Abstract
Herein, we report on the use of a tricyclic cytosine FRET pair, incorporated into DNA with different base pair separations, to study Z-DNA and B-Z DNA junctions. With its position inside the DNA structure, the FRET pair responds to a B- to Z-DNA transition with a distinct change in FRET efficiency for each donor/acceptor configuration allowing reliable structural probing. Moreover, we show how fluorescence spectroscopy and our cytosine analogues can be used to determine rate constants for the B- to Z-DNA transition mechanism. The modified cytosines have little influence on the transition and the FRET pair is thus an easily implemented and virtually non-perturbing fluorescence tool to study Z-DNA. This nucleobase analogue FRET pair represents a valuable addition to the limited number of fluorescence methods available to study Z-DNA and we suggest it will facilitate, for example, deciphering the B- to Z-DNA transition mechanism and investigating the interaction of DNA with Z-DNA binding proteins.
INTRODUCTION
Nucleic acids were first discovered as vectors of heredity in living organism and they are now also known to be involved in a variety of cellular processes like, for example, transcription, regulation and biocatalysis. The nucleic acid alphabet, composed of only five nucleobases (A, C, G and T or U), is however rather rudimentary and the complexity of the functions of DNA and RNA originates from their polymorphism. In vivo DNA is most commonly found as the canonical duplex right-handed helical structure known as B-DNA but DNA repeat-sequences can form several non-B DNA structures such as the G-quadruplex, the i-motif and Z-DNA (1–3). Since these structures are involved in important cellular processes, the regulation of which can be key in human diseases such as cancer (4), it is of utmost importance to understand their structural properties, thermodynamics and mechanisms of formation.
In this work we focus on Z-DNA, a left-handed duplex helix that can be formed in CpG (or GpC) dinucleotide repeats. Its crystal structure was first reported in 1979 by the team of Alexander Rich and its biological relevance has long been debated, but it is now admitted that left-handed DNA and RNA structures can be found in vivo and play a role in transcription regulation (5–7). Compared to B-DNA, Z-DNA is thermodynamically disfavored under normal cellular conditions and it forms only under specific conditions such as high salt concentrations or negative supercoiling, although the exact mechanism of its formation remains to be determined (8–10). To screen for the increased electrostatic repulsion of the phosphate-containing backbones that are closer to each other in Z-DNA than in B-DNA, high salt concentration is needed for in vitro studies. However, recent evidence indicates that anions can also influence B- to Z-DNA transition and so a pure electrostatic model fails to accurately explain the role of salts in Z-DNA formation (11). In vivo, Z-DNA is assumed to form as a result of mechanical constraints like negative supercoiling but also of Z-DNA binding proteins such as ADARI, a double-stranded RNA editing enzyme, which stabilizes DNA in its Z-conformation (12–14). The presence of Z-DNA amidst the widely predominant B-DNA structure is dependent on the formation of B-Z DNA junctions which require the presence of more weakly bound AT base pairs (15–18).
Monitoring of the B- to Z-DNA transition has up until now relied almost exclusively on circular dichroism (CD) measurements, since the switch from a right- to a left-handed helix results in a very distinctive inversion of the signs of the CD peaks (10,19–22). NMR and X-ray structural studies have elegantly complemented CD measurements and allowed the determination of high-resolution Z-DNA and B-Z DNA junction structures (5,16,22). However, NMR and CD both require large amounts of sample and X-ray diffraction is limited to crystallized, solid state structures and therefore makes it impossible to study the dynamics of the system in solution. A few other investigations of Z-DNA and B-Z DNA junctions have also been performed using fluorescence spectroscopy. Fluorescence is a very sensitive and versatile technique that allows monitoring of real-time processes and is, thus, particularly suited to study nucleic acids structure and dynamics (17,18). However, utilizing fluorescence spectroscopy requires adequate probes able to monitor the differences between the B- and Z-forms of DNA. The golden standard of fluorescent base analogues, 2-aminopurine (2-AP), has been used to study the base-pair extrusion at B-Z junctions, taking advantage of its environment-sensitive emission (23,24). While 2-AP remains one the most widely used fluorescent base analogues, it is a fairly poor fluorophore once incorporated into DNA and its use in the field of Z-DNA seems limited to the study of B-Z junctions and cannot be extended to a more comprehensive study of the Z-DNA structure. FRET-donor Cy3 and -acceptor Cy5 have also been used to study B-Z DNA junctions. This system can report on the B-Z DNA transition by following the corresponding change in FRET-efficiency at the single-molecule level (9,11,14). While such an investigation gives highly valuable insight on the system dynamics, single-molecule fluorescence is still not a routine experiment in most laboratories and is not easily implemented. Moreover, to maximize the change in FRET-efficiency, the Cy-dyes can only be positioned at the junction between B- and Z-DNA and, like 2-AP, this system is not suited to investigate the intrinsic Z-DNA part of the molecule. Hence there is a strong need for a versatile fluorescent tool that enables more flexible labelling of any part of the DNA sequence and that reliably monitors the B- to Z-DNA transition.
We previously reported on a FRET pair of modified tricyclic cytosines (tC) comprising a fluorescent donor, tCO and an acceptor, tCnitro, for quantitative FRET applications in DNA (25–27). On the contrary to conventional FRET systems where free dye rotation and an average orientation factor κ2 of 2/3 are assumed, the tCO/tCnitro probes, directly replacing native nucleobases, are firmly positioned inside DNA virtually inhibiting rotational diffusion. As a consequence, the FRET efficiency will not only depend on the distance between the acceptor and the donor but also significantly on their relative orientation. This makes it possible to use FRET quantitatively to resolve solution structures (28) and even small modifications of the DNA conformation may result in significant changes in the FRET efficiency for these probes. A similar orientation-dependence of energy transfer has been observed, for example, for a longer-range FRET pair involving Cy-dyes attached to the ends of DNA and RNA (29). This observation is an effect of significant π-stacking of the Cy-dyes to the terminal bases of the oligonucleotide. Since the relative orientation of the Cy-dyes’ transition dipole moments is less restricted, this end-labeling approach results in a slightly less pronounced orientation-dependence, compared to the case of the tCO/tCnitro FRET pair inside the base-stack (29).
With the knowledge of the properties of the tCO-tCnitro FRET pair in B-DNA at hand, we report herein on its application to monitor the salt-induced B- to Z-DNA transition by steady-state and time-resolved FRET. Two different DNA structures were investigated: a (GC)7 hairpin structure that completely converts to Z-DNA under high salt concentration and a mixed structure comprising a methylated or hemimethylated (GC)5 segment susceptible of forming a B-Z DNA junction (Figure 1). In the B-Z DNA junction sequence, the fluorescent donor tCO was positioned in the B-DNA part of the structure and the acceptor tCnitro at different positions of the GC segment undergoing the B- to Z-transition. Three different base pair separations (4, 6 and 8) were investigated for both structures. CD was used as a reference technique to evidence the B- to Z-DNA transition which was shown to be associated with substantial changes in FRET-efficiencies in both the Z-DNA hairpin and B-Z junction structures. Moreover, we show that the fluorescence intensity variation can be monitored over time to measure rate constants of the salt-induced B- to Z-DNA transition. Importantly, we find that despite their extended aromatic surfaces the modified tricyclic cytosines only have minor influence on the transition compared to unmodified DNA. This suggests that the tricyclic cytosines can be used efficiently and reliably in standard ensemble fluorescence experiments to monitor B- to Z-DNA transition virtually without interfering with the nucleic acid system structure and dynamics.
