Abstract
Ectopic expression of the double homeodomain transcription factor DUX4 causes facioscapulohumeral muscular dystrophy (FSHD). Mechanisms of action of DUX4 are currently unknown. Using immortalized human myoblasts with a titratable DUX4 transgene, we identify by mass spectrometry an interaction between the DUX4 C-terminus and the histone acetyltransferases p300/CBP. Chromatin immunoprecipitation shows that DUX4 recruits p300 to its target gene, ZSCAN4, displaces histone H3 from the center of its binding site, and induces H3K27Ac in its vicinity, but C-terminal deleted DUX4 does not. We show that a DUX4 minigene, bearing only the homeodomains and C-terminus, is transcriptionally functional and cytotoxic, and that overexpression of a nuclear targeted C-terminus impairs the ability of WT DUX4 to interact with p300 and to regulate target genes. Genomic profiling of DUX4, histone H3, and H3 modifications reveals that DUX4 binds two classes of loci: DNase accessible H3K27Ac-rich chromatin and inaccessible H3K27Ac-depleted MaLR-enriched chromatin. At this latter class, it acts as a pioneer factor, recruiting H3K27 acetyltransferase activity and opening the locus for transcription. In concert with local increased H3K27Ac, the strong H3K27Ac peaks at distant sites are significantly depleted of H3K27Ac, thus DUX4 uses its C-terminus to induce a global reorganization of H3K27 acetylation.
INTRODUCTION
Facioscapulohumeral muscular dystrophy (FSHD) is a devastating muscular dystrophy that affects more than 25 000 people in the United States. The disease is caused by altered chromatin on the D4Z4 macrosatellite repeats at 4q35.2 (1–4), which leads to transcription from these repeats. The altered chromatin is typically caused by unit array contractions reducing array size to <11 repeats (5,6), but in rare cases is caused by mutation of the SMCHD1 gene (7). The D4Z4 transcript encodes a double-homeodomain transcription factor, DUX4 (8,9). Although difficult to detect in human cells and tissue samples (10–12), DUX4 is nevertheless thought to be responsible for the pathology observed in FSHD, in part because FSHD only occurs when D4Z4 contractions occur on alleles that provide the DUX4-encoding RNA with a functional polyA signal (13), and in part because DUX4 has pathological effects on myogenic cells, inducing oxidative cell death at high levels of expression, and inhibiting MyoD and impairing myogenesis at low levels of expression (14).
The DUX4 protein has two conserved domains: the DNA-binding tandem homeodomains at the N-terminus of the protein which bind a sequence related to TAATCTAATCA (15,16), and the less conserved C-terminal domain (17) which confers transcriptional activation potential on Gal4 (18). Variants of DUX4 lacking the C-terminus are no longer cytotoxic (19), and lose the ability to upregulate DUX4 target genes (15), further suggesting that the C-terminus is a transcriptional activation domain. However, the mechanism of transcriptional activation by DUX4 is unknown.
The C-terminal domain contains acidic amino acid residues that are commonly found in other transactivation domains. However, apart from this, the sequence of DUX4 is not conserved with other known transactivators. Therefore, it is unclear whether DUX4 acts as a conventional transcription factor, recruiting co-activators or co-repressors to binding sites to alter chromatin structure, thereby regulating the activity of RNA pol II (20). DUX4 protein complexes or cofactors have not been described. In this study, we identify p300 and CBP, related co-activators with histone acetyltransferase activity (21), as factors that interact with DUX4 and are recruited to target genes at sites of DUX4 binding. We find H3K27 acetylation peaks flanking genomic DUX4 target loci within 6 hours of DUX4 expression, map the p300 interaction to the C-terminus of DUX4 and investigate interfering with this interaction as a way of inhibiting the transcriptional activation potential of DUX4.
