Abstract
Odorants are discriminated by hundreds of odorant receptor (OR) genes, which are dispersed throughout the mammalian genome. The OR genes are expressed in a highly specialized type of cell, the olfactory sensory neuron. Each one of these neurons expresses one of the 2 alleles from one single OR gene type. The mechanisms underlying OR gene expression are unclear. Here we describe recent work demonstrating that the olfactory sensory neuron shows a particular nuclear architecture, and that the genomic OR loci are colocalized in silencing heterochromatin compartments within the nucleus. These discoveries highlight the important role played by epigenetic modifications and nuclear genome organization in the regulation of OR gene expression.
Keywords: facultative heterochromatin, H3K27me3, nuclear architecture, odorant receptor gene choice, odorant receptor, olfactory sensory neuron
Abbreviations
- Odorant receptor
OR
Odorants are detected by a large family of odorant receptors (ORs) expressed by olfactory sensory neurons present in the nasal cavity.1 In mammals, the OR gene family is composed of more than a thousand genes, which are distributed throughout the genome2,3 (Fig. 1). Each olfactory sensory neuron, however, expresses only one of these OR genes. In addition, expression is monoallelical, that is, only one of the 2 homologous alleles of a given OR gene, the maternal or paternal, is expressed per neuron.4-7
Figure 1.
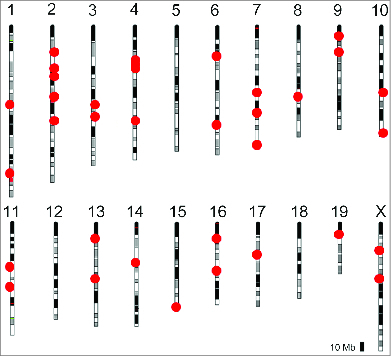
Distribution of the OR genes in the mouse genome. Approximately 1400 OR genes, including pseudogenes, are distributed in clusters throughout the chromosomes (based on Godfrey et al.2). Red dots represent loci containing OR genes. No OR gene is found in the Y chromosome.
The OR gene family is highly polymorphic, and a series of single nucleotide polymorphisms (SNPs) have been correlated with differences in odorant perception.8-10 Monoallelic expression therefore confers unique specificities to the olfactory neurons. Olfactory sensory neurons expressing the same type of OR project their axons to the same glomeruli in the olfactory bulb of the brain, and ORs are involved in axonal targeting.11 Odorant discrimination results from the activation of a combination of ORs,5 and therefore odorants are encoded by combinations of glomeruli. Thus, the OR type that is expressed in a given olfactory neuron, not only determines the range of odorants to which this neuron will respond, but it is also required for axonal targeting to specific glomeruli in the olfactory bulb. This organization of the olfactory system emphasizes the importance of the monogenic and monoallelic expression of OR genes.
OR genes need to be coordinately regulated in order to achieve mutually exclusive expression. What are the mechanisms that allow for this precise regulation if the OR genes are widespread in the genome? It has been shown that OR gene promoters are required for OR gene expression.12,13 Analysis of hundreds of OR gene promoters revealed that the majority of these regions share common cis-regulatory elements, which must play important roles in the regulation of OR gene transcription, but which do not explain monogenic or monoallelic expression.14,15 For example, the vast majority of OR genes contain O/E-like sites (EBF binding sites) and homeodomain-like sites.15,16 But the fact that OR genes share the same types of cis-regulatory elements, and that these elements are also present in the promoter region of other olfactory genes that are expressed in all mature olfactory neurons, such as the olfactory marker protein (OMP), Gαolf, and adenylyl cyclase III genes, indicates that these common promoter motifs are not sufficient for determining singular OR gene expression.
It is well known that the mammalian cell nucleus shows a highly organized architecture, where distinct nuclear processes, such as transcription and splicing, are compartmentalized.17-19 It is also clear now that genomes are nonrandomly arranged17,18 and that each chromosome occupies a distinct region in the nucleus (termed chromosome territories).20,21 Different cell types show different nuclear architectures, with different chromosome arrangements and distinct patterns of chromatin condensation.22,23 Differences in the nuclear organization must account for the differential gene expression.