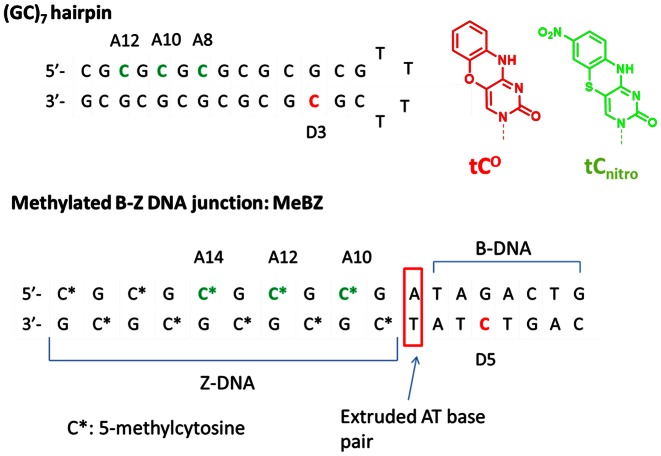
Figure 1.
DNA sequences investigated. Position of donor, tCO, in red and acceptor, tCnitro, in green. Cytosines marked by a star are methylated unless they are replaced by tCnitro. In the hemimethylated sequence the donor strand containing tCO is not methylated. The DX and AY notation of sequences reflect the position of the donor, D, and the acceptor, A, in the sequence. The red box marks expected position of the extruded base pair according to literature (16,17).
MATERIALS AND METHODS
Theoretical FRET-calculations
DNA models were generated using the UCSF Chimera package developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311) (30). For Z-DNA, an available crystal structure (PDB number 4LB5) was used, while the B-DNA was built up using the oligonucleotides build structure command of Chimera (Figure 2). The theoretical FRET efficiency values in B-DNA have already been reported elsewhere (25) and the new calculations were performed for Z-DNA only. The B-DNA structure is shown only for the purpose of comparison. The drawing of tCO and tCnitro structures was performed with ChemAxon Marvin Sketch (31). The structures of the modified cytosines were imported into Chimera and superimposed onto the native cytosines at the desired positions. To determine the orientation factor κ2 of the different FRET-pairs, the angles between the long axis of tCO and that of each tCnitro molecule were determined using atom coordinates, and angles (θ) between transition dipole moments were then inferred according to literature data (26,27).
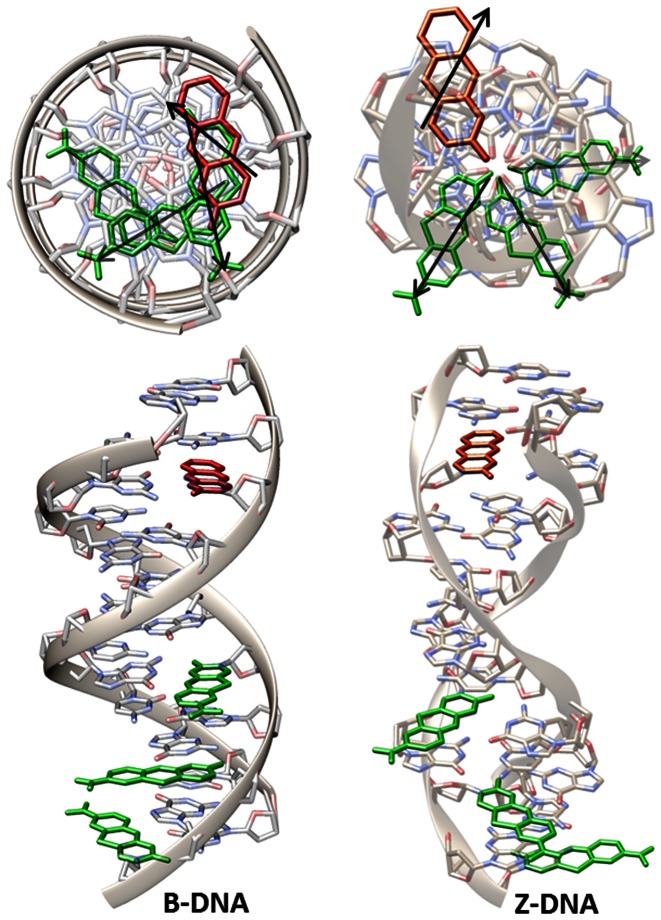
Figure 2.
Top and side view of the B- and Z-DNA structures showing the positions and orientations of tCO (red) and tCnitro (green) for different base-pair separations. Arrows in the top view indicate the direction of the transition dipole moments of the tCO/tCnitro chromophores. In the top view, transparency increases with the number of base pairs separating tCO and tCnitro.
To simplify the calculations, the bases were assumed to be in parallel planes, and thus, the distance d between the donor and acceptor planes and the orientation factor κ2 were calculated as follows:
- d = (n+1)r where n is the number of base-pairs separating donor and acceptor and r the rise per base pair (r = 3.7 Å for Z-DNA and 3.4 Å for B-DNA) (32)
- κ2 = cos2(θ) (simplified expression since the bases are assumed to be in parallel planes)
The Förster distance R0 (in Å) and the theoretical FRET efficiencies E were then calculated according to the following equations:
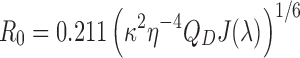 |
 |
Where η is the refractive index of the medium (η = 1.4 in DNA) and QD the quantum yield of the donor tCO (0.23). The overlap integral J(λ) has previously been calculated for the tCO/tCnitro pair and is equal to 1.3 × 1014 M−1 cm−1 nm4 (25).
Oligonucleotides
tCO and tCnitro were purchased from GlenResearch Corp. (Sterling VA, USA) and the oligonucleotides sequences were obtained from ATDBio (Southampton, UK). Single-stranded oligonucleotides were dissolved in mQ water at ∼500 μM and concentrations were measured by UV-Vis absorption measurements at 260 nm. Duplex- and hairpin-sequences were annealed at 100 μM in pH 7.5 25 mM PBS buffer with 100 mM NaCl by heating in a water bath at 95°C followed by slow cooling to 5°C over 12 h. B- to Z-DNA transition was induced by diluting the 100 mM NaCl B-DNA stock solution in a high salt (2–4.9 M NaCl) PBS buffer (10 μL of 100 μM stock solution into 990 μl of buffer for a final concentration of 1 μM or 20 μL of stock solution for a final concentration of 2 μM).
