Abstract
Purpose
We aimed to identify DNA methylation biomarkers of progression free survival (PFS) to platinum-based chemotherapy in high grade serous ovarian cancer (HGSOC) within biologically relevant ovarian cancer associated pathways.
Experimental Design
Association with PFS of CpG island (CGI) promoter DNA methylation at genes in the pathways Akt/mTOR, p53, redox and homologous recombination DNA repair was sought with PFS as the primary objective in a prospectively collected ovarian cancer cohort (n=150). Significant loci were validated for associations between PFS, methylation and gene expression in an independent TCGA data set of HGSOC (n=311).
Results
DNA methylation at 29 CGI loci linked to 28 genes was significantly associated with PFS, independent from conventional clinical prognostic factors (adjusted p<0.05). Of 17 out of the 28 genes represented in the TCGA data set, methylation of VEGFB, VEGFA, HDAC11, FANCA, E2F1, GPX4, PRDX2, RAD54L and RECQL4 was prognostic in this independent patient cohort (one-sided p<0.05, FDR<10%). A multivariate Cox model was constructed, with clinical parameters (age, stage, grade and histological type) and significant loci. The final model included NKD1, VEGFB and PRDX2 as the three best predictors of PFS (p=6.62x10-6, permutation test p<0.05). Focussing only on known VEGFs in the TCGA cohort showed that methylation at promoters of VEGFA, VEGFB and VEGFC was significantly associated with PFS.
Conclusions
A three loci model of DNA methylation could identify two distinct prognostic groups of ovarian cancer patients (PFS: HR=2.29, p=3.34×10-5; Overall Survival: HR= 1.87, p=0.007) and patients more likely to have poor response to chemotherapy (OR=3.45, p=0.012).
Keywords: Prognostic biomarkers, Ovarian Cancer, CpG island, DNA methylation
Introduction
There are an ever-increasing number of emerging novel agents being examined in clinical trials of ovarian cancer (1). However, debulking surgery with platinum-based chemotherapy remains the cornerstone of treatment at first presentation. Initial response rates are generally good (>75%), but patients relapse and will eventually develop resistant disease leading to treatment failure. Length of progression-free survival (PFS) of patients from primary presentation is an indication of whether patients will respond to second line platinum based chemotherapy (2). If robust biomarkers of poor PFS to platinum-based chemotherapy can be identified, then poor prognosis patients can potentially be stratified for novel treatment strategies. This may become particularly relevant for molecular-targeted therapies used in the maintenance setting, where those patients with high risk of relapsing earlier can be identified. DNA methylation has many advantages as a biomarker: its relative stability (in vivo and in vitro), functional links to gene expression and potential to be detected in cell-free DNA from body-fluids (3–5).
Targeted molecular therapies being clinically evaluated in ovarian cancer include angiogenesis inhibitors. Angiogenesis has been shown to be a crucial requirement for metastatic ovarian cancer and the development of ascites (6). Clinical trials using Bevacizumab, a humanised monclonal antibody targeting the pro-angiogenic vascular endothelial growth factor A (VEGFA), in combination with conventional chemotherapy have shown that Bevacizumab in combination with first line chemotherapy have improved PFS times in late stage ovarian cancer patients (7, 8). Prolonged PFS times have also been observed using Bevacizumab in addition to chemotherapy in recurrent chemo-resistant ovarian cancer (9, 10). However, pre-selection of patient subgroups based on their molecular and histological subtypes may be required in order for patients to optimally benefit from targeted agents beyond that achieved with conventional therapies.
In this study, we aimed at identifying the prognostic value of DNA methylation at GpG islands at the promoter of genes associated with known pathways involved in ovarian cancer development and progression. These were mainly genes and pathways as defined in Kyoto Encyclopedia of Genes and Genomes (KEGG) and included Akt/mTOR, p53, BRCA1/2, Redox and homologous recombination associated pathways. Previously we have shown that multiple CpG islands associated with the Wnt pathway significantly associate with PFS independently from clinical parameters (11). Together with the present study, we demonstrate the potential of DNA methylation biomarkers to be used for patient stratification for targeted care in clinical practice.
Methods & Materials
Patients
Tumour biopsies were prospectively collected in an ongoing Scottish Gynaecology Clinical Trial Group (SGCTG)/National Cancer Research Institute (NCRI) cohort study. Primary tumours included in the current study were restricted to those from patients with confirmed epithelial ovarian cancer (EOC) excluding clear cell and mucinous tumours, and treated with cytoreductive surgery followed by platinum-based chemotherapy. Biopsies of tumour were obtained at initial laparotomy or laparoscopic biopsy at the same time as diagnostic biopsy. All samples analysed were collected prior to chemotherapy.
The primary end-point of this study is to systematically examine any association between promoter methylation and PFS, defined as the time from the start of first-line chemotherapy to progressive disease or early death due to EOC or other causes. The secondary end points are the association of promoter methylation with response to platinum-based chemotherapy measured by RECIST 1.0 criteria and overall survival. Progression and survival status was assessed 2 monthly for the first two years post-treatment and subsequently 6 monthly to 5 years and annually thereafter as defined in the protocol. The study has been approved by MREC for Scotland (reference number 01/165). Genomic DNA was extracted from fresh frozen tumours for methylation analysis as previously described (12).
One hundred and seventy-nine EOC were used in this study. Twenty samples were excluded from subsequent analysis due to poor quality of signal intensities in over 10% of probes in the duplicates or methylation controls did not reach acceptance criteria. Nine samples with ovarian tumours only in the ovaries (Stage I) were further excluded. Therefore, the analysis was focused on 150 stage III and IV ovarian tumours. In a subsequent validation stage, data from 311 high-grade late-stage serous tumour samples profiled by The Cancer Genome Atlas (TCGA) project on HumanMethylation27 BeadChip using Illumina Infinium assay were analysed. Full details of clinical parameters are shown in Supplementary Table S1. Throughout this study we have followed the REMARK recommendations (13).
