. The structure is initially not well ordered. It becomes more of a standard β roll after 9 ns, causing the peptide to have more in-plane H-bonds than it had before 9ns. However, after 18 ns, the β strands start to rotate and form out-of-plane H-bonds, at the expense of in-plane H-bonds.
Figure 6.
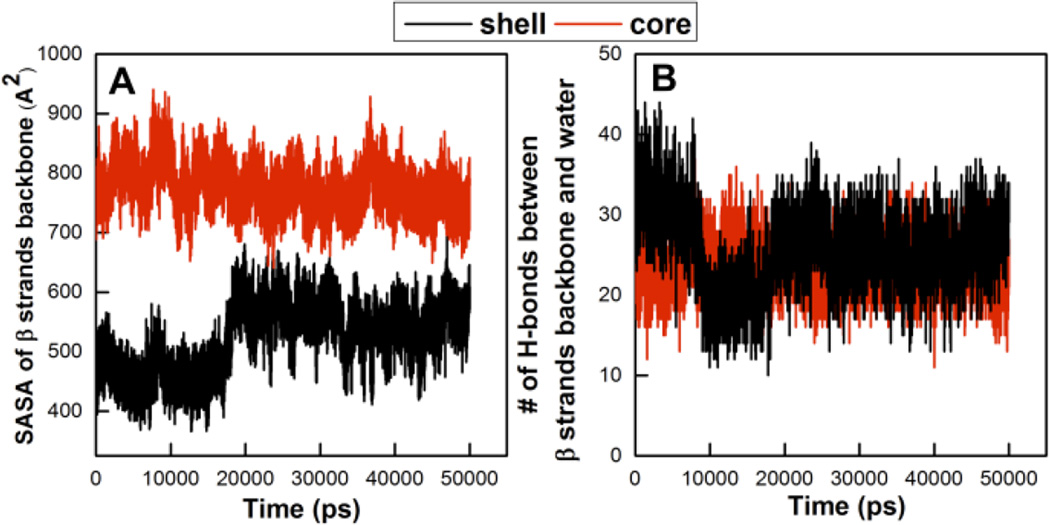
Solvent accessible surface area of the backbone of β strands (A) versus time and the number of H-bonds between backbone of β strands and water (B) versus time for both core (red) and shell (black) structures.
The hydration of the β strand backbone atoms in the core and shell structures was analyzed in an effort to learn why it is harder for the shell structure to maintain the standard β roll conformation and, hence, why the shell structure rotates its β strands. In Figure 6A, we plot the solvent accessible surface area (SASA) of the β strand backbone atoms in both the core (red) and shell (black) structures versus simulation time. There is a sharp increase in the SASA of the β strand backbone atoms for the shell structure after ~18 ns. This signifies that after 18 ns the β strand backbone atoms in the shell structure get more exposed to water, as it adopts a more favorable conformation. We therefore also plot the number of hydrogen bonds between the β strand backbone atoms and water molecules versus simulation time for both core and shell structures (Figure 6B). The shell structures exhibits three distinct behaviors over time. Initially, there are many bonds with solvent. The number of hydrogen bonds then decreases dramatically at ~ 9 ns as the shell structure adopts a more standard β roll structure with more hydrogen in-plane bonds than the structure before 9ns. The number of hydrogen bonds subsequently increases at 18 ns when the β strands rotate, meaning that the backbone atoms on the β strands in the shell structure get more exposed to water after 18 ns.
We also consider the energy of a dimer, i.e., a stack of two molecules. Table 1 lists (1) the internal energies of the two stacks, Epeptide-peptide, (2) the interaction energies between the peptides in each stack and surrounding water molecules, Epeptide-water, and (3) their sum. The internal energy includes both bonded and non-bonded energies. We see that the internal energy of the core stack is 135.79 kcal/mol higher than that of the shell stack. The difference mainly comes from the energy associated with dihedral angles; the core structure with type II β turns has a higher dihedral angle energy than the shell structure with type II’ β turns. This was apparently suspected by the authors of ref. 14, which made them decide not to consider the core structure any further. However, the interaction energy between the peptides in the core stack and water is 256.81 kcal/mol lower than that between the peptides in the shell stack and water. The peptides in the core stack have more interactions with water molecules than in the shell stack because their hydrophobic side chains are buried inside, leaving all of the backbone atoms exposed to water molecules. Finally, it is worth noting that the sum of the internal energy of the stacked peptides and the interaction energy between the peptides in the stack and water for the core stack is 121.02 kcal/mol lower than that for the shell stack, which means that the core stack is indeed more stable than the shell stack.