The olfactory sensory neurons are highly specialized cells which are continuously generated from a population of neuron progenitors located in the basal layer of the olfactory epithelium. During differentiation, the progenitor cells must develop a characteristic pattern of gene expression to acquire all of the specific properties of mature olfactory sensory neurons, including correct expression of OR genes. Recent experiments show that the organization of the olfactory nucleus is critical for regulating OR gene expression.
Spatial Organization of Heterochromatin in the Olfactory Nucleus
Studies of nuclear architecture showed that heterochromatin is organized in an unconventional fashion in the olfactory nucleus (Fig. 2A and B). Usually, in mammalian cells, heterochromatin is found at the nuclear periphery, whereas euchromatin is mostly localized in the nuclear center. In the olfactory nucleus, however, large blocks of constitutive heterochromatin are concentrated more centrally in the nucleus.24,25 There are 2 types of repressive heterochromatin: constitutive heterochromatin and facultative heterochromatin. Both types of heterochromatin are repressive, however, while constitutive heterochromatin is considered to be stably heterochromatized and transcriptionally silent, facultative heterochromatin can decondense and become transcriptionally active.26 It was recently shown that facultative heterochromatin is concentrated in a few domains which are localized around and close to the constitutive heterochromatin blocks in the olfactory nucleus24 (Fig. 2A and B). The other major cell types of the olfactory epithelium, the supporting cells and basal cells, do not show this same distribution of constitutive and facultative heterochromatin, indicating that this particular nuclear organization is characteristic of the olfactory sensory neurons.
Figure 2.
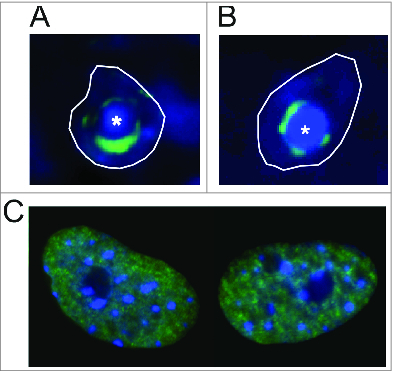
Distribution of constitutive and facultative heterochromatins in the nucleus of olfactory sensory neurons. Facultative heterochromatin as visualized by immunostaining for H3K27me3 (green) in the nuclei of female (A) or male (B) mouse olfactory sensory neurons and of mouse embryonic fibroblasts (MEFs) (C). Constitutive heterochromatin blocks (*) are densely stained by DAPI (blue). The white outline shows the boundary of a single nucleus. Nuclei are not shown to scale.
The organization of facultative and constitutive heterochromatin domains in the olfactory nuclei is striking, and not commonly observed in the nuclei of other cell types (Fig. 2C). The best known example of a nuclear facultative heterochromatin domain is the inactive X chromosome (Xi) in female mammalian cells. The inactive X chromosome forms a compact nuclear structure denominated the Barr body, which is enriched in both H3K9me3 (constitutive) and H3K27me3 (facultative) marks. Interestingly, it was shown that these 2 types of heterochromatin marks form spatially distinct domains in the interphase nucleus of human female cells, which are juxtaposed one to another within the Barr body.27 As mentioned above, the H3K27me3 and H3K9me3 domains in the olfactory nucleus are also distinct but closely associated one to another. Differently from the Barr body, which is only observed in female cells, these domains are observed in both female and male olfactory sensory nuclei (Fig. 2). These observations suggest that there is an interaction between the constitutive and facultative heterochromatin domains in these cells, which may contribute to the organization of inactivating heterochromatin and gene silencing over large genomic regions. It was for example shown that in the Barr body a protein named HBiX1 interacts with the H3K9me3 associated protein HP1, and with the Xist-H3K27me3 associated protein SMCHD1, and thereby bridges these 2 heterochromatin domains.27 Whether similar molecular interactions occur in the olfactory nucleus remains to be determined.
