Abstract
Bidirectional promoters are the major source of gene activation-associated noncoding RNA (ncRNA). PC12 cells offer an interesting model for understanding the mechanism underlying bidirectional promoter-mediated cell cycle control. Nerve growth factor (NGF)-stimulated PC12 cells elongate neurites, and are in a reversible cell-cycle-arrested state. In contrast, these cells irreversibly differentiate and cannot re-enter the normal cell cycle after NGF plus cAMP treatment. In this study, using directional RNA-seq, we found that bidirectional promoters for protein-coding genes with promoter-associated ncRNA (pancRNA) were enriched for cAMP response element consensus sequences, and were preferred targets for transcriptional regulation by the transcription factors in the cAMP-dependent pathway. A spindle-formation-associated gene, Nusap1 and pancNusap1 were among the most strictly co-transcribed pancRNA–mRNA pairs. This pancRNA–mRNA pair was specifically repressed in irreversibly differentiated PC12 cells. Knockdown (KD) and overexpression experiments showed that pancNusap1 positively regulated the Nusap1 expression in a sequence-specific manner, which was accompanied by histone acetylation at the Nusap1 promoter. Furthermore, pancNusap1 KD recapitulated the effects of cAMP on cell cycle arrest. Thus, we conclude that pancRNA-mediated histone acetylation contributes to the establishment of the cAMP-induced transcription state of the Nusap1 locus and contributes to the irreversible cell cycle exit for terminal differentiation of PC12 cells.
INTRODUCTION
Many long noncoding RNAs (lncRNAs) have been shown to be transcribed from the mammalian genome, and have emerged as key players of many cellular functions (1–4). The majority of non-coding RNAs (ncRNAs) involved in mRNA metabolism in mammals have been thought to downregulate the corresponding mRNA expression level in a pre- or post-transcriptional manner by forming ncRNA–mRNA duplex structures (2,5). However, several studies have shown that some lncRNAs function without forming RNA–RNA duplexes (6–9).
The transcripts derived from bidirectional promoters include not only protein-coding mRNAs, but also lncRNAs, and a significant proportion of such lncRNAs derived from bidirectional promoters are expressed in tissue-specific manners (10,11). We previously showed that functional polyA+, long (>200 bp) ncRNAs derived from bidirectional promoters, named promoter-associated ncRNAs (pancRNAs), are expressed in tissue-specific manners and function in the activation of their partner genes (7,8,11,12). For example, in mice, microinjection of siRNA against the abundant pancRNA partner of interleukin 17d (Il17d) mRNA at the 1-cell stage caused embryonic lethality, which was rescued by supplying IL17D protein in vitro at the 4-cell stage (7). Thousands of pancRNAs are generated by transcription of the antisense strand and exhibit expression changes coordinated with the expression of their cognate genes (11), making bidirectional promoters a major source of gene activation-associated ncRNA.
Transcriptional regulation by binding of transcription factors to the cAMP response element (CRE) downstream of cAMP signaling plays important roles in the cell differentiation process (13,14). Transcription factors that bind to CRE, such as CRE-binding protein (Creb) and CRE modulator (Crem), are activated by cAMP-dependent protein kinase (Pka)-mediated phosphorylation, and activate gene expression by means of recruitment of coactivator paralogs CREB-binding protein (Cbp) and p300 (15,16). Transcription factors that bind to CRE can act not only as transcriptional activators but also as transcriptional repressors. Inducible cAMP early repressor (Icer) is generated from an alternative intronic promoter of Crem, and acts as a transcriptional repressor (17). Icer lacks the activation and kinase-inducible domains, but retains the DNA-binding domain (18,19), and therefore is a potent repressor of cAMP-induced transcription. In a different context, CREB1 forms a complex with p53, and then inhibits p53-mediated transcriptional activation of the MDM2 gene in a human cell line during glucose deprivation (20). It is noteworthy that, using chromatin immunoprecipitation (ChIP) coupled with massively parallel sequencing (ChIP-seq), Impey et al. demonstrated that phosphorylated Creb1 (pCreb1)-bound regions are enriched in the bidirectional promoters of annotated transcript pairs that are organized in a head-to-head arrangement on opposite strands (15). Thus, comprehensive analysis of bidirectional promoters may identify a set of transcripts that are upregulated and downregulated by cAMP and that play a role in defining the cell state. We hypothesize that certain pancRNAs derived from bidirectional promoters are upregulated or downregulated by a cAMP-dependent mechanism.
In this context, rat adrenal phaeochromocytoma-derived PC12 cells offer an interesting model for studying cAMP-dependent pancRNA functions in cell cycle regulation associated with differentiation. Nerve growth factor (NGF)-stimulated PC12 cells elongate neurites and are reversibly arrested, that is, they can resume efficient cell cycling when they are put back under proliferative conditions (21). In contrast, when PC12 cells are stimulated simultaneously with NGF and a cAMP analog, dibutyryl cAMP (dbcAMP), they cannot re-enter the cell cycle even after the removal of NGF and dbcAMP (22). This shows that irreversible differentiation of PC12 cells can be induced by a cAMP-dependent mechanism. A previous in vivo study showed that pharmacological activation of the cAMP pathway rescued impairment of neuronal differentiation of neural progenitors caused by brain-specific knockout of the Nf1 gene in mice, suggesting that a cAMP-dependent mechanism is also required for the in vivo neuronal differentiation of neural progenitor cells (13). The cell cycle of terminally differentiated cells is repressed by a cAMP-dependent mechanism, but the underlying molecular mechanisms are unknown.
In this study, by comparing the transcriptome of NGF-differentiated (Ndiff) PC12 cells with that of NGF/cAMP-differentiated (NcAdiff) PC12 cells, we highlighted the critical importance of cell cycle regulation for the terminal differentiation of cells that cannot resume mitosis. We showed that a significant number of M-phase-associated genes were repressed in NcAdiff cells, compared to Ndiff cells. As expected, we found that a significant number of CREs were enriched in the thousands of newly identified bidirectional promoters for the expression of pancRNA–mRNA pairs. Here, we report that these bidirectional promoters were preferred targets for transcriptional regulation by the transcription factors in the cAMP-dependent pathway. Furthermore, among the pancRNA–mRNA pairs, a pancRNA (pancNusap1) at the Nusap1 locus, which encodes a spindle-formation-associated gene, and Nusap1 mRNA together play a functional role in the irreversible differentiation of PC12 cells. Artificial downregulation of the pancNusap1 expression level recapitulated the cAMP-triggered morphological features and epigenetic state of the irreversibly differentiated PC12 cells.
MATERIALS AND METHODS
PC12 culture and differentiation
PC12 cells were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM) (WAKO) containing 10% horse serum (HS; SAFC Biosciences), 5% fetal bovine serum (FBS; Biowest), 100 units/ml penicillin (PhytoTechnology Laboratories), and 100 μg/ml streptomycin (MP Biomedicals) at 37°C in 5% CO2. To induce differentiation, PC12 cells were placed on a collagen (Cellmatrix Type IV, Nitta Gelatin)-precoated dish at a density of 8000 cells/cm2, and were cultured in high-glucose DMEM containing 1% HS, 0.5% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 100 ng/ml NGF 2.5S (Millipore) or 50 ng/ml NGF 2.5S and 200 μM dibutyryl cAMP (SIGMA) for 7 days. During differentiation, cell culture medium was changed at day 3 and day 5. To observe the cell cycle resumption of differentiated PC12 cells, the differentiation medium was changed at day 7 to high-glucose DMEM containing 10% HS, 5% FBS and antibiotics. For doxycycline (Dox)-inducible pancRNA overexpression/knockdown (OE/KD) experiments, cells were incubated in the above medium containing 2 ng/ml Dox (Nacalai Tesque).
Immunocytochemistry
Immunocytochemistry was performed as follows: fixation with 4% paraformaldehyde (PFA) for 20 min in RT; washing with phosphate buffered saline (PBS) twice; permeabilization and blocking in PBS containing 0.1% Triton X-100 and 3% FBS in PBS for 30 min at RT; incubation with primary antibody diluted 1:500 in blocking solution for 2 h in RT; washing with PBS three times; incubation with Hoechst 33258 (Nacalai Tesque) and secondary antibody diluted 1:500 in PBS for 1 h in the dark at RT; washing with PBS three times. Imaging was performed using a Leica AF6000 microscope. As primary antibodies, chicken anti-GFP (AVES Labs), mouse anti-Ki67 (BD Biosciences) and rabbit anti-active Caspase 3 (R&D Systems) were used. As secondary antibodies, the following were used: CF647-conjugated anti-mouse IgG (Biotium) and CF647-conjugated anti-rabbit IgG (Biotium), FITC-conjugated anti-chick IgY (Biotium). For the EdU assay, cells were cultured with 10 μM EdU in Click-iT EdU Imaging Kits (Life Technologies) for 4 h, fixed with 4% PFA, permeabilized with 0.1% Triton-X and 3% FBS in PBS, and stained with Click-iT reaction buffer for 30 min at RT in the dark. After washing with PBS, primary and secondary staining were performed in the dark.
RNA analysis
To examine RNA expression, total RNA isolated with TRIzol reagent (Life Technologies) was treated with DNase I (Life Technologies) and reverse-transcribed with oligo dT primer using the SuperScriptIII First-Strand Synthesis System (Life Technologies). Synthesized cDNAs were subjected to qPCR using a KAPA SYBR Fast qPCR Kit (KAPA Biosystems). The primers used in these analyses are listed in Supplementary Table S1.
Directional RNA-seq library preparation
Directional RNA-seq libraries were prepared as follows. RNA with RNA integrity number above 9.6, calculated using total RNA Pico Bioanalyzer chip (Agilent), was used for Directional RNA-seq library preparation. polyA+ RNA was purified twice from 15 μg of total RNA of each PC12 sample by using Sera-mag Magnetic Oligo (dT) Beads (Thermo Scientific). In each polyA+ RNA sample, the fraction of rRNA was confirmed to be <2% by using a Total RNA Pico Bioanalyzer chip. The polyA+ RNA was fragmented by heating at 94°C for 2 min in 1 × fragmentation buffer (Affymetrix), and then the RNA was purified with two volumes of Agencourt RNAclean XP (Beckman Coulter). Fragmented RNA was decapped with 5 U TAP (Epicenter), and then subjected to phenol:chloroform:isoamylalcohol (PCI) extraction and ethanol precipitation. Fragmented and decapped RNA was 3′-dephosphorylated using Antarctic phosphatase (NEB). The RNA was 5′-phosphorylated using T4 polynucleotide kinase (NEB). The modified RNA was cleaned up with an RNeasy MinElute kit (Qiagen), was ligated to NEBNext Multiplex 3′ SR Adaptor with T4 RNA ligase 2 truncated K277Q (NEB) at 4°C overnight, and was further ligated to NEBNext Multiplex 5′ SR Adaptor with T4 RNA ligase (Illumina) at 20°C for 1 h. cDNA was synthesized with specific RT primer and the SuperScriptIII First-Strand Synthesis System. After RNase H treatment, unincorporated primers in each cDNA library sample were removed by using AMPure XP (Beckman Coulter). Each cDNA was independently amplified with KAPA Hifi HS polymerase (Kapa Biosystems) and NEBNext Index 1, 8, 10 and 11 primers in NEBNext Multiplex Oligos for Illumina (NEB) to produce four replicates for each sample. Thermal-cycling conditions were as follows: 30 s at 98°C, 12 cycles of 98°C for 15 s, 62°C for 30 s and 72°C for 30 s, followed by 5 min at 72°C. The polymerase chain reaction (PCR) product was purified twice with AMPure XP. Illumina HiSeq 2000 was used to perform 50-bp single-end sequencing according to the manufacturer's instructions.
