Summary
N-methyl-D-aspartate receptors (NMDARs) are glutamate-gated, calcium-permeable ion channels that mediate synaptic transmission and underpin learning and memory. NMDAR dysfunction is directly implicated in diseases ranging from seizure to ischemia. Despite its fundamental importance, little is known about how the NMDAR transitions between inactive and active states, and how small molecules inhibit or activate ion channel gating. Here we report electron cryo-microscopy structures of the GluN1-GluN2B NMDA receptor in an ensemble of competitive antagonist-bound states, an agonist-bound form, and a state bound with agonists and the allosteric inhibitor Ro25-6981. Together with double electron-electron resonance experiments, we show how competitive antagonists rupture the ligand binding domain (LBD) gating ‘ring’, how agonists retain the ‘ring’ in a dimer-of-dimers configuration, and how allosteric inhibitors, acting within the amino terminal domain, further stabilize the LBD layer. These studies illuminate how the LBD gating ‘ring’ is fundamental to signal transduction and gating in NMDARs.
Introduction
Ionotropic glutamate receptors (iGluRs) transduce transient glutamate release from presynaptic vesicles into postsynaptic neuronal excitation in synapses. iGluRs can be subdivided into three major families: N-methyl-D-aspartate (NMDA), AMPA and kainate receptors. Open NMDA receptors (NMDARs) not only generate an electrical signal, but they also mediate calcium influx, which in turn plays a pivotal role in synaptic transmission and plasticity during learning and memory (Traynelis et al., 2010). Hyper- and hypo-activation of NMDARs have been implicated in neurological disorders including stroke, schizophrenia, epilepsy, Alzheimer’s disease (Paoletti et al., 2013), and anti-NMDAR encephalitis (Dalmau et al., 2008). Moreover, human de novo and inherited NMDAR mutations are associated with neurodevelopmental disorders including mental retardation and epileptic aphasia (Endele et al., 2010; Lesca et al., 2013).
NMDARs are Hebbian-like coincidence detectors because their activation requires depolarization of the membrane potential to relieve voltage-dependent magnesium block (Mayer et al., 1984; Nowak et al., 1984), and binding of both glutamate and glycine (Johnson and Ascher, 1987). In contrast with the fast kinetics of AMPA and kainate receptors, NMDARs display slower gating kinetics, with activation occurring in approximately 10 ms (Dzubay and Jahr, 1996) and deactivation in tens to thousands of milliseconds (Attwell and Gibb, 2005; Traynelis et al., 2010). The deactivation time course depends largely on the GluN2 subunit composition (Vicini et al., 1998). Furthermore, NMDARs show relatively slow and incomplete desensitization compared to most AMPA or kainate receptors (Monyer et al., 1992; Vicini et al., 1998) and little is known about the molecular mechanism of NMDAR desensitization. NMDAR channel activity can be modulated by a variety of molecules such as endogenous zinc (Vergnano et al., 2014), protons (Traynelis et al., 1995), and polyamines (Mony et al., 2011), or by synthetic compounds such as the competitive antagonists 5,7-dichlorokynurenic acid (DCKA) (Baron et al., 1991) and D-(−)-2-amino-5-phosphonopentanoic acid (D-APV) (Evans et al., 1982), and by allosteric modulators such as Ro25-6981 (Fischer et al., 1997).
NMDARs are heterotetrameric assemblies typically composed of two GluN1 and two GluN2(A-D) subunits that are related in amino acid sequence and domain architecture (Monyer et al., 1992; Moriyoshi et al., 1991). Each subunit consists of an intracellular carboxyl-terminal domain (CTD), a transmembrane domain (TMD) and two extracellular domains: an amino-terminal domain (ATD) and a ligand-binding domain (LBD) that binds glycine in the GluN1 subunit and glutamate in the GluN2 subunit. Crystal structures of the full-length receptor in complex with agonist/modulator show a layered-domain architecture where the receptor assembles as a dimer-of-dimers, with a GluN1-2-1–2 arrangement, in which the ATDs and LBDs form pairs of 2-fold related dimers and the TMD forms a pseudo 4-fold pore (Furukawa et al., 2005; Karakas and Furukawa, 2014; Karakas et al., 2011; Lee et al., 2014). Functional studies suggest allosteric cooperativity between the extracellular ATDs and LBDs (Gielen et al., 2008; Zheng et al., 2001; Zhu et al., 2013), a feature apparently unique to NMDARs compared to non-NMDA iGluRs. In addition, the GluN2 subunits are the key determinants that dictate the biophysical and pharmacological properties of specific GluN1-GluN2(A-D) receptors (Paoletti et al., 2013), as controlled mainly by the distal ATD region of the GluN2 subunits (Gielen et al., 2009; Yuan et al., 2009). In spite of these advances, there is little structural understanding of how the ATD and LBD domains modulate receptor activity, and how this conformational information is transduced to the ion channel gating machinery. To capture structural information on the full-length NMDAR, we carried out single-particle cryo-electron microscopy (cryo-EM) (Cheng, 2015) combined with double electron-electron resonance (DEER) experiments (McHaourab et al., 2011).
Results and Discussion
Cryo-EM Structures in the Competitive Antagonist-Bound State
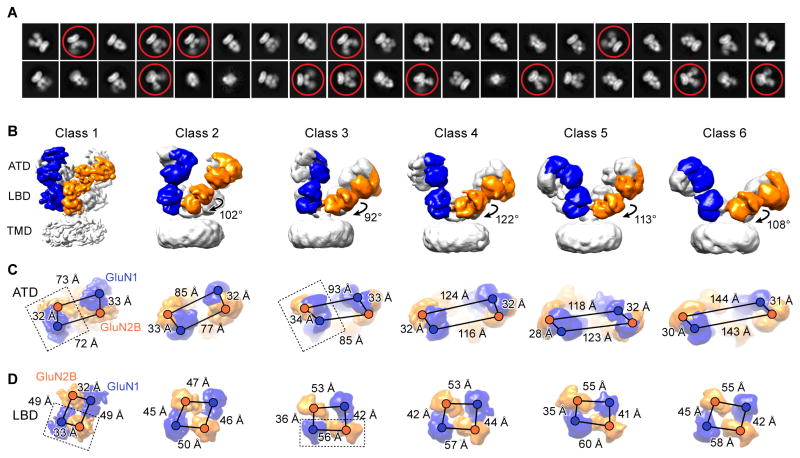
The previous crystal structures of the GluN1-GluN2B receptor exploited an engineered disulfide bond at residue K216C to reduce conformational mobility of the extracellular domains (Lee et al., 2014). Thus, we began by utilizing a receptor construct, deemed NMDAEM, in which we returned this residue to its wild-type identity, thus enabling the receptor to fully explore functionally relevant conformations (Figure S1A–S1C). To visualize how receptor rearrangement is coupled to antagonist binding, we then elucidated the structures of the full-length NMDAEM receptor in complex with GluN1 and GluN2B antagonists, DCKA and D-APV, respectively, by single particle cryo-EM (Figure S1E and S1G). The initial 2D class averages immediately revealed that the extracellular domains (ECDs) underwent large conformational changes relative to the previous crystal structures. In many classes, the 2D projections of the ATD layers show diffuse density (Figure 1A), indicating substantial conformational heterogeneity. After extensive 3D classification, the data yielded six different 3D models (Figure 1B) with well-sampled Euler angle distribution (Figure S2B). The final 3D refinement map of class 1 had an overall resolution of 10.5 Å, and the other five classes ranged in resolution from 13 to 15 Å (Figure S2 and Table S1).