MATERIALS AND METHODS
Plasmids
FUGW-rtTA was generated by ligating PCR amplified rtTA2(s)-m2-ires-GFP into BamH1/EcoR1 FUGW (22). The Dox-inducible DUX4 lentivector (pSam2-iDUX4-Flag-UBC-puro) was generated as follows: The SV40 ploy A signal was amplified from p2lox plasmid (23) and cloned into the Not1 site of pSAM2 (24), which contains a second generation tet-response element. The Pac1 to BsrG1 sequence of the plasmid was then replaced with a Pac1/BsrG1 fragment containing the Ubiquitin C promoter and EGFP from FUGW (22). Using In-Fusion HD cloning (Clontech), GFP was then replaced with a PCR fragment containing the puromycin resistance gene (N-acetyl-transferase, PAC) to generate pSam2-Ubc-Puro. An EcoRI/NotI PCR fragment encoding DUX4-flag was ligated into EcoR1/Not1 sites of pSam2-Ubc-puro plasmid to generate pSam2-iDUX4-Flag-Ubc-Puro. The dox-inducible C-terminally deleted DUX4 construct was constructed by replacing the EcoRI/NotI fragment with a PCR fragment encoding DUX4ΔC-Flag. The codon optimized DUX4-flag for bacterial expression was synthesized (Biomatik) and ligated into KpnI/NotI digested pMSCG10 (25). NLS-GFP-DUX4 fusion expression constructs were generated by ligating PCR amplified EGFP into NheI/EcoRI digested IRESpuro3 plasmid (Clontech), to generate pIRESpuro3-EGFP. DUX4-F1, DUX4-F2, and DUX4-F3 were generated by In-Fusion cloning (Clontech) and ligated into EcoRI/AgeI digested pIRESpuro3-EGFP. EGFP-NLS was amplified by PCR and replaced EGFP for control plasmid. To generate EGFP-NLS-DUX4-C98, the C-terminus of DUX4 was amplified by PCR and ligated to EcoRI/AgeI digested pIRESpuro3-EGFP. The regulatory elements of the ZSCAN4 gene were amplified by PCR and ligated into Nhe1/Xho1 digested pGL3 basic (Zscan4-luc). Primer sequences are provided in Supplemental Table S1.
Generation of doxycycline-inducible DUX4 cells
The immortalized human myoblast line LHCN-M2 (26), kindly provided by Vincent Mouly, were transduced with two lentivectors, the first (FUGW-rtTA) expressing the reverse tetracycline transactivator rtTA2m2 together with an IRES-EGFP from the human Ubiquitin C promoter. To generate lentiviral supernatants, 293T cells were transfected with vector plasmid, packaging (psPAX2) and envelope (pMD2.G) plasmids. Fourty eight hours post-transfection, media was collected and lentivirus was concentrated using ultracentrifugation, and cells were transduced in the presence of 10 μg/ml polybrene. GFP+ cells were sorted after the first transduction, then transduced with the second vector, which carried flag-tagged DUX4 under the control of the second-generation tetracycline response element (27) with a Puro-resistance gene (Figure 1A). After puromycin treatment, cells were individually sorted into 96-well dishes, and three independent clones were isolated and evaluated.
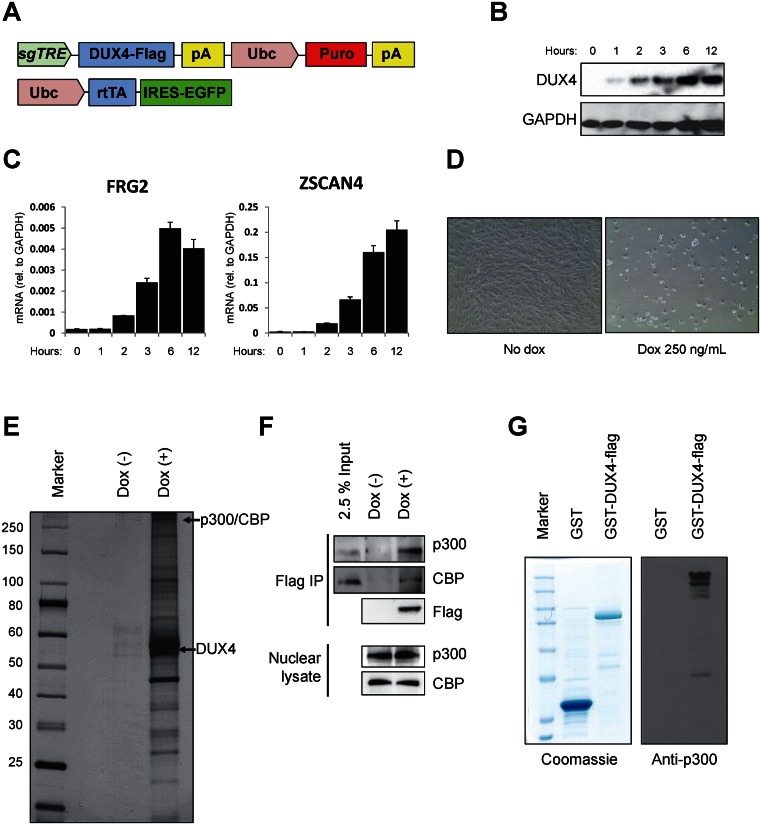
Figure 1.