Circular dichroism (CD)
Circular dichroism (CD) spectra of 2 μM DNA solutions were recorded on a Chirascan CD spectrophotometer at 20°C between 210 and 350 nm in a 0.5 cm path-length quartz cuvette. Spectra were an average of 4 scans at 2 nm s−1 and were corrected for background contribution.
Steady-state fluorescence
Fluorescence spectra of 1 μM DNA solutions were recorded in 1 cm path-length quartz cuvettes on a Cary Eclipse spectrophotometer between 370 and 600 nm with an excitation wavelength of 360 nm (excitation and emission slits set to 5 nm) and an integration time of 0.1 s. Spectra were corrected for variations in detector sensitivity with wavelength. Steady-state FRET-efficiencies were calculated according to the following equation:
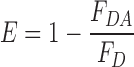 |
where FDA is the integrated fluorescence intensity of the DNA-sequence containing the donor tCO and the acceptor tCnitro and FD the integrated fluorescence intensity of the sequence containing the donor only.
Time-resolved fluorescence
Time-resolved fluorescence decays of 1 μM DNA solutions were measured using TCSPC (time-correlated single photon counting). The excitation source was a 377 nm laser diode pulsed at 10 MHz. The fluorescence emission was filtered by a monochromator with a resolution of 10 nm and detected by a microchannel plate photomultiplier Hamamatsu R3809U-50. The counts were fed into a multichannel analyzer with 2048 channels (Life-spec, Edinburgh Analytical Instruments) where a maximum of 10 000 counts were recorded in the top channel. All fluorescence decays were recorded in a time window of 100 ns. The data were convoluted with the instrument response function and fitted to mono- or bi-exponential functions using Fluofit Pro v.4 (PicoQuant GmbH). The average lifetimes were amplitude-weighed. Time-resolved FRET efficiencies were calculated according to the following equation:
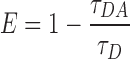 |
where τDA is the average lifetime of the system containing both donor and acceptor and τD the average lifetime of the corresponding donor only system. The values reported here are the average of two independent measurements.
Kinetics measurements
Kinetics of the B- to Z-DNA transition were measured by following the evolution of the CD signal at 285 nm or of the fluorescence at 460 nm immediately after the addition of B-DNA to the high salt buffer over a period of 900 to 2300 s. Given the considerable time scale of the transition, the time necessary to add the DNA into the buffer and to start the measurement (<5 s) was considered negligible. For CD measurements, an integration time of 100 μs/point was used and fluorescence parameters are the same as in the steady-state measurements.
RESULTS AND DISCUSSION
To evaluate the feasibility of using the tricyclic cytosine FRET-pair, we first calculated theoretical values of FRET-efficiencies for different base-pair separations and compared them to the corresponding, previously known values in B-DNA. Such calculations also provide us with reference values that can be compared with FRET experiments at high and low salt on the two DNA sequences, the hairpin and B-Z DNA junction (Figure 1). In this way the DNA conformations can be experimentally determined with steady-state and time-resolved fluorescence spectroscopy using the FRET pair and with CD as a reference method.
Theoretical estimates of FRET-values
Theoretical FRET-efficiencies of our tCO/tCnitro FRET-pair inside a Z-DNA were calculated using an available crystal structure of a (GC)6 duplex DNA in the Z-form (33). We used it to model the three different base-pair separations (4, 6 and 8) that we have in our (GC)7 hairpin. The transition from B- to Z-DNA induces a change in the distance between the donor and the acceptor since the rise per base pair is higher in Z-DNA than in the B-form (3.7 and 3.4 Å, respectively) (32) but more importantly, Figure 2 shows how the relative orientation of the tCO and the tCnitro chromophores is drastically altered due to the change from right to left-handed helicity. Whereas in most freely rotating FRET-pairs the orientation factor κ2 is assumed to be 2/3, it plays a very important role in the case of this internal FRET pair and, as a result, a distinct change in the FRET efficiencies is expected for each position upon transition from B- to Z-DNA (Table 1). As can be seen in Table 1, the high FRET anticipated for the four base-pair separation in B-form is not expected to change significantly due to the very short distance. On the other hand, both the six and eight base-pair configurations are expected to show large variations in FRET-efficiencies, more than tripling (from 13 to 50%) in the former case and decreasing 7-fold (from 42 to 6%) in the latter case (see Table 1).
Table 1. Theoretical κ2 and FRET-efficiencies (E) values.
| Bp separation | B-DNA | Z-DNA | ||||
|---|---|---|---|---|---|---|
| Distance (Å) | κ 2 | E | Distance (Å) | κ 2 | E | |
| 4 | 17 | 0.65 | 0.92 | 18.5 | 0.93 | 0.94 |
| 6 | 23.8 | 0.093 | 0.13 | 25.9 | 0.44 | 0.50 |
| 8 | 30.6 | 1 | 0.42 | 33.3 | 0.12 | 0.059 |
Monitoring right- and left-handed DNA-conformations using the tCO/tCnitro FRET-pair
(GC)7 hairpin sequence
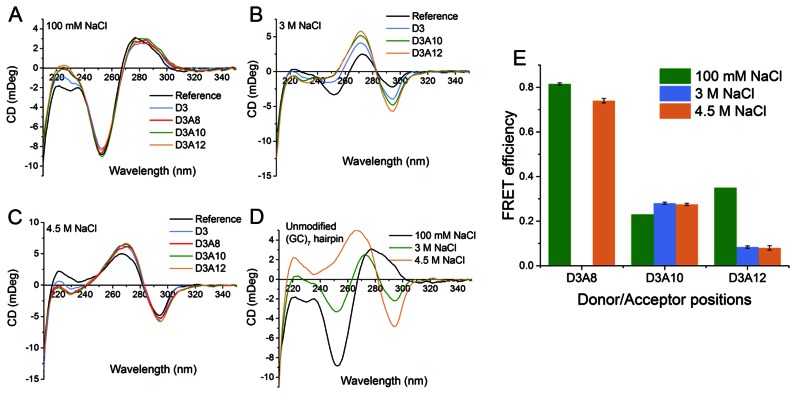
The hairpin sequence (Figure 1) built up by GpC repeats, with the exception of the 4 thymine loop, is expected to undergo a total conversion from B- to Z-conformation at high salt concentration. To study the effect of salt on the transition, two different high sodium chloride concentrations (3 and 4.5 M NaCl) were investigated and compared to the same sample at 100 mM NaCl. Since theoretical calculations suggested that the change in FRET-efficiency for the hairpin having the tCO/tCnitro FRET-pair separated by only four base-pairs will be negligible (Table 1), this sequence (D3A8; the DX and AY notation of sequences used throughout this study reflect the position of the donor, D, and the acceptor, A, in the sequence; the position numbering is indicated in Figure 1) was only studied at 4.5 M NaCl. As can be seen in Figure 3A the CD spectra at 100 mM Na+ show the characteristic features of B-DNA. A few minor differences in intensity were observed between the unmodified reference sequence and the corresponding modified ones, which most likely originate from the difference in absorptive properties between the native cytosines and the tricyclic cytosines that replace them. At 4.5 M NaCl, the CD spectra are essentially inverted (Figure 3C), with a negative peak at 295 nm and a positive one at 255 nm, which is characteristic of Z-DNA. Also at this salt condition the CD spectra are virtually identical for the native and the corresponding modified oligonucleotides. Hence the data strongly suggest that the hairpin sequence undergoes the expected B- to Z-DNA transition, whether the sequence contains our modified tricyclic cytosines or not. At 3 M NaCl, the CD spectra reveal incomplete transition and indicate different B-DNA:Z-DNA ratios for the unmodified and modified sequences (Figure 3B). Quantification of these ratios can be achieved using the CD spectra (see Table S2 in Supplementary Material) and shows that the reference sequence undergoes 57% transition to the Z-form compared to 73% for the D3, donor only, sequence and about 85% for the D3A10 and D3A12 sequences containing both donor tCO and acceptor tCnitro at 6 and 8 base-pairs separation configurations, respectively. A salt titration from 2 to 4.9 M NaCl was also performed on the reference and D3A10 sequences (see Figure S1 in the Supplementary Material), which shows that the transition is triggered at slightly lower salt concentration in the presence of the modified cytosines but that at 4.5 M NaCl they adopt the same Z-DNA conformation.
