Abstract
Friedreich ataxia, the most prevalent inherited ataxia, is caused by an expanded GAA triplet-repeat sequence in intron 1 of the FXN gene. Repressive chromatin spreads from the expanded GAA triplet-repeat sequence to cause epigenetic silencing of the FXN promoter via altered nucleosomal positioning and reduced chromatin accessibility. Indeed, deficient transcriptional initiation is the predominant cause of transcriptional deficiency in Friedreich ataxia. Treatment with 109, a class I histone deacetylase (HDAC) inhibitor, resulted in increased level of FXN transcript both upstream and downstream of the expanded GAA triplet-repeat sequence, without any change in transcript stability, suggesting that it acts via improvement of transcriptional initiation. Quantitative analysis of transcriptional initiation via metabolic labeling of nascent transcripts in patient-derived cells revealed a >3-fold increase (P < 0.05) in FXN promoter function. A concomitant 3-fold improvement (P < 0.001) in FXN promoter structure and chromatin accessibility was observed via Nucleosome Occupancy and Methylome Sequencing, a high-resolution in vivo footprint assay for detecting nucleosome occupancy in individual chromatin fibers. No such improvement in FXN promoter function or structure was observed upon treatment with a chemically-related inactive compound (966). Thus epigenetic promoter silencing in Friedreich ataxia is reversible, and the results implicate class I HDACs in repeat-mediated promoter silencing.
INTRODUCTION
Expanded triplet-repeat sequences are the cause of several human genetic diseases (1). Whereas the molecular mechanisms underlying disease pathogenesis can be varied, in some instances expanded triplet-repeats can serve as a source of repressive chromatin (2). In Friedreich ataxia (FRDA), the most prevalent inherited ataxia, patients are typically homozygous for expanded FXN alleles that contain 100–1300 GAA triplets in intron 1, with the majority of disease-causing alleles containing >400 GAA triplets (3). Repressive chromatin spreads from the expanded GAA triplet-repeat (GAA-TR) sequence (4–8) in intron 1 to the FXN promoter and results in epigenetic promoter silencing (6,8). The promoter of the FXN gene is rendered transcriptionally non-permissive, leading to severe deficiency of transcriptional initiation in FRDA (6,8,9). The magnitude of promoter silencing in FRDA and the resultant deficiency of transcriptional initiation is dependent on the length of the expanded GAA-TR sequence (10), which further substantiates the causal relationship between the expanded GAA-TR sequence and epigenetic promoter silencing. Presently, it remains unknown if the epigenetic promoter silencing caused by the expanded GAA-TR sequence in FRDA can be reversed.
In addition to epigenetic FXN promoter silencing in FRDA, there is also evidence of deficient transcriptional elongation through the expanded GAA-TR sequence. However, the magnitude of impedance of transcriptional elongation in FRDA is much less impressive in vivo than was previously suggested by in vitro studies (11–13). Indeed, measurement of steady-state transcript levels by quantitative RT-PCR did not reveal an appreciable FRDA-specific reduction in FXN transcript downstream (versus upstream) of the expanded GAA-TR sequence (6,10). Nevertheless, comparison of the level of FXN transcript upstream versus downstream of the expanded GAA-TR sequence, both via quantitative RT-PCR of metabolically labeled nascent transcript (8) and by comprehensive RNA-seq analysis (14), revealed ∼2-fold reduction in transcriptional elongation through the expanded GAA-TR containing intron 1. Consistent with this, Kumari et al. (6) detected ∼2-fold reduced level of the elongation-associated marker, H3K36me3, downstream of the expanded GAA-TR sequence in FRDA. In contrast, there is a 5–12-fold reduced level of transcriptional initiation in FRDA versus non-FRDA cells (8) and Silva et al. (9) estimated by RNA-FISH in single cells that the relative contribution of the transcriptional initiation defect to the total deficit of transcript level in FRDA is 6.3-fold greater than due to the defect in transcriptional elongation. Therefore, while there is a transcriptional elongation defect in FRDA, the predominant mechanism underlying the deficiency of FXN transcript in FRDA seems to be the reduced amount of transcriptional initiation due to promoter silencing.
Reversal of FXN transcriptional deficiency is expected to produce normal frataxin protein since the coding sequence of the FXN gene is unaffected in the vast majority of FRDA patients (3). Heterozygous carriers of the expanded GAA-TR sequence express ∼50% of normal levels of FXN transcript and remain asymptomatic, and patients typically express ∼15% of normal levels, therefore a 2–4-fold increase in FXN transcript levels would be expected to result in a clinically meaningful outcome. Furthermore, whereas the majority of people with FRDA are homozygous for the expanded GAA-TR sequence, a small number of patients (<5%) are compound heterozygous for one expanded GAA-TR sequence and another loss-of-function FXN mutation (15). This means that every FRDA patient has at least one expanded GAA-TR sequence with the capacity to cause epigenetic silencing of the FXN gene. For all of these reasons, reactivation of the epigenetically silenced FXN gene in FRDA is considered an attractive therapeutic strategy, and it is being widely explored (4,16–21). In fact, since deficient transcriptional initiation is the major cause of the deficiency of FXN transcript (6,8,9), reactivation of the epigenetically silenced FXN promoter is likely to be a particularly efficient strategy for restoring FXN transcript levels to therapeutically beneficial levels. 2-aminobenzamide derivatives that specifically inhibit class I histone deacetylases (HDACs) have been shown to increase FXN transcript levels in patient-derived cells (3); however, the precise molecular target in the FXN gene by which the increase in transcript levels is achieved remains unknown. One such molecule (109/RG2833) successfully completed a Phase 1b clinical trial (19) and is presently being developed as a therapeutic agent for FRDA. We sought to determine if 109 is able to reactivate the epigenetically silenced FXN promoter, thereby establishing the proof-of-principle of its reversibility, and potentially implicating class I HDACs in the establishment of repeat-mediated epigenetic promoter silencing in FRDA. Here we report that 109 acts by reversing the epigenetic promoter silencing in FRDA. It restores promoter structure as seen in individual chromatin fibers, and enhances promoter function as measured via in vivo labeling of nascent transcripts in patient-derived cells. Therefore, inhibition of class I HDACs reverses the major molecular defect in FRDA, and thus represents a rational molecular therapeutic strategy. Our data also implicate class I HDACs in the etiology of repeat-mediated epigenetic silencing.