Table 1.
Internal energy of the stack, Epeptide-peptide, and potential energy between stack and water, Epeptide-water, for core and shell stacks. Internal energy includes bonded (bond, angle and dihedral) and nonbonded (VdW and ELE) energies. The difference between the potential energy of the core stack and the shell stack is Ecore-Eshell.
| Potential Energy (kcal/mol) | Ecore | Eshell | Ecore-Eshell |
|---|---|---|---|
| Epeptide-peptide | −247.44±0.72 | −383.23±0.83 | 135.79 |
| Epeptide-water | −2193.40±1.19 | −1936.58±1.34 | −256.81 |
| Sum | −2440.84 | −2319.81 | −121.02 |
The stack of peptides in the core structure has more internal hydrogen bonds and more hydrogen bonds with water than that in shell structure. As shown in Table 2, the core stack has on average 5.73 more internal hydrogen bonds than the shell stack. In addition, the core stack has 12.98 more hydrogen bonds with water than the shell stack, which indicates that the core stack is better hydrated and is more stable in a water environment than the shell stack.
Table 2.
Number of H-bonds within the stack Npeptide-peptide, and between stack and water Npeptide-water for both the core and shell stacks. The difference between the number of H-bonds in the core and shell stack structures is Ncore-Nstack.
| Number of H-bonds | Ncore | Nshell | Ncore-Nshell |
|---|---|---|---|
| Npeptide-peptide | 72.42±0.11 | 66.69±0.11 | 5.73 |
| Npeptide-water | 141.34±0.14 | 128.37±0.14 | 12.98 |
| Sum | 213.42 | 195.06 | 18.71 |
The number of intramolecular hydrogen bonds for a single peptide increases when it is stacked with another peptide. In Table 3, the number of hydrogen bonds of a single peptide when it is in a stack and when it is not in a stack are calculated for both core and shell structure. We find that peptides have ~ 6 more hydrogen bonds when they are in a stack than when they are not. This indicates that the stacking has a stabilizing effect on the structure of each individual peptide in the stack.
Table 3.
The number of intramolecular hydrogen bonds within a single peptide when it is not in a stack, Nsingle-peptide and when it is in a stack, Nstacked-peptide and their difference, Nstacked-peptide - Nsingle-peptide.
| core | shell | |
|---|---|---|
| Nsingle-peptide | 27.57±0.07 | 24.74±0.06 |
| Nstacked-peptide | 34.05±0.08 | 31.00±0.05 |
| Nstacked-peptide- Nsingle-peptide | 6.48 | 6.26 |
Conclusions
The silk-like peptide (GAGAGAGX)n has been found in experiments to form fibrils by stacking one on top of another. The basic unit for these structures has been proposed to be a single peptide in a β roll structure with all hydrophobic side chains pointing out of the roll, what we called a hydrophobic shell structure. However, since most β solenoid peptides possess a hydrophobic core, i.e., the hydrophobic side chains lie inside the solenoid, we revisit this problem here. We construct a new type of β roll structure with the same dimensions as the hydrophobic shell structure, but with hydrophobic side chains pointing into the roll, what we call the hydrophobic core structure. We then compare the stability of these two structures using ground state analysis and atomistic molecular dynamics simulations and find that the core structure is a more probable structure than the shell structure.
By comparing secondary structures and Ramachandran plots, we find that peptides in the hydrophobic shell structure have type II’ β turns comprised of A-G-Q-G while peptides in the hydrophobic core have type II β turns comprised of G-Q-G-A. Our finding of type II β turns in the hydrophobic core structure matches well with a solid state NMR study18 conducted on sequence (GA)15 which found a type II β turn in the middle of the sequence. Moreover, theoretical predictions20 also support type II β turn for the sequence Q-G.