Data mining
To remove low quality reads and adaptor sequences (AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC), the FASTX tool kit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used as follows: Remove low quality reads (phred score <20); remove 3′-end multiplex adapter tag sequence; remove the reads shorter than 20 nt; remove low quality reads again. The raw reads from all PC12 samples resulted in a total of 517 million strand-specific reads. We mapped sequencing reads from each sample onto the rat Rn5 genome sequences except for random chromosome sequences using TopHat (v.2.0.8b, option: -g 1 –bowtie1) (23). RSeQC (24) was used for quality evaluation of the strandedness in our cDNA libraries. The read-enriched regions in each cell state were detected using MACS2 (v.2.0.10b, option: –nomodel –broad -g 2.57e9 -p 0.95) (25). All reads in biological replicates were merged and used as the input data for MACS2. Overlapping read-enriched regions between different cell states were merged to create one broad region using the mergeBed function of BEDTools (26). This resulted in 149 559 read-enriched regions (minimum length set to 300 nt). When a reference transcription start site (TSS) in the Rn5 genome assembly was overlapped with a read-enriched region, we defined the 5′-end of this region as the adjusted TSS in PC12 cell samples except for cases in which the 5′-end overlapped with another gene. The coding potentials of non-annotated transcripts and candidate-pancRNAs were estimated using the coding potential calculator (CPC) algorithm (27). For mRNA and ncRNA quantification, we counted the number of reads mapped to the exonic region of Ensembl protein-coding genes and read-enriched regions classified as non-annotated transcripts, respectively, followed by normalization using the iDEGES/edgeR methods and differential expression analyses using R package TCC (28).
For the correlation analysis in Figure 3C, Pearson correlation coefficients between the expression levels of novel transcripts and annotated protein-coding genes were calculated. In this analysis, randomly selected lncRNA and mRNA pairs were generated using the publicly available program (https://cell-innovation.nig.ac.jp/wiki/tiki-download_wiki_attachment.php?attId=5&download=y). For enrichment analysis of biological process ontology, differentially expressed genes were annotated using DAVID (29).
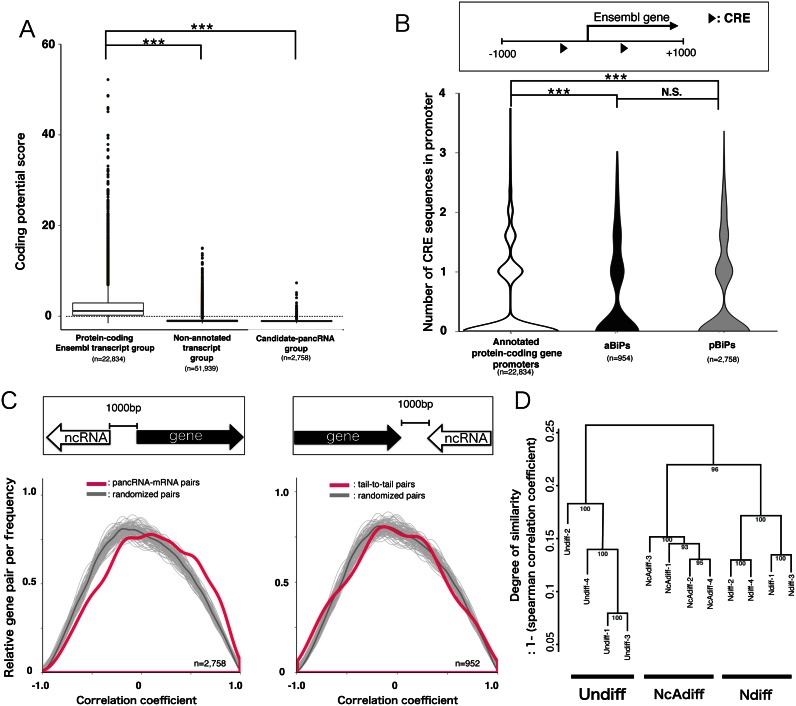
Figure 3.
The bidirectional promoters driving pancRNAs can be novel targets of cAMP signaling. (A) Distribution of coding potential scores for annotated protein-coding Ensembl transcripts, non-annotated transcripts and candidate-pancRNAs using the CPC algorithm. Negative scores indicate low coding potential. (B) Distribution of numbers of CREs located in the promoters (−1000 to +1000 from TSS) of all protein-coding genes, genes derived from aBiPs and pBiPs. (C) Distribution of Pearson correlation coefficients calculated based on the expression of protein-coding genes and nearest novel lncRNAs (magenta curve). The dark gray lines show the average correlation coefficient distribution based on 200 permutations of 2000 randomly selected pairs of lncRNAs and protein-coding gene. The pale gray lines represent 200 simulation results. Significance was calculated with the Mann–Whitney U test (left: P < 2.2e-16; right: P = 0.59). (D) Hierarchical clustering of sequence data sets based on pancRNA expression levels. The distance between samples was determined by clustering using group average methods and linkage criteria based on Spearman's rank coefficient of correlation. The numbers in tree diagrams are approximately unbiased P-values. Statistical analysis was performed using the Tukey-Kramer multiple comparison test in (A and B). N.S. = not significant. ***P < 0.001.
Enrichment analysis of CRE and Creb1 binding score in bidirectional promoter
For enrichment analysis of CREs, the locations of CREs were searched using the R package ChIPpeakAnno (option: min.score = ‘90%’) (30) using Creb1 binding sequence data that is registered in the JASPAR database (ID = MA0018.2) (31).
For enrichment analysis of Creb1 binding score, the mouse Mm9 genome-mapped Creb1 ChIP-seq dataset of E16.5 mouse cortical neurons that had been maintained in vitro for 7 days obtained from Gene Expression Omnibus (acc. no GSM530185) (32) was converted to the corresponding dataset in the rat Rn5 genome using the UCSC LiftOver tool (http://hgdownload.cse.ucsc.edu/admin/exe). Similarly, the rat rn3 genome-mapped pCreb1 ChIP-seq dataset of PC12 cells 15 min after cAMP signal activation by forskolin (15), deposited as a supplementary data of a serial report (33), was converted to the corresponding dataset in the rat Rn5 genome using the UCSC LiftOver tool.
Chromatin immunoprecipitation (ChIP) analysis
For ChIP analysis, we generated Dox-inducible short hairpin RNA (shRNA)-expressing or pancRNA-expressing PC12 cell lines, as described later. Cells (about 4 × 105) in a 10-cm Petri dish were fixed with 1% formaldehyde for 5 min at RT. Samples were placed on ice and washed twice with ice-cold PBS. Cells were harvested by scraping with 1 ml of resuspension solution (10% NaN3, 2% FBS in 1 × PBS) and counted, and were centrifuged at 2000 g for 5 min at 4°C. Cell pellets were washed with 1 ml of ice-cold PBS, resuspended in lysis buffer containing 1% sodium dodecyl sulphate (SDS), 50 mM Tris–HCl [pH 8.0], 10 mM ethylenediaminetetraacetic acid (EDTA) [pH 8.0] and 1% protease inhibitor cocktail (Nacalai Tesque) at a concentration of 1 × 105 cells per 100 μl, kept on ice for 10 min, then sonicated to an average size of 500 bp using a Bioruptor (Diagenode). After centrifugation for 15 min at 15 000 rpm at 4°C, supernatants containing sonicated chromatin were transferred to fresh tubes and diluted 1:10 with ChIP dilution buffer (1.1% Triton X-100, 50 mM Tris–HCl [pH 8.0], 167 mM NaCl, 0.11% sodium deoxycholate, 1% protease inhibitor cocktail). Immunoreaction was performed overnight at 4°C with rotation. For each sample preparation, 5 × 104 cells and 1 μl of each antibody (anti-mouse-H3K4me3, anti-mouse-H3K9me3, anti-mouse-H3K27me3, anti-mouse-H3K27ac or anti-mouse-H3K9ac; Cosmo Bio) or mouse-IgG (Santa Cruz) as a negative control were used. For ChIP for pCreb1 and Icer, 3 × 106 cells and 4 μg of each antibody (anti-rabbit-pCreb1(Ser133); Cell Signaling Technology, anti-rabbit-Crem(X-12); Santa Cruz) or rabbit-IgG (Santa Cruz) as a negative control were used. Immune complexes were captured with 10 μl of M-280 sheep anti-mouse IgG (Life Technologies) or 40 μl of M-280 sheep anti-rabbit IgG (Life Technologies) for 4 h at 4°C. After the beads were washed once with ice-cold RIPA buffer (0.1% SDS, 1% Triton X-100, 50 mM Tris–HCl [pH 8.0], 1 mM EDTA [pH 8.0], 150 mM NaCl, 0.1% sodium deoxycholate), once with ice-cold high-NaCl RIPA buffer (0.1% SDS, 1% Triton X-100, 50 mM Tris–HCl [pH 8.0], 1 mM EDTA [pH 8.0], 300 mM NaCl, 0.1% sodium deoxycholate) and twice with ice-cold TE (10 mM Tris–HCl [pH 8.0], 1 mM EDTA [pH 8.0]), formaldehyde cross-linking was reversed by overnight incubation with 200 μl of ChIP elution buffer (0.5% SDS, 10 mM Tris–HCl [pH 8.0], 5 mM EDTA [pH 8.0], 300 mM NaCl) at 65°C overnight. The immunoprecipitated samples and the same amount of chromatin fragments without IP (input) were treated with proteinase K for 1 h at 55°C, further treated with RNase A (Thermo Scientific) for 30 min at 37°C, and purified by PCI extraction and ethanol precipitation. DNA was resuspended in 20 μl of sterile water and used as a template for quantitative PCR with specific primers (see Supplementary Table S1). The results from three independent experiments were averaged.
Bisulfite sequencing
To determine the DNA methylation profiles of the Nusap1 promoter regions, genomic DNAs were subjected to the bisulfite reaction using a MethylCode Kit (Life Technologies) according to the manufacturer's instructions. Each bisulfite-treated genome was amplified using AmpliTaq Gold 360 Master Mix (Life Technologies) or LA Taq (TaKaRa) with the primers listed in Supplementary Table S3. Each PCR product was cloned into pGEM T-easy (Promega) and 8 randomly picked clones were sequenced.
Plasmid construction and lentivirus production
Lentiviral vectors pLLX and pLEMPRA (34) were generously provided by Drs Z. Zhou and M. E. Greenberg.
For knockdown experiments, pLLX was used to enable the simultaneous expression of the shRNA and EGFP RNA under the control of U6 and ubiquitin-C promoters, respectively. The sh-RNA oligos were inserted into the HpaI and XhoI sites of pLLX (see Supplementary Table S2).