MATERIALS AND METHODS
Cell lines
Lymphoblastoid cell lines from two healthy subjects [GM22647 (CNTR-1) and GM22671 (CNTR-2)], and 12 individuals with FRDA [GM16216, GM16207, GM16204, GM16230, GM16223, GM16244 (FRDA-17), GM16210 (FRDA-10), GM16203 (FRDA-6), GM16243, GM16798, GM16197, GM14518] were obtained from Coriell Cell Repositories. All individuals with FRDA were homozygous for expanded GAA-TR alleles that ranged in size from 200 to 1122 triplets (shorter allele range: 200–925 triplets; longer allele range: 500–1122 triplets).
Drug treatment
HDAC inhibitors, 109 [N-(6-(2-aminophenylamino)-6-oxohexyl)-4-methybenzamide] and 966 [(E)-N-(2-amino-4-flurophenyl)-3-(1-cinnamyl-1H-pyrazol-4-yl)acrylamide] (22) were synthesized by Repligen Corporation (Waltham, MA, USA). Drugs were dissolved in dimethyl sulfoxide (DMSO). Cells were treated with drug (or only DMSO) for 24 h at a final concentration of 10 μM each. For all drug (and no drug) treatments the final concentration of DMSO in the cell culture medium was 0.1% (23). Drug treatment was carried out in low passage cells that were maintained <400 000 cells/ml.
Quantitative measurement of FXN transcript
This was done by reverse transcription followed by quantitative polymerase chain reaction (quantitative RT-PCR), using protocols and primer sequences as previously described (8). Briefly, reverse transcription was performed either with a mixture of random hexamers and oligo dT primers (for the Exon 3–4 amplicon (4)) or with a strand-specific RT primer (for the Exon 1 amplicon) using the QuantiTect® reverse transcription kit (Qiagen). Transcript levels were quantified by real-time polymerase chain reaction (PCR) relative to expression of the control TBP gene (forward primer: 5′-CCACTCACAGACTCTCACAAC-3′, reverse primer: 5′-CTGCGGTACAATCCCAGAACT-3′) using the ΔΔCt method on the Eppendorf RealPlex-4 Mastercycler with SsoAdvanced™ SYBR® green supermix (BioRad) or power SYBR green supermix (Life Technologies). Primary FXN transcript was quantified in intron 1 at upGAA (see Figure 1A) (4) by strand-specific reverse transcription (RT primer: 5′- CTTTTAAGCACTGGCAACCAATC-3′) using the QuantiTect® reverse transcription kit (Qiagen) followed by real-time PCR (forward primer: 5′-GAAACCCAAAGAATGGCTGTG-3′, reverse primer: 5′-TTCCCTCCTCGTGAAACACC-3′) (4) using the ΔΔCt method on the Roche LightCycler® 96 System with power SYBR green supermix (Life Technologies).
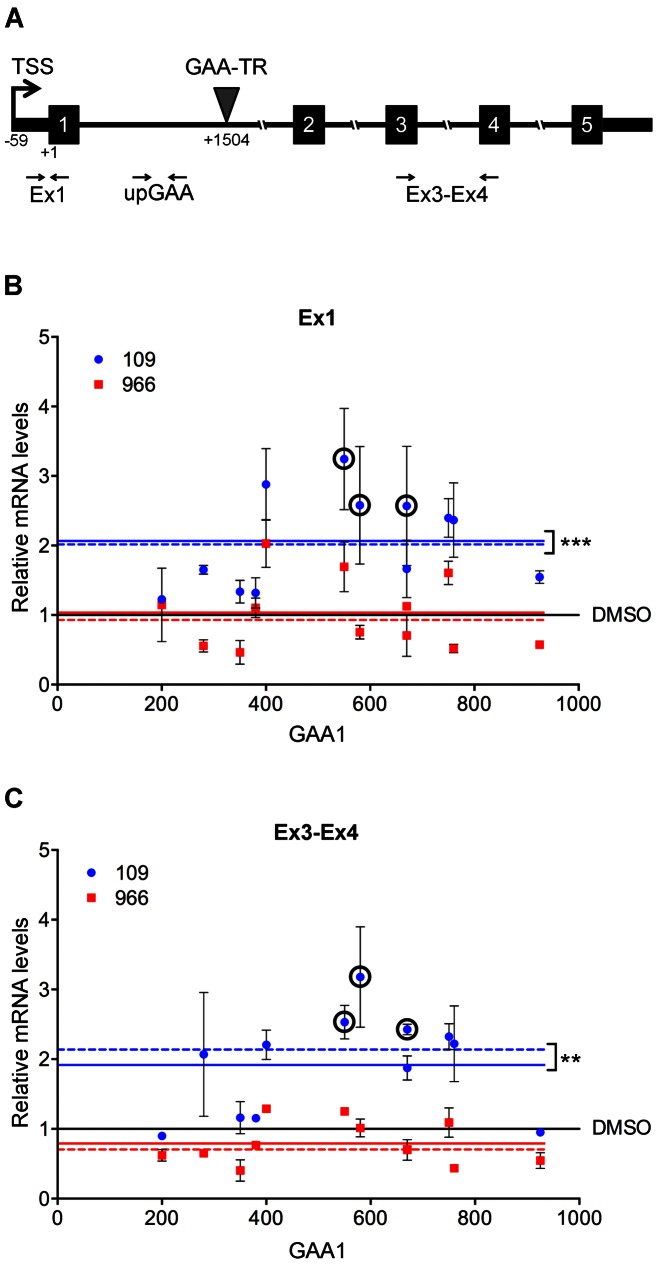
Figure 1.
Treatment with 109 significantly increases FXN transcript levels in FRDA both upstream and downstream of the expanded GAA-TR sequence. (A) Schematic representation of the FXN gene showing the location of the expanded GAA-TR sequence in intron 1, the FXN transcription start site (TSS, at −59, relative to the ‘A’ in the translational start codon as +1), and the amplicons, Ex1 and Ex3–Ex4, used for quantitative RT-PCR to assay FXN transcript levels upstream and downstream of the GAA-TR sequence, respectively. The intronic amplicon, upGAA, is also shown, which was used for quantitative RT-PCR to assay primary FXN transcript levels. (B and C) Quantitative RT-PCR showing relative levels of FXN transcript (Y-axis) in 12 FRDA cell lines, distributed (X-axis) by the size of the shorter of the two expanded GAA-TR alleles (GAA1, triplets), upon treatment with 109 (blue circles) and 966 (red squares), upstream (Ex1) and downstream (Ex3–Ex4) of the expanded GAA-TR sequence in FRDA. Relative to treatment of each cell line with only DMSO (indicated by the black line at 1.0), treatment with 109 resulted in a statistically significant ∼2-fold increase in FXN transcript at both upstream (B) and downstream (C) locations; mean and median of transcript levels in response to 109 in all 12 cell lines is indicated by solid and interrupted blue lines, respectively. No such increase was seen upon treatment with 966, either upstream (B) or downstream (C) of the expanded GAA-TR sequence in FRDA; mean and median of transcript levels in response to 966 in all 12 cell lines is indicated by solid and interrupted red lines, respectively. The variable response to 109 was not correlated with GAA1 repeat length (R2 = 0.13 for Ex1 and R2 = 0.08 for Ex3–Ex4). All subsequent mechanistic studies were performed using the three ‘circled’ FRDA cell lines (FRDA-6, FRDA-10, FRDA-17) that showed consistent and robust increase in FXN transcript levels. The graphs represent cumulative data from two complete experiments using 12 lymphoblastoid cell lines from individuals who are homozygous for the expanded GAA-TR sequence, treated with 109, 966 or no drug (only DMSO), each assayed in triplicate. FXN transcript levels are shown relative to the expression of the housekeeping RPS29 gene using the ΔΔCt method. Error bars represent +/−SEM. Medians were analyzed with the Mann–Whitney test, and mean values were analyzed using the two-tailed Student's t-test. ***P < 0.001; **P < 0.01.