Figure 3.
CD spectra and FRET efficiencies calculated from time-resolved emission measurements of the (GC)7 hairpin sequence at different salt concentrations. CD spectra of modified (GC)7 hairpins at (A) 100 mM, (B) 3 M and (C) 4.5 M NaCl, the reference hairpin being the corresponding unmodified sequence with only natural nucleobases. To facilitate comparison, the CD spectra of the reference structures at the three salt conditions are displayed together in panel (D). (E) FRET efficiencies calculated from time-resolved measurements of the (GC)7 hairpin structures at the three salt conditions. Measurements performed at room temperature in pH 7.5 buffer (25 mM PBS).
The intrinsic fluorescence of tCO increases upon B- to Z-DNA transition and this is accompanied by a change of fluorescence lifetime from 3.36 ns in the B-form to 4.68 ns in the Z-conformation (for spectra and lifetimes values see Figure S3 and Table S3 in Supplementary Material). These two parameters can readily be used to probe the B- to Z-DNA transition without resorting to FRET-measurements and calculations but as we will discuss below, FRET presents many advantages. Recently, a fluorescent guanosine analogue was reported that, in a similar fashion, was able to monitor B- to Z-DNA transitions with a fluorescence intensity increase (34). A similar enhancement of the fluorescence in Z-DNA has previously also been observed for 2-AP and was attributed to the disruption of the continuous base stacking of B-DNA, as the Z-DNA architecture forms a discreet 4-base stack (35,36). This increase in emission, thus, appears to be an intrinsic property of Z-DNA regardless of the fluorophore. FRET experiments have significant advantages compared to intensity-based measurements since environmental (solvents, cosolutes, oxygen) and instrumental factors (lamp fluctuations, detector sensitivity) influencing fluorescence intensity, in particular when working in complex samples and media, can be handled more reliably. In FRET experiments the fluorescence of the donor alone is measured as a reference in exactly the same conditions as those of the donor/acceptor sample. This means that the additional donor intensity change upon addition of the acceptor is a change strictly associated with the FRET mechanism and not to other external factors. Importantly, the change in fluorescence associated with a conformational change as a result of energy transfer can be predicted through Förster theory, as has been done here for the hairpin structure (Table 1), giving a good indication of what change of emission to expect when performing the actual FRET-experiment. The advantage of FRET is particularly large when using time-resolved emission, since measurements of fluorescence lifetimes are independent of concentration and therefore also avoid such experimental errors that can accompany intensity-based studies.
The time-resolved FRET efficiencies measured here were calculated with high reproducibility in duplicate from the measured average lifetime values and are shown in Figure 3E and reported in Table 2 (see Table S3 in Supplementary Material for fluorescence decay fittings and lifetime values). FRET-efficiencies calculated from steady-state emission spectra are virtually similar to those from the time-resolved measurements but experimental errors are higher (Figure S4 and Table S1 in Supplementary Material). As can be seen in Figure 3E we find a distinct change in FRET-efficiency for all donor/acceptor configurations. For the 8 base-pair separation sample this change is most pronounced, with a significant drop from 35% in the B-form to 8% in the Z-form while the 6 base pair separation sample gives a smaller but still significant change from 23% to 28% going from B- to Z-DNA (Table 2). The general trend and the values correlate well with the theoretical calculations (Table 1), except for the four base-pair separation sequence (sample D3A8), where an unexpected change in FRET efficiency is observed. It is possible that, due to the short distance between the donor and acceptor in this configuration, the energy transfer occurs through several different pathways and thus will not strictly obey the Förster theory. The larger variation in the theoretical estimates of the FRET-change in the 6 base-pair separation case compared to experiment could be an effect of the very high responsiveness to small structural variations around such base-pair separations (25).
Table 2. Time-resolved FRET efficiencies of the (GC)7 hairpin sequences at 100 mM, 3 M and 4.5 M NaCl. Values are the average of two independent measurements.
| Sequence | 100 mM NaCl | 3 M NaCl | 4.5 M NaCl |
|---|---|---|---|
| D3A8 | 0.82 ± 0.01 | n.d. | 0.74 ± 0.01 |
| D3A10 | 0.23 ± 0.01 | 0.28 ± 0.01 | 0.28 ± 0.01 |
| D3A12 | 0.35 ± 0.01 | 0.084 ± 0.005 | 0.080 ± 0.010 |
As expected from the calculations the transition from B- to Z-DNA results in significant changes in the FRET-efficiencies of the tCO/tCnitro FRET pair. Taking into account only the distance change and an average κ2 of 2/3, a decrease in the FRET efficiency from 71 to 60% for the 6 base-pair separation and from 35 to 25% for the 8 base-pair separation would be expected. With our FRET-pair in this system, the orientation dependence of the FRET results in larger variations with an increase or decrease of the FRET efficiencies. This results in a specific FRET pattern, finger-print, allowing the unambiguous identification of Z-DNA. A minor stabilizing effect of the Z-form of DNA by the tC-bases was observed but it does not interfere with the measurements when the salt effect is strong enough to induce complete conversion to the Z-form (4.5 M NaCl). However, at 3 M NaCl where the unmodified sequence is present in a mixture of B- and Z-form DNA, the additional stabilization of the tC-bases in the modified oligonucleotides is clearly visible (Figure 3B). The FRET-efficiency results at 3 M NaCl are essentially the same as those at 4.5 M NaCl but should be considered with a slight caution since the CD-spectra and our calculations in the Supplementary Materials mentioned above (Supplementary Table S2) indicate that the DNA sequences containing the donor and those containing both donor and acceptor do not have the same distribution of DNA-forms (more B-form in the donor-only sequence D3 compared to the donor–acceptor systems D3A10 and D3A12, vide supra).