Figure 1. Cryo-EM Structures of Antagonist-Bound Receptors.
(A) Representative 2D class average images of the DCKA/D-APV-bound state. Conformational heterogeneity with highly mobile extracellular domains are circled. (B) Six distinct classes of three-dimensional reconstruction density maps of DCKA/D-APV-bound GluN1-GluN2B (one subunit of each highlighted in blue and orange, respectively), showing antagonist-induced conformational changes of the ECDs. Rotation angles based on the inertial axis of mass weighting of the GluN2B-LBDs between the class 1 and the other five classes are indicated. (C–D) Measurements of the distances from the center of masses (shown as spheres) between GluN1 and GluN2B subunits are indicated by scale bars for the classes represented above. (C) The distances between the heterodimers in the ATD layer increase, while the distances within the dimer remain relatively unchanged. (D) The distance changes result in a transition from a twofold to a pseudo-fourfold symmetry on the LBD layer. The dimer units are highlighted by dashed box in class 1 and class 3. Views are ‘top down’ in (C) and (D).
The density map of class 1 shows secondary structure features, particularly for the majority of α-helices within the ATD, LBD and TMD layers, and also some of the ATD-LBD and LBD-TMD linkers (Figure 2A). We improved the extracellular domain density by additional rounds of refinement using an ECD mask (see Experimental procedure; Figure S2C). The final resolution of the ECD-masked map was 9.3 Å, which was sufficient to fit the R1 and R2 lobes of the ATDs, and D1 and D2 lobes of the LBDs as rigid bodies. Although the general domain arrangement in class 1 is similar to the full-length crystal structure (Lee et al., 2014), there are striking differences, as indicated by the relatively high RMSD of 5.4 Å following superposition of the ECDs. First, the ATDs are more widely separated compared to the disulfide crosslinked crystal structure (Figure S3). Second, the GluN2B-ATDs undergo an opening and untwisting motion, likely as a response to antagonist binding on the LBD layer (Figure 2B and Figure S4B). Third, comparison of the DCKA-bound cryo-EM structure and the glycine-bound structures reveals that the GluN1-LBD clamshell is more open in the antagonist-bound state (Figure 2B and S4C), as predicted by the studies from isolated domains (Furukawa and Gouaux, 2003; Jespersen et al., 2014). Interestingly, the EM-derived GluN2B-LBD conformation does not show opening of the clamshell in comparison to the agonist-bound crystal structure (Figure 2B and S4D), perhaps because either not all particles are fully occupied by antagonist or because the conformational changes induced by D-APV binding are too small to be discerned at this resolution.
Figure 2. Competitive Antagonist Binding Induces Clamshell Conformational Changes.
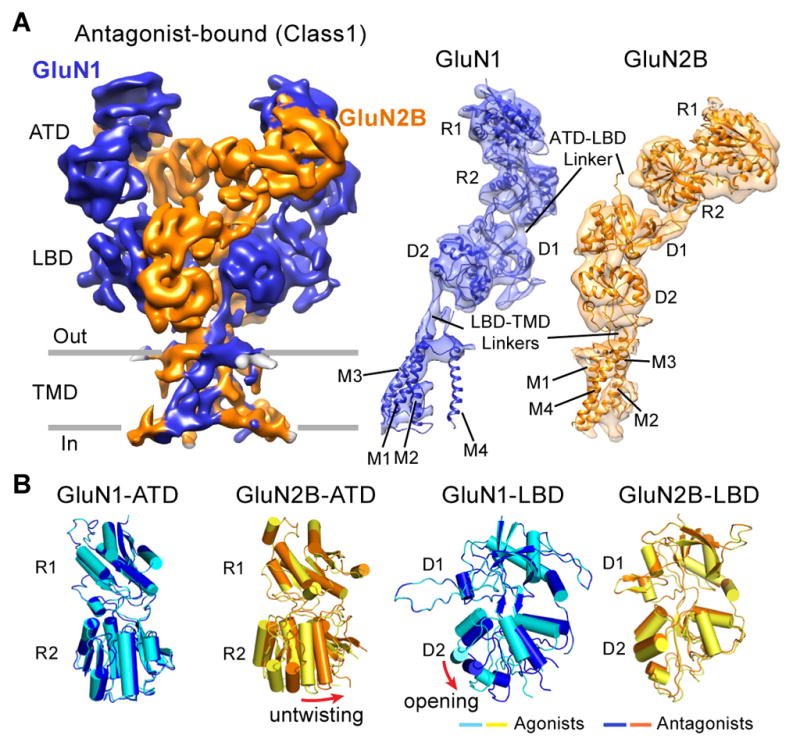
(A) The 3D reconstruction density map of the DCKA/D-APV-bound GluN1-GluN2B receptor (in blue and orange, respectively), referring to class 1 in figure 1. View is perpendicular to the overall 2-fold axes of symmetry. Right panels are corresponding coordinate fits into the reconstruction map of the R1/R2 lobes of ATD, D1/D2 lobes of LBD, and TMD of the GluN1 or GluN2B subunits, independently. (B) Superimposition of the R1 lobes of the antagonist- and agonist-bound GluN1-ATD and GluN2B-ATD models (left panels), showing the untwisting motions of the GluN2B R2 lobe in the antagonist-bound state. Superimposition of the D1 lobes of the antagonist- and agonist-bound states of the GluN1-LBD and GluN2B-LBD models (right panels), showing the opening of the GluN1 D2 lobe in the DCKA-bound state.
The most striking conformational changes were extracted from the five additional classes, which represent the majority of particles in this condition. To better interpret these structural changes, we performed rigid body fitting using individual clamshell ATD and LBD domains, and measured distances between the centers-of-mass (COMs) of the ATDs or LBDs (Figure 1C and 1D). The first major change involves rupture of the D1-D1 LBD dimer interface. The GluN2B-LBD clamshells “swing” clockwise by nearly 110° in the horizontal plane (Figure 1B), which cleaves the LBD dimer assembly in the agonist-bound states (Karakas and Furukawa, 2014; Lee et al., 2014) and leads to an arrangement with pseudo 4-fold symmetry (Figure 1D). Due to the LBD dimer separation, the characteristic domain swapping feature of the NMDAR crystal structure is no longer observed in these antagonist-bound classes (Figure 1B). Second, there is an unambiguous separation between the ATD dimers from ~70 Å to ~140 Å, whereas the distance within the ATD dimer remains unaltered, around 30 Å. Specifically, the R2 lobes of GluN2B-ATD rotate away from the 2-fold axis and the two ATD dimers progressively move apart, while the R1-R1 heterodimeric interface is retained in the complex (Figure 1B and 1C). Third, the relative geometry of ATD and LBD clamshell orientation in the GluN1 subunits remains similar to the agonist-bound structures, while there is a clear separation between the ATD and LBD interface in the GluN2B subunits (Figure 1B).