DUX4 interacts with p300/CBP. (A) Schematic diagram of the lentiviral constructs carrying doxycycline-inducible DUX4 and rtTA. sgTRE: second generation tet-response element for doxycycline-induced expression. (B) Immunoblot showing doxycycline dose-dependent inducible expression of DUX4-flag in LHCN-M2 cells. (C) Time course of induction of DUX4 downstream target genes, FRG2 and ZSCAN4. (D) Cell death of immortalized human LHCN-M2 myoblasts 48 hours after DUX4 induction with 250 ng/ml doxycycline. (E) Silver stained SDS-PAGE gel of DUX4-flag associated proteins, 6 h after 250 ng/ml doxycycline treatment. (F) Immunoblots of DUX4-associated factors, p300 and CBP, in immunoprecipitates with Flag-DUX4. (G) Direct interaction of DUX4 with p300. Left panel: Coomassie staining of GST and GST-DUX4 proteins purified from E. coli. Right panel: immunoblot for recombinant p300 after GST-pull down, showing interaction with GST-DUX4 but not GST alone.
Immunoprecipitation and mass spectrometry
109 LHCN-M2-iDUX4-flag cells were prepared in the absence or presence of doxycycline (500 ng/ml) for 6 h. Cells were washed twice with PBS, harvested, and nuclear extract was prepared as described (Dignam et al., 1983). Briefly, cells were lysed in hypotonic solution, low salt buffer, and high salt buffer and nuclear extract was treated with Benzonase nuclease (Sigma) (van den Berg et al., 2010). Nuclear extract was dialyzed in BC300 and precleared with mouse IgG-agarose beads (Sigma). Pre-cleared nuclear lysate was immunoprecipitated with Anti-flag M2-magnetic beads (Sigma) and extensively washed with BC300 (20 mM Tris, 20% glycerol, 0.2mM EDTA, 300mM KCl) containing 0.1% NP-40. Immunoprecipitates were eluted with flag peptides (Sigma) and 5% of eluted samples were tested by SDS-PAGE and visualized with silver staining (Pierce). The remaining samples were used for mass spectrometry analysis performed at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School. To validate DUX4 interacting proteins, western blot analysis was performed with following antibodies : DUX4 (abcam; ab124699), Flag (Sigma; F1804), Flag-HRP (Sigma; A8592), p300 (Millipore 05-257 and Santa Cruz; sc-584), CBP (Santa Cruz; sc-369), GAPDH-HRP (GenScript; A00192), and GFP (Santa Cruz; sc-8334).
DUX4 protein purification and GST-pull down
Codon optimized GST-TEV-DUX4-Flag in pMCSG10 was expressed in Rosetta2 pLysS strain E. coli. First, C-terminal flag epitope tagged DUX4 was purified using flag peptide elution (Gu and Roeder, 1997). Briefly, 3 h after 0.1 mM IPTG induction, E. coli was lysed in lysis buffer (500 mM NaCl, 20 mM Tris, 10% glycerol, 0.2 mM EDTA). DUX4-Flag was immunoprecipitated with anti-flag M2-magnetic beads (Sigma) and washed extensively with BC300 containing 0.1% NP-40. DUX4-flag was eluted with flag peptide (Sigma) in BC100 (20 mM Tris, 20% glycerol, 0.2 mM EDTA, 100 mM KCl) solution. GST-only control or GST-DUX4-flag was purified again with glutathione magnetic beads (Pierce) and GST or GST-DUX4-flag and a recombinant p300 protein (Millipore) were incubated in NETN buffer (100 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl, pH8.0, and containing 0.5% detergent NP-40) (Kim et al., 2003). After washing, bound proteins were analyzed by immunoblot with anti-p300 antibody.