B-Z DNA junction sequence
Next, we carried out similar experiment for the duplexes susceptible of forming a B-Z DNA junction, in their unmethylated (BZ), hemimethylated (hMeBZ) and methylated (MeBZ) forms. The study of B-Z DNA junctions is of high importance to understand the biological role of Z-DNA and its transition mechanism, since such junctions are required for Z-DNA (or Z-RNA) to form in vivo amidst the widely predominant B-DNA (or A-RNA) (17).
The sequence is comprised of a methylated or hemimethylated (GC)5 repeat segment susceptible of forming Z-DNA and of an 8 bp segment expected to remain in the B-form even at high salt conditions (Figure 1). The B-DNA part of the sequence was selected randomly, except for the three AT base pairs which have been proven to be necessary for the formation of B-Z junctions as they are weaker and, thus, can induce the flexibility necessary for the DNA sequence to switch from a right- to a left-handed helix (15–17). The transition is less prone to occur in such a B-Z DNA junction sequence than in the GC-hairpins discussed in the previous paragraph and methylation was necessary to induce the B to Z-DNA transition (Figure 4D). The short distance sample, D5A10, was only studied in its methylated form since, for similar reason as in the hairpin structure, at this base-pair separation it is not expected to give useful data in the FRET measurements.
Figure 4.
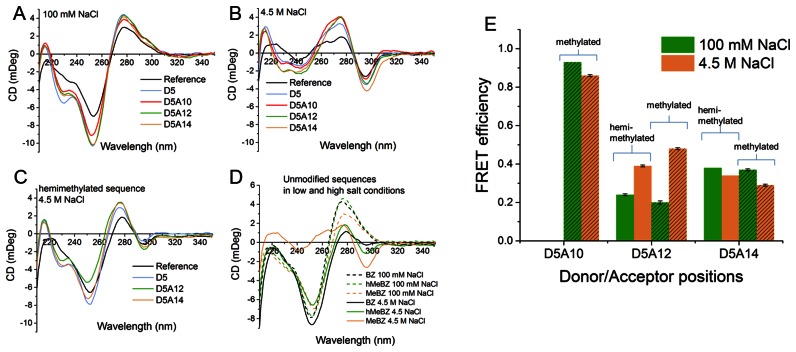
CD spectra and time-resolved FRET efficiencies for the methylated and hemimethylated B-Z DNA junction sequences at low and high salt concentrations. CD spectra of the methylated B-Z DNA junction sequences (MeBZ) in (A) 100 mM and (B) 4.5 M NaCl. (C) CD spectra of the hemimethylated B-Z DNA junction sequences (hMeBZ) at 4.5 M NaCl. (D) CD spectra of the reference methylated (MeBZ), hemimethylated (hMeBZ) and unmethylated (BZ) B-Z DNA junction sequences at 100 mM and 4.5 M NaCl. (E) Time-resolved FRET efficiencies of the methylated (patterned bars) and hemimethylated duplexes at 100 mM and 4.5 M NaCl. The reference sample in panels A to C is the sequence containing no tricyclic cytosine molecules. Measurements performed at room temperature in pH 7.5 buffer (25 mM PBS).
The methylated sequences display typical B-DNA CD spectra at 100 mM NaCl (Figure 4A) whereas at high salt concentration a negative peak at 295 nm characteristic of Z-DNA is formed (Figure 4B). However, the ellipticity at 255 nm remains negative at 4.5 M NaCl which, unlike in the Z-hairpin structure, suggests the expected formation of a mixture of B- and Z-DNA with a B-Z DNA junction. The differences between the CD-spectra of the reference and the modified sequences at both low and high salts are larger than in the case of the hairpin sequence (Figure 3A, B). Also here the variations most likely originate from differences in absorption between normal C and the tricyclic cytosines. Additionally, unlike for the hairpin structures, excess single strand can here contribute to the CD and increase the differences. Due to the significant differences in the CD spectra, salt titrations from 2 to 4.9 M NaCl were performed to monitor for differences in DNA conformation. Comparison of the variations of the CD intensities at 295 nm for the reference MeBZ sequence and the MeBZ-D5A12 sequence shows that they undergo the same transition with a transition mid-point around 3 M NaCl and saturation at 4 M NaCl (see Figure S2 in Supplementary Material). These results confirm that despite the differences in intensity, the reference and modified sequences undergo a similar change of conformation and end up with the same BZ junction structure.
The standard CD results are validated by the FRET measurements performed using the tCO/tCnitro pair where a variation of the FRET-efficiencies subsequent to the transition is observed for all sequences (Figure 4E and Table 3). Again, the FRET-efficiencies calculated from steady-state emission spectra are virtually similar to those from the time-resolved measurements but experimental errors are higher (Figure S4 and Table S1 in Supplementary Material). Moreover, similar steady-state FRET results were observed for the methylated sequence D5A12 at a sample amount 333 times lower than for the CD experiments, without changing the measurement conditions (data not shown). This highlights the advantage of fluorescence over CD: a satisfactory CD spectrum requires a significantly higher sample amount (the amount used in the CD-experiments presented in this study is approaching the limit) and a longer acquisition time (in our case we used 12 min for CD versus 3 min for fluorescence).
Table 3. Time-resolved FRET efficiencies with mean errors for the hemimethylated and methylated BZ-junction sequences at two different salt concentrations. Values are average of two independent measurements.
| Sequence | Hemimethylated form (hMeBZ) | Methylated form (MeBZ) | ||
|---|---|---|---|---|
| 100 mM NaCl | 4.5 M NaCl | 100 mM NaCl | 4.5 M NaCl | |
| D5A10 | n.d. | n.d. | 0.93 ± 0.01 | 0.86 ± 0.01 |
| D5A12 | 0.24 ± 0.01 | 0.39 ± 0.01 | 0.20 ± 0.01 | 0.48 ± 0.01 |
| D5A14 | 0.38 ± 0.01 | 0.34 ± 0.01 | 0.37 ± 0.01 | 0.29 ± 0.01 |
It is worth noting that the intrinsic fluorescence intensity and lifetime of tCO remain much more stable during this transition than in the case of the hairpin (for spectra and lifetimes values see Figure S3, Table S4 and S5 in Supplementary Material). These two observations constitute further evidence that a B-Z DNA junction and not a pure Z-DNA is formed and that, as expected, tCO is located in the B-DNA segment and its environment, therefore, is kept unchanged. This results in a different relative orientation of tCO and tCnitro, and thus in a different orientation factor and consequently FRET efficiency, in the B-Z DNA junction compared to in the hairpin (vide supra) where the structure is all Z-form. The trend in the FRET efficiency variation is similar to that of the hairpin, with an increase for the 6 bp separation duplex and a decrease for the 8 bp separation duplex. However, in this case the larger variation is observed for the 6 bp separation with a change in efficiency from 20 to 48% and, on the contrary to the Z-DNA hairpin structure, the 8 bp separation undergoes a smaller variation in FRET efficiency from 37 to 29% (Table 3).