Quantitative analysis of FXN promoter activity
This was done by metabolic labeling of nascent RNA using the Click-iT® Nascent RNA Capture Kit (Life Technologies) as previously described (8). Briefly, lymphoblastoid cell lines were incubated with 5-ethynyl uridine (EU) at the end of 24 h treatment with drug (or only DMSO). Following 0.5–4 h of incubation, RNA was extracted and used for biotinylation by Click reaction. Biotinylated RNA bound to streptavidin beads was reverse transcribed using the SuperScript® VILO™ cDNA synthesis kit (Life Technologies). Mature FXN transcript was measured at Ex1 and Ex3–Ex4, and primary FXN transcript was measured at upGAA in intron 1 (see Figure 1A) (4), as described above. Due to the differential drug-dependent effects on the expression of house-keeping genes (23), nascent FXN transcript levels were quantified by real-time PCR relative to expression of the control TBP gene (for 109 treated samples) or the OAZ1 gene (for 966 treated samples) from total biotinylated RNA using the ΔΔCt method on the Eppendorf RealPlex-4 Mastercycler with SsoAdvanced SYBR green supermix (BioRad).
Quantitative measurement of FXN transcript stability
FXN mRNA stability was analyzed by the pulse-chase method to measure the rate of decay of metabolically labeled transcript using the Click-iT® Nascent RNA Capture Kit (Life Technologies). As outlined in Figure 4A, 1.2 million cells were treated with either 109 or no drug (DMSO only) for 24 h. After 20 h of drug/DMSO treatment (i.e. 4 h prior to the end of the 24 h treatment period), cells were pulsed with 0.2 mM EU for 4 h. Cells were then re-suspended in fresh RPMI medium supplemented with 109 or DMSO. Total RNA was isolated from cells harvested following 1, 2, 4, 8 and 16 h in non-EU containing medium, and was subjected to biotinylation by Click reaction. EU-labeled-biotinylated RNA bound to streptavidin beads was reverse transcribed using the SuperScript® VILO™ cDNA synthesis kit (Life Technologies). The mature metabolically-labeled FXN mRNA was quantified at Ex1 (plotted as ‘% mRNA remaining’ in Figure 4B) by real-time PCR and normalized to total RNA by measuring the control OAZ1 gene using the ΔΔCt method on the Roche LightCycler® 96 System with SsoAdvanced SYBR green supermix (BioRad). The one-phase exponential decay constants and half-lives were solved by nonlinear regression of the percentage of FXN mRNA remaining versus time, using GraphPad Prism version 6.0 h for Mac OS X (GraphPad Software, La Jolla, CA, USA).
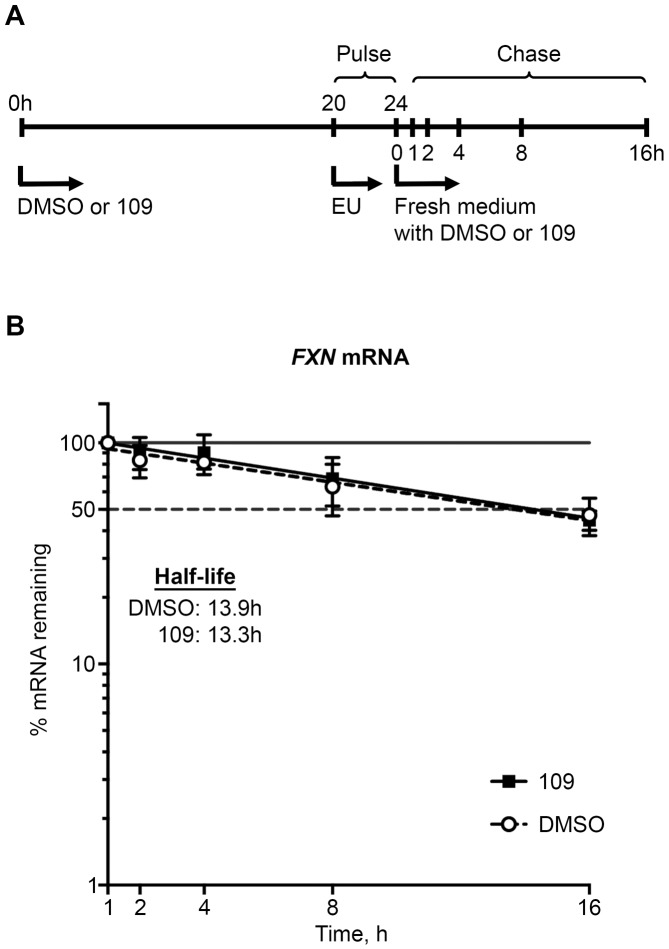
Figure 4.
Treatment with 109 does not alter the stability of FXN transcript in FRDA. (A) Scheme of pulse-chase experiment performed to measure the half-life of FXN transcript in patient-derived lymphoblastoid cells. Treatment with 109 (or only DMSO) was carried out for 24 h, with the last 4 h in the presence of EU to label nascent transcripts. The decay of labeled FXN transcript was measured by quantitative RT-PCR over the subsequent 16 h, in the absence of EU. (B) The temporal decay profile of newly labeled FXN transcript (% mRNA remaining, plotted using a log scale on the Y-axis), assayed in triplicate (twice using FRDA-6 and once using FRDA-10), indicates that the half-life of FXN transcript remained unchanged at ∼13.5 h following treatment with 109.