ECD Conformational Changes Detected by DEER
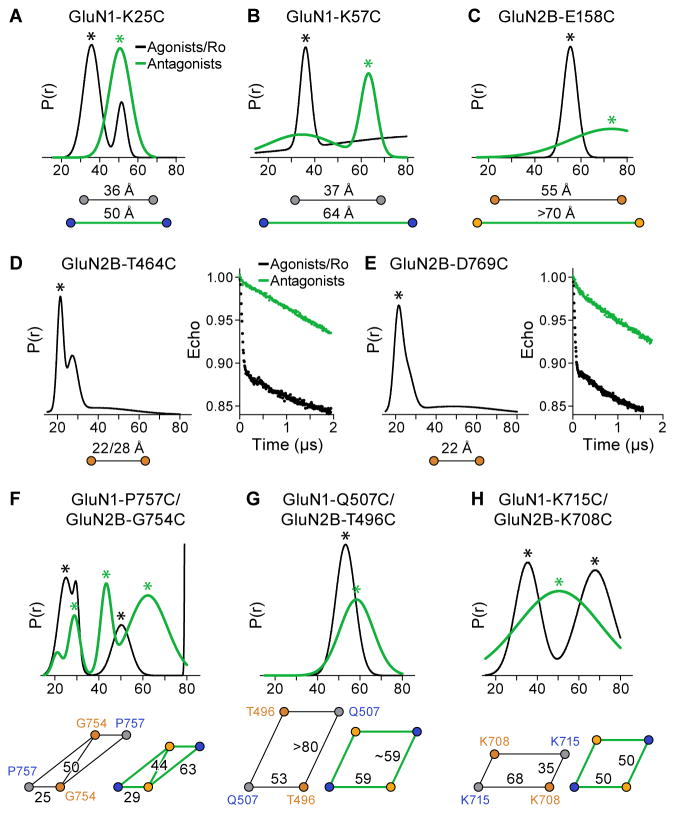
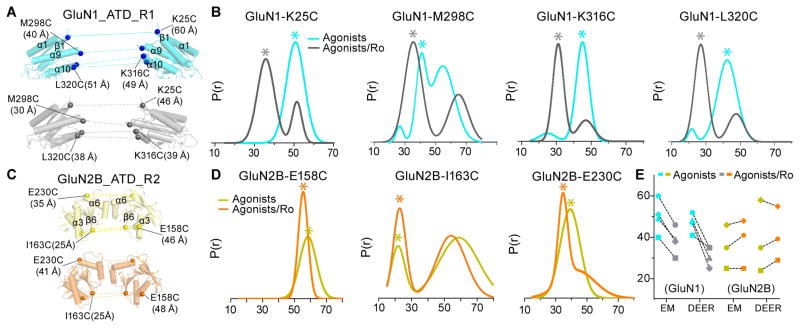
We carried out DEER spectroscopy (McHaourab et al., 2011) at selected ATD or LBD positions by first designing cysteine substitutions on the GluN1-ATD R1 lobe (K25 and K57 on β-sheet 1 and β-sheet 2, respectively), and on the GluN2B-ATD R2 lobe (E158 on α-helix 3, and E230 on α-helix 6). We labeled the cysteine substitutions with MTSSL, and measured probability distributions of distances in antagonist-bound or agonist/Ro25-6981-bound states. The agonist/Ro25-6981condition was included as a control because the crystal structures were solved in the presence of both agonists and Ro25-6981 (Karakas and Furukawa, 2014; Lee et al., 2014). The extent of closure in the antagonist-bound state can then be compared to the agonist/Ro25-6981 condition using the depth of modulation, i.e. the relative depths of the decays. Therefore, a smaller depth of modulation corresponds to a less homogenous closure of the labeled domain.
The DEER distance distribution of spin-labeled GluN1-K25C receptors shows only a single peak around 50 Å in the antagonist-bound state, whereas a major peak around 36 Å is observed under the agonist/Ro25-6981 condition (Figure 3A and Figure S5A). For the spin-labeled GluN1-K57C receptors, the DEER result shows a major peak at 64 Å in the antagonist state and at 37 Å in the agonist/Ro25-6981 state (Figure 3B and Figure S5B). On the other hand, the DEER spectra of the spin-labeled GluN2B-E158C receptors in the antagonist-bound state shows a shift to a distance longer than 70 Å, compared to 55 Å in the agonist/Ro25-6981 state (Figure 3C and Figure S5C). Not surprisingly, the spin-labeled GluN2B-E230C receptors exhibit little depth of modulation with the antagonists, which is likely due to ATD domain fluctuation dynamics (Figure S5D). Taken together, these results corroborate the large separation of ATD dimers seen in the cryo-EM structures when the LBDs are bound to the antagonists.
Figure 3. Conformational Changes in the Antagonist-Bound State Monitored by DEER.
(A–C) Probability distributions of DEER distances determined from DEER decays of MTSSL labeled antagonist- and agonist/Ro25-6981-bound states of GluN1-K25C (A), GluN1-K57C (B) and GluN2B-E158C (C) receptors. Distance changes for agonist/Ro25-6981 (in black) and antagonist (in green) are indicated. (D–E) DEER data of MTSSL-labeled GluN2B-T464C (D) and GluN2B-D769C (E). Peak-normalized echo decays and fits are shown on right. Measurements for agonist/Ro25-6981 are shown in black. (F–G) Probability distributions of DEER distances determined from DEER decays of MTSSL labeled antagonist- and agonist/Ro25-6981-bound states of GluN1-P757C/GluN2B-G754C (F), GluN1-Q507C/GluN2B-T496C (G), and GluN1-K715C/GluN2B-K708C (H) receptors. The asterisks indicate putative distances of the spin-labels. The diagrams below each distribution reflect the distances extracted from the peaks of DEER distribution.