We conducted a ground state analysis in which each peptide was assumed to be in a perfect β roll structure. The ground state analysis suggested that the hydrophobic core stack is more stable than the hydrophobic shell stack and that the difference becomes larger as the peptides get longer. Interestingly, the relative stability of the core stack compared to the shell stack was independent of the number N of molecules in the stack, since it mainly comes from the fact that a stack of shell structure features two unfavorable hydrophobic faces, one at either end.
Atomistic simulations of single peptide molecules with core and shell structures show that the hydrophobic shell structure is less stable than the core structure. During the simulation, a molecule in the shell structure tends to rotate its β strands to form hydrogen bonds between top and bottom layers, which then enhances exposure of their backbones to water. In contrast, a single molecule in the hydrophobic core structure remains stable in the system throughout the simulation. Further simulations of two stacked peptides in both core and shell structures reveal that the core stack has lower interaction energy with water than the shell stack. We also observe that the core stack has a higher number of intra-molecular hydrogen bonds and a higher number of hydrogen bonds between stack and water than the shell stack. Moreover, each peptide in the stack has more intra-molecular hydrogen bonds than when isolated. The stacking of peptides clearly helps to stabilize the folded structure. This is probably relevant for the mechanism by which the molecules change configuration from a less ordered state in solution to a more ordered one in the stack; it seems likely that this occurs in contact with a folded molecule sitting at the (growing) end of a stack of pre-folded molecules, i.e., the roll forms after docking.
In summary, we reported here that the hydrophobic core structure is more probable than the shell structure for the silk-like sequence (GAGAGAGX). This was accomplished through comprehensive analyses of simulations of a single (GAGAGAGQ)10 molecule and a stack of two (GAGAGAGQ)10 in both the hydrophobic core and hydrophobic shell structure. In the future, we plan to investigate the mechanism by which the peptide docks onto the template and folds on top of it using a combination of molecular dynamics simulation and accelerated molecular dynamics.
Method
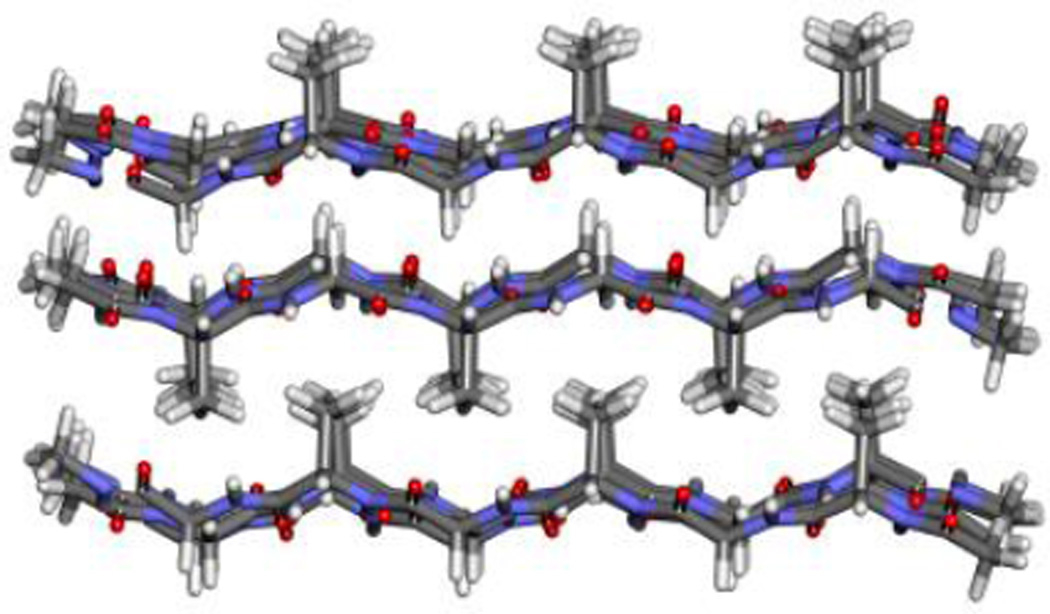
We performed explicit-solvent, atomistic molecular dynamics simulations on (GAGAGAGQ)10 using Amber 12 with the ff12SB force field22. The PDB for the initial configuration of the hydrophobic shell structure was provided by the Bolhuis14 group. The initial configuration for the hydrophobic core structure was constructed using the 2SLK silk block in the pdb bank [16]. The β strands in the silk block, AGAGAG, form three layers of anti-parallel β sheets as shown in Figure 7.