Nucleosome occupancy and Methylome sequencing (NOMe-Seq)
NOMe-Seq (24) was performed according to the manufacturer's protocol (Active Motif) with few modifications, as previously described (8). Briefly, cells were cross-linked and chromatin was treated with GpC methyltransferase (GpC-MT) and subjected to bisulfite conversion. A total of 20–50 ng of each bisulfite-treated DNA was used for PCR amplification (forward primer: 5′-ATCTCCCTTAAATCAAAAATCCTAA-3′, reverse primer: 5′-GCTGCGGGATTCGGGTGAGGGTTTGG-3′) using AmpliTaq® Gold DNA polymerase (Applied Biosystems). PCR products were TA-cloned using the TOPO TA Cloning® Kit (Invitrogen), transformed into competent Escherichia coli (DH5α) and individual clones were sequenced. Cytosines at GpC sites that are accessible to GpC-MT (i.e. not within a nucleosome) are methylated and therefore not converted by bisulfite treatment, i.e. read as C (denoted as white boxes in Figure 5A). Conversely, inaccessible cytosines, which remain unmethylated after GpC-MT treatment are converted via bisulfite treatment to uracil, i.e. read as T (denoted as black boxes in Figure 5A). The discriminatory capacity of the assay relies on the efficiency of bisulfite conversion, therefore only clones that demonstrated complete conversion efficiency at all non-GpC (and non-CpG) sites were used for NOMe-Seq analysis (>98% conversion of all non-GpC sites was attained for the clones used in Figure 5A). The Shapiro–Wilk test revealed that the percent open boxes (i.e. percent accessibility) for the three test groups (non-FRDA controls, and FRDA cells treated with 109 or DMSO-only) was not normally distributed so the Wilcoxon–Mann–Whitney test was used in pairwise comparisons of the medians. We used n = 17 in each of the three test groups (control, FRDA + 109 and FRDA + DMSO) for each of the 17 GpC sites in the nucleosome depleted region.
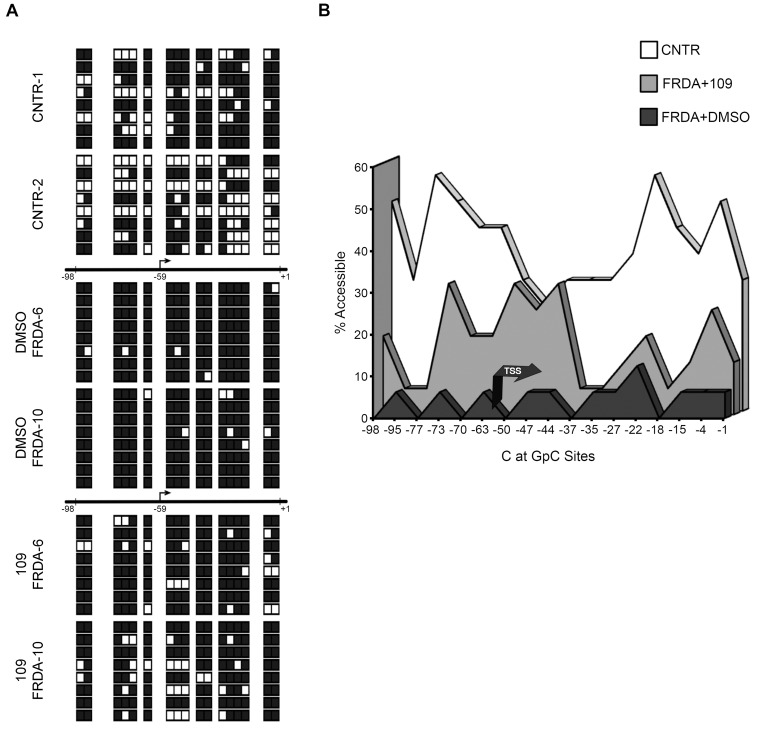
Figure 5.
Treatment with 109 significantly improves FXN promoter structure and chromatin accessibility in FRDA. (A) NOMe-Seq data, measuring the nucleosomal footprint in vivo, for two non-FRDA cell lines (CNTR1 and 2), and two FRDA cell lines either treated with 109 or with no drug (DMSO) are shown. The horizontal lines depict the 100-bp region analyzed, which represents the nucleosome depleted region flanking the FXN–TSS at −59 (relative to the ‘A’ in the translational start codon as +1). NOMe-Seq data of eight chromatin fibers per cell line, i.e. n = 16 fibers for each of the three conditions is depicted. For each of the 48 fibers are shown 17 white/black boxes, which represent accessible/inaccessible GpC sites, respectively. (B) The cumulative NOMe-Seq data of all 48 chromatin fibers, measured at each of 17 GpC dinucleotides within the nucleosome depleted region, are shown with promoter accessibility (%) plotted on the Y-axis. In non-FRDA controls (CNTR) the FXN-TSS is located within a nucleosome depleted region (median accessibility = 37.5%), which is obliterated in FRDA patients (FRDA + DMSO; median accessibility = 6.25%; P < 0.0001 compared with CNTR). Treatment with 109 (FRDA + 109) resulted in a significant, ∼3-fold increase in chromatin accessibility of the nucleosome depleted region (median accessibility = 18.8%; P < 0.0001 compared with FRDA + DMSO).
RESULTS
Treatment with 109 significantly increases steady-state levels of FXN transcript levels in FRDA
Deficient transcriptional initiation in FRDA results in transcriptional deficiency that extends both upstream and downstream of the expanded GAA-TR sequence in intron 1 of the FXN gene. We performed quantitative RT-PCR to measure steady-state levels of FXN transcript both upstream (exon 1 [Ex1 in Figure 1A]) and downstream (spliced product of exons 3–4 [Ex3–Ex4 in Figure 1A]) of the expanded GAA-TR sequence. Relative FXN transcript levels following treatment of patient-derived lymphoblastoid cell lines (n = 12; homozygous for expanded alleles containing 200–1122 GAA triplets) with 109 (10 μM for 24 h) or only DMSO are shown in Figure 1B (Ex1) and Figure 1C (Ex3–Ex4). Relative to treatment of each cell line with only DMSO (indicated by the solid black line at 1.0), treatment with 109 resulted in a statistically significant ∼2-fold increase in FXN transcript at both upstream (Figure 1B) and downstream (Figure 1C) locations (mean/median of transcript levels in all 12 cell lines is indicated by solid/interrupted blue lines, respectively).