Cell viability assay
Cell viability assay was performed as described (Bosnakovski et al., 2014). Cells were treated with doxycycline for 48 h and cell viability was measured using the CellTiter-Glo luminescent assay based on quantitation of ATP (Promega). Briefly, the plates were equilibrated at room temperature and media was removed. CellTiter-Glo reagent diluted (1:1) in PBS was added and the plates were read in a Cytation3 plate reader (BioTek).
Chromatin immunoprecipitation and ChIP-qPCR/ChIP-seq analysis
ChIP assays were performed according to previously described protocol with slight modifications (Boyer et al., 2005). The sonicated chromatin was incubated with the indicated antibodies overnight at 4°C followed by incubation with 30 μl of Magna ChIP Protein A+G magnetic beads (Millipore) for 2 h. For ChIP and ChIP-seq experiments, the following antibodies were used: Flag (Sigma; F1804), p300 (Santa Cruz; sc-584), H3 (abcam; ab1791), H3K18Ac (abcam; ab1191), H3K27Ac (abcam; ab4729), and H3K4me3 (abcam; ab8580). Beads were washed with RIPA buffer and TE buffer containing 50 mM NaCl. The immunoprecipitated complexes were eluted from the beads by heating at 65°C and reverse crosslinked by overnight incubation at 65°C. Immunoprecipitated DNA was treated with RNase A and proteinase K. The immunoprecipitated DNA was purified using a PCR purification kit (Qiagen) and quantified by qPCR using SYBR Premix Ex Taq (Clontech) according to the manufacturer's instructions. All primers for ChIP-qPCR are shown in Supplemental Table S1. All reactions were performed in a two-step PCR condition (95°C for 15 s and 60°C for 30 s) using Applied Biosystems 7500 Real-time PCR system (Applied Biosystems) and the quantification cycle (Cq) was determined by default settings. Each PCR reaction was tested with a melting curve analyses to confirm the specificity of the amplification and the absence of primer dimers. All data were converted to relative values based on a standard curve and normalized to 1% input values of the same sample. Primers are listed in Supplemental Table S1.
For ChIP-seq, inputs and immunoprecipitates were used to create libraries as follows: Blunt end repair of ChIPed DNA was performed using End-it DNA repair kit (Epicentre, ER0720) and ‘A’ base was added to 3′ ends using Klenow Fragment (3′→5′ exo-) (NEB, M0212). NEXTflex™ DNA Barcodes (BIOO Scientific, 514103) were ligated to DNA fragments using LigaFast™ Rapid DNA Ligation System (Promega, M8221) and 200–500 base pair DNA was selected using Agencourt AMPpure XP (Beckman Coulter, A63881). 15 cycles of PCR amplification were performed using Phusion High-Fidelity PCR Master Mix with HF Buffer (ThermoFisher Scientific, F531S). Fifty base pairs of DNA sequence were read from a single end on a HiSeq 2000/2500 (Illumina) by the University of Minnesota Genomics Center. Read sequences were trimmed (Trimmomatic 0.32), mapped to the hg19 genome (Bowtie 2) and sorted (Samtools 1.2) at the University of Minnesota Supercomputing Institute. Peaks were identified (MACS 1.4) and analyzed with custom R scripts (28) available at https://github.com/micahgearhart/dux4. DNase-Seq data for LHCN-M2 cells was obtained from the ENCODE project accession ENCFF001BVR. Sequencing data have been deposited in the Gene Expression Omnibus repository under accession code GSE78158.