As expected, the structures and FRET-efficiencies at 100 mM are virtually identical for the methylated and hemimethylated sequences. However, under high salt conditions the CD-spectra of the hemimethylated sequences show that the transition occurs to a significantly smaller extent than for the fully methylated one (Figure 4C and Table 3). The negative band at 295 nm is indeed less pronounced than for the fully methylated sequence. This emphasizes the well-known ability of 5-methylcytosines to facilitate the B-Z transition. The effect of methylation is clearly shown by comparing the CD-spectra of the unmethylated, hemimethylated and fully methylated unmodified sequences at different salt concentration (Figure 4D). The unmethylated B-Z sequence shows almost no transition to the Z-form and the extent of Z-DNA in the B-Z DNA junction sequence increases with the extent of the methylation of the cytosines (see also Figure S2 in Supplementary Material). The large decrease in intensity visible in the long-wavelength part of the CD spectrum of the unmethylated sequence when increasing the salt concentration (BZ sequence, Figure 4D) has been observed previously and seems to be a consequence of the presence of high salt B-DNA structures (37). Importantly, the absence of significant change in CD signal in the 250 nm band for the same sample is a clear indication that no Z-DNA is formed (37). The need for cytosine methylation and the absence of B- to Z-transition at 3 M NaCl (data not shown) shows that the transition is thermodynamically more disfavored compared to the same transition in the full Z-DNA hairpin due to the necessity of forming a B-Z DNA junction.
At high salt the variation of the efficiencies of the hemimethylated samples follows the same trend as for their methylated counterpart but with a lower magnitude. For instance, it changes from 24% in the B-form to only 39% in the Z-form (instead of 48% for the methylated counterpart) for the 6 base-pair separation sample (Figure 4E), which is consistent with a partial transition to Z-DNA as was also found using CD measurements. Despite the slight stabilizing effect of the tC-bases observed in the hairpin structure, the hemimethylated sequences all seem to display similar amount of Z-DNA regardless of the modifications, although exact quantification is not possible due to the mixture of B- and Z-DNA. Along with the salt titrations performed on the methylated sequences (see Supplementary Material Figure S1 and S2), this demonstrates that the stabilizing effect of the tC bases observed in the case of the hairpin is negligible compared to that of cytosine methylation. This property is necessary if the tC bases should serve as virtually non-perturbing probes for Z-DNA and was not obvious considering that the extended ring system of the tricyclic cytosines represents a much larger structural modification to the cytosine scaffold than methylation. For instance, C8-arylguanine have been shown to drastically stabilize Z-DNA, thus enabling the transition to occur under physiological conditions and a similar effect of the tC-probes would not have been surprising (38,39). The fact that the tCO/tCnitro bases seem to behave essentially as normal cytosines in this respect allows their use to quantitatively measure the extent of Z-DNA in a partially transitioned methylated DNA sequence. This was not the case for the unmethylated GC-hairpin investigated above. For both the hemimethylated D5A12 and D5A14 sequences, the variation of FRET efficiency is about half that of the fully methylated one. Assuming that the transition is complete in the GC-segment for the fully methylated sequence, this means that a mixture of 50% B-DNA and 50% B-Z junction DNA is present in the hemimethylated state. The quantification of Z-DNA in a B-Z junction sequence is not straightforward using CD-spectra (11), and we envision that our FRET-system can be used with an advantage for this purpose.
Kinetics of the B- to Z-DNA transition
After demonstrating the ability of our FRET pair to correctly discriminate between the B- and Z-forms of DNA, we set out to study the kinetics of the B- to Z-DNA transition in the (GC)7 hairpin sequence by following the evolution of the fluorescence signal at the two high salt conditions (4.5 and 3 M NaCl, Figure 5). Despite being thermodynamically more disfavored, the transition in the B-Z DNA junction sequence occurred much faster, possibly due to the shorter GC-repeat segment, and was not possible to measure accurately without using stopped-flow techniques (data not shown). Since following the change in the intrinsic fluorescence of tCO during the transition (vide supra) is enough to obtain kinetics curves, the presence of our FRET acceptor tCnitro is not mandatory here. However, studying the kinetics for the sequences containing our complete FRET pair was also interesting in order to evaluate the effect of the modified nucleobases on the B- to Z-DNA transition. Also, the signal variation during the transition is significantly higher in the case of the 8 base-pair separation FRET-sequence than the intrinsic variation of tCO fluorescence intensity, thus affording more reliable experimental data.
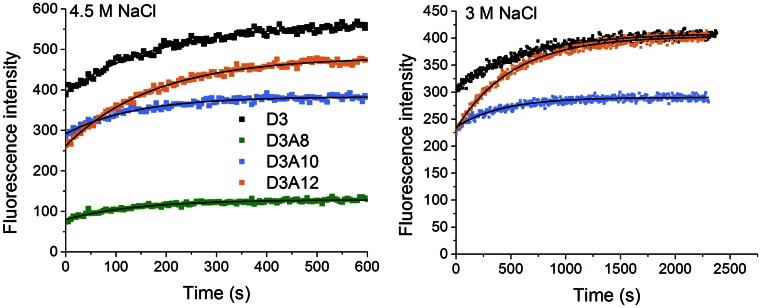
Figure 5.
Kinetics of B- to Z-DNA transition of (GC)7 hairpin structure. Measurements were performed at 4.5 M NaCl (left) and 3 M NaCl (right) and 20°C by monitoring the fluorescence intensity at 460 nm (data points). Lines are mono-exponential fits of the experimental data.
In all cases a significant variation in fluorescence was observed due to the change in FRET efficiency or as an effect of the change of the intrinsic fluorescence of tCO (D3). The kinetics curves could be well fitted to a mono-exponential function (Figure 5), suggesting first-order kinetics for the B- to Z-transition of the hairpin. The fits yielded rate constants, k, between 5 × 10−3 and 8 × 10−3 s−1 (Table 4) with a good reproducibility (5.1% error on average for the four sequences). The values we find are about one order of magnitude lower than the transition rate constants that have been measured by single molecule fluorescence means for a B-Z DNA junction sequence with a (GC)6 segment in the presence of the Zα binding domain (14). Even though strict comparison is impossible since the rate of transition will depend on the length of the GC-segment and on the conditions used to induce the transition, the values found herein are reasonable when comparing to the previously obtained results.