We also tested 966, a drug with a structure similar to 109, but which is known to not increase FXN transcript levels in patient-derived cell lines (19), as a negative control. Compound 966 is a potent inhibitor of HDAC3 (22), but unlike 109 it does not increase histone acetylation at the FXN locus in FRDA (19). As expected, similar treatment with 966 (10 μM for 24 h) did not result in increased FXN transcript levels either upstream (Figure 1B) or downstream (Figure 1C) of the expanded GAA-TR sequence in FRDA (mean/median of transcript levels in all 12 cell lines is indicated by solid/interrupted red lines, respectively).
There was considerable inter-individual variability in the response to 109, ranging from only slight increases over no drug treatment to ∼3-fold increase in levels of steady-state FXN transcript (Figure 1B and C), which is consistent with previous findings (20). However, the response to 109 did not correlate with repeat length (R2 = 0.13 for Ex1 and R2 = 0.08 for Ex3–Ex4; using the shorter of the two expanded alleles, i.e. the allele best correlated with transcriptional deficiency (10)). Interestingly, despite the variability in response to 109 treatment, the same magnitude of response was noted at both upstream and downstream locations in each of the 12 cell lines tested (Figure 1B and C), indicating that some individuals respond more robustly to 109 treatment than others. In subsequent experiments, which were designed to characterize the precise mechanism of transcriptional upregulation by 109, we used the three cell lines that showed consistent and robust (2.4–3.2-fold) increases in FXN transcript levels (circled in Figure 1B and C). These cell lines carry expanded GAA-TR alleles that are in the ‘typical’ size range for FRDA mutations; FRDA-6 [670/830 triplets], FRDA-10 [580/580 triplets] and FRDA-17 [550/920 triplets].
Treatment with 109 significantly increases FXN promoter function in FRDA
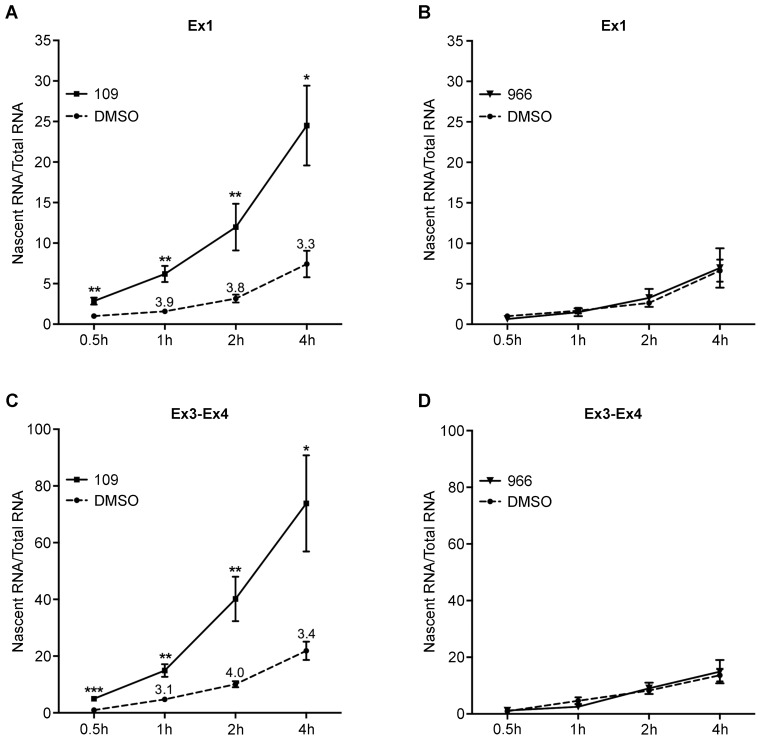
To directly test if treatment with 109 increases FXN promoter activity in FRDA, FXN transcriptional initiation was measured via metabolic labeling of nascent transcripts in patient-derived lymphoblastoid cell lines. Nascent transcripts were metabolically labeled with EU, to which biotin was subsequently added via click chemistry, thus permitting a quantitative, dynamic, in vivo analysis of newly synthesized FXN transcript. This is similar to a nuclear run-on assay, with the added advantage of measuring nascent transcripts in live cells. Quantitative RT-PCR was performed to measure FXN transcript levels immediately downstream of the FXN transcriptional start site [FXN-TSS, at −59; ‘Ex1’ in Figure 1A]). Three patient-derived cell lines (FRDA-6, FRDA-10, FRDA-17) were treated with either 109 or 966 (10 μM for 24 h) and no drug, i.e. DMSO only, followed by labeling with EU for 0.5, 1, 2 and 4 h. Treatment with 109 resulted in 3.3–3.9-fold increased production of newly synthesized FXN transcript compared with no drug (P < 0.05; Figure 2A). No such increase in FXN promoter function was noted with 966 (Figure 2B). Thus, 109 directly results in significantly improved function of the epigenetically silenced FXN promoter in FRDA. Quantitative RT-PCR was also performed downstream of the expanded GAA-TR sequence [Ex3–Ex4 in Figure 1A], which showed that 109 resulted in a similar 3.1–4.0-fold increased production of newly synthesized FXN transcript compared with no drug (P < 0.05; Figure 2C). Again, no such increase in FXN transcript was noted with 966 (Figure 2D). The similarity in magnitude of accumulation of nascent FXN transcript at both the upstream and downstream locations, especially at the 1–4 h time points (3–4-fold increase) where the reaction is more reliably quantitative, suggests that 109 acts mainly via improving transcriptional initiation in FRDA.
Figure 2.
Treatment with 109 significantly increases FXN promoter function in FRDA. Treatment with 109 (A and C), but not 966 (B and D), resulted in >3-fold increased accumulation of nascent FXN transcript at various time points assayed (0.5–4 h) after initiation of metabolic labeling. Quantitative RT-PCR was performed both immediately downstream of the transcription start site (i.e. upstream of the expanded GAA-TR sequence; Ex1 in Figure 1A) and downstream of the expanded GAA-TR sequence (Ex3–Ex4 in Figure 1A), with both locations showing a similar >3-fold increase in nascent FXN transcript accumulation in response to 109 treatment. Nascent transcript levels were quantified by real-time PCR relative to expression of the control TBP gene (for 109 treated samples [A and C]) or the control OAZ1 gene (for 966 treated samples [B and D]) from total biotinylated RNA using the ΔΔCt method. Graphs represent cumulative data from two complete experiments using three lymphoblastoid cell lines from individuals with FRDA who are homozygous for the expanded GAA-TR sequence, treated with 109, 966 or no drug (only DMSO), each assayed in triplicate. Error bars represent +/−SEM. ***P < 0.001; **P < 0.01; *P < 0.05.