Design of Agilent customised promoter CpG island microarray
Genes involved in Akt/mTOR pathway, p53 pathway, BRCA1/2 pathway, redox pathway, homologous recombination (HR) were mainly collected from Kyoto Encyclopedia of Genes and Genomes (KEGG). Promoter CGIs of those genes were identified as previously described (11). The genomic locations of the targets are specified by Human Mar. 2006 (NCBI36/hg18) assembly. In total, 51 genes represented by 78 loci in the Akt/mTOR pathway, 68 genes by 140 loci in the p53 pathway, 64 genes by 101 loci in the BRCA1/2 pathway, 48 genes by 63 loci in the Redox pathway, 35 genes by 45 loci in homologous recombination were examined in this study. A full list of genes and the genomic location of the promoter regions targeted on the array are in Supplementary Table S2.
The 60mer-oligos/probes targeting those regions were mainly selected from Oligome™ (Oxford Genome Technology, UK). The selected probes were uploaded to eArray (Agilent Technologies, UK) and the 60-mer microarray was fabricated using Agilent SurePrint Technology. The customised array was further evaluated in Differential Methylation Hybridisation (DMH) assay using 0% and 100% methylated samples (Millipore, UK). Only 3% of probes were identified as non-informative either due to lack of McrBC recognition sites within the loci or having low log2 transformed DMH ratios in 100% methylated samples compared to 0% methylated sample (Supplementary Figure S1).
Differential methylation hybridisation (DMH)
Methylation levels of our targets were measured by DMH in duplicates and microarray data pre-processing was done as previously described (11). In brief, DNA was digested with MseI, ligated to an end-linker and divided into two aliquots. One aliquot was mock-treated, the other aliquot was digested with the methylation-sensitive restriction enzyme McrBC (14, 15), followed by PCR amplification. The amplicons labeled with Cy3 or Cy5 were then competitively hybridized to the customized microarrays. Labeling of DNA, array hybridization and image scanning was done according to the standard Agilent aCGH protocol. DMH ratio is the ratio of the signals from McrBC mock digested and McrBC digested samples. The DMH dataset is available at GEO (accession ID: GSE23240). The quality of DMH assay was assessed as previously described (11).
The Cancer Genome Atlas (TCGA) dataset
The level 2 expression dataset on Affymetrix HGU133A microarrays and level 3 methylation dataset on Illumina HumanMethylation27 Beadchip of serous tumours were obtained from TCGA data portal (16). We limited the analysis in late-stage tumours with methylation and expression data, therefore, 311 HGSOC were included in the study.
The expression microarray data have been pre-processed and normalized across the samples, and methylation data have been summarized as β value, which was calculated as M/(M+U), where M is the signals of methylation bead type and U is the signals of unmethylation bead type of the targeted CpG site. The poor quality probes have been excluded by TCGA (16).
Bisulphite pyrosequencing
Bisulphite pyrosequencing was performed in a panel of n=142 HGSOC from the SGCTG cohort to validate the prognostic value of promoter methylation at VEGFA, VEGFB and VEGFC as previously described (11). In brief, 1 µg of genomic DNA was bisulfite modified using the EpiTect Bisulfite Kit (Qiagen, West Sussex, UK) according to the manufacturer's instructions. Pyrosequencing primer sets and PCR conditions are listed in Supplementary Table S3. The methylation was quantified as the percentage of methylated cytosine over the sum of methylated and unmethylated cytosines using Pyro Q-CpG™ software (Biostage, Uppsala, Sweden). The methylation level of the three genes in each sample was calculated by using the average percentages of methylation across all targeted CpG sites in duplicates, respectively, and subsequently, was used as a continuous variable in the Cox model in the survival analysis.
Statistical power estimation
The initial screening set consisted of DMH data from 150 tumours, with 133 (89%) patients having disease progression. To estimate approximate statistical power of this screening set prior to analysis, we assumed 5% of the loci examined in each pathway were true positives and split patients into two groups based on the upper quartile of methylation level at each locus following what we have observed in previous methylation profiling study (11). With a hazard ratio at 2 and false discovery rate (FDR) (17) less than 10%, we estimated the average power of the screening study to be 75% (Supplementary Method 1). In the subsequent analysis methylation levels have been treated as a continuous variable, meaning we are underestimating the study power.
Survival analysis
All the survival analysis was done in R (version 2.10.1) using survival package. The DMH ratios of multiple probes targeting the same locus (MseI fragment) were averaged. The mean value of methylation at the locus in duplicates was then standardized to Z score (Z~N(0, 1)). The Z scores were used as a continuous variable in the Cox model. The proportional hazards were examined before the association between methylation and PFS was examined by univariate Cox model. The hazard ratio was then adjusted by conventional prognostic factors (FIGO stage, grade, histology and age) in multivariate Cox model. The significance of estimated hazard ratios was calculated by Score test in univariate analysis, and Wald test in multivariate analysis. External validation of prognostic value of biomarkers identified from SGCTG cohort was done in TCGA cohort using methylation level (β value) as a continuous variable in univariate analysis.
To determine the best predictors of PFS in patients with late-stage (stage III and IV) ovarian cancer, a multivariate Cox model was constructed using the forward stepwise method based on likelihood ratio statistics with a probability of 0.05 for entry and 0.10 for removal. Among the variables including clinical parameters and validated, independent methylation markers identified in current study and previous study (11) only three methylation markers meet the entry criteria, thus selected into the model in this study (see Results). Subsequently, a methylation index (MI) was calculated using the selected covariates from this model. Permutation test involved in the same process as our modelling procedure including feature selection in the univariate and multivariate analysis adjusted by clinical parameters as well as model construction in the SGCTG cohort was performed 100 times to evaluate the significance of the final multivariate Cox model.