For rescue experiments, pLEMPRA was used to enable the simultaneous expression of the shRNA and EGFP-IRES-Nusap1 under the control of U6 and ubiquitin-C promoters, respectively. A FLAG-tagged rat Nusap1 expression lentiviral vector was constructed by inserting Nusap1 cDNA between the EcoRI and AscI sites, and inserting the sh-pancNusap1 sequence between the HpaI and XhoI sites of pLEMPRA (see Supplementary Table S2).
For conditional pancNusap1-OE or -KD experiments, we employed the lentivirus-based pSLIK_Neo vector system (Addgene), which allows tight Dox-inducible, RNA polymerase II (RNAPII)-mediated transcription of a gene of interest, and co-expression of a Neomycin selection gene (35). The pancNusap1 fragments (−1685 to −323 relative to the TSS of Nusap1), isolated by genomic PCR with specific primers (see Supplementary Table S2), were inserted into the SacII and XbaI sites of the entry vector pEN_TmiRc3 (Addgene). The annealed sh-pancNusap1 oligo (see Supplementary Table S2) was inserted between the HpaI and XhoI sites of pEN_TmiRc3. Recombination of pEN_TmiRc3 and pSLIK_Neo was performed using the LR Clonase Enzyme Mix (Life Technologies) to obtain pancNusap1- or sh-pancNusap1-expressing vectors according to manufacturer's instructions. We confirmed that Dox-inducible expression of sh-pancNusap1 successfully caused a reduction of pancNusap1 expression in the PC12 cell line (Supplementary Figure S1).
HEK 293T cells were used as producers of lentiviruses, and were cultured in high-glucose DMEM (Nacalai Tesque) containing 10% FBS (Biowest) and antibiotics. The expression vector was transfected along with third-generation lentiviral packaging and pseudotyping plasmids (36) using PEI-MAX (COSMO BIO). Ten micrograms of pSLIK, pLLX or pLEMPRA plasmid, 3 μg of the packaging plasmid pCAG-HIVgp and 3 μg of pCMV-VSV-G-RSV-Rev were diluted in 500 μl of Opti-MEM (Gibco) and used for cells in a 10-cm-diameter dish. The medium was replaced after 12 h of transfection. The virus-containing supernatant was collected 48 h later, and the virus was concentrated by centrifugation at 6000 g overnight at 4°C, and used for the manipulation of pancNusap1 or Nusap1 expression in PC12 cells.
RESULTS
Cell cycle arrest was observed in the irreversible differentiation of NGF/cAMP-differentiated PC12 cells
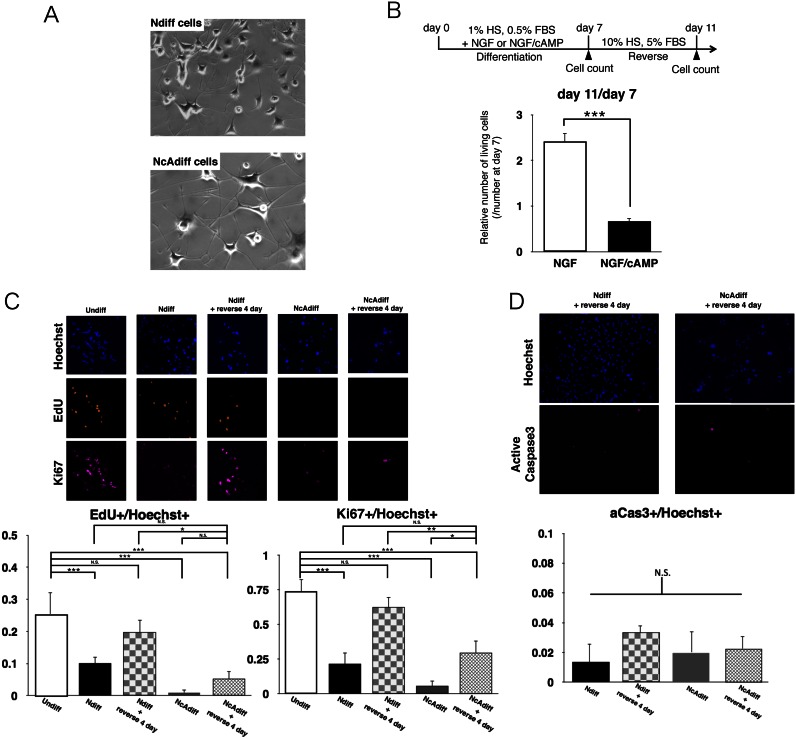
Clear morphological differences were observed here between Ndiff and NcAdiff cells, as previously documented in the literature (22). NcAdiff cells could be distinguished from Ndiff cells by the thickening of neurites and increase in cell soma size of the former (Figure 1A). To ascertain whether Ndiff and NcAdiff cells showed a difference in the reversibility of their differentiation after removal of these agents, we counted the number of cells after 7 days of differentiation in the presence of NGF or NGF/cAMP followed by 4 days of culture in proliferative conditions without NGF or cAMP (Figure 1B). After the NGF-containing differentiation medium was changed back to the proliferation medium, Ndiff cells resumed proliferation. In contrast, NcAdiff cells did not resume proliferation after such a change. Thus, 7 days of differentiation in the presence of NGF/cAMP induced irreversible differentiation of a fraction of PC12 cells, whereas the differentiation induced NGF alone was reversible. To monitor the cell cycle progression in each cell state, we immunostained incorporated 5-ethynyl-2′-deoxyuridine (EdU) and the proliferation marker Ki67 (Figure 1C). In Ndiff cells, both the EdU+ and the Ki67+ cells decreased by about 40%, compared to undifferentiated (Undiff) PC12 cells. After the NGF-containing differentiation medium was changed back to the proliferation medium, the fractions of both the EdU+ and the Ki67+ cells increased to levels comparable to those of Undiff cells. In contrast, the fractions of both the EdU+ and the Ki67+ cells were very low in NcAdiff cells. Even after the NGF/cAMP-containing differentiation medium was changed back to the proliferation medium, the fractions of both the EdU+ and the Ki67+ cells remained low. Thus, cells continued to proliferate marginally in the NGF-containing differentiation medium, whereas NcAdiff cells almost completely stopped progressing through the cell cycle. To investigate whether apoptosis occurred in each cell state, we immunostained an apoptosis marker, activated Caspase 3 (aCas3; Figure 1D). The fractions of aCas3+ cells were continuously very low, and did not differ markedly among all the samples examined. Taken together, these results showed that cell cycle arrest, but not apoptosis, occurred in the differentiated PC12 cells after the removal of cAMP.
Figure 1.
Combination of NGF and cAMP stimulations caused cell cycle arrest in differentiated PC12 cells. (A) Morphologies of PC12 cells that grew during 7 days of differentiation in the presence of NGF (upper) or NGF/cAMP (lower). (B) Relative number of living cells after removal of NGF or of NGF and cAMP. Upper diagram shows experimental scheme. After 7 days of differentiation of PC12 cells, NGF and cAMP were removed to allow proliferation. Number of living cells was evaluated by counting trypan blue staining-negative cells. Number of cells in each differentiation state at day 7 was set as 1. (C) Cell proliferation monitored by EdU incorporation and Ki67 staining in undifferentiated (Undiff), NGF-differentiated (Ndiff) and NGF/cAMP-differentiated (NcAdiff) cells, and in Ndiff and NcAdiff cells after 4 days of incubation in proliferative condition (Ndiff + reverse 4 day, NcAdiff + reverse 4 day, respectively). (D) Proportion of apoptotic cells before/after removal of NGF or NGF and cAMP, detected by active Caspase 3 (aCas3) staining. In (B–D), values are mean ± SD (n = 3). Statistical analysis was performed using Student's two-tailed t-tests, or the Tukey-Kramer multiple comparison test. N.S. = not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
A significant number of M-phase-associated genes were specifically repressed in NcAdiff cells
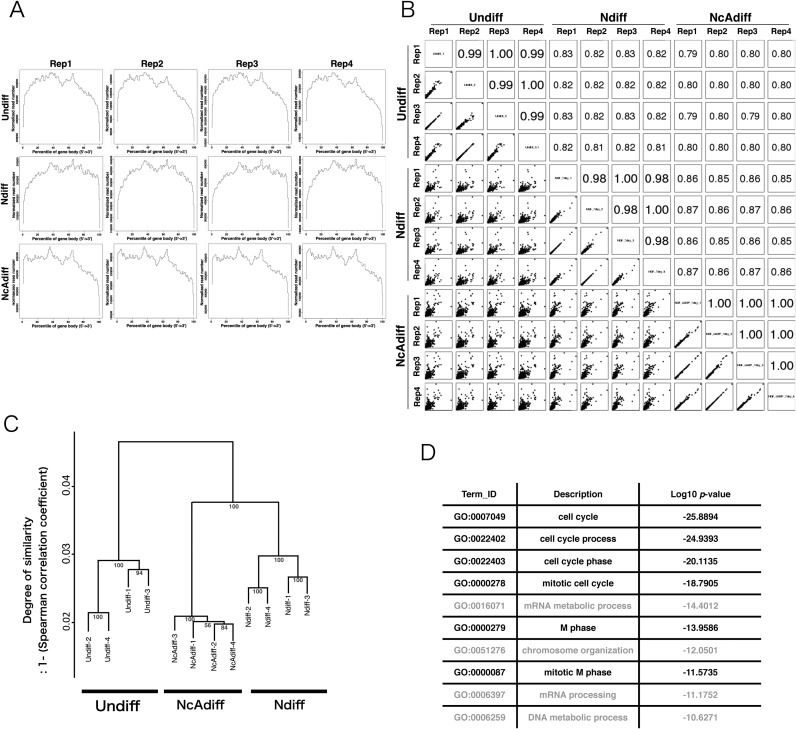
To identify RNAs whose expression dynamics differed between reversibly and irreversibly differentiated PC12 cells, we performed high-throughput directional sequencing of Undiff, Ndiff and NcAdiff cells using Illumina Hiseq2000. The data contained over 148 million raw reads per sample, with a total of 517 million reads after removing low quality reads (Supplementary Table S4). These reads were mapped to the rat Rn5 reference genome using TopHat2 (see ‘Materials and Methods’ section). The average percentage of uniquely mapped reads among the valid reads was 81.7% (Supplementary Table S4). Our RNA-seq data showed little 5′-3′ mapping bias for annotated protein-coding genes (Ensembl gene annotation, version 76: Figure 2A) and robust reproducibility among four replicates (Pearson correlation coefficient, R > 0.98: Figure 2B). Hierarchical clustering analysis of the sequenced datasets based on the expression levels of protein-coding genes revealed that the expression profiles of protein-coding genes were cell-state-specific and highly reproducible among replicates (Figure 2C). In order to validate the strandedness of our directional RNA-seq data, we mapped the reads to the annotated RefSeq genes and calculated the proportion that mapped on the correct strand, and thereby found that more than 99% of the reads were mapped to the correct strands (Supplementary Table S5). In Ndiff cells, 3346 protein-coding genes showed significantly higher expression levels compared to those in NcAdiff cells (q-value < 0.05). Gene Ontology analysis of these Ndiff PC12-specific protein-coding genes revealed a strong enrichment in the Gene Ontology biological process terms associated with cell cycle, especially terms related to G2/M-phase progression (e.g., mitotic cell cycle, M phase and mitotic M phase: Figure 2D), which is consistent with our finding that cell cycle arrest was observed much more frequently in NcAdiff cells than in Ndiff cells (Figure 1C).