Table 4. Rate constants of the B- to Z-DNA transition of the (GC)7 hairpin structure calculated by fitting the fluorescence kinetics curves to a mono-exponential function. The rate constants are an average of two independent measurements.
| Sequence | k (10−3 s−1) | |
|---|---|---|
| 4.5 M NaCl | 3 M NaCl | |
| D3 | 5.3 ± 0.1 | 1.7 ± 0.1 |
| D3A8 | 6.9 ± 0.1 | -a |
| D3A10 | 7.8 ± 0.5 | 2.6 ± 0.2 |
| D3A12 | 6.3 ± 0.6 | 2.1 ± 0.2 |
aAs was the case for time-resolved fluorescence measurements, sequence D3A8 was only studied at 4.5 M NaCl (vide supra).
We compared our fluorescence kinetics measurements to investigations made by CD which currently is the most common method to follow the B- to Z-DNA transition kinetics (10). This also allowed us to measure the kinetics of the unmodified DNA and compare it to our tC-modified oligonucleotides (see Figure S5 in Supplementary Materials for CD kinetics curves).
The kinetics curves obtained using CD could also be fitted to a mono-exponential function (Table 5). At 4.5 M NaCl, the rate constants are all in good agreement with those measured using fluorescence. Moreover, the rate constant of the unmodified hairpin sequence measured by CD is the same as that of the sequence containing only tCO (D3). At 3 M only the unmodified sequence was measured as a reference (Table 5) and once again its rate constant is essentially the same as that of the D3 sample measured using fluorescence (Table 4). The sequences containing both tCO and tCnitro display slightly higher rate constants but they remain in the same order of magnitude showing that the incorporation of the tricyclic cytosine probes has only a slight impact on the kinetics of the transition. Importantly, we find that our fluorescence-based technique gives rate constants that well match the ones obtained using the more conventional CD-method on non-modified DNA, making it an excellent new tool to follow the kinetics of the B- to Z-DNA transition in real-time at low concentrations.
Table 5. Rate constants of the B- to Z-DNA transition of the (GC)7 hairpin structure calculated by fitting the CD kinetics curves to a mono-exponential function. The rate constants are an average of two independent measurements.
| Sequence | k (10−3 s−1) | |
|---|---|---|
| 4.5 M NaCl | 3 M NaCl | |
| Unmodified hairpin | 5.75a | 2.2a |
| D3 | 5.75a | - |
| D3A8 | 7.8a | - |
| D3A10 | 7.55a | - |
| D3A12 | 5.6a | - |
aErrors are <10%.
CONCLUSIONS
Fluorescence spectroscopy is one of the most sensitive technique routinely available in the research laboratory today, and our new reliable FRET-based method to study B- to Z-DNA transitions and the formation of B-Z DNA junctions is a valuable and straightforward tool to gain insight into the mechanism and kinetics of the transition. Variation in the fluorescence intensity and lifetime of tCO can be used to qualitatively probe the B- to Z-DNA transition but the identification of Z-DNA using FRET is more reliable due to the distinct characteristic responses for each donor/acceptor configuration. Moreover, the variation of the FRET efficiencies can be used to quantify the extent of the B- to Z-DNA transition in the methylated B-Z DNA junction structure or to follow the kinetics of the transition. Overall, and despite a minor stabilizing effect that has been shown to be negligible compared to that of 5-methylcytosine, the tCO/tCnitro FRET pair is a reliable fluorescent tool to monitor Z-DNA with little influence on the thermodynamics and the kinetics of the transition and, thus, represents a valuable alternative to the less sensitive and less versatile CD measurements.
Among the available fluorescence methods to monitor Z-DNA, the non-perturbing tCO/tCnitro FRET pair presents the advantage of being more quantitative than simple fluorescence intensity-based assay and the tricyclic cytosine molecules can be positioned at different position inside the DNA sequence, for instance inside the Z-DNA segment or across a B-Z DNA junction. Our straightforward FRET-based method is also more easily implemented than single-molecule experiments, although the latter can in certain cases give deeper insights compared to ensemble measurements. A recent report on the use of modified nucleobases in single-molecule fluorescence show that increasingly sensitive instrumentation and state-of-the-art methods may render the use of the tricyclic cytosine molecules in single-molecules experiments possible in the near future (40). This would make the tC nucleobases even more valuable for B- to Z-DNA transition mechanism investigation, since this would enable measuring single-molecule fluorescence and real-time dynamics of a probe positioned directly inside the Z-DNA segment and/or across a B-Z DNA junction.
Supplementary Material
FUNDING
Swedish Research Council [2013–4375]; Olle Engkvist Byggmästare Foundation. Funding for open access charge: The Swedish Research Council (VR) [2013-4375].
Conflict of interest statement. None declared.
REFERENCES
- 1.Choi J., Majima T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011;40:5893–5909. doi: 10.1039/c1cs15153c. [DOI] [PubMed] [Google Scholar]
- 2.Belotserkovskii B.P., Mirkin S.M., Hanawalt P.C. DNA sequences that interfere with transcription: Implications for genome function and stability. Chem. Rev. 2013;113:8620–8637. doi: 10.1021/cr400078y. [DOI] [PubMed] [Google Scholar]
- 3.Doluca O., Withers J.M., Filichev V. V. Molecular engineering of guanine-rich sequences: Z-DNA, DNA triplexes, and G-quadruplexes. Chem. Rev. 2013;113:3044–3083. doi: 10.1021/cr300225q. [DOI] [PubMed] [Google Scholar]
- 4.Bacolla A., Wells R.D. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 5.Wang A., Quigley G., Kolpak F., Crawford J., van Boom J.H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 6.Rich A., Zhang S. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 7.Liu R., Liu H., Chen X., Kirby M., Brown P.O., Zhao K. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 8.Pohl F.M., Jovin T.M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC) J. Mol. Biol. 1972;67:375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- 9.Lee M., Kim S.H., Hong S.-C. Minute negative superhelicity is sufficient to induce the B-Z transition in the presence of low tension. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4985–4990. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González V.M., Fuertes M. A., Pérez J.M., Alonso C. Kinetics of the salt-induced B- to Z-DNA transition. Eur. Biophys. J. 1998;27:417–423. [Google Scholar]