Logistic regression analysis
The correlation between response and promoter methylation in AKT/mTOR pathway, p53 pathway, BRCA1/2 pathway, Redox pathway and HR pathway was tested by logistic regression. Patients were classified as responders (complete or partial response) or non-responders (stable disease or progressive disease or not evaluable response generally due to the poor physical condition of the patients) according to RECIST 1.0 criteria. The analysis was restricted to patients with measurable disease at baseline level. Methylation level was used as a continuous variable as well as a categorical variable in SGCTG cohort and as a categorical variable in TCGA cohort, where the categorical variable was used, the top 20% of the patients with high methylation level at the biomarker examined were categorised into the ‘high methylation group’, otherwise, they were included in the ‘low methylation group’.
We constructed a multivariate logistic regression model incorporating multiple methylation biomarkers selected by forward stepwise likelihood ratio algorithm in SGCTG cohort. The prediction value of these biomarkers was further evaluated in TCGA cohort (Supplementary method 2).
Results
DNA methylation and association with PFS
We have systematically profiled CpG island (CGI) promoter DNA methylation at genes in pathways implicated in ovarian cancer, including p53, AKT, redox and DNA repair pathways (Table 1). Association with PFS was sought in 150 Stage III/IV ovarian tumours prospectively collected through a cohort study (SGCTG cohort). Mucinous and clear cell cancers were excluded due to their different clinical outcome from more common serous and endometrioid EOC (18, 19). Thirty eight loci were identified as significantly associated with PFS (p<0.01 and FDR<10%) (see univariate PFS analysis in SGCTG cohort in Table 1 for a summary and univariate PFS analysis in Supplementary Table S4 for details).
Table 1. A summary of the progression-free survival analysis in 5 key signalling pathways.
| Pathway/family | Source | Total # Genes | Total # CGIs | # loci (130–6000bp) | SGCTG cohort (n=150) | TCGA cohort (n=311) | Genes | |
|---|---|---|---|---|---|---|---|---|
| # loci Univariate PFS analysis P<0.01 and FDR<10% |
# loci Multivariate PFS analysis p<0.05 |
# validated loci/ # loci on the array |
||||||
| Akt/mTOR | hsa041501 | 51 | 51 | 78 | 6 | 4 | 2/3 | VEGFA, VEGFB* |
| p53 | hsa041151 | 68 | 87 | 140 | 14 | 10 | 1/4 | SESN2 |
| BRCA1/2 | Biocompare2 | 64 | 72 | 101 | 10 | 7 | 3/5 | E2F1, FANCA, HDAC11 |
| Redox | Manually curated | 48 | 44 | 63 | 5 | 5 | 2/3 | PRDX2, GPX4* |
| HR | ko034401 | 35 | 31 | 45 | 3 | 3 | 2/2 | RECQL4, RAD54L* |
Kyoto Encyclopaedia of Genes and Genomes (KEGG)
Biocompare website: http://www.biocompare.com/
The locus is significantly correlated with response measured by RECIST criteria (version 1.0) (logistic regression, p<0.05)
The hazard ratios of 38 loci identified as having p<0.01 and FDR<10% in univariate analysis were adjusted by age, stage, grade and histological type and the patients were stratified into three groups who either received platinum alone (n=42), combination of platinum and taxane (n=85), or other platinum-based treatment (n=16). We found hypermethylation at 29 loci linked to 28 genes was associated with increased hazard of disease progression independent from conventional clinical prognostic factors: CGIs at VEGFA, AKT1, ULK2, VEGFB, TP73, CD82, SMARCA2, SESN2, RRM2, LRDD, CCND1, SERPINB5, BAX, BID, GTSE1, HDAC11, HDAC7, FANCA, HDAC5, E2F1, BACH1, TR2IT1, FOXO1, GPX4, PRDX2, RAD54L, RECQL4 and EME (adjusted p<0.05) (see multivariate PFS analysis in Supplementary Table S4).
To validate in a further tumour set the prognostic value of the methylation biomarkers that are independent from clinical parameters, we further analysed associations between progression/relapse-free survival and methylation at these loci in data from an independent cohort (n=311) of HGSOC available through The Cancer Genome Atlas (TCGA) study established by the NCI and NHGRI. The methylation data was generated using a different method of DNA methylation analysis: hybridisation of bisulphite modified DNA to Illumina HumanMethylation27 BeadChip. Given that the batch effect of methylation profiling in Illumina Infinium assay could potentially influence the survival analysis, our study was limited to 16,587 CpG sites with small variation in technical replicates. Only 17 out of 28 genes identified from the SGCTG cohort study are represented on the BeadChip either within the same regions or within the promoter CGI linked to the loci identified by DMH assay. Among these 17 genes examined, methylation of promoter regions of VEGFB, VEGFA, HDAC11, FANCA, E2F1, GPX4, PRDX2, RAD54L and RECQL4 were still prognostic in this independent patient cohort (one-sided p<0.05, FDR<10%, n=311), and methylation of SESN2 shows marginal correlation with PFS in this patient cohort (one-sided p=0.0458, FDR=18%) (see univariate PFS analysis in TCGA cohort in Table 1 for a summary and Supplementary Table S5 for details).