Figure 2.
A significant number of M-phase associated genes were not stringently repressed in Ndiff cells. (A) Density plots of directional RNA-seq reads mapped to the RefSeq genes to evaluate 5′-3′ potential mapping bias across genes. Each sample category consisted of four replicates (Rep1–4). (B) Scatter plots of gene expression levels and Pearson correlation coefficients among all sequenced samples. The expression levels of genes were calculated based on the number of reads mapped to each Ensembl protein-coding gene normalized by iDEGES/edgeR methods. (C) Hierarchical clustering of sequence data sets based on the expression levels of Ensembl protein-coding genes. The distance between samples was determined by clustering using group average methods and linkage criteria based on Spearman's rank coefficient of correlation. The numbers in tree diagrams are approximately unbiased P-values. (D) The top 10 most significantly enriched GO terms for 3346 Ndiff-specific protein-coding genes. The P-value is the Benjamini–Hochberg corrected P-value.
pancRNA and mRNA pairs were possible downstream targets of cAMP signaling pathway
Impey et al. reported that more than half of total pCreb1-binding sites are located adjacent to genes, and in particular, pCreb1-binding regions are significantly enriched in bidirectional promoters that produce annotated transcripts in both directions (annotated bidirectional promoters or aBiPs) (15). Recent studies have revealed that antisense noncoding transcripts are derived from upstream of many protein-coding genes in mammals (7,11,37). We therefore hypothesized that not only aBiPs, but also protein-coding gene promoters that simultaneously produce pancRNAs (pancRNA-associated bidirectional promoters or pBiPs) were potential targets of transcriptional regulation by a CRE-mediated mechanism.
To test our hypothesis, first, we adjusted the annotated TSSs of genes according to our directional RNA-seq data in order to improve the accuracy of prediction of pBiPs (see ‘Materials and Methods’ section). Next, because MACS2 showed more sensitivity and accuracy for detection of lncRNA than transcript models reconstructed using ab initio transcript assembly tools (e.g. Cufflinks and Trinity; Supplementary Figure S2), we identified the read-enriched regions using MACS2 software after parameter setting, and classified these regions according to their positions relative to those of annotated transcripts. A total of 51 939 regions were classified as regions of non-annotated transcripts. Of these, 2758 regions were classified as regions of candidate-pancRNAs, which were overlapped with the 1-kb region upstream of the TSS of an annotated protein-coding gene in an antisense direction. The average length of the candidate-pancRNAs was 1928 bp, and the average distance between the 5′ ends of candidate-pancRNAs and that of its partnered gene was 167 bp (Supplementary Figure S3). As expected, based on coding potential estimation using the CPC algorithm (27), 92.5% of the non-annotated transcripts and 96.9% of candidate-pancRNAs exhibited negative coding potential scores, suggesting that most of these regions can be regarded as templates of pancRNAs (Figure 3A). We regarded the annotated protein-coding gene promoters producing pancRNAs as pBiPs. As a result, we identified 3235 bidirectional promoters in PC12 cells. Of these, 948 genes derived from 474 out of 477 aBiPs, 2718 pancRNAs and their paired genes derived from 2718 out of 2758 pBiPs were transcribed in Undiff, Ndiff or NcAdiff cells.
Then, we investigated the frequency of CRE occurrence in aBiPs and pBiPs. There were one or more CRE sequences in 27.6% of all protein-coding gene promoters (−1000 to +1000 bp from the TSS) and in 36.1% of aBiPs and 32.3% of pBiPs. Thus, in agreement with our hypothesis, CRE sequences were highly enriched not only in aBiPs, but also in pBiPs compared to all protein-coding gene promoters (Figure 3B). To further validate our hypothesis, we analyzed a publicly available pCreb1 ChIP-seq dataset of PC12 cells immediately after cAMP signal activation by forskolin (15). Investigation of the frequency of overlap of pCreb1-binding at protein-coding gene promoters showed that pCreb interacted with 59% of aBiPs, 45% of pBiPs and 23% of all protein-coding gene promoters. Similarly, we analyzed a publicly available Creb1 ChIP-seq dataset of E16.5 mouse cortical neurons that had been maintained in vitro for 7 days (32). Creb1 binding signals were significantly enriched not only in aBiPs, but also in pBiPs (Supplementary Figure S4). These results supported our hypothesis that not only aBiPs but also pBiPs were preferred downstream targets of cAMP signaling during the irreversible differentiation of PC12 cells.
Next, because the expression of pancRNA and the corresponding annotated protein-coding gene are positively correlated in both mouse and human brain (11,38), we investigated whether the expression dynamics of pancRNA–mRNA pairs were also positively correlated during the differentiation of PC12 cells. First, we calculated the correlation coefficient of the expression level of each RNA pair during PC12 differentiation, and then classified them into three groups. The first group was composed of antisense lncRNAs that overlapped with the 1-kb region upstream from the 5′ ends of mRNAs and the partner mRNAs (pancRNA–mRNA pairs). The second group was composed of antisense lncRNAs that overlapped with the 1-kb region downstream from the 3′ ends of mRNAs and the partner mRNAs (tail-to-tail pairs). The third group was composed of randomly selected RNA transcripts (randomized pairs). We found that the distribution of the correlation coefficients of the pancRNA–mRNA pairs was significantly different from that of the tail-to-tail pairs (Bonferroni-corrected Mann–Whitney U test, P < 2.1e-9) and that of randomized pairs (P < 2e-16; Figure 3C left). However, no significant difference was observed between the distribution of the correlation coefficients of the tail-to-tail pairs and that of randomized pairs (P = 0.99; Figure 3C right). These statistical analyses highlighted the positive correlation of pancRNA–mRNA expression during PC12 differentiation. The hierarchical clustering analysis of sequenced datasets based on candidate-pancRNA expression levels revealed that the expression profiles of candidate-pancRNAs were cell-state-specific and highly reproducible among replicates (Figure 3D). These genome-wide analyses supported the notion that pancRNAs are physically associated with cAMP targets for the partner gene activation.
After irreversible differentiation, both Nusap1 and pancNusap1 expression levels, and the histone acetylation level of the Nusap1 promoter, were dramatically decreased
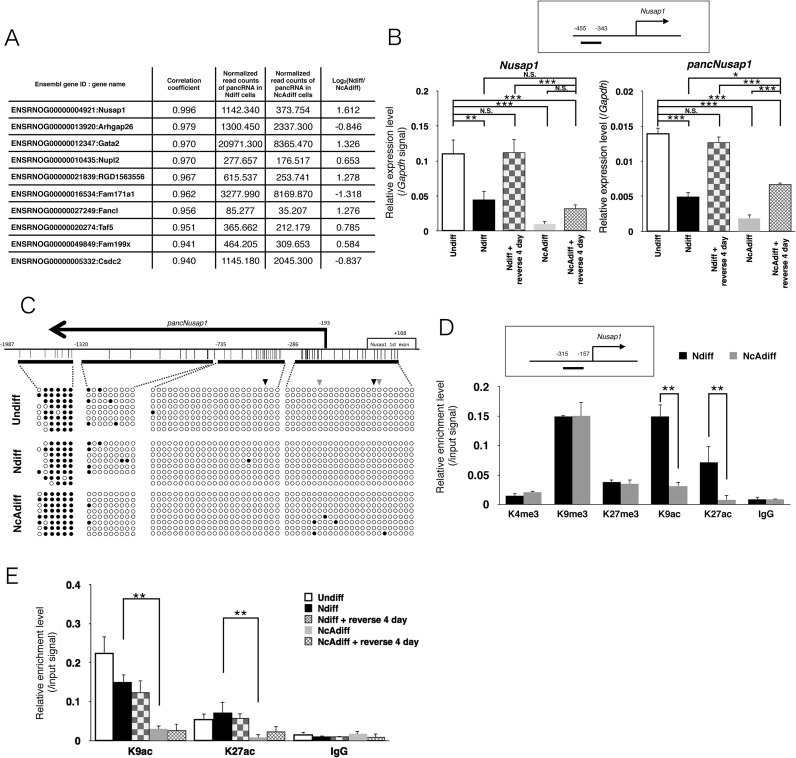
Among candidate-pancRNAs, we then focused on a pancRNA from the promoter of a spindle-formation-associated gene Nusap1 in an attempt to elucidate possible functions of this pancRNA. This pancRNA and its paired annotated protein-coding gene pair exhibited the highest Pearson correlation coefficient among all pancRNA–mRNA pairs during PC12 differentiation (P = 0.995: Figure 4A). However, the RNA-seq data showed that there was no coordination of the expression changes of any Nusap1-neighboring genes with that of Nusap1 and its pancRNA (Supplementary Figure S5). We named this pancRNA pancNusap1. Its expression as estimated by RNA-seq analysis was lowest in NcAdiff cells among all of the cell samples examined. TGACG, one of the previously identified non-canonical CREs (15), was symmetrically located at −12 and −380 bp relative to the TSS of Nusap1 (Figure 4C, black arrowheads). Similarly, CGCCA (33) was located at +31 and −230 bp relative to the TSS of Nusap1 (Figure 4C, gray arrowheads, Supplementary Figure S6). These non-canonical CREs also appeared near the Nusap1 TSS in the mouse and human genomes (Supplementary Figure S6). We then examined the expression patterns of pancNusap1 and Nusap1 in each differentiation state of PC12 cells. As shown in Figure 4B, the expression dynamics of pancNusap1 as determined by RT-qPCR seemed to be positively correlated with those of Nusap1. We confirmed that the expression levels of both Nusap1 and pancNusap1 were significantly lower in NcAdiff cells than in Ndiff cells. Interestingly, these lower levels of pancNusap1 and Nusap1 expression seemed to continue after NcAdiff cells were put back under proliferative conditions. That is, after the NGF-containing differentiation medium was changed back to the proliferation medium, the expression levels of both Nusap1 and pancNusap1 in Ndiff cells recovered to a level similar to that in Undiff cells, whereas the expression levels of both Nusap1 and pancNusap1 in NcAdiff cells failed to fully recover after 4 days in proliferative condition.
Figure 4.
Both Nusap1 and pancNusap1 were epigenetically repressed in NcAdiff cells. (A) List of the top 10 candidate-pancRNA/mRNA pairs showing highest correlation of expression during PC12 differentiation. The expression levels of pancRNAs normalized with the iDEGES/edgeR method are also shown. (B) RT-qPCR analysis of pancNusap1 and Nusap1 in Undiff, Ndiff and NcAdiff cells, before and after NGF and cAMP factor deprivation. Upper diagram shows the location of the primer used for RT-qPCR of pancNusap1. Gapdh was used as a control. (C) DNA methylation levels of the Nusap1 promoter region in Undiff, Ndiff and NcAdiff cells. Upper diagram shows the genomic structure of the rat Nusap1 promoter. Thick horizontal lines indicate the regions analyzed by bisulfite sequencing. Vertical lines mark the location of CpG dinucleotides. Primer positions are numbered relative to TSS of the Nusap1 gene. Filled and empty circles indicate methylated and unmethylated cytosines, respectively. Black and gray arrowheads indicate locations of the previously identified non-canonical CRE, TGACG and CGCCA, respectively. (D and E) Histone modification status of the Nusap1 promoter in each differentiation state of PC12 cells. Upper diagram shows the location of the primers used in the ChIP assay. Status of histone H3 lysine 4 tri-methylation (H3K4me3), lysine 9 tri-methylation (H3K9me3), lysine 27 tri-methylation (H3K27me3), histone H3 lysine 9 acetylation (H3K9ac) and lysine 27 acetylation (H3K27ac) was evaluated. Normal mouse IgG (IgG) was used as a negative control. The same amount of chromatin fragments as used for each immunoprecipitation was also subjected to PCR without IP as a positive control (Input). In (B), (D) and (E), values are mean ± SD (n = 3). Statistical analysis was performed using Student's two-tailed t-tests, or the Tukey-Kramer multiple comparison test. N.S. = not significant. **P < 0.01; ***P < 0.001.