- 11.Bae S., Son H., Kim Y.-G., Hohng S. Z-DNA stabilization is dominated by the Hofmeister effect. Phys. Chem. Chem. Phys. 2013;15:15829–15832. doi: 10.1039/c3cp52047a. [DOI] [PubMed] [Google Scholar]
- 12.Kim Y.G., Lowenhaupt K., Maas S., Herbert A, Schwartz T., Rich A. The zab domain of the human RNA editing enzyme ADAR1 recognizes Z-DNA when surrounded by B-DNA. J. Biol. Chem. 2000;275:26828–26833. doi: 10.1074/jbc.M003477200. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y.-M., Kim H.-E., Lee E.-H., Seo Y.-J., Lee A.-R., Lee J.-H. NMR investigation on the DNA binding and B-Z transition pathway of the Zα domain of human ADAR1. Biophys. Chem. 2013;172:18–25. doi: 10.1016/j.bpc.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Bae S., Kim D., Kim K.K., Kim Y.G., Hohng S. Intrinsic Z-DNA is stabilized by the conformational selection mechanism of Z-DNA-binding proteins. J. Am. Chem. Soc. 2011;133:668–671. doi: 10.1021/ja107498y. [DOI] [PubMed] [Google Scholar]
- 15.Bothe J.R., Lowenhaupt K., Al-Hashimi H.M. Sequence-specific B-DNA flexibility modulates Z-DNA formation. J. Am. Chem. Soc. 2011;133:2016–2018. doi: 10.1021/ja1073068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ha S.C., Lowenhaupt K., Rich A., Kim Y.-G., Kim K.K. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 17.Kim D., Reddy S., Kim D.Y., Rich A., Lee S., Kim K.K., Kim Y.-G. Base extrusion is found at helical junctions between right- and left-handed forms of DNA and RNA. Nucleic Acids Res. 2009;37:4353–4359. doi: 10.1093/nar/gkp364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bothe J.R., Lowenhaupt K., Al-Hashimi H.M. Incorporation of CC steps into Z-DNA: interplay between B-Z junction and Z-DNA helical formation. Biochemistry. 2012;51:6871–6879. doi: 10.1021/bi300785b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Knop R., Cohen J. Adriamycin Inhibits the B to Z Transition of Poly(dGm5dC).Poly(dGm5dC) Biochemistry. 1983;22:5468–5471. [Google Scholar]
- 20.Sheardy R.D., Levine N., Marotta S., Suh D., Chaires J.B. A thermodynamic investigation of the melting of B-Z junction forming DNA oligomers. Biochemistry. 1994;33:1385–1391. doi: 10.1021/bi00172a014. [DOI] [PubMed] [Google Scholar]
- 21.Sheardy R.D., Suh D., Kurzinsky R., Doktycz M.J., Benight A.S., Chaires J.B. Sequence dependence of the free energy of B-Z junction formation in deoxyoligonucleotides. J. Mol. Biol. 1993;231:475–488. doi: 10.1006/jmbi.1993.1295. [DOI] [PubMed] [Google Scholar]
- 22.Sheardy R.D., Winkle S.A. Temperature-dependent CD and NMR studies on a synthetic oligonucleotide containing a B-Z junction at high salt. Biochemistry. 1989;28:720–725. doi: 10.1021/bi00428a046. [DOI] [PubMed] [Google Scholar]
- 23.Ward D.C., Reich E., Stryer L. Fluorescence studies of nucleotides and polynucleotides: I. formycin, 2-aminopurine riboside, 2, 6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 24.Wilhelmsson L.M. Fluorescent nucleic acid base analogues. Q. Rev. Biophys. 2010;43:159–183. doi: 10.1017/S0033583510000090. [DOI] [PubMed] [Google Scholar]
- 25.Börjesson K., Preus S., El-Sagheer A.H., Brown T., Albinsson B., Wilhelmsson L.M. Nucleic acid base analog FRET-pair facilitating detailed structural measurements in nucleic acid containing systems. J. Am. Chem. Soc. 2009;131:4288–4293. doi: 10.1021/ja806944w. [DOI] [PubMed] [Google Scholar]
- 26.Preus S., Börjesson K., Kilså K., Albinsson B., Wilhelmsson L.M. Characterization of nucleobase analogue FRET acceptor tCnitro. J. Phys. Chem. B. 2010;114:1050–1056. doi: 10.1021/jp909471b. [DOI] [PubMed] [Google Scholar]
- 27.Sandin P., Börjesson K., Li H., Mårtensson J., Brown T., Wilhelmsson L.M., Albinsson B. Characterization and use of an unprecedentedly bright and structurally non-perturbing fluorescent DNA base analogue. Nucleic Acids Res. 2008;36:157–167. doi: 10.1093/nar/gkm1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preus S., Kilså K., Miannay F.-A., Albinsson B., Wilhelmsson L.M. FRETmatrix: a general methodology for the simulation and analysis of FRET in nucleic acids. Nucleic Acids Res. 2013;41:e18. doi: 10.1093/nar/gks856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iqbal A., Arslan S., Okumus B., Wilson T.J., Giraud G., Norman D.G., Ha T., Lilley D.M.J. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 2008;105:11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettersen E.F., Goddard T.D., Huang C.C., Cough G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.MarvinSketch version 15.3.16. ChemAxon. 2015. http://www.chemaxon.com http://www.chemaxon.com
- 32.Fuertes M.A., Cepeda V., Alonso C., Pérez J.M. Molecular mechanisms for the B-Z transition in the example of poly[d(G-C) x d(G-C)] polymers. A critical review. Chem. Rev. 2006;106:2045–2064. doi: 10.1021/cr050243f. [DOI] [PubMed] [Google Scholar]
- 33.De Rosa M., Zacarias S., Athanasiadis A., Grande R.Q. Structural basis for Z-DNA binding and stabilization by the zebrafish Z-DNA dependent protein kinase PKZ. Nucleic Acids Res. 2013;41:9924–9933. doi: 10.1093/nar/gkt743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park S., Otomo H., Zheng L., Sugiyama H. Highly emissive deoxyguanosine analogue capable of direct visualization of B-Z transition. Chem. Commun. 2014;50:1573–1575. doi: 10.1039/c3cc48297a. [DOI] [PubMed] [Google Scholar]
- 35.Tashiro R., Sugiyama H. A nanothermometer based on the different π stackings of B- and Z-DNA. Angew. Chemie Int. Ed. 2003;42:6018–6020. doi: 10.1002/anie.200352752. [DOI] [PubMed] [Google Scholar]
- 36.Tashiro R., Sugiyama H. Biomolecule-based switching devices that respond inversely to thermal stimuli. J. Am. Chem. Soc. 2005;127:2094–2097. doi: 10.1021/ja044138j. [DOI] [PubMed] [Google Scholar]
- 37.Kypr J., Kejnovská I., Renciuk D., Vorlícková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vongsutilers V., Phillips D.J., Train B.C., McKelvey G.R., Thomsen N.M., Shaughnessy K.H., Lewis J.P., Gannett P.M. The conformational effect of para-substituted C8-arylguanine adducts on the B/Z-DNA equilibrium. Biophys. Chem. 2011;154:41–48. doi: 10.1016/j.bpc.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Train B.C., Bilgesü S.A., Despeaux E.C., Vongsutilers V., Gannett P.M. Single C8-arylguanine modifications render oligonucleotides in the Z-DNA conformation under physiological conditions. Chem. Res. Toxicol. 2014;27:1176–1186. doi: 10.1021/tx5000798. [DOI] [PubMed] [Google Scholar]
- 40.Alemán E.A., de Silva C., Patrick E.M., Musier-Forsyth K., Rueda D. Single-molecule fluorescence using nucleotide analogs: a proof-of-principle. J. Phys. Chem. Lett. 2014;5:777–781. doi: 10.1021/jz4025832. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.