LHCN-M2 RNA-Seq
Total RNA was harvested from 6 independent cultures of LHCN-M2iDUX4 cells, three of which were treated with 250 ng/ml dox for 6hrs. Libraries were created by the University of Minnesota Genomics Center from the polyA+ mRNAs using the TruSeq RNA library preparation kit. Fifty bp paired-end sequence was obtained and trimmed as above. Paired-end reads were mapped to GRCh37 genome (Star 2.4) and uniquely mapped reads were assigned to genes using the Ensembl version 75 annotation. Differential expression was determined with DESeq2 (29) using custom R scripts available at https://github.com/micahgearhart/dux4.
DUX4 reporter assay
293T cells were transfected using TansIT-LT1 (Mirus) according to manufacturer instructions.
Briefly, 293T cells were transfected with a DUX4 reporter construct, Renilla luciferase ZSCAN4-Luc (firefly luciferase under the control of the regulatory elements of the ZSCAN4 gene) together with pCI-neo empty vector (EV) or pCI-neo-DUX4 in combination with the indicated DUX4 fragments. Cells were lysed 24 h post-transfection in Passive Lysis Buffer (Promega). Luciferase activities were quantified using reagents from the Dual-Luciferase Reporter Assay System (Promega) following manufacturer's instructions. Light emission was measured using on a Cytation3 plate reader (BioTek). The ratio of firefly/Renilla luciferase data was calculated and given as the averages ± SD of at least triplicates.
Gene expression analysis
293T-iDUX4 cells were transiently transfected with control or the indicated DUX4 fragments and 24 h post-transfection cells were treated with doxycycline for 6 h to induce WT DUX4. Immortalized myoblast cells expressing dox-inducible DUX4-ΔC or DUX4 (three independent clones of each) were also treated with doxycycline for 6 h. Total RNA was extracted from around 1 × 106 fresh cells using RNeasy mini kit (Qiagen) and genomic DNA was removed by RNase-free DNase (Qiagen). Reverse transcription (RT) was performed using the Vero cDNA synthesis kit (Thermo Scientific) and quantitative polymerase chain reactions (PCR) were performed using Premix Ex Taq™,ROX Plus (Clontech) and hydrolysis (Taqman) probes (ZSCAN4; Hs00537549_m1, MYF5; Hs00271574_m1, MYOD1; Hs00159528_m1, CCNA1; Hs00171105_m1, GAPDH; Hs99999905_m1) (Applied Biosystems). Reactions were performed as described above for ChIP-qPCR experiments. The data were normalized to the reference gene GAPDH and relative mRNA expression was calculated by the ΔCq method. Gene expression data are given as the averages ± SD of at least triplicates.
RESULTS
Doxycycline-inducible DUX4-expressing human myoblasts
In order to study the activity of DUX4 in human myogenic cells, we engineered an immortalized myoblast line (26) with lentivectors for doxycycline (dox)-inducible expression of flag-tagged DUX4 (Figure 1A). In these cells, DUX4 is expressed within 2 h (Figure 1B), target genes are strongly upregulated by 6 h (Figure 1C), and cells die within 24 h of 250 ng/ml dox (Figure 1D), similar to what has been observed in the murine system (14,30).
DUX4 interacts with p300 and CBP
To identify proteins that interact with DUX4, we immunoprecipitated DUX4 from nuclear extracts prepared from cells exposed to dox (500 ng/ml) for 6 hours or control cells not exposed to dox. Flag immunoprecipitated extract contained DUX4 and multiple dox-dependent (DUX4-specific) proteins (Figure 1E). We performed mass-spectrometry and identified potential interacting proteins. Of these, CBP was the protein with the greatest number of unique reads, and its homologue p300 had the third most reads, with no other coactivators in the top 10 most-read sequences (Supplemental Figure S1 shows sequence coverage of p300 and CBP). We then confirmed both p300 and CBP interactions by independent co-immunoprecipitation experiments (Figure 1F). To further confirm the interaction of DUX4 with p300, we tested the ability of GST-DUX4 versus GST control to pull down recombinant p300 protein (Millipore). This revealed a specific interaction between p300 and GST-DUX4 (Figure 1G).