OR Gene Positioning in the Nucleus
Even though the OR genes are distributed over almost all chromosomes (Fig. 1), they are located close together in the 3-dimensional organization of the genome in the olfactory nucleus. DNA-FISH experiments using a probe that recognizes the complete repertoire of mouse OR genes showed that these genes are aggregated in the centrally located constitutive heterochromatin blocks.25 The same is not observed for the olfactory marker protein (OMP) gene, which is biallelically expressed in all mature olfactory sensory neurons.24 Although the OMP gene is located close to the S6 and P2 OR genes on chromosome 7, in the olfactory nucleus, the OMP gene alleles are located further away from the central repressive blocks when compared to the OR gene alleles.24 Accordingly, chromatin immunoprecipitation (ChIP) experiments showed that OR genes are marked with H3K9me3 and H4K20me3, which are typical marks for constitutive heterochromatin.28
Recent DNA-FISH experiments where the 2 alleles from different OR genes were analyzed in great detail indicated that the alleles of a given OR gene are segregated in the nucleus: while one allele is localized in the constitutive heterochromatin blocks, the homologous allele is located further away and in or next to the facultative heterochromatin domains mentioned above.24 These results could explain monoallelic expression: while one of the alleles would be ‘permanently’ inactive’, the remaining one would be available for transcription. The fact that OR genes show asynchronous replication29 further suggests that the alleles are differentially marked, with one being more prone for activation than the other.
Differently from what has been shown for the constitutive heterochromatin H3K9me3 and H4K20me3 marks, there is no evidence so far from ChIP experiments that OR genes are marked with H3K27me3, the typical facultative heterochromatin mark.28 H3K27me3 marks have been previously identified in a variety of genomic positions, such as gene promoters, enhancers, coding regions, intergenic regions or in large genomic blocks.30 It remains to be determined, which are the genomic regions that are marked with H3K27me3 and account for the facultative heterochromatin compartments present in the olfactory nucleus.
OR Gene Activation
The OR genes are therefore globally confined in repressive compartments in the olfactory nucleus. It is not clear when exactly during neuronal differentiation these epigenetic marks are deposited. Genetic experiments showed that the P2 OR locus shows a low permissiveness for transcription, which is consistent with a repressive OR chromatin.31 Younger neurons show, however higher permissiveness, indicating that the fully repressive state of the OR chromatin occurs only after OR gene choice is stabilized.31
It is also not clear yet, what are the precise mechanisms that result in the expression of a single allele per neuron. In order to achieve singular expression of OR genes one single OR gene allele must escape repression. One possibility is that all OR genes are marked with the constitutive heterochromatin H3K9me3 marks, and these silencing marks would be removed later by the H3K9me2 demethylase (LSD1) from a single OR gene allele to assure monogenic and monoallelic OR gene expression.32 Another enzyme, yet to be identified, would be required to demethylate H3K9me3 first. Once one single OR allele is activated, production of adenylyl cyclase III by mature olfactory sensory neurons leads to LSD1 downregulation, preventing activation of a second OR allele.32 The fact that in new born mice where LSD1 is knocked out in progenitor neurons, there is a reduction of the total OR transcript level in the MOE, indicates that this enzyme is involved in OR gene regulation.32 Also, in zebrafish embryos, an inhibitor of LSD1 leads to a decreased number of olfactory cells expressing endogenous odorant receptors, suggesting that H3K9me3 demethylation is also required for proper OR gene expression in this organism.33
Another possibility, is that the OR alleles which are associated with facultative heterochromatin, are the ones that retain the potential to be transcribed, and repression would have to be relieved from a single allele by a H3K27me3 demethylase, such as UTX (also named KDM6A) or JMJD3 (also named KDM6B).34 The involvement of H3K27me3 modifying enzymes, however, was not experimentally addressed yet. Curiously, it was shown that in the zebrafish olfactory epithelium, signaling through Gβγ is both necessary and sufficient to suppress the expression of odorant receptor genes through heterochromatin deposition in these loci.33 When Gβγ activity is blocked, the number of olfactory sensory neurons expressing an individual OR is increased, and the expression of KDM6B is upregulated while expression of ehmt2, a histone H3K9 methyltrasferase is downregulated, probably resulting in a decrease in repressive H3K27 and H3K9 histone marks.33
Alternatively, it is possible that the repressive facultative marks are deposited as a result of the feedback signal that is elicited by an OR gene which was expressed first, to prevent expression of additional OR genes and contribute in this way to the stability of OR gene choice. Future experiments will be required to unravel the exact role played by facultative heterochromatin in OR gene expression.