Since we reported that the expression changes of pancRNAs caused a regional alteration in the epigenetic states in the pBiPs (8), next we examined the epigenetic state of the Nusap1 promoter by means of bisulfite sequencing and ChIP-qPCR assay. The DNA methylation states were not changed upon differentiation in PC12 cells (Figure 4C). However, the levels of active histone modifications, such as histone H3 lysine 9 acetylation (H3K9ac) and lysine 27 acetylation (H3K27ac), were lower in NcAdiff cells than in Undiff and Ndiff cells (Figure 4D and E). The levels of signals of H3K9ac and H3K27ac on the Nusap1 promoter in NcAdiff cells remained very low after the differentiation medium was changed back to the proliferation medium (Figure 4E). These kinetics of the histone acetylation levels were concordant with the pancNusap1 expression pattern, raising the possibility that pancNusap1 is required for local open chromatin formation, and cessation of the expression of pancNusap1 causes histone deacetylation at the Nusap1 promoter. Histone methylation levels, such as the active histone modification histone H3 lysine 4 tri-methylation (H3K4me3), and the repressive histone modifications histone H3 lysine 9 tri-methylation (H3K9me3) and lysine 27 tri-methylation (H3K27me3), were not different between Ndiff and NcAdiff cells (Figure 4D). Taken together, these results indicate that pancNusap1 expression is associated with local histone acetylation specifically in a cAMP-minus condition.
Knockdown of pancNusap1 induced a closed chromatin structure in the Nusap1 promoter, resulting in cell cycle arrest
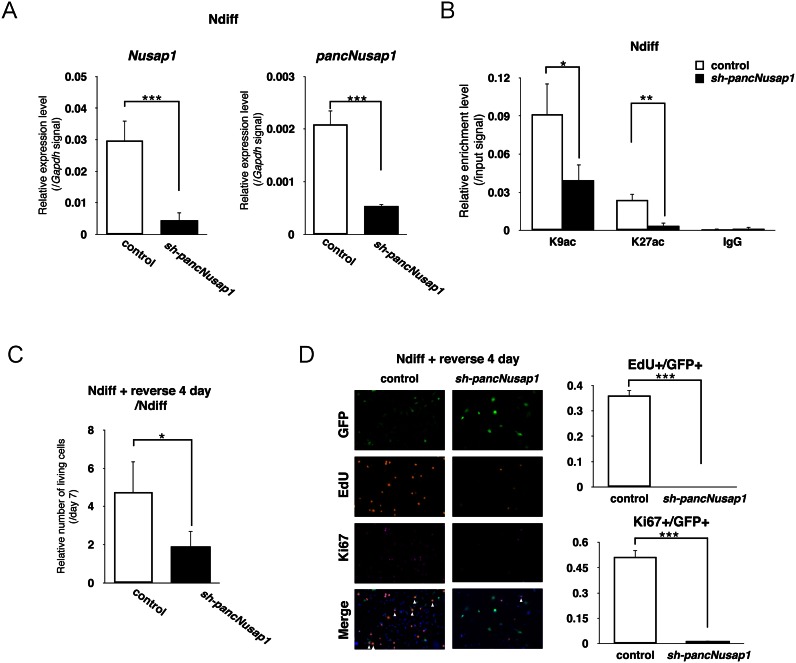
To test the hypothesis that the regulation of pancNusap1 is critical for irreversible differentiation, first we examined the effects of pancNusap1 KD on the expression levels of Nusap1 in the cells under reversible differentiation condition (see ‘Materials and Methods’ section). Both of the tested shRNAs for pancNusap1 downregulated the expressions of not only pancNusap1 but also Nusap1 in Ndiff cells (Figure 5A and Supplementary Figure S7). We confirmed that neither of the shRNAs for pancNusap1 showed off-target effects (Supplementary Figure S8). Next, we investigated the effect of pancNusap1-KD on the histone acetylation profile in the Nusap1 promoter. ChIP-qPCR analysis revealed that pancNusap1-KD significantly decreased the signals of both H3K9ac and H3K27ac in the Nusap1 promoter without affecting the signal levels in the Gapdh promoter (Figure 5B and Supplementary Figure S9). Then we examined whether pancNusap1-KD affected the proliferation of Ndiff cells after they were returned to the proliferation medium. Counting the living cells revealed that pancNusap1-KD inhibited the proliferation of Ndiff cells after the NGF-containing differentiation medium was changed to the proliferation medium (Figure 5C). Moreover, consistent with our hypothesis, immunostaining showed that pancNusap1-KD decreased both the EdU+ and the Ki67+ cells even after the NGF-containing differentiation medium was changed back to the proliferation medium (Figure 5D). Thus, repression of pancNusap1 mimics the epigenetic features of irreversibly differentiated PC12 cells.
Figure 5.
Knockdown (KD) of pancNusap1 mimicked irreversible differentiation of PC12 cells induced by cAMP (A) The effect of pancNusap1 KD on the expression level of Nusap1 in Ndiff cells. Gapdh was used as a control. Empty vector-introduced PC12 cells were used as a negative control. (B) Histone modification status of the Nusap1 promoter in pancNusap1-KD PC12 cells under reversible differentiation condition. The expression of sh-pancNusap1 was induced by Dox at day 5. Empty vector-infected PC12 cells were used as a control. (C) The effects of pancNusap1 KD on cell proliferation in Ndiff cells. The numbers of living cells were counted. (D) Proportion of the EdU+ and the Ki67+ cells in pancNusap1-KD PC12 cells after NGF deprivation. After 7 days of differentiation, NGF was removed to allow proliferation. White arrowheads indicate locations of proliferating infected cells (triple-positive for Ki67, EdU and GFP). In (A–C), viral vectors were introduced at day 3. In (A–D), values are mean ± SD (n = 3). Statistical analysis was performed using Student's two-tailed t-tests, or the Tukey-Kramer multiple comparison test. N.S. = not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Overexpression of pancNusap1 induced cell cycle progression with an open chromatin formation in the Nusap1 promoter
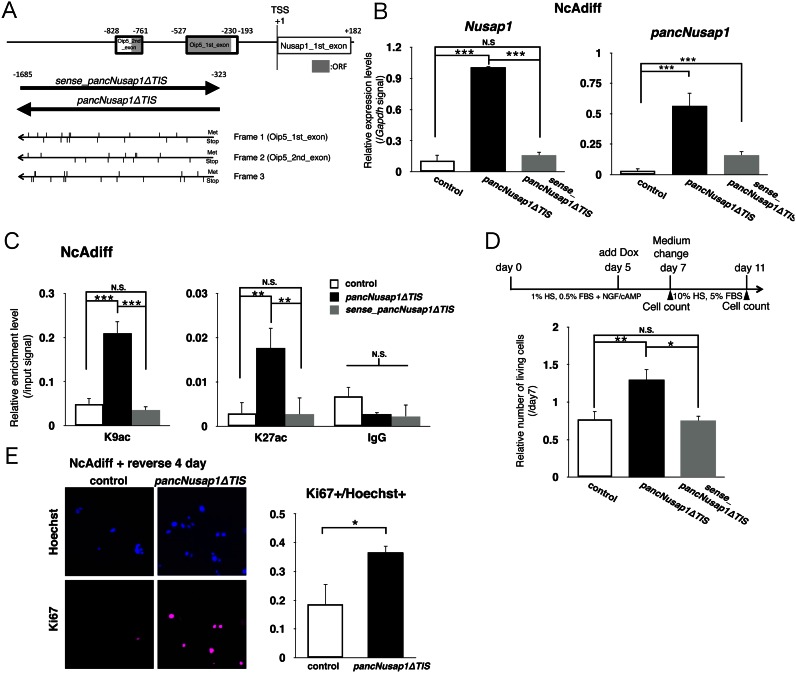
To further test our hypothesis that regulation of pancNusap1 is critical for irreversible differentiation, we examined the effects of pancNusap1 OE on the expression levels of Nusap1 in NcAdiff cells. Since pancNusap1 contained regions highly homologous with mouse Oip5 (Figure 6A), it seemed possible that this pancRNA might need to be translated in order for it to function. However, when we used a genomic region lacking the predicted translation initiation site (pancNusap1ΔTIS), we still found upregulation of Nusap1 (Figure 6B). Because we discovered previously that several pancRNAs function in a strand-specific manner (8), we also prepared PC12 cells in which the strand opposite pancNusap1 (sense_pancNusap1ΔTIS) was exogenously expressed (Figure 6A), and found that the introduction of sense_pancNusap1ΔTIS could not upregulate the expression of Nusap1 (Figure 6B). This indicates that the direction of transcription of this pancRNA was important for its function as a local gene activator. We also investigated the effect of pancNusap1ΔTIS-OE and sense_pancNusap1ΔTIS-OE on the histone acetylation profile of the Nusap1 promoter in NcAdiff cells, and found that pancNusap1ΔTIS-OE, but not sense_pancNusap1ΔTIS-OE, increased the signals of both H3K9ac and H3K27ac at the Nusap1 promoter in parallel with Nusap1 upregulation (Figure 6C). Then, to investigate the effects of pancNusap1-OE on the phenotypic irreversibility of PC12 cells, we immunostained PC12 cells with antibody against Ki67 and counted the number of living cells before/after the NGF/cAMP-containing medium was changed back to the proliferation medium, and found that the number of cells was increased by pancNusap1ΔTIS-OE, but not by sense_pancNusap1ΔTIS-OE (Figure 6A and D). Immunostaining revealed an increase of the Ki67+ cells after pancNusap1ΔTIS-OE (Figure 6E), indicating that re-proliferation was enhanced by pancNusap1ΔTIS-OE. These data showed that pancNusap1ΔTIS-OE expression could induce morphological and epigenetic features characteristic of the reversibly differentiated state of PC12 cells in irreversibly differentiated cells.