Methylation index identifies two prognostic groups in late-stage ovarian tumours
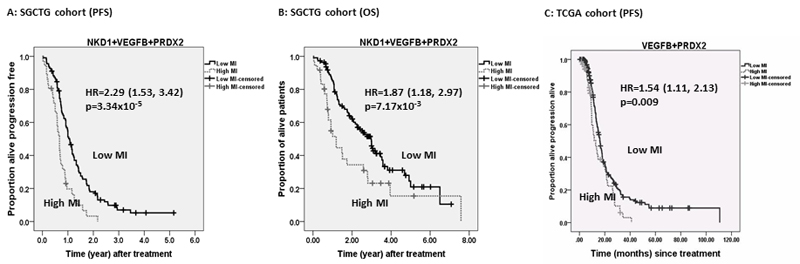
To identify the best methylation predictors of PFS, we constructed a multivariate Cox model from the SGCTG cohort data. Covariates were selected by likelihood ratio (LR) forward stepwise algorithm from clinical parameters (age, stage, grade and histological type) and promoter methylation at VEGFA, VEGFB, HDAC11, FANCA, E2F1, GPX4, PRDX2, RAD54L and RECQL4 identified in the SGCTG cohort and validated in TCGA cohort, as well as methylation at 7 gene promoters (FZD4, DVL1, NKD1, ROCK1, LRP5, AXIN1 and NFATC3) that have been shown to be significantly associated with PFS in SGCTG cohort in a previous study of promoter CGI methylation profiling in the Wnt pathway in EOCs using identical platforms and statistical analysis plan (11). The final model included NKD1 (HR = 1.26; 95% CI 1.02-1.45; p=0.025), VEGFB (HR =1.22, 95% CI 1.04-1.44; p=0.015) and PRDX2 (HR=1.22, 95% CI 1.02-1.45; p=0.029) as the three best predictors of PFS (p=6.62x10-6, permutation test p=0.05). The hazard ratio (HR) represents the relative risk per unit increase in Z score. The remaining variables including clinical parameters and remaining 14 methylation biomarkers were not selected into the model because they did not provide additional prognostic information beyond that provided by NKD1, VEGFB and PRDX2. A methylation index (MI) calculated from this model (, Z denotes Z score) could identify two distinct prognostic groups using the third quartile of the index as the cut-off (PFS: HR=2.29, 95% CI 1.53-3.42, log rank test p=3.34x10-5; OS: HR= 1.87, 95% CI 1.18-2.97, log rank test p=0.007) (Figure 1). The patients with increased MI were also more likely to have poor response to chemotherapy (OR=3.45 95% CI 1.31-9.08, p=0.012; PR+CR vs. PD+SD+NE: 30% vs. 70%).
Figure 1. Kaplan-Meier plots of progression free survival and overall survival in SGCTG cohort.
The patients were separated into two groups with high/low level of methylation index (MI) estimated from a multivariate Cox model incorporating NKD1, VEGFB and PRDX2 promoter methylation. The cut-off was determined by the third quartile of MI across all the patients (n=150). A) Progression free survival curve; B) overall survival curve.
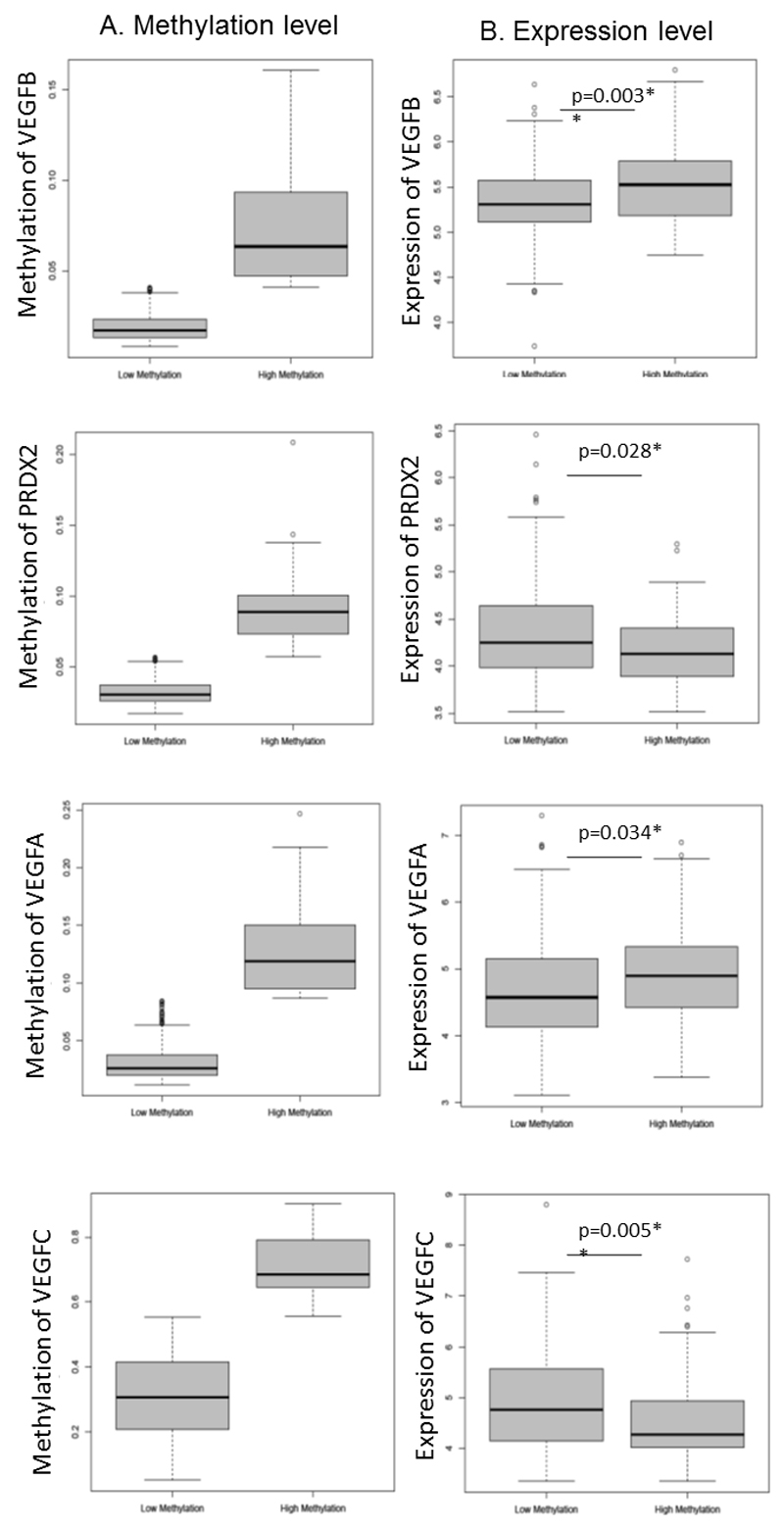
Subsequently, we further evaluated the multivariate model incorporating VEGFB and PRDX2 in terms of association with PFS in the TCGA cohort data (the locus linked to NKD1 is absent on Illumina BeadChip and so NKD1 could not be included in this model). Consistent with the result found in SGCTG cohort, the association between MI estimated from this model and progression/relapse-free survival as well as response to chemotherapy remained significant in this independent patient cohort (PFS: log-rank test p=0.009; response: logistic regression analysis adjusted OR=4.11, 95% CI 1.21-13.96, p=0.024). However, the association with overall survival did not stand in this cohort (OS: log rank test p=0.291). By combining with the expression dataset from TCGA, patients with higher methylation of VEGFB had increased expression of VEGFB, while patients with higher methylation of PRDX2 had reduced expression of PRDX2 (Figure 2).