Figure 7.
2SLK silk block structure from the pdb bank.
The initial configuration for the hydrophobic core structure was created by placing the bottom two layers of the silk block next to each other since these have all the hydrophobic side chains between them. The strands in one layer are then connected with the strands going in the opposite direction in the other layer via turns that contain the missing residues G-Q-G-A, leading to a single β roll structure with the sequence (GAGAGAGQ)10. The peptide was solvated in a box with explicit TIP3P water molecules23. The initial configurations for the two-molecule stack were made by stacking the peptides on top of each other. All the simulations were conducted at 300 K.
Each simulation consists of the following steps. A 10,000-step solvent minimization was performed using the steepest descent method with the peptide constrained by a force of 200 kcal/mol. The system was then gradually heated over 10 ps to 300 K with all the atoms in the peptide restrained via a force of 200 kcal/mol. A short 40 ps NPT ensemble MD run was performed with the peptide restrained by a force of 200 kcal/mol, after which another 10,000-step minimization was conducted with a 25 kcal/mol restraint, followed by another 20 ps NPT ensemble MD run via a force of 25 kcal/mol. Next another 10,000-step unconstrained minimization was performed, after which there was a reheating over 40ps at a constant volume to 300K. Finally, an NPT ensemble MD run was performed to ensure uniform solvent density. The simulation temperature was maintained by using Berendsen thermostat24. Partial mesh Ewald (PME) summation25 was used to calculate the long-ranged electrostatic interactions and a 9 Å cut-off radius and a 1E-5 tolerance for the Ewald convergence were used to calculate the non-bonded interaction. The SHAKE algorithm26 was used to constrain bonds involving hydrogen atoms. The run times for the simulations were 50 ns for systems with single peptide, and 180 ns for systems with stacked peptides. The last 20 ns of the trajectories from simulations with a single molecule and the last 30 ns of the trajectories from simulations with stacked peptides were considered for statistical analysis. Ambertools, VMD, NAMD and in-house scripts were used for statistical analysis. Secondary structures were determined using the DSSP algorithm27, which has been widely used as a standard method for assigning secondary structures to amino acids.
Acknowledgments
M.A. CS acknowledges financial support from ERC Advanced Grant 267254 (BioMate) for his stay at NCSU which enabled him to carry out this work. CKH acknowledges support by the Research Triangle MRSEC under grant DMR-1121107 and by the National Institutes of Health USA under grant EB006006.
Appendix: detailed ground state analysis
Let εHB denote the energy for a single hydrogen bond. In a perfect hydrophobic core β roll structure, each octamer consists of a β strand (G-A-G-A) that can form a β sheet via hydrogen bonding with its neighboring strand and a β turn (G-Q-G-A) that can connect two consecutive β strands. In a perfect roll, each β strand (G-A-G-A) can form 8 hydrogen bonds through N-H and C=O groups with its neighbors on both sides, except for the 4 strands at both terminals which only have neighboring strands on one side. So the total number of hydrogen bonds contributed by strands with neighboring strands on both sides is (8(n−4))/2=4(n−4). (It is divided by two because each hydrogen bond is counted twice.) The number of hydrogen bonds contributed by strands with a neighboring strand on only one side is 4*4/2=8. Adding these two numbers together and multiplying by εHB gives us the hydrogen bonding energy contributed by the β sheets, 4(n−2)εHB.