We then investigated conformational changes within the LBD layer. To quantify the diagonal separation of the LBDs, we measured inter-GluN2B subunit distances between two marker residues, GluN2B-T464 on helix B and GluN2B-D769 on helix J. For both spin-labeled receptors, only under the agonist/Ro25-6981 condition does the DEER distance distribution give a short distance peak. However, they both exhibit little depth of modulation under the antagonist-bound state (Figure 3D–3E and Figure S5E–S5F), probably due to the large separation between the two D1 lobes of GluN2B-LBDs. We next introduced two pairs of cysteine substitutions at the LBD dimer interface: GluN1-P757C/GluN2B-G754C and GluN1-Q507C/GluN2B-T496C. Both pairs are situated on top of the D1 lobes, with the first pair proximal and second pair distal to the dimer interface. The labeled GluN1-P757C/GluN2B-G754C receptors in the presence of the agonists and Ro25-6981 show three major peaks in the distribution profiles: the shortest is ~25 Å, indicative of the distance within the LBD heterodimer, the longest is more than 80 Å, indicative of the distance between adjacent GluN1 and GluN2B LBDs in separate dimers, while the middle peak likely corresponds to the distance between the diagonal two GluN1 domains. Interestingly, with DCKA/D-APV, the shortest peak lengthens to 29 Å, the longest peak shortens to 63 Å, and the middle peak also shortens to 44 Å (Figure 3F and Figure S5G). We propose that these distance changes reflect the separation of D1-D1 lobes upon disruption of the LBD intra-dimer interface and that upon rupture of the dimer interface, the former inter dimer interactions become closer in the antagonist-bound ensemble of conformations. Furthermore, the distance distribution obtained from the DEER data of GluN1-Q507C/GluN2B-T496C receptors showed a sharp peak close to 53 Å in the agonist/Ro25-6981 condition, and a peak with moderate width around 59 Å in the antagonist-bound condition (Figure 3G and Figure S5H). This indicates that the GluN1-Q507C and GluN2B-T496C sites move apart in the DCKA/D-APV bound state, which reflects the increase of intra-dimer distance upon binding of competitive antagonists.
To monitor conformational change of the D2-D2 lobes, we engineered cysteines at the sites of GluN1-K715C and GluN2B-K708C. Under the condition with agonist/Ro25-6981, the short distance peak of 35 Å most likely reflects dipolar interactions between labels attached to the adjacent ‘inter dimer’ GluN1 and GluN2 LBDs, whereas the longer distances of the larger peak of 58 Å may arise from dipolar interactions of spin-labels within a GluN1-GluN2B dimer. However, the antagonist-bound receptors exhibit only one broad distribution with the peak centered at 50 Å (Figure 3H and Figure S5I). We speculate that upon binding of antagonists, the D2-D2 lobes rotate from the 2-fold to pseudo 4-fold symmetry. The DEER data is in general agreement with the cryo-EM structures and supports a model whereby domain separation within LBD dimers causes separation of the dimer-of-dimers assembly on the ATD layer in the antagonist-bound state.
NMDAR in the Agonist-Bound Inactive State
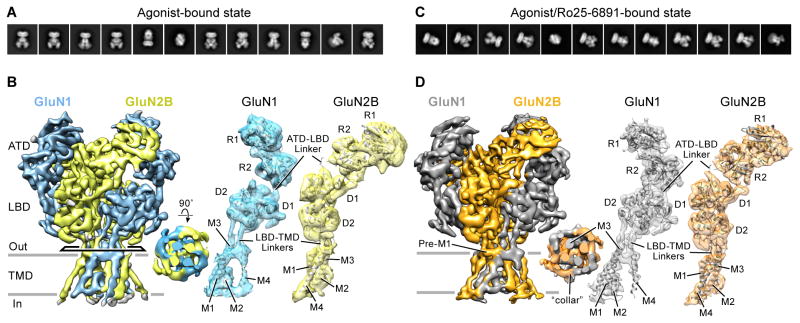
Given the unexpected observation that competitive antagonist binding leads to the disassociation and rearrangement of the LBD and ATD layers, we sought to determine the cryo-EM structure of NMDAR in the presence of glycine and glutamate (Figure S1F). The 2D class averages indicate particle homogeneity of this agonist-bound condition (Figure 4A). The final 3D reconstruction had a resolution of 7.0 Å (Figure S6C), allowing visualization of the majority of α-helices (Figure 4B). Local resolution estimation suggested that the LBD layer was at a higher resolution than the ATD and TMD layers (Figure S6D).
Figure 4. Cryo-EM Structures of Agonist- and Agonist/Ro25-6981-Bound Receptors.
(A and C) Representative 2D class average images of the agonist-bound (A) and agonist/Ro25-6981-bound (C) states. (B and D) The three-dimensional reconstruction density maps of the GluN1-GluN2B receptors in the agonist-bound (GluN1 in cyan and GluN2B in yellow, respectively) and agonist/Ro25-6981-bound states (GluN1 in grey and GluN2B in orange, respectively). Top-down views of the TMD are inserted in the middle panel. Right panels are corresponding coordinate fits into the reconstruction maps of the R1/R2 lobes of ATD, D1/D2 lobes of LBD, and TMD of the GluN1 or GluN2B subunits, independently. View is perpendicular to the overall 2-fold axes of symmetry.
To improve the resolution of the ECD domains, we used a mask to remove the TMD region (see Experimental procedures). The resulting ECD density map yielded a final resolution of 6.7 Å (Figure S6C–S6D). Overall, the agonist-bound structure shows that the ATD and LBD assemble as a dimer-of-dimers. On the “top” layer, the GluN1 and GluN2B ATDs associate through the R1-R1 interface (Figure 4B) with two GluN2B proximal and two GluN1 distal positions to the 2-fold axis (Figure S3). Among the layers, there is a remarkable domain swapping, where pairing of domains between the ATD and LBD layers involves different subunits (Figure 4B). The densities for the ATD-LBD and LBD-TMD linkers are visible (Figure 4B). The differences in the structure of these linkers could contribute to the non-equivalent conformation between the two types of subunits.
The resolution of the full-length density map allows us to reveal detailed features in the TMD region. Pre-M1 helices are parallel to the membrane and form a “collar” around the ion channel core assembled by the M3 helices ((Sobolevsky et al., 2009), Figure 4B). The M1 and M4 helices cross the membrane forming the outer periphery of the ion channel. The M2 segment forms a short re-entrant helix and an extended region, defining the selectivity filter (Figure 4B). Upon comparison of the x-ray crystal structure with the structure derived from the EM density map, we find that the LBDs superimpose well (RMSD=1.9 Å), whereas the ATDs superimpose less well (Figure S3, RMSD=3.9 Å). Because the ion channel gate is closed in our agonist-bound EM structure, we speculate that the receptor is trapped in an agonist-bound desensitized or pre-open state.
Experiments probing the binding of agonists to non-NMDA iGluRs are consistent with the concept that agonists induce rapid receptor desensitization concomitant with disruption of the D1-D1 LBD dimer interface and rearrangement of the ATD and LBD layers (Armstrong et al., 2006; Chaudhry et al., 2009; Durr et al., 2014; Meyerson et al., 2014; Sun et al., 2002; Weston et al., 2006). In contrast, our full-length agonist-bound NMDA receptor structure reveals that the D1-D1 interface remains intact. Indeed, a recent study of isolated LBDs shows that agonist binding stabilizes the GluN1-GluN2 LBD heterodimer (Cheriyan et al., 2015). In light of the ensemble of antagonist-bound NMDAR structures, we propose that binding of glutamate and glycine stabilizes the LBD heterodimer interfaces and gating ring, setting the structural stage for receptor activation (Movie S1).