Figure 2. Methylation and expression of VEGFB, PRDX2, VEGFA and VEGFC in TCGA cohort.
The top 20% of the patients with high methylation level at the biomarker examined were categorised into the ‘high methylation group’, otherwise, they were included in the ‘low methylation group’. Left panel (A): methylation level of candidate biomarker; right panel (B): expression level of candidate biomarker. Mann-Whitney U test (two sided) was used to examine the significant difference between two groups. *p<0.05, **p<0.01.
Surgical debulk status is correlated with survival in advanced ovarian cancer (20). In the SGCTG and TCGA cohorts studied in the present study, patients with any microscopic tumour after surgery have shorter overall and progression-free survival than those without any detectable tumour (SGCTG: HR=1.98, 95% CI 1.1-3.58, p=0.023; TCGA cohort: HR=1.67, 95% CI 1.06-2.65, p=0.029). We therefore further assessed the association between PFS and the MI estimated from our study adjusted by debulk status by forcing both variables into the multivariate Cox model. The MI remained significance after adjustment (SGCTG: adjust HR=2.56, 95% CI 1.54-4.26, p=2.91x10-4, n=89; TCGA: adjust HR=2.87, 95% CI 1.17-7.06, p=0.021, n=228), showing that the MI has independent prognostic value from surgical outcome.
DNA methylation correlates with response to platinum-based chemotherapy
DNA methylation at the 29 loci associated with poor PFS was assessed for any relationship with patients’ response to first line platinum-based chemotherapy. Increased methylation at VEGFB, RRM2, CD82, TR2IT1, GPX4, RAD54L and EME2 was associated with poor response in SGCTG cohort (Supplementary Table S6). Out of these 7 biomarkers, methylation of VEGFB and GPX4 are the most significant and independent biomarkers of response identified by multivariate logistic regression analysis using forward stepwise LR algorithm (Supplementary Method 2). 78% (29/37) patients with increased methylation either at VEGFB or at GPX4 had poor response to chemotherapy (OR=6.18, 95% CI 2.47-15.43, p=0.0001). Consistently, patients with methylation either at VEGFB or GPX4 from TCGA cohort were more likely to have poor response to chemotherapy (OR=1.99, 95% CI 0.93-4.25, p=0.078). This trend became clearer after correction for batch effect (adjusted OR=3.56, 95% CI 1.09-11.62, p=0.036) (Supplementary Method 2).
Association between disease progression and methylation at VEGFs in EOCs
Anti-angiogenesis treatment has shown clinical benefit to ovarian cancer patients when combined with chemotherapy (7, 8). The vascular endothelial growth factors (VEGFs) are prime regulators of pathological angiogenesis. Given that two biomarkers identified in this study are associated with VEGFA and VEGFB, we systematically examined methylation at promoter regions and expression of VEGFs in the TCGA cohort for correlation with progression/relapse free survival and response to chemotherapy where the majority of the patients received platinum-based chemotherapy. Expression data showed that patients with increased methylation at VEGFB and VEGFA had elevated expression level, while patients with increased methylation at VEGFC had reduced expression of this gene (Figure 2). Among 5 members of VEGFs including VEGFA, VEGFB, VEGFC, VEGFD and PIGF, we found methylation at the promoter region of VEGFA, VEGFB and VEGFC was significantly relevant to disease progression (TCGA cohort in Table 2).
Table 2. Association between PFS and methylation of VEGFA, VEGFB and VEGFC.
| TCGA cohort (HumanMethylation27 beadchip) |
|||||
|---|---|---|---|---|---|
| Genes | Genomic location | HR | 95% CI | two-sided p value | N |
| VEGFA | chr6: 43845984-43845985 | 1.35 | (0.99, 1.84) | 0.056 | 311 |
| VEGFB | chr11:63758874-63758875 | 1.95 | (1.12,3.42) | 0.018* | 311 |
| VEGFC | chr4: 177951450-177951451 | 0.92 | (0.86, 0.99) | 0.021* | 311 |
| SGCTG cohort (Bisulfite Pyrosequencing) |
|||||
| Genes | Genomic location | HR | 95% CI | one-sided p value | N |
| VEGFA | chr6: 43845984-43845985 | 1.17 | (0.94, 1.44) | 0.08+ | 141 |
| VEGFB | chr11: 63758262-63758287 | 1.12 | (0.99, 1.28) | 0.044* | 126 |
| VEGFC | chr4: 177951442-177951520 | 0.99 | (0.98, 1.00) | 0.03* | 142 |
Methylation of VEGFA, VEGFB and VEGFC was then quantified by bisulfite pyrosequencing in the SGCTG cohort. Consistent with previous findings, patients with increased methylation of VEGFB and decreased methylation of VEGFC have increased risk of tumour progression (one-sided p<0.05) (SGCTG cohort in Table 2). Methylation at VEGFA shows the trend to be correlated with PFS in this analysis, though the methylation level at promoter region of this gene is extremely low (about 1%).
Methylation of VEGFB is a strong predictor of response to chemotherapy in both SGCTG and TCGA cohort (SGCTG cohort: OR=5.92, 95% CI 1.85-18.94, p=0.003; TCGA cohort: OR=2.13, 95% CI 0.97-4.68, p=0.059). Expression level of VEGFB also showed a significant association with response in TCGA cohort (OR=2.57, 95% CI 1.22-5.43, p=0.013) (Table 3).