Treatment with 109 significantly increases levels of primary FXN transcript in FRDA
To further test if 109 acts by improving FXN promoter function, we assessed if treatment with 109 would also result in an increase in the level of primary FXN transcript. Firstly, quantitative RT-PCR was performed to measure steady-state levels of FXN transcript in intron 1 (upGAA in Figure 1A) (4). Given that bidirectional transcription is known to occur in this part of the FXN gene (25), we performed reverse transcription using a strand-specific primer to ensure that we specifically measured transcription of the sense strand (see ‘Materials and Methods’ section). Treatment of three patient-derived lymphoblastoid cell lines (FRDA-6, FRDA-10, FRDA-17) with 109 (10 μM for 24 h) resulted in ∼3.5-fold increase (P < 0.05) in levels of steady-state primary FXN transcript compared with no drug (i.e. only DMSO), and no such increase was seen upon treatment with 966 (Figure 3A). Next we measured metabolically-labeled nascent primary FXN transcript by quantitative RT-PCR at upGAA following 0.5, 1, 2 and 4 h of labeling with EU in the same three cell lines. The measurement of metabolically labeled primary transcript, wherein reverse transcription is performed using a purified (‘pull down’) template consisting only of nascent RNA, has the added advantage of minimizing the potentially confounding effects of ‘endogenous’ priming, which intronic regions are sometimes prone to. Treatment with 109 resulted in >4-fold increased production of newly synthesized primary FXN transcript compared with no drug (P < 0.05 at each of the four time points; Figure 3B). As expected, given the much lower stability of primary transcripts, the primary FXN transcript attained steady-state levels prior to what we had observed with the mature FXN transcript; steady-state levels were reached in <1 h for the primary transcript (Figure 3B), but this was not attained by 4 h for the mature transcript (Figure 2A and C). The increase in both nascent primary and mature FXN transcript further supports the conclusion that 109 has a direct effect on improving transcriptional initiation in FRDA.
Figure 3.
Treatment with 109 significantly increases levels of primary FXN transcript in FRDA. (A) Quantitative RT-PCR showing relative levels of primary FXN transcript assayed via strand-specific RT-PCR at upGAA in intron 1 (see Figure 1A) in three patient-derived lymphoblastoid cell lines, following treatment with 109, 966 or only DMSO. Treatment with 109, but not 966, resulted in >3-fold increase in levels of steady-state primary FXN transcript compared with no drug (i.e. only DMSO). Graphs represent cumulative data from four complete experiments using three lymphoblastoid cell lines from individuals with FRDA who are homozygous for the expanded GAA-TR sequence, each assayed in triplicate. Error bars represent +/−SEM. *P < 0.05. (B) Quantitative RT-PCR of metabolically-labeled nascent primary FXN transcript assayed at upGAA in intron 1 following 0.5, 1, 2 and 4 h of labeling with 5-ethynyl uridine (EU) in the same three cell lines used above. Treatment with 109 resulted in >4-fold increased production of newly synthesized primary FXN transcript compared with no drug at each of the four time points. Graphs represent cumulative data from two complete experiments using three lymphoblastoid cell lines from individuals with FRDA, each assayed in triplicate. Error bars represent +/−SEM. *P < 0.05; **P < 0.01.
Treatment with 109 does not alter the stability of FXN transcript in FRDA
It has been previously shown that stability of the FXN transcript remains unchanged in FRDA (5). However, it is not known if the increase in FXN transcript levels caused by treatment with 109 stems, even partly, from increased transcript stability. A pulse-chase experiment was performed to measure the half-life of the FXN transcript in lymphoblastoid cells (see experimental scheme in Figure 4A). Treatment with 109 (or only DMSO) was carried out for 24 h, with the terminal 4 h in the presence of EU to label nascent transcripts. The decay of labeled FXN transcript was measured by quantitative RT-PCR over the subsequent 16 h. The temporal decay profile of newly labeled FXN transcript, assayed in triplicate (twice using FRDA-6 and once using FRDA-10), revealed that the half-life of FXN transcript remained unchanged at ∼13.5 h following treatment with 109 (Figure 4B). These data indicate that the increase in FXN transcript following 109 treatment must derive largely from increased synthesis of FXN transcript, and further supports the conclusion that 109 acts mainly via reactivation of the epigenetically silenced promoter in FRDA.
Treatment with 109 substantially improves FXN promoter structure and chromatin accessibility in FRDA
The FXN gene has a typical CpG island promoter, with a well-positioned +1 nucleosome that is preceded by a nucleosome depleted region (8). The FXN promoter is normally pre-associated with RNA pol II, which initiates FXN transcription within the nucleosome depleted region (at position −59 in lymphoblastoid cells (6)). In FRDA the nucleosome depleted region is obliterated by altered nucleosome positioning at the FXN-TSS which leads to severely deficient transcriptional initiation (6,8,9). We used Nucleosome Occupancy and Methylome Sequencing (24) (NOMe-Seq; Active Motif), a high-resolution in vivo footprint assay to detect nucleosome occupancy in individual chromatin fibers based on accessibility of GpC methyltransferase. Specifically, we probed the nucleosome depleted region at the FXN-TSS in order to test for any beneficial effect of treatment with 109 on the structure of the FXN promoter. A 100-bp window flanking the FXN-TSS at −59 that is relatively depleted of nucleosomes and contains 17 GpC dinucleotides was selected for structural analysis of the FXN promoter (8). We reasoned that the ability of 109 to improve FXN promoter function would stem from its ability to restore access to the FXN-TSS. In the NOMe-Seq assay (24), chromatin is treated with a GpC methyltransferase, subjected to bisulfite conversion and individual clones are sequenced to determine nucleosome occupancy in single chromatin fibers. Cytosines at GpC sites that are accessible to the GpC methyltransferase (i.e. not within a nucleosome) are methylated and therefore not converted by bisulfite treatment, i.e. read as C (denoted as white boxes in Figure 5A). Conversely, inaccessible cytosines, which remain unmethylated after GpC methyltransferase treatment are converted via bisulfite treatment to uracil, i.e. read as T (denoted as black boxes in Figure 5A). NOMe-Seq was performed using lymphoblastoid cell lines from two non-FRDA controls (CNTR-1, CNTR-2) and two FRDA patients (FRDA-6, FRDA-10), either treated with 109 (10 μM for 24 h) or only DMSO. Individual chromatin fibers (n = 8) were sequenced for each cell line/condition, and the NOMe-Seq data of all 48 chromatin fibers are depicted in Figure 5A and cumulative data are plotted in Figure 5B. Compared to non-FRDA controls, where the FXN-TSS is located within a nucleosome depleted region (median accessibility = 37.5% [25th–75th% = 31.3, 50.0]), FRDA patients (‘DMSO’) showed dramatic loss of chromatin accessibility at the FXN-TSS (median accessibility = 6.25% [25th–75th% = 0, 6.25]; P < 0.0001 using the Wilcoxon–Mann–Whitney test for pairwise comparison of medians), with almost no accessible GpC dinucleotides (Figure 5B). Treatment of FRDA cells with 109 resulted in a significant, 3-fold increase in accessibility of the nucleosome depleted region; median accessibility was 18.8% [25th–75th% = 6.25, 25.0] in FRDA cells treated with 109 compared with 6.25% in cells treated only with DMSO (Figure 5B; P < 0.0001 using the Wilcoxon–Mann–Whitney test). While not quite restoring promoter accessibility to the levels seen in non-FRDA cells, these data indicate that 109 results in significant reversal of the abnormal structural organization of the epigenetically silenced FXN promoter in FRDA, concomitant with the ∼3-fold increase seen in promoter function.