NMDAR in the Agonist/Modulator-Bound State
By performing TEVC recordings, we confirmed that the Ro25-6981 inhibition is maintained in the NMDAEM construct (Figure S1D). Cryo-EM analysis of NMDAEM construct with glycine, glutamate and Ro25-6981 revealed well-defined 2D class averages, similar to the agonist-bound dataset (Figure 4C and Figure S6E). The final 3D reconstruction was determined to overall resolution of 7.5 Å (Figure S6G). Secondary structures of the extracellular ATD and LBD domains, and also the α-helices of TMDs and LBD-TMD linkers, were well-resolved (Figure 4D), and the ion channel gate is closed. Further refinement with masking of the TMD obtained a map of ECD to a final resolution of 7.0 Å. By superimposing this map with the agonist-bound map, we observed that the ATD layer undergoes conformational changes upon Ro25-6981 binding (see next section). On the LBD layer, the two heterodimers are shifted slightly counterclockwise, but the D1-D1 dimer interface is retained (Figure S3).
To test whether the action of ATD inhibitors requires disruption of the LBD D1-D1 interface, we engineered two pairs of disulfide bridges to crosslink adjacent D1-D1 LBD dimer interfaces: N519C and L775C mutants on GluN1, and E514C and L778C on GluN2B, homologous positions according to the GluN1-GluN2A LBD structure (Furukawa et al., 2005). TEVC recording reveals that the Ro25-6981 inhibition is maintained in this LBD crosslinked receptor (Figure S1D). By comparison to the antagonist-bound structures, we propose that the negative allosteric modulator and competitive antagonists have different inhibition mechanisms. Early electrophysiological studies suggested that phenylethanolamines antagonize the channel gating in an activity-dependent manner. Ifenprodil exhibits >10-fold higher apparent binding affinity in the active or desensitized state in comparison to the resting or agonist-unbound state; furthermore, the binding of ifenprodil increases receptor affinity for glutamate (Kew et al., 1996). Our structural data is in line with these binding experiments, supporting the notion that, in GluN2B receptors, Ro25-6981 inhibition is associated with intact LBD D1-D1 heterodimer interfaces.
ATD Conformational Rearrangement upon Ro25-6981 Binding
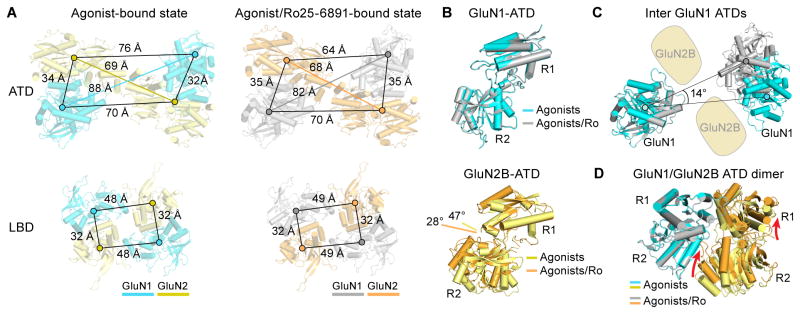
To understand the molecular basis of ATD ligand binding and signal transduction, we analyzed and compared the conformational state of ATDs in the absence or presence of ATD ligand. We generated the model by fitting the R1 or R2 lobes as independent rigid bodies into the agonist/Ro25-6981-bound or agonist-bound ECD density maps. A counterclockwise movement viewed from the top was observed upon Ro25-6981 binding, as well as an unforeseen inward movement of the two R1 lobes of the distal GluN1 subunits (Figure 5 and Movie S2). To quantify the distance changes between the ATD protomers, we measured the distances of the center of masses (COMs) between individual clamshells. An ~6 Å contraction and 14° counterclockwise rotation is observed in the GluN1-ATDs with the binding of Ro25-6981 (Figure 5A and 5C). In contrast, between the two conditions only a small ~1 Å COM distance difference for the two GluN2B-ATDs was observed (Figure 5A).
Figure 5. Conformational Changes Effected by Allosteric Inhibitor.
(A) Agonist- and agonist/Ro25-6981-bound GluN1-GluN2B receptor models derived from the density maps are shown in cyan/yellow and grey/orange, respectively. Distances from the center of mass (spheres) between subunits for the ATD (top panels) and LBD (bottom panels) are indicated by scale bars. (B) Superimposition of the R1 lobes of the agonist- and agonist/Ro25-6981-bound states of the GluN1-ATD (top panel) and GluN2B-ATD (bottom panel) domains, showing the 20° opening (measured by dihedral angles connecting the Cα atoms of E101, Q147, E230 and Y277) of the GluN2B clamshell in the agonist-bound state. (C) Superimposition of one GluN1 for the agonist- and agonist/Ro25-6981-bound states, showing the intersubunit rotation of the other GluN1 clamshell-liked domain.(D) Superimposition of the GluN1-ATD of the agonist- and agonist/Ro25-6981-bound states within the ATD heterodimer, showing relative movement of the R1 and R2 lobes of GluN2B ATD.
Within each ATD, superimposing the R1 lobes of the Ro25-6981-bound and non-Ro25-6981 GluN2B-ATDs illustrates the GluN2B ATD adopts a clamshell conformation that is ~20° more open in the absence of Ro25-6981 compared with the structure in its presence (Figure 5B and Movie S2). In contrast, the conformation of the GluN1 ATD remains similar regardless of the presence or absence of Ro25-6981 (Figure 5B and Figure S4A). When superimposing the GluN1-ATDs, we observed that the R1 lobe of the GluN2 moves outward, away from the dimer interface (Figure 5D). Furthermore, Ro25-6981 induces a major structural change, in that the R2 lobe of the GluN2B-ATD undergoes a large degree of closure and forms a dimeric interface with the GluN1 R1 lobe. This new GluN1-R1 and GluN2B-R2 interface uniquely exists in NMDARs (Karakas et al., 2011; Mayer, 2011). Interestingly, previous functional experiments have demonstrated that several pairs of ionic interactions at this interface directly contribute to “lock” the GluN2 ATD in the closed state (Zhu et al., 2014). Based on the agonist-bound structure, we show that this interface is not formed in the absence of the Ro25-6981 molecule (Figure 5D). Thus, we provide evidence for the “open” GluN2B-ATD conformation, and also demonstrate that the open and closed states of the GluN2-ATDs exist in equilibrium, as proposed by former functional studies (Gielen et al., 2009; Yuan et al., 2009). The presence of Ro25-6981 shifts this equilibrium toward the closed state of GluN2-ATDs. Taken together, these results indicate that Ro25-6981 binding to the ATD dimer interface induces rearrangement of the entire ATD tetramer (Figure S3).