Table 3. Association between response and methylation/expression of VEGFA, VEGFB and VEGFC.
| TCGA cohort (HumanMethylation27) (CR+PR: 174 vs. SD+PD 39) |
||||||
|---|---|---|---|---|---|---|
| Genes | Methylation level$ | OR | 95% CI | p value | # PR+CR | # PD+SD |
| VEGFA | High methylation | 1.61 | (0.66, 3.92) | 0.29 | 24 | 8 |
| Low methylation | 1 | 150 | 31 | |||
| VEGFB | High methylation | 2.13 | (0.97, 4.68) | 0.059+ | 30 | 12 |
| Low methylation | 1 | 144 | 27 | |||
| VEGFC | High methylation | 1.03 | (0.43, 2.43) | 0.955 | 35 | 8 |
| Low methylation | 1 | 139 | 31 | |||
| TCGA cohort (Affymetrix HGU133A) (CR+PR: 174 vs. SD+PD 39) |
||||||
| Genes | Expression level$$ | OR | 95% CI | p value | # PR+CR | # PD+SD |
| VEGFA | High expression | 1.15 | (0.48, 2.72) | 0.759 | 32 | 8 |
| Low expression | 1 | 142 | 31 | |||
| VEGFB | High expression | 2.57 | (1.22, 5.43) | 0.013* | 34 | 15 |
| Low expression | 1 | 140 | 24 | |||
| VEGFC | High expression | 2.2 | (0.73, 6.61) | 0.159 | 139 | 35 |
| Low expression | 1 | 35 | 4 | |||
20% of the patients with highest level of methylation were categorised in the ‘high methylation group’, otherwise, they were include in the ‘low methylation group’
If expression was positively correlated with methylation, 20% of the patients with highest level of expression were categorised in the “high expression group”, otherwise, they were included in the “low expression group”. If expression was reversely correlated with expression, 20% of the patients with lowest level of expression were categorised in the “low expression group”, otherwise, they were in the “high expression group”
Discussion
Novel biomarkers of disease progression and response to chemotherapy are needed to guide current treatment strategies thereby potentially optimising patient benefit and clinical trial design. We have systematically profiled CGI DNA methylation at genes in pathways previously implicated in ovarian cancer development and progression and identified validated association of methylation at multiple CGI with clinical outcomes in high grade serous ovarian cancer. Aberrant DNA methylation frequently occurs in cancer, particularly at CGIs which are generally unmethylated in normal cells. CGIs often co-localise with the promoters of genes and promoter hypermethylation is associated with repression of gene transcription (21). Several studies have shown that CGI methylation has potential as a biomarker for monitoring tumour progression (22, 23) and is associated with platinum-based chemoresistance in EOC (24, 25). However, many of these studies are either limited by small sample size or lack of validation of the methylation biomarker as an independent prognostic marker. By building on our previously reported DNA methylation model derived from loci at genes in the Wnt pathway (11) we present here a novel multivariate Cox model including three DNA methylation-dependent loci (NKD1, VEGFB, PRDX2) that can separate late-stage ovarian cancer patient subgroups with distinct PFS and overall survival (OS), and shows improved prognostic value when compared to conventional clinical parameters. Alongside our previously identified Wnt-associated NKD1 locus, this model includes loci from further cancer-related pathways VEGFB and PRDX2. Furthermore, MI estimated from this model is associated with response to the first-line platinum-based chemotherapy, suggesting these pathways might be involved in the chemosensitivity in EOCs.
The identification of VEGFA, VEGFB and VEGFC as associated with clinical outcome in ovarian cancer patients is particularly of interest given the clinical trials showing improvement of PFS in ovarian cancer patients treated with the VEGF targeted agent Bevacizumab in addition to standard chemotherapy (Carboplatin and Paclitaxel). It has been proposed that Bevacizumab activity might be limited to a small proportion of patients (26), and hence it is important to identify patient subsets likely to benefit from Bevacizumab. Thus the prognostic and predictive value of DNA methylation at VEGF loci in such Bevacizumab trials now needs to be assessed.
In contrast to VEGFA, which has a defined role of in angiogenesis and has recently been shown to fulfil a direct, tumour-promoting effect (27), the function of VEGFB remains largely elusive (28). A number of studies have reported conflicting observations and relatively little is known about the role of VEGFB in tumourigenesis and progression (29). In brain and retinal neuron apoptosis models VEGFB can potently inhibit apoptosis and promote survival (30). Although the role of VEGFB may largely depend on tumour context, an anti-apoptotic role for VEGFB could provide a rational for increased methylation of VEGFB and increased VEGFB expression being associated with worse PFS and poor response to platinum-based chemotherapy in ovarian cancer. Although speculative, VEGFB methylation may indicate a subset of patients which may particularly benefit from Bevacizumab treatment in not only abrogating tumour angiogenesis but also targeting increased tumour growth, while being more resistant to platinum induced DNA apoptosis.
We initially identified two out of five VEGF-associated genes, including VEGFA and VEGFB, as having prognostic value in high grade serous carcinoma. Systematic analysis of VEGF family members from the TCGA dataset showed that methylation of VEGFC was further associated with PFS of ovarian cancer patients. These observations highlight the prognostic significance of VEGF biomarkers for ovarian cancer. Furthermore, methylation of VEGFB, in particular, emerged as being a predictor of response in two independent patient cohorts. It is interesting that patients with increased VEGFB methylation at the promoter region have elevated VEGFB expression in the TCGA cohort. We looked for transcription factor binding around the PFS-associated CpG site and found a consensus binding site for the transcription factor ZEB1 (AREB6/TCF8) close to this genomic site. ZEB1 is a negative transcriptional regulator of angiogenesis (31). It has been reported that increased methylation close to the ZEB transcription binding site impairs ZEB transcription factor binding at TP73 thereby upregulating gene expression in ovarian cancer cell lines (32), however, it remains unclear whether this is also the case at VEGFB.