DISCUSSION
The deficiency of mature FXN transcript in FRDA is known to be caused by at least two mechanisms: deficient transcriptional initiation and inefficient transcriptional elongation through the expanded repeat in intron 1. Various assays, including measurement of steady-state transcript levels, whole transcriptome analysis, metabolic labeling of nascent transcripts in living cells and RNA-FISH in single cells have shown that FRDA cells exhibit a major defect in transcriptional initiation and a relatively minor defect in transcriptional elongation (6,8,9,14). Indeed, it is estimated that defective transcriptional initiation is about 5–6-fold more severe than inefficient transcriptional elongation in the etiology of FXN transcriptional deficiency in FRDA (8,9). Therefore, the direct targeting and significant improvement of the abnormal structural and functional properties of the FXN promoter, as seen with 109, indicates that class I HDAC inhibition represents a rational and efficient therapeutic strategy in FRDA.
The ‘openness’ of the nucleosome depleted region in CpG island promoters is a reliable indicator of promoter activity (24,26), and our data indicate that inhibition of class I HDACs via 109 treatment achieves ∼3-fold improvement in both the openness and activity of the epigenetically silenced FXN promoter in FRDA (Figure 6). Patients who have at least one shorter-than-average expanded allele (containing <400 triplets) typically have a milder phenotype associated with a later age of onset (27) and slower disease progression (28,29). These individuals have 2-fold higher promoter activity compared with typical FRDA patients who are homozygous for expanded alleles containing >400 GAA triplets (10). Therefore, in addition to its sound molecular rationale, the use of class I HDAC inhibitors such as 109 that improve promoter function in FRDA by ∼3-fold is also likely to be clinically meaningful.
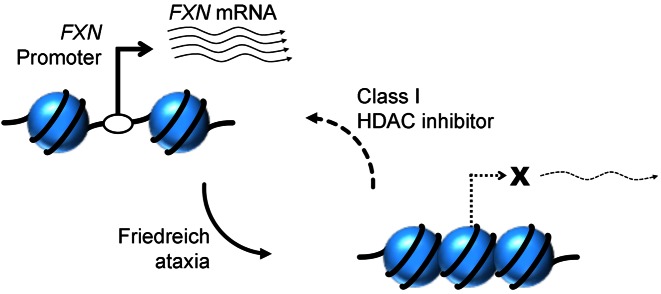
Figure 6.
Model of reversal of epigenetic FXN promoter silencing in FRDA by a class I HDAC inhibitor. The FXN promoter, which normally has a transcription start site within a nucleosome depleted region, is rendered inaccessible in FRDA because of increased nucleosome density. This results in a severe deficiency of transcriptional initiation in FRDA. Treatment with 109, a class I HDAC inhibitor, results in significantly improved FXN promoter accessibility and promoter activity in FRDA.
Compound 109 is a potent inhibitor of class I HDACs HDAC1 and HDAC3 (21), and 966 predominantly inhibits HDAC3 (22). Indeed, Soragni et al. (30) elegantly showed that simultaneous inhibition of HDAC1 and HDAC3 with compounds such as 109 that exhibit a slow-on/slow-off kinetic profile are most effective at reversing the transcriptional repression in FRDA. Taken together, our data support the simultaneous roles of HDAC1 and HDAC3 in mediating the abnormal structural organization and functional silencing of the FXN promoter in FRDA.
A potential caveat of our study is that it explores the corrective role of class I HDAC inhibition in the context of lymphoblastoid cells, a cell type that is not involved in disease pathogenesis. However, class I HDAC inhibition via 2-aminobenzamides (including 109) has been shown to increase FXN transcript levels in many human and murine cell types (4,18–21,31) and, moreover, the underlying mechanism of gene silencing in FRDA does not seem to be tissue-specific (32). The analysis of lymphoblastoid cell lines therefore represents the use of a tractable and well-characterized model system to perform detailed mechanistic studies of drug action on FXN promoter structure and function.
We conclude that repeat-mediated epigenetic promoter silencing in FRDA is mediated by class I HDACs, and it is reversible via treatment with specific inhibitors. It is noteworthy that the correction of both the structural and functional defects of the FXN promoter in FRDA, albeit partial, occurs in its natural genomic context, i.e. while in continued physical proximity to the cis-acting expanded GAA-TR sequence. These features bode well for the development of class I HDAC inhibitors as a rational therapeutic modality for FRDA.
Acknowledgments
We thank Michael Michalopulos for technical assistance. We thank James R. Rusche, Ph D of Repligen Corporation (Waltham, MA, USA) and Joel Gottesfeld, Ph D (Scripps Research Institute, La Jolla, CA, USA) for generously providing the HDAC inhibitor drugs, 109 (RG2833) and 966 (RGFP966).
FUNDING
National Institutes of Health [R01 NS072418]; Muscular Dystrophy Association [MDA344862]; Translational Neuroscience Research program of the Oklahoma Center for Neuroscience (to S.I.B., Y.K.C.); Million Dollar Bike Ride Grant Program of the Orphan Disease Center at University of Pennsylvania Postdoctoral Research Fellowship [MDBR-15-212-FA (to Y.K.C.)]. Funding for open access charge: National Institutes of Health [R01 NS072418].
Conflict of interest statement. None declared.