Monitoring ATD Interdomain Distances by DEER
To further investigate modulator-dependent structural rearrangement of the ATDs, we carried out DEER spectroscopy on spin-labels at various positions on the ATD layer. We introduced cysteines at four positions on GluN1-ATD based on the NMDAEM construct: K25C as aforementioned, M298C on α-helix 9, K316C at the beginning of α-helix 10, and L320C in the middle of the α-helix 10 (Figure 6A); and at three positions on GluN2B-ATD: E158C and E230C as aforementioned, and I163C in the beginning of the β-sheet 6 (Figure 6C). DEER distance probability distributions calculated from the echo decays of the spin-labeled cysteine mutant receptors with or without compound Ro25-6981 are shown in Figure 6B and 6D, respectively.
Figure 6. Separation of GluN1 ATDs, but not GluN2 ATDs Detected by DEER.
(A and C) Corresponding models from coordinate fits into the three-dimensional EM reconstruction ECD maps with side views of the R1 lobes of inter-GluN1 ATDs (A) and R2 lobes of inter-GluN2B ATDs (C) in complex with agonist (cyan in A; yellow in C) and agonist/Ro25-6981 (grey in A; orange in C). Distance between Cα atoms for GluN1 (Lys 25, Met 298, Lys 316 and Leu 320) and GluN2B (Glu 158, Ile 163 and Glu 230) are indicated. (B and D) Probability distributions of DEER distances determined from DEER decays of MTSSL labeled agonist- and agonist/Ro25-6981-bound GluN1-GluN2B of indicated receptor constructs. Asterisk indicates putative distances from the spin-labels under the given condition. (E) Summary of the distance changes in the agonist- and agonist/Ro25-6981-bound GluN1-GluN2B receptor states observed from the cryo-EM structures and DEER measurements.
To better characterize the physiological relevance of ATD movement, we compared the distance distribution measured from the DEER experiments and the Cα-Cα distances measured from agonist-bound and agonist/Ro25-698-bound EM structures (Figure 6E). Among the four GluN1 sites tested, the distance probabilities between the pairs of spin labels in the Ro25-6981-bound receptors are consistently shorter than those in the agonist-bound receptors (Figure 6B and Figure S7A–S7D). These results are consistent with measurements from the cryo-EM structures, showing that Ro25-6981 binding to the dimer interface shortens the distance between the two GluN1 ATDs, and brings the ATDs closer to each other (Figure 6A). However, with the three sites on the GluN2B-ATD, both DEER distributions and measurements from EM models show only minor movements between the two GluN2B-ATDs (Figure 6C–6E and Figure S7E–S7G).
Differential Signal Transduction
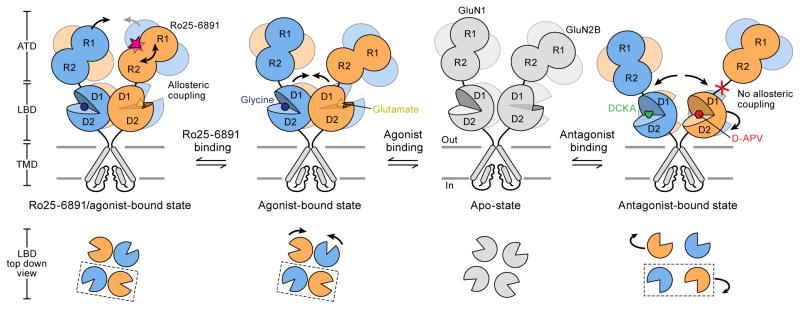
Combined with preexisting functional data, we propose the following mechanism for NMDAR activity (Figure 7). First, the binding of glutamate and glycine stabilizes the LBD heterodimers and the LBD gating ring. Second, the binding of phenylethanolamines to the ATD heterodimer interface induces GluN2-ATD clamshell closure and also shortens the distance of inter-GluN1 ATDs. We speculate that the ATD allosteric inhibitor binding further strengthens the LBD dimer interaction and enhances conformational coupling between ATD and LBD layers. Third, upon binding of competitive antagonists, the GluN2B LBDs move apart and “flip” and, as a consequence, the GluN2 ATDs are displaced by the horizontal rotation of the LBDs, disrupting the interaction between the ATD and LBD layers and entirely decoupling agonist-binding from ion channel gating. This motion of the GluN2B subunit may explain why the GluN2 subunit composition in the tetramer is the key determinant for biophysical and pharmacological diversity.
Figure 7. Schematic Summary of ECD Conformational Changes in NMDAR.
Shown are the conformational changes of the extracellular ATD and LBD layers associated with the transition from the apo state to the antagonist-bound state (right), the agonist-bound state (middle) and the agonist/Ro25-6981-bound (left). The bottom panel shows top-down views of various LBD conformations at different states. At present there is no structure for an apo-state and thus the schematic is grey.
Our studies also uncover structural insights into the disparate mechanisms of NMDA receptor inhibition by LBD competitive antagonists in comparison with the ATD allosteric modulator. Competitive antagonist binding disrupts the LBD heterodimers and likely affects the gating ring through loss of tension in the LBD-TMD linkers, while ATD modulators such as the phenylethanolamines preserve interactions within the LBD dimer, likely stabilizing the receptor in an agonist-bound but desensitized or desensitized-like state. We speculate that the effects of competitive antagonists and phenylethanolamine-like compounds exhibit negative cooperativity, such that application of competitive antagonists diminishes the inhibitory effect of phenylethanolamines, and vice versa. Furthermore, comparison to another ATD allosteric inhibitor, zinc, reveals yet another distinctive inhibition mechanism. Specifically, the phenylethanolamine binding pocket is at the ATD dimer interface involving both GluN1 and GluN2B (Karakas et al., 2011), while the zinc binding site is at the outer end of the GluN2 interlobe cleft (Karakas et al., 2009). In previous functional studies of the LBD cysteine crosslinked GluN1-GluN2A receptors, zinc sensitivity is largely reduced (Gielen et al., 2008). Thus, phenylethanolamine allosteric inhibition of GluN2B receptors probably acts through a different ATD rearrangement in comparison to the zinc inhibition in GluN2A receptors.
By comparison with antagonist-bound or agonist-bound desensitized AMPA and kainate receptors, we note a fundamental difference in the conformational rearrangement during agonist-binding or antagonist-action between the NMDA and non-NMDA iGluRs. The agonist-bound desensitized non-NMDA iGluRs exhibit remarkable D1-D1 LBD dimer interface separation, accompanied with the LBD conformational transition from 2-fold to 4-fold symmetry (Durr et al., 2014; Meyerson et al., 2014; Schauder et al., 2013), somewhat reminiscent of our structures of the antagonist-bound NMDARs. By contrast, the full-length AMPA GluA2 crystal (Sobolevsky et al., 2009) and cryo-EM structures (Meyerson et al., 2014) in complex with a competitive antagonist, display intact LBD D1-D1 interfaces. Taken together, our work provides fundamental insights into mechanisms of activation and inhibition of NMDARs and emphasizes important distinctions between AMPA and kainate receptors, and NMDARs.