Next we calculate the hydrogen-bonding energy contributed by the β turns in the core structure. Since there are n octamers, the number of β turns should be n−1. Each β turn has a hydrogen bond within the turn between residue i and i+4, so the number of hydrogen bonds inside the β turns is n−1. Each β turn can also form hydrogen bonds with its neighboring turns on both sides, except for the 4 turns at the terminals. The number of hydrogen bonds formed between turns and their neighbors on both sides is 2(n−1−4)/2=n−5. Additionally, the number of hydrogen bonds formed between the 4 turns at each terminal and their neighbor on just one side is 4/2=2. Thus the total number of hydrogen bonds formed by β turns is (n−1)+(n−5)+2=2n−4. Thus in a perfect β roll with n octamers, the hydrogen bonding energy contributed by β turns is (2n−4)εΗB. Therefore the hydrogen bonding energy of a perfect hydrophobic core β roll structure, EHB, is
| (1) |
The hydrophobic interaction is calculated as follows. εAA denotes the interaction energy between an alanine hydrophobic side chain and its closest alanine side chain neighbor on the other plane. For the hydrophobic energy in the hydrophobic core structure, we need to consider three types of hydrophobic side chains, those that have four neighbors, those that have two neighbors and those that have one neighbor. We ignore interactions between alanine side chains on the same plane, because these interactions are the same in both the core or shell structures. Each repeating unit has two alanine residues in the β strand segment and one alanine residue in the β turn segment. So a roll structure with n octamers has a total number of 3n alanine residues. Alanine side chains in the β strand segment (except for the one at each terminal) have 4 neighbors, so the number of side chains that have four neighbors is 2(n−2). Thus the total number of interactions attributed to side chains with 4 neighbors is 4[2(n−2)]/2=4(n−2). As shown in Figure 2 (C), alanine side chains with just 1 neighbor are located at the upper left and the lower right, so there are 2 of them. Thus the total number of interactions contributed by side chains with 1 neighbor is 2*1/2=1. If we subtract the number of side chains with 4 neighbors and the number of side chains with 1 neighbor from the total number of side chains, we get the number of alanine side chains that have 2 neighbors, which is 3n−2(n−2)−2=n+2. It follows that the total number of interactions contributed by side chains with 2 neighbors is 2(n+2)/2=n+2. The hydrophobic interaction energy of a perfect hydrophobic core β roll structure, EHP, is
| (2) |
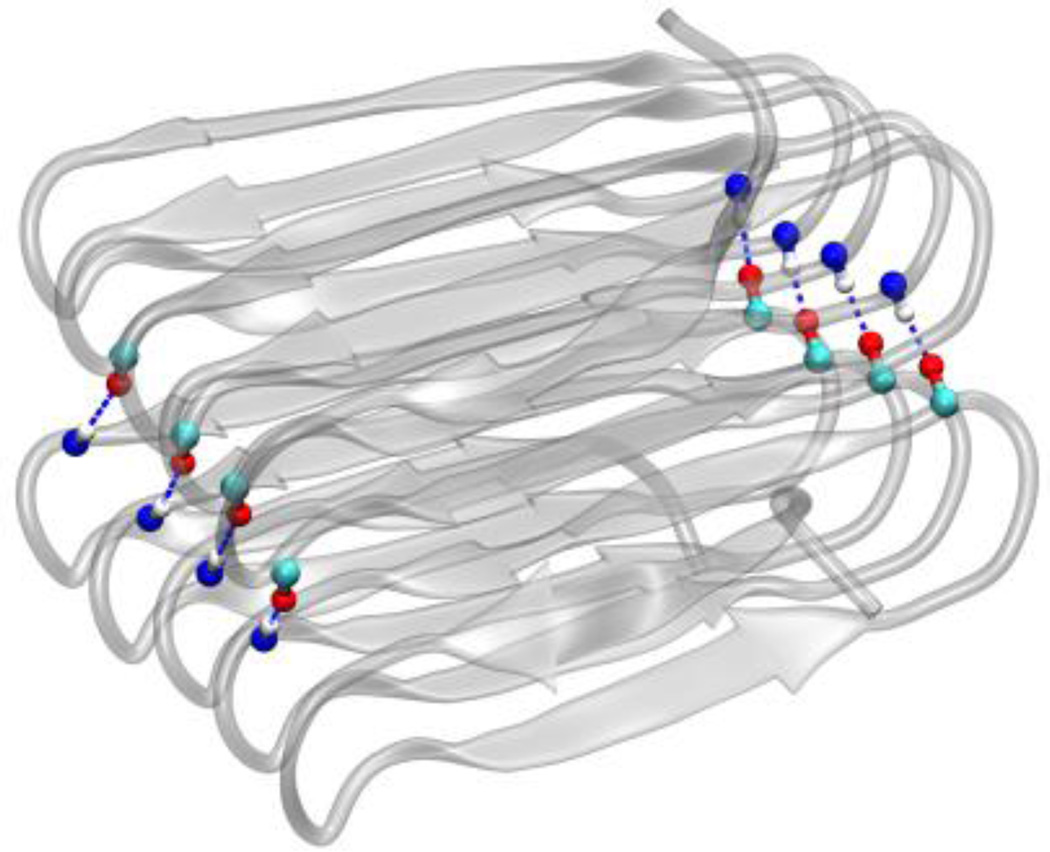
Interactions also occur between neighboring molecules when they form a stack of n molecules. No hydrophobic interactions occur within the interface between two molecules of hydrophobic core β roll structure, because all the hydrophobic side chains are buried within the β roll structure. However, each β turn has an N-H and a C=O group pointing out of the turn structure, which enables the two molecules to form hydrogen bonds on both sides through the turns. As is shown in Figure 8, a β roll structure with 10 octamers can form 8 hydrogen bonds with its nearest molecules when stacking. Therefore, a β roll structure with n octamers can form n−2 hydrogen bonds with its neighboring molecule within their interface. A stack of N molecules with the core structure has (N−1) interfaces. The interaction energy between N hydrophobic core molecules at the interface, , is