The C-terminus of DUX4 is required for interaction with p300 and transcriptional upregulation
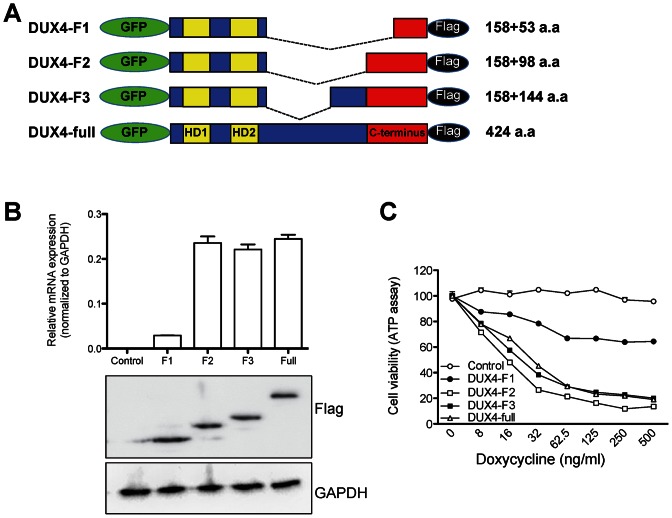
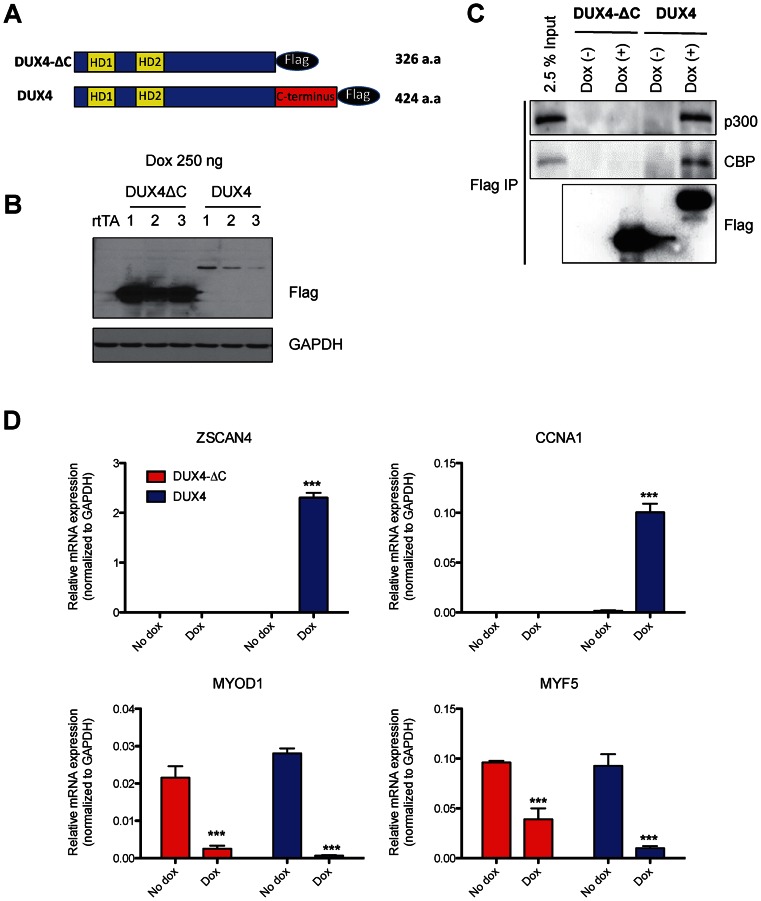
The DUX4C gene, which encodes a version of DUX4 with an alternate C-terminus replacing the C-terminal 82 amino acids, does not induce cell death when overexpressed (19) and a short form of DUX4 that does not contain C-terminus did not have the ability to upregulate DUX4 target genes (15), suggesting that the C-terminus of DUX4 is the transactivation domain, and perhaps the domain that interacts with p300. First, to definitively delineate the transactivation domain(s) of DUX4, we created internal deletions of various sizes, the largest retaining only the last 53 amino acids (Figure 2A), which is the unique region of conservation with DUXC family members (17). We tested the transcriptional activity of each deletion mutant by measuring expression of ZSCAN4, a strong DUX4 target gene (15) (Figure 2B). The smallest construct with only 52 amino acids, DUX4-F1, only weakly induced the DUX4 reporter, while DUX4-F2 and DUX4-F3 which contain 98 and 144 amino acids of the C-terminus, respectively, induce ZSCAN4 expression just as well as full length DUX4. The toxicity of these constructs correlated with their activation potential, i.e. constructs with 98 amino acids or more of C-terminal sequence were highly and equally toxic (Figure 2C). Thus, a minigene containing the homeodomains and the C-terminal 98 amino acids of DUX4 is functionally equivalent to full length DUX4. To further examine the role the C-terminus, we generated immortalized myoblast clones expressing dox-inducible DUX4-ΔC, which lacks the C-terminal 98 amino acids (Figure 3A). We compared expression levels of DUX4 and DUX4-ΔC (three independent clones of each) and found that DUX4-ΔC was expressed at much higher levels than DUX4 (Figure 3B), an effect that we attribute to a post-transcriptional mechanism. While FL DUX4 precipitated both p300 and CBP, DUX4-ΔC did not (Figure 3C), indicating that the C-terminus of DUX4 is essential for interaction with these coactivators, and providing an explanation for the lack of transcriptional activity of C-terminal deletion forms of DUX4. To verify the lack of transcriptional activation, we examined the expression level of several DUX4 target genes in DUX4 and DUX4-ΔC myoblasts 6 h after doxcycline