Experimental Procedures
More detailed experimental procedures are described in the Supplemental Experimental Procedures.
Plasmid Construction, Expression and Purification
The construct used for cryo-EM and DEER studies was derived from the previously reported Xenopus laevis NMDA Δ2 construct (Lee et al., 2014), with the K216C mutation reverted to the wild-type lysine residue (deemed NMDAEM). The DEER constructs were designed with cysteine substitutions introduced into the NMDAEM construct. For the scintillation proximity assay (SPA), a streptavidin-binding peptide (SBP) tag was introduced to the GluN2B subunit of NMDAEM after the 3C protease tag. The NMDA receptor constructs were expressed in HEK293S GnTI- cells by baculovirus-mediated gene transduction of mammalian cells and the resulting protein purified as previously described (Lee et al., 2014).
Spin Labeling and DEER Experiments
Single cysteine mutations of the NMDAEM construct were reacted with the spin label 1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl-methanethiosulfonate (MTSSL). Electron paramagnetic resonance (EPR) spectra were obtained using continuous wave EPR as described previously (Mishra et al., 2014). DEER experiments were performed using a standard four-pulse protocol (Jeschke et al., 2002) and the resulting signals were analyzed assuming that the distance distributions, P(r), consist of a sum of Gaussians (Durr et al., 2014; Stein et al., 2015).
Cryo-EM Grid Preparation and Data Acquisition
Detergent-solubilized NMDA receptor protein was mixed with D-APV and DCKA for the antagonist-bound state, glutamate and glycine for the agonist-bound state; and Ro25-6981, glutamate and glycine for the agonist/modulator-bound state, all at final concentrations of 1 mM. Vitrified specimens were prepared by adding a 2.5 μl protein sample at the concentration of 4.5 mg/ml to R1.2/1.3 200 mesh gold holey carbon grids glow discharged for 120 s. Grids were blotted for 5 s, then plunge-frozen under conditions of 18°C and 100% humidity.
The agonist-bound dataset was collected on a Titan Krios cryo-electron microscope operated at 300 kV at the Electron Imaging Center for Nanomachines at UCLA. Images were recorded on a Gatan K2 Summit direct-detector using electron counting mode at a magnification of 130000x (corresponding to a calibrated pixel size of 1.07 Å). The beam intensity on the camera was set to 8.7 e−/pixel/s. Total exposure time was 8 s fractionated over 40 frames. Nominal defocus values ranged from −2.5 to −4.0 μm. The agonist/Ro25-6981- and antagonist-bound data sets were collected on Titan Krios I at the Janelia Research Campus. Krios I is equipped with a CETCOR Cs Corrector and a Gatan Image Filter (GIF). A 30 eV energy slit as well as a 70 μm objective aperture (corresponding to a 2 Å cutoff) was used during data collection. The Cs was turned to 0.01 mm. Both data sets were collected using a K2 in super-resolution mode, then 2 Χ 2 binned in Fourier space, resulting in a pixel size of 1.35 Å. The dose rate was set to 10 e−/pixel/s. The total exposure time was 15 s for the agonist/Ro25-6981-bound state and 9.6 s for the antagonist-bound state. Nominal defocus values ranged from −1.5 to −3.5 μm. The dose-fractionated images were recorded using the automated image acquisition programs Leginon (Suloway et al., 2005) at UCLA and SerialEM (Mastronarde, 2005) at Janelia.
Image Processing and 3D Refinement
The down sampled frames were motion-corrected using Unblur (Grant and Grigorieff, 2015). After motion correction, the summed images were processing in RELION (Scheres, 2012). Defocus values were estimated using CTFFIND3 (Mindell and Grigorieff, 2003). For each dataset, approximately 3000 particles were manually picked and processed using reference-free 2D classification to generate 5–8 classes for template-based particle picking. Auto-picked particles were further cleaned-up with several additional rounds of 2D classification. An initial 3D model was generated from a previous crystal structure of the GluN1-GluN2B receptor (PDB code: 4TLM) (Lee et al., 2014) and low-pass filtered to 50 Å. No symmetry was applied during 3D classification and auto-refinement. Due to the substantial conformational heterogeneity in the antagonist-bound dataset, an initial 3D classification was carried out yielding six (out of ten) classes that was carried to further 3D refinement. Fourier shell coefficient (FSC) was generated in RELION using the gold standard FSC=0.143 criteria (Scheres and Chen, 2012) and final maps were made using soft masks and B-factor estimation and sharpening as implemented in RELION’s post-processing step. To improve the density of the ECDs, an additional round of auto-refinement was run using masks to reduce the effect of heterogeneity on alignment in the TMD. These masks were created using the Volume Eraser feature in Chimera (Pettersen et al., 2004) to remove the TMD, followed by low-pass filtering to 20 Å, application of a binary threshold, and a mask extension of ~8 Å with a 5 Å soft edge.
Model Fitting and Structural Analysis
Fittings of the PDB coordinates into the ECD density maps were carried out using UCSF Chimera (Pettersen et al., 2004). The separated ATD R1 or R2 lobe (PDB: 3QEL) and LBD D1 or D2 lobe (PBD: 2A5T) were each fitted as independent rigid bodies. For antagonist-bound classes (class 2–6), individual ATD or LBD clamshells were fitted. After model fitting, the homology model of the Xenopus laevis GluN1-GluN2B receptor was generated based on the PDB coordinate files of separated domains using the SWISS-MODEL Automated Mode (Biasini et al., 2014). All structural measurements were carried out based on the PDB coordinates from ECD density maps. The movie figures were generated in Chimera using the map interpolation feature to animate the transition between density maps, and the morph conformation feature to interpolate and animate the movement between each of the rigid-body domain fittings.
Two-Electrode Voltage-clamp Electrophysiology
Recombinant NMDA receptors were expressed in Xenopus laevis oocytes after nuclear injection of RNAs encoding GluN1 and GluN2 subunits (at 100 ng/μL concentration, ratio 1:1). 2–3 days after injection, currents in response to agonists, antagonists and allosteric modulator Ro25-6981 were recorded at room temperature using two-electrode voltage-clamp at the holding potential of −60 mV.
Ligand Binding Assays
The binding constant of 3H-glycine was measured in the absence of glutamate, and of 3H-glutamate in the absence of glycine. Non-specific binding was determined by the addition of 1 mM DCKA (for 3H-glycine) or 1 mM D-APV (for 3H-glutamate). The inhibition constant of DCKA was determined in the presence of 1 μM glycine (10% 3H-glycine), and of D-APV in the presence of 1 μM glutamate (10% 3H-glutamate). The data were analyzed and fitted in GraphPad Prism using a one-site binding model. All data are represented as mean ± SD, calculated from 2–3 independent trials performed in triplicate.