REFERENCES
- 1.Brouwer J.R., Willemsen R., Oostra B.A. Microsatellite repeat instability and neurological disease. Bioessays. 2009;31:71–83. doi: 10.1002/bies.080122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saveliev A., Everett C., Sharpe T., Webster Z., Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 3.Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 4.Herman D., Jenssen K., Burnett R., Soragni E., Perlman S.L., Gottesfeld J.M. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 5.Punga T., Buhler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari D., Biacsi R.E., Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J. Biol. Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim E., Napierala M., Dent S.Y. Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich's ataxia. Nucleic Acids Res. 2011;39:8366–8377. doi: 10.1093/nar/gkr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chutake Y.K., Costello W.N., Lam C., Bidichandani S.I. Altered nucleosome positioning at the transcription start site and deficient transcriptional initiation in Friedreich ataxia. J. Biol. Chem. 2014;289:15194–15202. doi: 10.1074/jbc.M114.566414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva A.M., Brown J.M., Buckle V.J., Wade-Martins R., Lufino M.M. Expanded GAA repeats impair FXN gene expression and reposition the FXN locus to the nuclear lamina in single cells. Hum. Mol. Genet. 2015;24:3457–3471. doi: 10.1093/hmg/ddv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chutake Y.K., Lam C., Costello W.N., Anderson M., Bidichandani S.I. Epigenetic promoter silencing in Friedreich ataxia is dependent on repeat length. Ann. Neurol. 2014;76:522–528. doi: 10.1002/ana.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidichandani S.I., Ashizawa T., Patel P.I. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohshima K., Montermini L., Wells R.D., Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J. Biol. Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto N., Ohshima K., Montermini L., Pandolfo M., Wells R.D. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J. Biol. Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Lu Y., Polak U., Lin K., Shen J., Farmer J., Seyer L., Bhalla A.D., Rozwadowska N., Lynch D.R., et al. Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum. Mol. Genet. 2015;24:6932–6943. doi: 10.1093/hmg/ddv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bidichandani S.I., Delatycki M.B. Friedreich ataxia. In: Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Dolan CR, Fong CT, Smith RJH, editors. GeneReviews(R) Seattle: University of Washington; 2014. pp. 1993–2016. [Google Scholar]
- 16.Lufino M.M., Silva A.M., Nemeth A.H., Alegre-Abarrategui J., Russell A.J., Wade-Martins R. A GAA repeat expansion reporter model of Friedreich's ataxia recapitulates the genomic context and allows rapid screening of therapeutic compounds. Hum. Mol. Genet. 2013;22:5173–5187. doi: 10.1093/hmg/ddt370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahdeo S., Scott B.D., McMackin M.Z., Jasoliya M., Brown B., Wulff H., Perlman S.L., Pook M.A., Cortopassi G.A. Dyclonine rescues frataxin deficiency in animal models and buccal cells of patients with Friedreich's ataxia. Hum. Mol. Genet. 2014;23:6848–6862. doi: 10.1093/hmg/ddu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libri V., Yandim C., Athanasopoulos S., Loyse N., Natisvili T., Law P.P., Chan P.K., Mohammad T., Mauri M., Tam K.T., et al. Epigenetic and neurological effects and safety of high-dose nicotinamide in patients with Friedreich's ataxia: an exploratory, open-label, dose-escalation study. Lancet. 2014;384:504–513. doi: 10.1016/S0140-6736(14)60382-2. [DOI] [PubMed] [Google Scholar]
- 19.Soragni E., Miao W., Iudicello M., Jacoby D., Mercanti S., Clerico M., Longo F., Piga A., Ku S., Campau E., et al. Epigenetic therapy for Friedreich ataxia. Ann. Neurol. 2014;76:489–508. doi: 10.1002/ana.24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plasterer H.L., Deutsch E.C., Belmonte M., Egan E., Lynch D.R., Rusche J.R. Development of frataxin gene expression measures for the evaluation of experimental treatments in Friedreich's ataxia. PLoS One. 2013;8:e63958. doi: 10.1371/journal.pone.0063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rai M., Soragni E., Chou C.J., Barnes G., Jones S., Rusche J.R., Gottesfeld J.M., Pandolfo M. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich's ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malvaez M., McQuown S.C., Rogge G.A., Astarabadi M., Jacques V., Carreiro S., Rusche J.R., Wood M.A. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soragni E., Xu C., Cooper A., Plasterer H.L., Rusche J.R., Gottesfeld J.M. Evaluation of histone deacetylase inhibitors as therapeutics for neurodegenerative diseases. Methods Mol. Biol. 2011;793:495–508. doi: 10.1007/978-1-61779-328-8_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly T.K., Liu Y., Lay F.D., Liang G., Berman B.P., Jones P.A. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012;22:2497–2506. doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Biase I., Chutake Y.K., Rindler P.M., Bidichandani S.I. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns B.R. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 27.Durr A., Cossee M., Agid Y., Campuzano V., Mignard C., Penet C., Mandel J.L., Brice A., Koenig M. Clinical and genetic abnormalities in patients with Friedreich's ataxia. N. Engl. J. Med. 1996;335:1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 28.Regner S.R., Wilcox N.S., Friedman L.S., Seyer L.A., Schadt K.A., Brigatti K.W., Perlman S., Delatycki M., Wilmot G.R., Gomez C.M., et al. Friedreich ataxia clinical outcome measures: natural history evaluation in 410 participants. J. Child Neurol. 2012;27:1152–1158. doi: 10.1177/0883073812448462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metz G., Coppard N., Cooper J.M., Delatycki M.B., Durr A., Prospero N.A., Giunti P., Lynch D.R., Schulz J.B., Rummey C., et al. Rating disease progression of Friedreich's ataxia by the International Cooperative Ataxia Rating Scale: analysis of a 603-patient database. Brain. 2013;136:259–268. doi: 10.1093/brain/aws309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soragni E., Chou C.J., Rusche J.R., Gottesfeld J.M. Mechanism of action of 2-aminobenzamide HDAC inhibitors in reversing gene silencing in Friedreich's ataxia. Front. Neurol. 2015;6:44. doi: 10.3389/fneur.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandi C., Pinto R.M., Al-Mahdawi S., Ezzatizadeh V., Barnes G., Jones S., Rusche J.R., Gottesfeld J.M., Pook M.A. Prolonged treatment with pimelic o-aminobenzamide HDAC inhibitors ameliorates the disease phenotype of a Friedreich ataxia mouse model. Neurobiol. Dis. 2011;42:496–505. doi: 10.1016/j.nbd.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chutake Y.K., Costello W.N., Lam C.C., Parikh A.C., Hughes T.T., Michalopulos M.G., Pook M.A., Bidichandani S.I. FXN promoter silencing in the humanized mouse model of Friedreich ataxia. PLoS One. 2015;10:e0138437. doi: 10.1371/journal.pone.0138437. [DOI] [PMC free article] [PubMed] [Google Scholar]