Figure 6.
pancNusap1 overexpression (OE) caused cells subjected to the irreversibly differentiation condition to have the morphological and epigenetic characters of reversibly differentiated cells. (A) Genomic structure of the rat Nusap1 promoter. Bold-lined boxes show the region homologous to mouse Oip5. Gray boxes indicate predicted locations composing the Oip5 open reading frame (ORF). Lower thick arrows indicate the region and transcribed strands used for OE-experiments of ncRNA. Upper and lower vertical lines show the positions of methionines (Met) or stop codons (Stop), respectively, in each translational frame. (B) The effect of Dox-induced sense or antisense pancNusap1 OE on the Nusap1 expression level in NcAdiff cells. Gapdh was used as a control. Empty vector-introduced PC12 cells were used as a negative control. (C) Histone modification status of the Nusap1 promoter in pancNusap1-overexpressing PC12 cells under irreversible differentiation condition. (D) The effect of Dox-induced sense or antisense pancNusap1 OE on cell proliferation in NcAdiff cells. Upper diagram shows experimental scheme. Number of living cells was counted. After 7 days of differentiation of PC12 cells, NGF/cAMP were removed to allow proliferation. (E) Proportion of Ki67+ cells in antisense pancNusap1-overexpressing PC12 cells after NGF/cAMP deprivation. In (B–E), values are mean ± SD (n = 3). Statistical analysis was performed using Student's two-tailed t-tests or the Tukey-Kramer multiple comparison test. N.S. = not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Overexpression of Nusap1 rescued pancNusap1-KD-induced cell cycle arrest
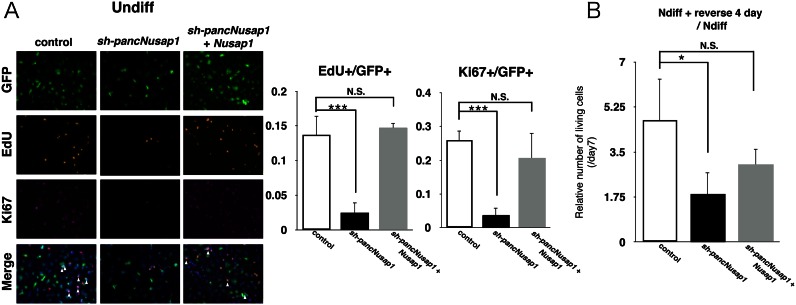
To test the epigenetic effects of pancNusap1-KD and -OE in a single gene expression setting, we performed rescue experiments of pancNusap1-KD cells with Nusap1-OE, and monitored the proliferation of Undiff or Ndiff cells after introduction of sh-pancNusap1 and Nusap1. In Undiff cells, the reductions of both the EdU+ and the Ki67+ cells by pancNusap1-KD were rescued by the simultaneous introduction of Nusap1-OE (Figure 7A and Supplementary Figure S10). Counting the number of living cells after 4 days of culture in re-proliferative condition further confirmed that, after the NGF-containing differentiation medium was changed back to the proliferation medium, the reduction of the number of cells by pancNusap1-KD was successfully rescued by simultaneous Nusap1-OE expression (Figure 7B). These results clearly indicate that pancNusap1 acts to upregulate the transcription of Nusap1.
Figure 7.
pancNusap1 regulates irreversibility of PC12 differentiation through transcriptional regulation of Nusap1. (A) Proportion of the EdU+ and the Ki67+ cells in pancNusap1-KD and Nusap1-OE Undiff cells. White arrowheads indicate locations of the proliferating infected cells (triple-positive for Ki67, EdU and GFP). (B) The effect of Nusap1-OE on cell proliferation in the pancNusap1-KD cells. Number of living cells was counted. In (A and B), values are mean ± SD (n = 3). Statistical analysis was performed using the Tukey-Kramer multiple comparison test. N.S. = not significant. *P < 0.05; ***P < 0.001.
The activities of pBiPs, including that for the pancNusap1-Nusap1 pair, were regulated by cAMP signaling
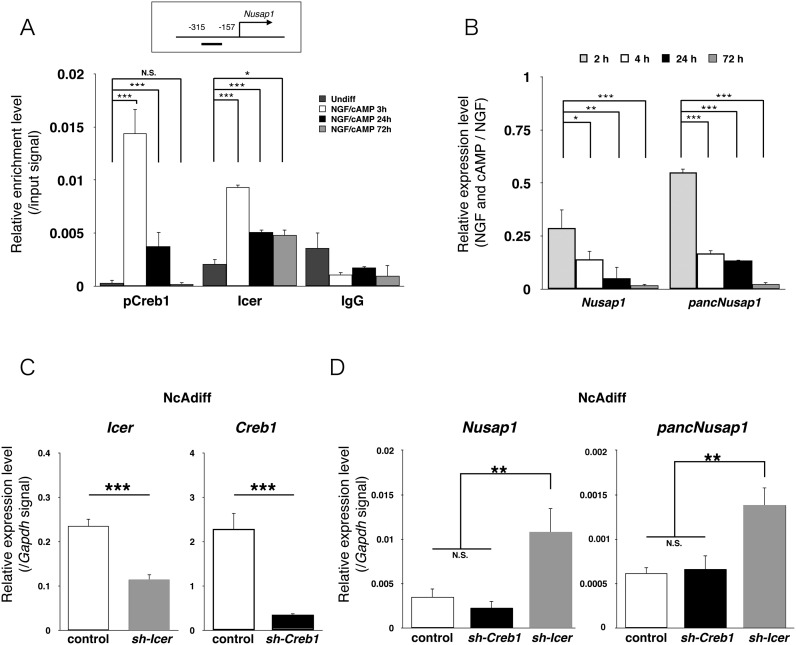
By using the publicly available pCreb1 ChIP-seq dataset of PC12 cells (15), we found that pCreb1 interacted with the Nusap1 promoter immediately after cAMP signal activation by forskolin. However, because pancNusap1 and Nusap1 were silenced in response to cAMP stimulation in NcAdiff cells (Figure 4B), we tried to experimentally confirm the pCreb1-binding at the Nusap1 promoter and to identify the factors that shut down cAMP-dependent activation of the pancNusap1–Nusap1 pair as well. Some reports showed that transcription of a dominant-negative form of Crem, Icer (Supplementary Figure S11A), is activated by binding of pCreb1 after cAMP treatment in PC12 cells (19,39), and therefore we investigated the transcriptional dynamics of Creb1, Crem and Icer using our RNA-seq data. Consistent with previous reports, we found that transcription of Icer was dramatically activated in NcAdiff cells (Supplementary Figure S11B). We confirmed these results by RT-qPCR (Supplementary Figure S11C). Next, we performed time-course ChIP experiments to examine the binding of pCreb1 and Icer to the Nusap1 promoter after NGF/cAMP treatment (Figure 8A). We found that pCreb1-binding was upregulated at 3 h and then diminished by 72 h after the NGF/cAMP treatment. In contrast, Icer-binding was upregulated at 3 h but then partially maintained until 72 h after the NGF/cAMP treatment. These ChIP-qPCR data agree well with the RT-qPCR data, in which both Nusap1 and pancNusap1 transcripts were reduced to the basal levels by 72 h (Figure 8B). To further validate the differential roles of pCreb1 and Icer in the transcriptional regulation of the Nusap1 promoter during PC12 differentiation, we examined the effects of Icer and Creb1 KD on the expression levels of Nusap1 and pancNusap1. As expected, our shRNAs specifically targeted Icer or Creb1, resulting in their significant downregulation (Figure 8C and Supplementary Figure S12). KD of Icer upregulated the expression levels of both pancNusap1 and Nusap1 in NcAdiff cells (Figure 8D). In contrast, KD of Creb1 did not affect the expression levels of either Nusap1 or pancNusap1 in NcAdiff cells (Figure 8D). Taking these results all together, we concluded that cAMP signaling represses pancNusap1 and Nusap1 through Icer, leading ultimately to the reduction of Nusap1 expression via pancNusap1 downregulation.
Figure 8.
The transcriptional activity of pancNusap1-Nusap1 pair was regulated by Icer after cAMP stimulation. (A) pCreb1- and Icer-binding status at the Nusap1 promoter after 3, 24 and 72 h of NGF/cAMP treatment. Normal rabbit IgG (IgG) was used as a negative control. (B) The expression dynamics of pancNusap1 and Nusap1 after 2, 4, 24 and 72 h of NGF and/or cAMP treatment. (C and D) The effects of sh-Icer and sh-Creb OEs on the expression levels of their target genes (C) and the pancNusap1-Nusap1 pair (D) in NcAdiff cells. Viral vectors were introduced at day 3. In (A–D), values are mean ± SD (n = 3). Statistical analysis was performed using the Tukey-Kramer multiple comparison test. N.S. = not significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Finally, to investigate whether pBiPs other than the Nusap1 promoter were transcriptionally regulated by downstream transcription factors of the cAMP signaling pathway, we examined the effects of Icer and Creb1 KD on the expression levels of two genes listed next to Nusap1: Arhgap26 and Gata2 (Figure 4A, Supplementary Table S6). We found that in NcAdiff cells, KD of Icer significantly upregulated the expression level of Arhgap26, and KD of Creb1 significantly downregulated the expression level of Gata2 (Figure S13). These results indicated that Icer and Creb1 were involved in the transcriptional regulation of Arhgap26 and Gata2, respectively, and supported our notion that pBiPs are preferred targets of downstream transcription factors of cAMP signaling during the irreversible differentiation of PC12 cells.
DISCUSSION
In this study, we utilized PC12 cells as a model for understanding the bidirectional promoter-mediated mechanism underlying cAMP-triggered irreversible differentiation. Genome-wide analysis of our directional RNA-seq dataset and the publicly available pCreb1 ChIP-seq dataset of PC12 cells (15) revealed that bidirectional promoters for the expression of pancRNA–mRNA pairs are preferred downstream targets of cAMP signaling during the irreversible differentiation of PC12 cells. In irreversibly differentiated PC12 cells, the activities of representative bidirectional promoters for the expression of pancRNA–mRNA pairs were regulated by transcription factors in the cAMP-dependent pathway (Figure 8 and Supplementary Figure S13). To examine whether such transcriptional regulation of pancRNA–mRNA pairs contributes to the cAMP-triggered irreversible differentiation, we focused on pancNusap1 and Nusap1, the pancRNA–mRNA pair showing the highest Pearson correlation coefficients of the expression during PC12 differentiation. KD and OE experiments showed that pancNusap1 positively regulated the Nusap1 expression in a sequence-specific manner, which was accompanied by acetylation of histone H3 lysine 9 and 27 at the promoter. Both the pancNusap1 and the Nusap1 expression levels and the histone acetylation level of the Nusap1 promoter decreased concomitantly, most likely as part of the regulation responsible for irreversible differentiation, as shown by our finding that the epigenetic silencing of Nusap1 via KD of pancNusap1 recapitulated the cAMP effect on cell cycle arrest during the process of irreversible differentiation of PC12 cells.
Downstream transcription factors of cAMP signaling regulate the expression of hundreds of gene activation-associated ncRNAs derived from bidirectional promoters
A previous ChIP-seq study showed that a quarter of pCreb1-target genes are derived from aBiPs, suggesting that such head-to-head promoter structures may provide adaptive advantages for cAMP functioning in cells (15). In this study, we showed that annotated protein-coding genes were expressed from 474 aBiPs, and from 2718 pBiPs, in rat PC12 cells. Genome-wide analysis, supported by representative experimental tests, revealed that not only aBiPs but also pBiPs were enriched for CRE consensus sequences, and were preferential targets for regulation by the transcription factors in the cAMP-dependent pathway (Figures 3B and 8 and Supplementary Figure S13). We reported previously that transcription of pancRNAs from pBiPs epigenetically activates partner tissue-specific genes (7,8,11). Consistent with previous reports that pancRNAs contribute to the establishment of cell identity via epigenetic regulation of tissue-specific gene expression, the expression levels of pancRNAs derived from pBiPs tended to be positively correlated with the expression levels of their partner mRNAs during differentiation of PC12 cells (Figure 3C). Furthermore, epigenetic activation via pancRNAs was confirmed here by the manipulation of the pancNusap1 levels in PC12 cells (Figures 5–7). Thus, the present results provide the new insight that transcriptional regulation by means of pancRNAs provides adaptive advantages for CRE-mediated gene regulation. Considering that the number of pBiPs is almost 6-fold greater than the number of aBiPs, the impacts of pBiPs as important effectors of cAMP signals might be greater than those of aBiPs for coordinated activation of a fraction of cAMP-responsive genes.