In contrast to VEGFB where increased methylation increases risk of disease progression, methylation at the VEGFC locus reduces risk of progression. In tumours, VEGFC fulfils a role in promoting lymphangiogenesis and metastasis via the lymphatic system (33). In ovarian cancer, VEGFC protein expression and serum levels have been correlated with lymphatic metastasis and high VEGFC correlates with poor prognosis (34, 35). It has been suggested that induction of VEGFC by the transcription factor LEDGF/p75 could be a potential strategy in tumours to escape VEGFA targeted therapy and to sustain tumour progression (36). In this respect, it remains to be seen whether patients with increased risk of progression due to low VEGFC methylation and high VEGFC expression may benefit from targeted therapies such as anti-angiogenesis agents.
Taken together, by using systematic profiling of DNA methylation at CGI promoters of pathways relevant to ovarian carcinogenesis, we have identified three DNA methylation biomarkers (NKD1, VEGFB, PRDX2) that give rise to a methylation index capable of predicting PFS in ovarian cancer patients independently from known clinical prognostic feature. These biomarkers could aid in the identification of patients with suboptimal benefit from standard platinum-based chemotherapy. Patients with an increased methylation index may especially benefit from being stratified for more targeted therapies. In addition, methylation at individual VEGF family members associates with differential risk of disease progression and further evaluation of the predictive value of methylation and expression at VEGFB and VEGFC as biomarkers for patient’s response to targeted therapies such as Bevacizumab is warranted.
Supplementary Material
Figure S1: Distribution of of McrBC recognition sites
Log2 transformed DMH ratios in 100% methylated samples compared to 0% methylated sample
Translational relevance.
Debulking surgery with platinum-based chemotherapy remains the primary treatment for high grade serous ovarian cancer. Independent biomarkers of progression free survival to platinum-based chemotherapy could aid in identifying patients prone to early relapse who could particularly benefit from novel treatments. We have evaluated DNA methylation at promoter GpG islands of genes in multiple key pathways implicated in epithelial ovarian cancer, including Akt/mTOR, p53, Redox and homologous recombination DNA repair associated pathways. We have identified nine loci whose methylation is associated with progression-free survival independent from conventional clinical parameters. Together with our previous study of the Wnt pathway, 3 loci (NKD1, VEGFB and PRDX2), when combined in a multivariate Cox model, are strong predictors of progression-free survival independent from known clinical factors (PFS: HR=2.3 p=3.3×10-5; Overall Survival: HR= 1.9, p=0.007). These loci have the potential to aid in stratifying patients for targeted therapy in ovarian cancer.
Acknowledgements
We thank Gail Anderson for her initial help with pyrosequencing assay optimization. External validation of our candidate genes used expression and methylation data generated by The Cancer Genome Atlas (TCGA) Pilot Project established by the NCI and NHGRI. We would like to thank members of SGCTG and patients for their help in the study. We would also like to thank Liz Anne Lewsley and Jan Graham for help with patient recruitment and data management.
Grant support: The study is supported by Cancer Research UK (C536/A13086 to RB), Imperial Experimental Cancer Medicines Centre, Imperial Biomedical Research Centre, Imperial Cancer Research UK Centre and Ovarian Cancer Action.
Footnotes
Disclosure of potential conflicts of interest
There are no relationships that the authors believe could be construed as resulting in an actual, potential, or perceived conflict of interest with regards to this manuscript.
Conflict of Interest Statement:
No potential conflicts of interest are disclosed.
References
- 1.Hasan J, Jayson G. Novel Anti-angiogenic Therapies in Ovarian Cancer. In: Kaye S, Brown R, Gabra H, Gore M, editors. Emerging Therapeutic Targets in Ovarian Cacner. New York: Springer; 2011. pp. 51–72. [Google Scholar]
- 2.Parmar MK, Ledermann JA, Colombo N, du Bois A, Delaloye JF, Kristensen GB, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361:2099–106. doi: 10.1016/s0140-6736(03)13718-x. [DOI] [PubMed] [Google Scholar]
- 3.Laird PW. The power and the promise of DNA methylation markers. Nature reviews Cancer. 2003;3:253–66. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 4.Widschwendter M, Jones A, Teschendorff AE. Epigenetics makes its mark on women-specific cancers--an opportunity to redefine oncological approaches? Gynecologic oncology. 2013;128:134–43. doi: 10.1016/j.ygyno.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Nogueira da Costa A, Herceg Z. Detection of cancer-specific epigenomic changes in biofluids: powerful tools in biomarker discovery and application. Molecular oncology. 2012;6:704–15. doi: 10.1016/j.molonc.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown MR, Blanchette JO, Kohn EC. Angiogenesis in ovarian cancer. Baillieres Best Pract Res Clin Obstet Gynaecol. 2000;14:901–18. doi: 10.1053/beog.2000.0134. [DOI] [PubMed] [Google Scholar]
- 7.Perren TJ, Swart AM, Pfisterer J, Ledermann JA, Pujade-Lauraine E, Kristensen G, et al. A phase 3 trial of bevacizumab in ovarian cancer. N Engl J Med. 2011;365:2484–96. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 8.Burger RA, Brady MF, Bookman MA, Fleming GF, Monk BJ, Huang H, et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N Engl J Med. 2011;365:2473–83. doi: 10.1056/NEJMoa1104390. [DOI] [PubMed] [Google Scholar]