| (3) |
Figure 8.
Both core and shell stacks have hydrogen bonds between the two molecules in the interface on both sides of the β roll structure. Carbon is in cyan, oxgen is in red and hydrogen is in white. Both the N-H and C=O group belongs to the residues in β turns.
For a stack of N molecules in the core structure, the total potential energy of the stack Ecore-stack consists of the internal energy (hydrogen bonding and hydrophobic interaction energy) EHB+EHP within each molecule, and the interaction energy at the interfaces between the neighboring molecules. Ecore-stack has the following form,
| (4) |
By plugging equation (1), (2) and (3) into equation (4), we arrive at the final expression for the total potential energy of a stack consists of n molecules with perfect hydrophobic core β roll structures, Ecore-stack, as
| (5) |
The energy Eshell-stack of a stack of molecules with hydrophobic shell structures can be derived by following the same steps as for Ecore-stack. In a perfect hydrophobic shell β roll structure, each repeating unit consists of β strand (A-G-A-G) that can form β sheets via hydrogen bonding with its neighboring strands. The strands are connected with β turns (A-G-Q-G). Even though a hydrophobic shell molecule has its β strands and β turns at a slightly different position, it still has the same number of backbone hydrogen bonds as a hydrophobic core molecule. Thus the hydrogen bonding energy in a single hydrophobic shell molecule, EHB, is the same as in equation (1).
The hydrophobic shell structure has all of its hydrophobic side chains pointing out, so there are no interactions between hydrophobic side chains on the different planes within a single molecule. Thus the hydrophobic interaction energy between these side chains is zero. The hydrophobic interaction of a hydrophobic shell structure only occurs when one molecule stacks with another molecule. Therefore, the total energy of a stack of N molecules with perfect hydrophobic shell β roll structure, Eshell-stack, consists of the internal energy of each molecule, EHB, and the interaction energy of the interfaces between neighboring molecules, ,
| (6) |
The interface between two hydrophobic shell molecules comprises both hydrogen bonding interactions and hydrophobic interactions. The hydrogen bonding interactions at the interface between two shell molecules is the same as that between two core molecules, and can be expressed, as in equation (3), as (N−1)(n−2)εHB for a stack of N molecules with (N−1) interfaces. Hydrophobic interactions between side chains at the interface between two shell molecules have the same expression as equation (2). Therefore, the hydrophobic energy at interfaces for a stack of N molecules should be (N−1)(5n−5)εAA. Thus the interaction energy between two shell molecules at the interface, , is
| (7) |
By plugging equation (1) and (7) in equation (6), we get the final expression for the total potential energy of a stack consisting N molecules with perfect hydrophobic shell β roll structures, Eshell-stack as
| (8) |
Finally, we subtract Eshell-stack from Ecore-stack to get the energy difference between the two types of stacked structures, as shown in the following equation
| (9) |
Since εAA is less than zero, Δ is also less than zero, which means that the core stack has a lower potential energy than the shell stack. The value of Δ depends linearly on the number of octamers n.