The silencing of M-phase progression contributes to the establishment of an irreversibly differentiated state: implications for neuronal differentiation
We showed that regulation of cell-cycle-associated genes plays crucial roles in the process of irreversible differentiation of PC12 cells (Figures 5–7). The critical importance of maintaining the cell-cycle-arrested state in neurons is supported by studies on somatic cell nuclear transfer. When nuclei of adult cortical neurons are used as donors, the majority of reconstructed mouse oocytes are arrested at the first mitotic cleavage with abnormal chromosome condensation (40), whereas reconstructed oocytes cloned from nuclei of other types of differentiated somatic cells, such as cumulus cells (41) and tail tip fibroblasts (42), show much greater developmental potential. Because the cell cycle of neurons is arrested at G0 phase, many studies have focused on the relationship between neuronal differentiation and inhibition of G1/S-phase progression (43,44). For example, neuronal differentiation of mouse neural stem cells is enhanced by double knockout of G1/S-phase-progression-associated genes Cdk2 and Cdk4 (45). Similarly, Ndiff PC12 cells have been widely used as an in vitro model for investigating the relationship between G1/S-phase inhibition and neurite outgrowth. In fact, like the Cdk2 protein level, the level of Pcna and phosphorylated retinoblastoma protein, also known as G1/S-phase-progression-associated proteins, are downregulated in Ndiff PC12 cells (46–48), suggesting that G1/S-phase inhibition also occurs in reversibly differentiated Ndiff PC12 cells.
In addition, several lines of evidence indicate that inhibition of M-phase-associated genes is involved in the functional differentiation of neurons (49,50). For example, reconstructed mouse oocytes cloned from nuclei of adult cortical neurons undergo abnormal cell cycle arrest at the first mitotic cleavage (40). In mouse neural progenitors, KD of an M-phase-progression-associated gene, doublecortin-like kinase, causes neuronal differentiation (51). Therefore, coordinated regulation of G1/S-phase-progression-associated genes and M-phase-progression-associated genes seems to be important for neuronal differentiation. In this context, it is interesting to note that the expression of cell-cycle-associated genes, especially those related to G2/M-phase progression, remained high in Ndiff PC12 cells (Figure 2) and epigenetic silencing of M-phase-associated Nusap1 could convert reversibly differentiated cells into irreversibly differentiated cells (Figure 5). Considering that inhibition of G1/S-phase progression is observed in reversibly differentiated Ndiff PC12 cells (Supplementary Figure S14), NGF and cAMP signalings appear to be strongly involved in G1/S- and G2/M-inhibition, respectively, for completing the differentiation of neurons. Our results suggested that irreversible/reversible PC12 differentiation could be a good model for further addressing the lncRNA-mediated mechanisms that link cell cycle regulation to irreversible differentiation.
The pancRNA repertoires are differentially utilized from the zygotic to the terminally differentiated stages
In this study, the RNA-seq method was utilized to test whether pancRNA-mediated gene activation mechanisms function in the terminal differentiation of the cells on a genome-wide scale, and the results confirmed this hypothesis (Figures 3, 5 and 6). Indeed, pancRNAs have been detected from more than a thousand promoter regions at a very early stage of life called zygotic gene activation (7). Previous reports have also shown that thousands of pancRNAs are transcribed in ES cells and various tissues (11,52). Thus, a non-negligible number of pancRNAs seem to be expressed in various contexts. Since pancRNAs and mRNAs exhibit coordinated expression changes not only in totipotent/pluripotent cells but also in terminally differentiated cells (Figure 3C), pancRNAs may be commonly utilized for gene activation from the zygotic to the terminally differentiated stage. Although we focused on a study of functional pancRNAs from the polyA+ and >200 bp ncRNA fraction in this study, other fractions of ncRNAs (e.g. polyA-, <200 bp) derived from the promoter of protein-coding genes may also function in transcriptional activation of their partner genes (37,53). The general usage of the pancRNA system over the course of life sheds light on the importance of gene-activation-associated ncRNAs as counterparts of gene-silencing ncRNAs, such as microRNA, which work at the post-transcriptional level. Studies of possible coordinated regulation with lncRNAs derived from enhancer regions, called eRNAs (32,54–59) would be the next step in elucidating the sequence-specific gene activation mechanisms in which ncRNA structurally recognizes genomic DNA.
Transcriptional silencing of Nusap1 in response to extracellular differentiation stimuli is an important step of cAMP signaling toward terminal differentiation of PC12 cells
Nusap1 protein has a well-conserved microtubule-binding domain in its COOH terminus and a DNA-binding SAP domain in its NH2 terminus, and promotes the stabilization and crosslinking of microtubules near chromosomes in metaphase/anaphase (60,61). Nusap1 is highly expressed in proliferative tissues, and essential for M-phase progression. In cultured cell lines, KD of Nusap1 causes severe mitotic defects with defective anaphase and cytokinesis (60), and knockout of Nusap1 induces early embryonic lethality in mice (62). Like other cell cycle regulators, the protein level of Nusap1 is under the regulation of a multisubunit E3 ubiquitin ligase, anaphase promoting complex/cyclosome, in proliferating cells (63). However, the mechanisms of transcriptional regulation of Nusap1 in terminally differentiated cells are still unknown. In this study, we showed that reduced transcription of Nusap1 via downregulation of pancNusap1 in response to cAMP stimulation functions in the terminal differentiation of PC12 cells (Figures 5–8). These findings indicated that transcriptional silencing of Nusap1 is a key mediator of cAMP signaling for terminal differentiation of PC12 cells. The epigenetic silencing by the concomitant reduction of pancNusap1 by cAMP may further support the complete shut-down of neuronal cell division.
Possible molecular mechanism underlying pancNusap1-mediated epigenetic modification
We and others have reported that pancRNA promotes the assembly of an open chromatin conformation that includes histone acetylation, H3K4 methylation, H3K9/K27 demethylation and active DNA demethylation (7,8,12,64). Differently from these previous results, the present data indicated that the activation and reduction of pancNusap1 expression increased and decreased the histone acetylation levels of the Nusap1 promoter, respectively, but not the levels of histone methylation or DNA methylation (Figures 4–6, and Supplementary Figure S1). There are many reports indicating that functional lncRNAs recruit epigenetic modification complexes, such as histone methyltransferase myeloid/lymphoid or mixed-lineage leukemia (MLL) complex and the BER components at specific genomic locations (1,56,65–67). However there are no reports that prove the interaction of lncRNAs with histone acetyltransferase complex. Recently, nuclease-null Cas9-based transcriptional activation technology has revealed that artificial transcription initiation at an intergenic locus elevates H3K27ac levels (68). Furthermore, single living cell analysis has shown that recruitment of RNAPII occurs within a few minutes after histone acetylation at the glucocorticoid receptor gene locus (69). Given these reports, one possible scenario is that transcription initiation of pancNusap1 primary causes histone acetylation at the Nusap1 promoter and thus enhances specific gene expression there.
Interestingly, we and others have reported that only antisense pancRNAs are associated with chromatin, and pancRNA-mediated transcriptional activation occurs in a strand-specific manner (8,64). Consistent with these reports, the transcriptional direction of pancNusap1 is important for pancNusap1-mediated histone acetylation (Figure 6). In this context, it is worthwhile to note that pancNusap1 has a G-rich sequence (Supplementary Figure S15). Because guanine (G)-rich RNA that hybridizes with single-stranded cytosine (C)-rich complementary DNA forms a thermodynamically stable DNA:RNA hybrid structure, known as an R-loop structure (70,71), the sequence-dependent stability of R-loops may explain the increase of histone acetylation by pancNusap1. Further studies will be needed to unravel the molecular mechanisms that determine the sequential epigenetic changes induced by pancRNA-mediated histone acetylation.
In conclusion, pancRNA-mediated histone acetylation contributes to the establishment of PC12 cells’ stimulation-induced transcription status during the irreversible differentiation process.
DATA AVAILABILITY
RNA-seq data have been deposited in the DDBJ Sequence Read Archive (DRA) under accession number DRA004148.
Supplementary Material
Acknowledgments
We thank Osamu Nishimura and Masahiro Uesaka for providing advice about transcriptome analysis; Nobuhiko Hamazaki and Hirofumi Noguchi for scientific insights and suggestions throughout the study; Katsuhide Igarashi, Hideyuki Nakashima, Tsukasa Sanosaka and Sayako Katada for experimental advice; and Elizabeth Nakajima for proofreading the manuscript.
Footnotes
Present address: Naoki Yamamoto, Life Science Tokyo Advanced Research center (L-StaR), Hoshi University School of Pharmacy and Pharmaceutical Science, Japan.
FUNDING
Japan Society for the Promotion of Science [15H04603; 24380158 to T. I., 241544 to N. Y.]; Global COE program [A06 to Kyoto University]; Scientific Research on Innovative Areas ‘Genome Science’ from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) [221S0002 to T.I.]. Funding for open access charge: Japan Society for the Promotion of Science [15H04603, 24380158 to T. I., 241544 to N. Y.]; Global COE program [A06 to Kyoto University]; Scientific Research on Innovative Areas ‘Genome Science’ from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) [221S0002].
Conflict of interest statement. None declared.