Supplementary Material
Acknowledgments
We are grateful to Z.H. Yu, R. Huang and C. Hong (Janelia Campus), Z. Hong Zhou and I. Atanasov (UCLA) for assistance with data collection and to R. Stites, M. Hakanson and A. Trzynka (OHSU) for computational support. Microscopy at Oregon Health & Science University (OHSU) was performed at the Multiscale Microscopy Core (MMC) with technical support from the OHSU-FEI Living Lab, Intel and the OHSU Center for Spatial Systems Biomedicine (OCSSB). We thank L. Vaskalis for help with illustrations, K.L. Durr and H. Owen for proofreading, L. Wei for help with data processing, and to all the Gouaux lab members for helpful discussions. H.S.M. and R.A.S. were supported by grants U54-GM087519 and pulsed EPR instrumentation was acquired through a shared instrumentation grant S10 RR027091. This work was supported by the National Institute of Health (5R37NS038631). E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Accession Numbers
The density maps and structural models have been deposited in the Electron Microscopy Data Bank under ID codes EMD-8097, 8098, 8101, 8102, 8103, 8104, 8105 and 8106 and Protein Data Bank under ID codes 5IOU, 5IOV, 5IPQ, 5IPR, 5IPS, 5IPT, 5IPU, 5IPV, respectively.
Author Contributions
S.Z. and E.G. designed the project, S.Z. performed protein preparation, cryo-EM data collection and data analysis, and electrophysiological recordings, C.Y. participated in the cryo-EM data analysis, R.A.S. and H.S.M. carried out the DEER experiment, A.G. helped with the molecular cloning for the DEER constructs, C.L collected the agonist-bound dataset. S.Z. and E.G. wrote the manuscript with input from all co-authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Attwell D, Gibb A. Neuroenergetics and the kinetic design of excitatory synapses. Nature reviews Neuroscience. 2005;6:841–849. doi: 10.1038/nrn1784. [DOI] [PubMed] [Google Scholar]
- Baron BM, Siegel BW, Slone AL, Harrison BL, Palfreyman MG, Hurt SD. [3H]5,7-dichlorokynurenic acid, a novel radioligand labels NMDA receptor-associated glycine binding sites. European journal of pharmacology. 1991;206:149–154. doi: 10.1016/0922-4106(91)90023-b. [DOI] [PubMed] [Google Scholar]
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer ML. Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization. The EMBO journal. 2009;28:1518–1530. doi: 10.1038/emboj.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheriyan J, Mezes C, Zhou N, Balsara RD, Castellino FJ. Heteromerization of ligand binding domains of N-methyl-D-aspartate receptor requires both coagonists, L-glutamate and glycine. Biochemistry. 2015;54:787–794. doi: 10.1021/bi501437s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmau J, Gleichman AJ, Hughes EG, Rossi JE, Peng X, Lai M, Dessain SK, Rosenfeld MR, Balice-Gordon R, Lynch DR. Anti-NMDA-receptor encephalitis: case series and analysis of the effects of antibodies. The Lancet Neurology. 2008;7:1091–1098. doi: 10.1016/S1474-4422(08)70224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, McHaourab HS, Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzubay JA, Jahr CE. Kinetics of NMDA channel opening. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1996;16:4129–4134. doi: 10.1523/JNEUROSCI.16-13-04129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endele S, Rosenberger G, Geider K, Popp B, Tamer C, Stefanova I, Milh M, Kortum F, Fritsch A, Pientka FK, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nature genetics. 2010;42:1021–1026. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- Evans RH, Francis AA, Jones AW, Smith DA, Watkins JC. The effects of a series of omega-phosphonic alpha-carboxylic amino acids on electrically evoked and excitant amino acid-induced responses in isolated spinal cord preparations. British journal of pharmacology. 1982;75:65–75. doi: 10.1111/j.1476-5381.1982.tb08758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G, Mutel V, Trube G, Malherbe P, Kew JN, Mohacsi E, Heitz MP, Kemp JA. Ro 25-6981, a highly potent and selective blocker of N-methyl-D-aspartate receptors containing the NR2B subunit. Characterization in vitro. The Journal of pharmacology and experimental therapeutics. 1997;283:1285–1292. [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. The EMBO journal. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G, Koch A, Jonas U, Godt A. Direct conversion of EPR dipolar time evolution data to distance distributions. J Magn Reson. 2002;155:72–82. doi: 10.1006/jmre.2001.2498. [DOI] [PubMed] [Google Scholar]
- Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. Structural insights into competitive antagonism in NMDA receptors. Neuron. 2014;81:366–378. doi: 10.1016/j.neuron.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325:529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- Karakas E, Furukawa H. Crystal structure of a heterotetrameric NMDA receptor ion channel. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. The EMBO journal. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew JN, Trube G, Kemp JA. A novel mechanism of activity-dependent NMDA receptor antagonism describes the effect of ifenprodil in rat cultured cortical neurones. The Journal of physiology. 1996;497(Pt 3):761–772. doi: 10.1113/jphysiol.1996.sp021807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Lu W, Michel JC, Goehring A, Du J, Song X, Gouaux E. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014;511:191–197. doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesca G, Rudolf G, Bruneau N, Lozovaya N, Labalme A, Boutry-Kryza N, Salmi M, Tsintsadze T, Addis L, Motte J, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nature genetics. 2013;45:1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of structural biology. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mayer ML. Emerging models of glutamate receptor ion channel structure and function. Structure. 2011;19:1370–1380. doi: 10.1016/j.str.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Mishra S, Verhalen B, Stein RA, Wen PC, Tajkhorshid E, McHaourab HS. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife. 2014;3:e02740. doi: 10.7554/eLife.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. The EMBO journal. 2011;30:3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nature reviews Neuroscience. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Schauder DM, Kuybeda O, Zhang J, Klymko K, Bartesaghi A, Borgnia MJ, Mayer ML, Subramaniam S. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nature methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein RA, Beth AH, Hustedt EJ. A Straightforward Approach to the Analysis of Double Electron-Electron Resonance Data. Methods in enzymology. 2015;563:531–567. doi: 10.1016/bs.mie.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Hartley M, Heinemann SF. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacological reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnano AM, Rebola N, Savtchenko LP, Pinheiro PS, Casado M, Kieffer BL, Rusakov DA, Mulle C, Paoletti P. Zinc dynamics and action at excitatory synapses. Neuron. 2014;82:1101–1114. doi: 10.1016/j.neuron.2014.04.034. [DOI] [PubMed] [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. Journal of neurophysiology. 1998;79:555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nature structural & molecular biology. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F, Erreger K, Low CM, Banke T, Lee CJ, Conn PJ, Traynelis SF. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nature neuroscience. 2001;4:894–901. doi: 10.1038/nn0901-894. [DOI] [PubMed] [Google Scholar]
- Zhu S, Riou M, Yao CA, Carvalho S, Rodriguez PC, Bensaude O, Paoletti P, Ye S. Genetically encoding a light switch in an ionotropic glutamate receptor reveals subunit-specific interfaces. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:6081–6086. doi: 10.1073/pnas.1318808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nature structural & molecular biology. 2013;20:477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.