treatment. Genes that are upregulated by DUX4 (ZSCAN4 and CCNA1) were not upregulated by DUX4-ΔC (Figure 3D). On the other hand, genes that are downregulated by DUX4 (MYF5 and MYOD1) were also downregulated by DUX4-ΔC, indicating that the C-terminus participates in transcriptional activation but not transcriptional repression. These studies define the last 98 amino acids of DUX4 as necessary for interaction with p300 and sufficient for full upregulation of DUX4 target genes.
Figure 2.
The central unstructured domain of DUX4 is dispensable for transcriptional activation and cytotoxicity. (A) Scheme of DUX4 deletion mutants. HD: homeodomain. GFP was added to visualize DUX4 fragments. The number of DUX4 amino acids in the N-terminal and C-terminal parts of each construct are indicated. (B) RT-qPCR analysis of RNA of the target gene ZSCAN4 after transient transfection of each construct. Protein expression levels of DUX4 deletion mutants are shown by immune blot, below. (C) Viability of C2C12 cells expressing different levels of each DUX4 deletion mutant. All constructs were targeted into the same genomic inducible locus. Cell viability (ATP content) was measured 24 h after treatment with doxycycline.
Figure 3.
The C-terminal domain of DUX4 is required for p300 interaction and DUX4-mediated transcriptional activation, but not for repression of target genes. (A) Schematic diagram of DUX4-ΔC and full length DUX4. The C-terminal 98 amino acids were deleted in DUX4-ΔC. (B) Western blots indicating the level of DUX4-ΔC and DUX4 expression in three independent LHCN-M2 clonal cell lines. Whole cell lysates were prepared after 6 h of dox treatment. Note that the C-terminal deletion is expressed much more abundantly. (C) p300/CBP binds to the C-terminal domain of DUX4. DUX4-ΔC-flag and DUX4-flag were immunoprecipitated with flag antibody 6 h after 250 ng dox treatment. Endogenous p300 and CBP were coprecipitated with DUX4-flag, but not with DUX4-ΔC-flag. (D) RT-qPCR analysis of two upregulated genes (ZSCAN4 and CCNA1), and two downregulated genes (MYOD1 and MYF5) in DUX4-ΔC or DUX4 inducible LHCN-M2 cells 6 h after 250 ng/ml dox treatment. Expression is normalized to GAPDH (n = 3, three independent clones, error bars represent SEM, t-test: ***P < 0.001 versus no dox group). Note that the C-terminus is necessary for activation, but dispensable for downregulation of MYOD1 and MYF5.