REFERENCES
- 1.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G., Young G., Lucas A.B., Ach R., Bruhn L., et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrieri C., Cimatti L., Biagioli M., Beugnet A., Zucchelli S., Fedele S., Pesce E., Ferrer I., Collavin L., Santoro C., et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature. 2012;491:454–457. doi: 10.1038/nature11508. [DOI] [PubMed] [Google Scholar]
- 3.Ng S.-Y., Bogu G.K., Soh B.S., Stanton L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Herzog V.A., Lempradl A., Trupke J., Okulski H., Altmutter C., Ruge F., Boidol B., Kubicek S., Schmauss G., Aumayr K., et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat. Genet. 2014;46:973–981. doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ting A.H., Schuebel K.E., Herman J.G., Baylin S.B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.-C., Hung T., Argani P., Rinn J.L., et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamazaki N., Uesaka M., Nakashima K., Agata K., Imamura T. Gene activation-associated long noncoding RNAs function in mouse preimplantation development. Development. 2015;142:910–920. doi: 10.1242/dev.116996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomikawa J., Shimokawa H., Uesaka M., Yamamoto N., Mori Y., Tsukamura H., Maeda K.-I., Imamura T. Single-stranded noncoding RNAs mediate local epigenetic alterations at gene promoters in rat cell lines. J. Biol. Chem. 2011;286:34788–34799. doi: 10.1074/jbc.M111.275750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sigova A.A., Abraham B.J., Ji X., Molinie B., Hannett N.M., Guo Y.E., Jangi M., Giallourakis C.C., Sharp P.A., Young R.A. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uesaka M., Nishimura O., Go Y., Nakashima K., Agata K., Imamura T. Bidirectional promoters are the major source of gene activation-associated non-coding RNAs in mammals. BMC Genomics. 2014;15:35. doi: 10.1186/1471-2164-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imamura T., Yamamoto S., Ohgane J., Hattori N., Tanaka S., Shiota K. Non-coding RNA directed DNA demethylation of Sphk1 CpG island. Biochem. Biophys. Res. Commun. 2004;322:593–600. doi: 10.1016/j.bbrc.2004.07.159. [DOI] [PubMed] [Google Scholar]
- 13.Hegedus B., Dasgupta B., Shin J.E., Emnett R.J., Hart-Mahon E.K., Elghazi L., Bernal-Mizrachi E., Gutmann D.H. Neurofibromatosis-1 regulates neuronal and glial cell differentiation from neuroglial progenitors in vivo by both cAMP- and Ras-dependent mechanisms. Cell Stem Cell. 2007;1:443–457. doi: 10.1016/j.stem.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Zahir T., Chen Y.F., MacDonald J.F., Leipzig N., Tator C.H., Shoichet M.S. Neural stem/progenitor cells differentiate in vitro to neurons by the combined action of dibutyryl cAMP and interferon-gamma. Stem Cells Dev. 2009;18:1423–1432. doi: 10.1089/scd.2008.0412. [DOI] [PubMed] [Google Scholar]
- 15.Impey S., McCorkle S.R., Cha-Molstad H., Dwyer J.M., Yochum G.S., Boss J.M., McWeeney S., Dunn J.J., Mandel G., Goodman R.H. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 16.Cha-Molstad H., Keller D.M., Yochum G.S., Impey S., Goodman R.H. Cell-type-specific binding of the transcription factor CREB to the cAMP-response element. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13572–13577. doi: 10.1073/pnas.0405587101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molina C.A., Foulkes N.S., Lalli E., Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 18.Klejman A., Kaczmarek L. Inducible cAMP early repressor (ICER) isoforms and neuronal apoptosis in cortical in vitro culture. Acta Neurobiol. Exp. (Wars) 2006;66:267–272. doi: 10.55782/ane-2006-1615. [DOI] [PubMed] [Google Scholar]
- 19.Borlikova G., Endo S. Inducible cAMP early repressor (ICER) and brain functions. Mol. Neurobiol. 2009;40:73–86. doi: 10.1007/s12035-009-8072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okoshi R., Kubo N., Nakashima K., Shimozato O., Nakagawara A., Ozaki T. CREB represses p53-dependent transactivation of MDM2 through the complex formation with p53 and contributes to p53-mediated apoptosis in response to glucose deprivation. Biochem. Biophys. Res. Commun. 2011;406:79–84. doi: 10.1016/j.bbrc.2011.01.114. [DOI] [PubMed] [Google Scholar]
- 21.Gunning P.W., Landreth G.E., Layer P., Ignatius M., Shooter E.M. Nerve growth factor-induced differentiation of PC12 cells: evaluation of changes in RNA and DNA metabolism. J. Neurosci. 1981;1:368–379. doi: 10.1523/JNEUROSCI.01-04-00368.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel P.P., Vyas S., Agid Y. Synergistic differentiation by chronic exposure to cyclic AMP and nerve growth factor renders rat phaeochromocytoma PC12 cells totally dependent upon trophic support for survival. Eur. J. Neurosci. 1995;7:251–260. doi: 10.1111/j.1460-9568.1995.tb01061.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Wang S., Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong L., Zhang Y., Ye Z.-Q., Liu X.-Q., Zhao S.-Q., Wei L., Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun J., Nishiyama T., Shimizu K., Kadota K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., Green M.R. ChIPpeakAnno: a bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.-Y., Chou A., Ienasescu H., et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesiak A., Pelz C., Ando H., Zhu M., Davare M., Lambert T.J., Hansen K.F., Obrietan K., Appleyard S.M., Impey S., et al. A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PLoS One. 2013;8:e64658. doi: 10.1371/journal.pone.0064658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Rocca R., Fulciniti M., Lakshmikanth T., Mesuraca M., Ali T.H., Mazzei V., Amodio N., Catalano L., Rotoli B., Ouerfelli O., et al. Early hematopoietic zinc finger protein prevents tumor cell recognition by natural killer cells. J. Immunol. 2009;182:4529–4537. doi: 10.4049/jimmunol.0802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shin K.-J., Wall E.A., Zavzavadjian J.R., Santat L.A., Liu J., Hwang J.-I., Rebres R., Roach T., Seaman W., Simon M.I., et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. U.S.A. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zufferey R., Dull T., Mandel R.J., Bukovsky A., Quiroz D., Naldini L., Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J. Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu H.Y., He L., Khaitovich P. Deep sequencing reveals a novel class of bidirectional promoters associated with neuronal genes. BMC Genomics. 2014;15:457. doi: 10.1186/1471-2164-15-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang J.H., Vuppalanchi D., van Niekerk E., Trepel J.B., Schanen N.C., Twiss J.L. PC12 cells regulate inducible cyclic AMP (cAMP) element repressor expression to differentially control cAMP response element-dependent transcription in response to nerve growth factor and cAMP. J. Neurochem. 2006;99:1517–1530. doi: 10.1111/j.1471-4159.2006.04196.x. [DOI] [PubMed] [Google Scholar]
- 40.Osada T., Kusakabe H., Akutsu H., Yagi T., Yanagimachi R. Adult murine neurons: their chromatin and chromosome changes and failure to support embryonic development as revealed by nuclear transfer. Cytogenet. Genome Res. 2002;97:7–12. doi: 10.1159/000064037. [DOI] [PubMed] [Google Scholar]
- 41.Wakayama T., Perry A.C., Zuccotti M., Johnson K.R., Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature. 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 42.Wakayama T., Yanagimachi R. Cloning of male mice from adult tail-tip cells. Nat. Genet. 1999;22:127–128. doi: 10.1038/9632. [DOI] [PubMed] [Google Scholar]
- 43.Lee E.Y., Hu N., Yuan S.S., Cox L.A., Bradley A., Lee W.H., Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 44.Galderisi U., Jori F.P., Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22:5208–5219. doi: 10.1038/sj.onc.1206558. [DOI] [PubMed] [Google Scholar]
- 45.Lim S., Kaldis P. Loss of Cdk2 and Cdk4 induces a switch from proliferation to differentiation in neural stem cells. Stem Cells. 2012;30:1509–1520. doi: 10.1002/stem.1114. [DOI] [PubMed] [Google Scholar]
- 46.Yan G.Z., Ziff E.B. NGF regulates the PC12 cell cycle machinery through specific inhibition of the Cdk kinases and induction of cyclin D1. J. Neurosci. 1995;15:6200–6212. doi: 10.1523/JNEUROSCI.15-09-06200.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan G.Z., Ziff E.B. Nerve growth factor induces transcription of the p21 WAF1/CIP1 and cyclin D1 genes in PC12 cells by activating the Sp1 transcription factor. J. Neurosci. 1997;17:6122–6132. doi: 10.1523/JNEUROSCI.17-16-06122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobashi Y., Shoji M., Kitagawa M., Noguchi T., Kameya T. Simultaneous suppression of cdc2 and cdk2 activities induces neuronal differentiation of PC12 cells. J. Biol. Chem. 2000;275:12572–12580. doi: 10.1074/jbc.275.17.12572. [DOI] [PubMed] [Google Scholar]
- 49.Hayes T.E., Valtz N.L., McKay R.D. Downregulation of CDC2 upon terminal differentiation of neurons. New Biol. 1991;3:259–269. [PubMed] [Google Scholar]
- 50.Okano H.J., Pfaff D.W., Gibbs R.B. RB and Cdc2 expression in brain: correlations with 3H-thymidine incorporation and neurogenesis. J. Neurosci. 1993;13:2930–2938. doi: 10.1523/JNEUROSCI.13-07-02930.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shu T., Tseng H.-C., Sapir T., Stern P., Zhou Y., Sanada K., Fischer A., Coquelle F.M., Reiner O., Tsai L.-H. Doublecortin-like kinase controls neurogenesis by regulating mitotic spindles and M phase progression. Neuron. 2006;49:25–39. doi: 10.1016/j.neuron.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 52.Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., Almada A.E., Lin C., Sharp P.A., Giallourakis C.C., et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermüller J., Hofacker I.L., et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 54.Melgar M.F., Collins F.S., Sethupathy P. Discovery of active enhancers through bidirectional expression of short transcripts. Genome Biol. 2011;12:R113. doi: 10.1186/gb-2011-12-11-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hah N., Murakami S., Nagari A., Danko C.G., Kraus W.L. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C., et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai F., Orom U.A., Cesaroni M., Beringer M., Taatjes D.J., Blobel G.A., Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Léveillé N., Melo C.A., Rooijers K., Díaz-Lagares A., Melo S.A., Korkmaz G., Lopes R., Akbari Moqadam F., Maia A.R., Wijchers P.J., et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat. Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raemaekers T., Ribbeck K., Beaudouin J., Annaert W., Van Camp M., Stockmans I., Smets N., Bouillon R., Ellenberg J., Carmeliet G. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 2003;162:1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ribbeck K., Raemaekers T., Carmeliet G., Mattaj I.W. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 2007;17:230–236. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- 62.Vanden Bosch A., Raemaekers T., Denayer S., Torrekens S., Smets N., Moermans K., Dewerchin M., Carmeliet P., Carmeliet G. NuSAP is essential for chromatin-induced spindle formation during early embryogenesis. J. Cell. Sci. 2010;123:3244–3255. doi: 10.1242/jcs.063875. [DOI] [PubMed] [Google Scholar]
- 63.Li L., Zhou Y., Sun L., Xing G., Tian C., Sun J., Zhang L., He F. NuSAP is degraded by APC/C-Cdh1 and its overexpression results in mitotic arrest dependent of its microtubules’ affinity. Cell. Signal. 2007;19:2046–2055. doi: 10.1016/j.cellsig.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 64.Boque-Sastre R., Soler M., Oliveira-Mateos C., Portela A., Moutinho C., Sayols S., Villanueva A., Esteller M., Guil S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5785–5790. doi: 10.1073/pnas.1421197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K.C., Yang Y.W., Liu B., Sanyal A., Corces-Zimmerman R., Chen Y., Lajoie B.R., Protacio A., Flynn R.A., Gupta R.A., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D., Zhang J., et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puc J., Kozbial P., Li W., Tan Y., Liu Z., Suter T., Ohgi K.A., Zhang J., Aggarwal A.K., Rosenfeld M.G. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–380. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hilton I.B., D'Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stasevich T.J., Hayashi-Takanaka Y., Sato Y., Maehara K., Ohkawa Y., Sakata-Sogawa K., Tokunaga M., Nagase T., Nozaki N., McNally J.G., et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 70.Ratmeyer L., Vinayak R., Zhong Y.Y., Zon G., Wilson W.D. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 71.Roberts R.W., Crothers D.M. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the DDBJ Sequence Read Archive (DRA) under accession number DRA004148.