Notes and references
- 1.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17342. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Protein Sci. 2008;17:1467. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnhart MM, Chapman MR. Annu. Rev. Microbiol. 2006;60:131. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Der-Sarkissian A, Jao CC, Chen J, Langen R. J Biol. Chem. 2003;278:37530. doi: 10.1074/jbc.M305266200. [DOI] [PubMed] [Google Scholar]
- 6.Middleton AJ, Marshall CB, Faucher F, Bar-Dolev M, Braslavsky I, Campbell RL, Walker VK, Davies PL. J Mol. Biol. 2012;416:713–724. doi: 10.1016/j.jmb.2012.01.032. [DOI] [PubMed] [Google Scholar]
- 7.Wimley WC. Curr. Opin. Struct. Biol. 2003;13:404. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 8.Hennetin J, Jullian B, Steven AC, Kajava AV. J Mol. Biol. 2006;358:1094. doi: 10.1016/j.jmb.2006.02.039. [DOI] [PubMed] [Google Scholar]
- 9.Kajava AV, Steven AC. Fibrous Proteins: Amyloids, Prions and Beta Proteins. Elsevier; London: 2006. [Google Scholar]
- 10.Mita K, Ichimura S, James TC. J Mol. Evol. 1994;38:583. doi: 10.1007/BF00175878. [DOI] [PubMed] [Google Scholar]
- 11.Krejchi MT, Cooper SJ, Deguchi Y, Atkins E. Biophys. J. 1997;30:5012. [Google Scholar]
- 12.Beun LH, Beaudoux XJ, Kleijn JM, de Wolf FA, Cohen Stuart MA. ACS Nano. 2012;6:133. doi: 10.1021/nn203092u. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez-Garcia A, Kraft DJ, Janssen AFJ, Bomans PHH, Sommerdijk NAJM, Thies-Weesie DME, Favretto ME, Brock R, de Wolf FA, Werten MWT, van der Schoot P, Stuart MC, de Vries R. Nature Nanotechnology. 2014;9:698. doi: 10.1038/nnano.2014.169. [DOI] [PubMed] [Google Scholar]
- 14.Schor M, Martens AA, deWolf FA, Cohen Stuart MA, Bolhuis PG. Soft Matter. 2009;5:2658. [Google Scholar]
- 15.Martens AA, Portale G, Werten MWT, de Vries RJ, Eggink G, Cohen Stuart MA, de Wolf FA. Macromolecules. 2009;42:1002. [Google Scholar]
- 16.Johnson KG, Jackson JBC, Budd AF. Science. 2008;319:1521. doi: 10.1126/science.1152197. [DOI] [PubMed] [Google Scholar]
- 17.Fossey SA, Némethy G, Gibson KD, Scheraga HA. Biopolymers. 1991;31:1529. doi: 10.1002/bip.360311309. [DOI] [PubMed] [Google Scholar]
- 18.Gullion T, Kishore R, Asakura T. J Am. Chem. Soc. 2003;125:7510. doi: 10.1021/ja0342345. [DOI] [PubMed] [Google Scholar]
- 19.Rose GD, Glerasch LM, Smith JA. Advances in Protein Chemistry. London: Elsevier; 1985. [DOI] [PubMed] [Google Scholar]
- 20.Ting D, Wang G, Shapovalov M, Mitra R, Jordan MI, Dunbrack RL. PLoS Comput Biol. 2010;6:e1000763–e1000721. doi: 10.1371/journal.pcbi.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson EG, Thornton JM. Protein Sci. 1994;3:2207. doi: 10.1002/pro.5560031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Götz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER 12. San Francisco: University of California; 2012. [Google Scholar]
- 23.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem. Phys. 1983;79:926. [Google Scholar]
- 24.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem. Phys. 1984;81:3684. [Google Scholar]
- 25.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem. Phys. 1995;103:8577. [Google Scholar]
- 26.Ryckaert J-P, Ciccotti G, Berendsen HJC. J Comput. Phys. 1977;23:327. [Google Scholar]
- 27.Kabsch W, Sander C. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]