Conclusion
OR genes are spread throughout the genome, however, in the 3-dimensional organization of the olfactory nucleus they are clustered together in centrally located repressive heterochromatin compartments. This particular nuclear architecture, which differs from other cell types, appears to be essential for the expression of a single OR gene per olfactory neuron. Future challenges will be to determine the molecular mechanisms that lead to OR gene activation as well as the contributing roles played by these 2 types of heterochromatin.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the lab for comments and suggestions.
Funding
We would like to thank FAPESP and CNPq for financial support.
References
- 1. Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 1991; 65:175-87; PMID:1840504; http://dx.doi.org/ 10.1016/0092-8674(91)90418-X [DOI] [PubMed] [Google Scholar]
- 2. Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci U S A 2004; 101:2156-61; PMID:14769939; http://dx.doi.org/ 10.1073/pnas.0308051100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang X, Zhang X, Firestein S. Comparative genomics of odorant and pheromone receptor genes in rodents. Genomics 2007; 89:441-50; PMID:17303377; http://dx.doi.org/ 10.1016/j.ygeno.2007.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell 1994; 78:823-34; PMID:8087849; http://dx.doi.org/ 10.1016/S0092-8674(94)90562-2 [DOI] [PubMed] [Google Scholar]
- 5. Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell 1999; 96:713-23; PMID:10089886; http://dx.doi.org/ 10.1016/S0092-8674(00)80581-4 [DOI] [PubMed] [Google Scholar]
- 6. Serizawa S, Ishii T, Nakatani H, Tsuboi A, Nagawa F, Asano M, Sudo K, Sakagami J, Sakano H, Ijiri T, et al. Mutually exclusive expression of odorant receptor transgenes. Nat Neurosci 2000; 3:687-93; PMID:10862701; http://dx.doi.org/ 10.1038/76641 [DOI] [PubMed] [Google Scholar]
- 7. Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara S, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in the mouse. Science 2003; 302:2088-94; PMID:14593185; http://dx.doi.org/ 10.1126/science.1089122 [DOI] [PubMed] [Google Scholar]
- 8. Keller A, Zhuang H, Chi Q, Vosshall L, Matsunami H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007; 449:468-72; PMID:17873857; http://dx.doi.org/ 10.1038/nature06162 [DOI] [PubMed] [Google Scholar]
- 9. Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, et al. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci 2014; 17:114-20; PMID:24316890; http://dx.doi.org/ 10.1038/nn.3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Menashe I, Abaffy T, Hasin Y, Goshen S, Yahalom V, Luetje C, Lancet D. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol 2007; 5:e284; PMID:17973576; http://dx.doi.org/ 10.1371/journal.pbio.0050284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mori K, Sakano H. How is the olfactory map formed and interpreted in the mammalian brain? Annu Rev Neurosci 2011; 34:467-99; PMID:21469960; http://dx.doi.org/ 10.1146/annurev-neuro-112210-112917 [DOI] [PubMed] [Google Scholar]
- 12. McIntyre J, Bose S, Stromberg A, McClintock T. Emx2 stimulates odorant receptor gene expression. Chem Senses 2008; 33:825-37; PMID:18854508; http://dx.doi.org/ 10.1093/chemse/bjn061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vassalli A, Rothman A, Feinstein P, Zapotocky M, Mombaerts P. Minigenes impart odorant receptor-specific axon guidance in the olfactory bulb. Neuron 2002; 35:681-96; PMID:12194868; http://dx.doi.org/ 10.1016/S0896-6273(02)00793-6 [DOI] [PubMed] [Google Scholar]