- 9.O’Malley DM, Richardson DL, Rheaume PS, Salani R, Eisenhauer EL, McCann GA, et al. Addition of bevacizumab to weekly paclitaxel significantly improves progression-free survival in heavily pretreated recurrent epithelial ovarian cancer. Gynecologic oncology. 2011;121:269–72. doi: 10.1016/j.ygyno.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Aghajanian C, Blank SV, Goff BA, Judson PL, Teneriello MG, Husain A, Sovak MA, Yi J, Nycum LR. OCEANS: a randomized, double-blind, placebo-controlled phase III trial of chemotherapy with or without bevacizumab in patients with platinum-sensitive recurrent epithelial ovarian, primary peritoneal, or fallopian tube cancer. J Clin Oncol. 2012;30:2039–45. doi: 10.1200/JCO.2012.42.0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai W, Teodoridis JM, Zeller C, Graham J, Hersey J, Flanagan JM, et al. Systematic CpG islands methylation profiling of genes in the wnt pathway in epithelial ovarian cancer identifies biomarkers of progression-free survival. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4052–62. doi: 10.1158/1078-0432.CCR-10-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strathdee G, MacKean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18:2335–41. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 13.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–72. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 14.Stewart FJ, Raleigh EA. Dependence of McrBC cleavage on distance between recognition elements. Biol Chem. 1998;379:611–6. [PubMed] [Google Scholar]
- 15.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. Journal of molecular biology. 1992;225:327–48. doi: 10.1016/0022-2836(92)90925-a. [DOI] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 18.Rosen DG, Yang G, Liu G, Mercado-Uribe I, Chang B, Xiao XS, et al. Ovarian cancer: pathology, biology, and disease models. Frontiers in bioscience : a journal and virtual library. 2009;14:2089–102. doi: 10.2741/3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama T, Kamura T, Kigawa J, Terakawa N, Kikuchi Y, Kita T, et al. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer. 2000;88:2584–9. [PubMed] [Google Scholar]
- 20.Bristow RE, Tomacruz RS, Armstrong DK, Trimble EL, Montz FJ. Survival effect of maximal cytoreductive surgery for advanced ovarian carcinoma during the platinum era: a meta-analysis. J Clin Oncol. 2002;20(5):1248–1259. doi: 10.1200/JCO.2002.20.5.1248. [DOI] [PubMed] [Google Scholar]
- 21.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahapatra S, Klee EW, Young CY, Sun Z, Jimenez RE, Klee GG, et al. Global methylation profiling for risk prediction of prostate cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2882–95. doi: 10.1158/1078-0432.CCR-11-2090. [DOI] [PubMed] [Google Scholar]
- 23.Watts GS, Futscher BW, Holtan N, Degeest K, Domann FE, Rose SL. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC medical genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gifford G, Paul J, Vasey PA, Kaye SB, Brown R. The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:4420–6. doi: 10.1158/1078-0432.CCR-03-0732. [DOI] [PubMed] [Google Scholar]
- 25.Zeller C, Dai W, Steele NL, Siddiq A, Walley AJ, Wilhelm-Benartzi CS, et al. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene. 2012;31:4567–76. doi: 10.1038/onc.2011.611. [DOI] [PubMed] [Google Scholar]
- 26.Jubb AM, Harris AL. Biomarkers to predict the clinical efficacy of bevacizumab in cancer. Lancet Oncol. 2010;11:1172–83. doi: 10.1016/S1470-2045(10)70232-1. [DOI] [PubMed] [Google Scholar]
- 27.Cao Y, E G, Wang E, Pal K, Dutta SK, Bar-Sagi D, et al. VEGF exerts an angiogenesis-independent function in cancer cells to promote their malignant progression. Cancer research. 2012;72:3912–8. doi: 10.1158/0008-5472.CAN-11-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Kumar A, Zhang F, Lee C, Tang Z. Complicated life, complicated VEGF-B. Trends Mol Med. 2012;18:119–27. doi: 10.1016/j.molmed.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 29.Albrecht I, Kopfstein L, Strittmatter K, Schomber T, Falkevall A, Hagberg CE, et al. Suppressive effects of vascular endothelial growth factor-B on tumor growth in a mouse model of pancreatic neuroendocrine tumorigenesis. PLoS One. 2010;5:e14109. doi: 10.1371/journal.pone.0014109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Zhang F, Nagai N, Tang Z, Zhang S, Scotney P, et al. VEGF-B inhibits apoptosis via VEGFR-1-mediated suppression of the expression of BH3-only protein genes in mice and rats. J Clin Invest. 2008;118:913–23. doi: 10.1172/JCI33673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inuzuka T, Tsuda M, Tanaka S, Kawaguchi H, Higashi Y, Ohba Y. Integral role of transcription factor 8 in the negative regulation of tumor angiogenesis. Cancer research. 2009;69:1678–84. doi: 10.1158/0008-5472.CAN-08-3620. [DOI] [PubMed] [Google Scholar]
- 32.Ibrahim N, He L, Leong CO, Xing D, Karlan BY, Swisher EM, et al. BRCA1-associated epigenetic regulation of p73 mediates an effector pathway for chemosensitivity in ovarian carcinoma. Cancer research. 2010;70:7155–65. doi: 10.1158/0008-5472.CAN-10-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plate K. From angiogenesis to lymphangiogenesis. Nat Med. 2001;7:151–2. doi: 10.1038/84579. [DOI] [PubMed] [Google Scholar]
- 34.Cheng D, Liang B, Li Y. Serum Vascular Endothelial Growth Factor (VEGF-C) as a Diagnostic and Prognostic Marker in Patients with Ovarian Cancer. PLoS One. 2013;8:e55309. doi: 10.1371/journal.pone.0055309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda M, Hung YC, Terai Y, Kanda K, Kanemura M, Futakuchi H, et al. Vascular endothelial growth factor-C expression and invasive phenotype in ovarian carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:3225–32. doi: 10.1158/1078-0432.CCR-04-1148. [DOI] [PubMed] [Google Scholar]
- 36.Cohen B, Addadi Y, Sapoznik S, Meir G, Kalchenko V, Harmelin A, et al. Transcriptional regulation of vascular endothelial growth factor C by oxidative and thermal stress is mediated by lens epithelium-derived growth factor/p75. Neoplasia. 2009;11:921–33. doi: 10.1593/neo.09636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.