- 14. Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, Lomvardas S. High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res 2011; 21:1249-59; PMID:21705439; http://dx.doi.org/ 10.1101/gr.120162.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michaloski J, Galante P, Malnic B. Identification of potential regulatory motifs in odorant receptor genes by analysis of promoter sequences. Genome Research 2006; 16:1091-8; PMID:16902085; http://dx.doi.org/ 10.1101/gr.5185406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Michaloski JS, Galante PA, Nagai MH, Armelin-Correa L, Chien MS, Matsunami H, Malnic B. Common promoter elements in odorant and vomeronasal receptor genes. PLoS One 2011; 6:e29065; PMID:22216168; http://dx.doi.org/ 10.1371/journal.pone.0029065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 2007; 8:104-15; PMID:17230197; http://dx.doi.org/ 10.1038/nrg2041 [DOI] [PubMed] [Google Scholar]
- 18. Misteli T. Beyond the sequence: cellular organization of genome function. Cell 2007; 128:787-800; PMID:17320514; http://dx.doi.org/ 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- 19. Rajapakse I, Groudine M. On emerging nuclear order. J Cell Biol 2011; 192:711-21; PMID:21383074; http://dx.doi.org/ 10.1083/jcb.201010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories-a functional nuclear landscape. Curr Opin Cell Biol 2006; 18:307-16; PMID:16687245; http://dx.doi.org/ 10.1016/j.ceb.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 21. Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature 2007; 445:379-781; PMID:17251970; http://dx.doi.org/ 10.1038/445379a [DOI] [PubMed] [Google Scholar]
- 22. Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol 2000; 1:137-43; PMID:11253366; http://dx.doi.org/ 10.1038/35040083 [DOI] [PubMed] [Google Scholar]
- 23. Takizawa T, Meshorer E. Chromatin and nuclear architecture in the nervous system. Trends Neurosci 2008; 31:343-52; PMID:18538423; http://dx.doi.org/ 10.1016/j.tins.2008.03.005 [DOI] [PubMed] [Google Scholar]
- 24. Armelin-Correa LM, Gutiyama LM, Brandt DY, Malnic B. Nuclear compartmentalization of odorant receptor genes. Proc Natl Acad Sci U S A 2014; 111:2782-7; PMID:24550308; http://dx.doi.org/ 10.1073/pnas.1317036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clowney EJ, Legros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 2012; 151:724-37; PMID:23141535; http://dx.doi.org/ 10.1016/j.cell.2012.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 2007; 28:1-13; PMID:17936700; http://dx.doi.org/ 10.1016/j.molcel.2007.09.011 [DOI] [PubMed] [Google Scholar]
- 27. Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol 2013; 20:566-73; PMID:23542155; http://dx.doi.org/ 10.1038/nsmb.2532 [DOI] [PubMed] [Google Scholar]
- 28. Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell 2011; 145:555-70; PMID:21529909; http://dx.doi.org/ 10.1016/j.cell.2011.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A. Coordination of the random asynchronous replication of autosomal loci. Nat Genet 2003; 33:339-41; PMID:12577058; http://dx.doi.org/ 10.1038/ng1102 [DOI] [PubMed] [Google Scholar]
- 30. Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343-9; PMID:21248841; http://dx.doi.org/ 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fleischmann A, Abdus-Saboor I, Sayed A, Shykind B. Functional interrogation of an odorant receptor locus reveals multiple axes of transcriptional regulation. PLoS Biol 2013; 11:e1001568; PMID:23700388; http://dx.doi.org/ 10.1371/journal.pbio.1001568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell 2013; 154:325-36; PMID:23870122; http://dx.doi.org/ 10.1016/j.cell.2013.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferreira T, Wilson SR, Choi YG, Risso D, Dudoit S, Speed TP, Ngai J. Silencing of odorant receptor genes by G protein bg signaling ensures the expression of one odorant receptor per olfactory sensory neuron. Neuron 2014; 81:847-59; PMID:24559675; http://dx.doi.org/ 10.1016/j.neuron.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell 2007; 131:29-32; PMID:17923085; http://dx.doi.org/ 10.1016/j.cell.2007.09.026 [DOI] [PubMed] [Google Scholar]