FL DUX4 recruits p300 and drives epigenetic changes at ZSCAN4
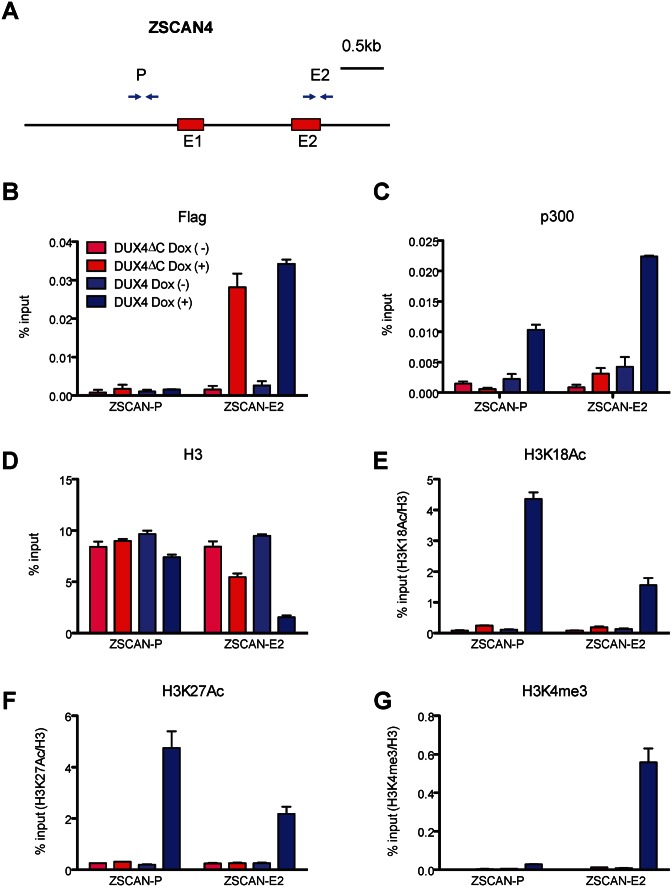
Because p300 has histone acetyltransferase activity, its recruitment by DUX4 would be expected to result in increased H3K18Ac and H3K27Ac marks in regions near DUX4 binding sites (31). We therefore tested for presence of DUX4, p300, and a variety of histone H3 marks in different regions of one of the most responsive DUX4 target genes, ZSCAN4. Previous ChIP-seq data revealed a DUX4 peak in the annotated second exon of ZSCAN4 (15). We prepared chromatin from both DUX4- and DUX4-ΔC-inducible immortalized myoblasts 6 h after dox treatment, performed ChIP with antibodies to Flag (DUX4) and p300, and assayed the annotated promoter region as well as exon 2 of ZSCAN4 (Figure 4A). As expected, we found that the exon 2 site was robustly pulled down by Flag (DUX4) antibodies. We did not observe enrichment of DUX4 at the annotated ZSCAN4 promoter. Similarly, DUX4-ΔC was enriched specifically at the exon 2 site and not at the promoter (Figure 4B). We then evaluated p300, and found robust enrichment at both the promoter region and at exon 2, but only in the inducible DUX4 cells, not in the DUX4-ΔC cells (Figure 4C). These data show that when FL DUX4 binds, p300 is recruited, but when DUX4ΔC binds, p300 is not recruited, in agreement with the data above, demonstrating that the C-terminus is the p300 interaction domain.
Figure 4.
p300 is recruited to the DUX4 binding site in ZSCAN4. (A) Location of PCR amplicons in the ZSCAN4 gene used for ChIP analyses. The previously-identified DUX4-binding site is at +1430, in exon 2. (B–G) ChIP assays with various antibodies at the indicated ZSCAN4 loci (ZSCAN4-P: promoter region, ZSCAN4-E: exon 2 region). LHCN-M2 cells were treated with 250 ng/ml dox for 6 h to express DUX4 or DUX4-ΔC and subjected to ChIP analyses.% input of H3K18Ac, H3K27Ac, and H3K4me3 were normalized with% input of H3 (n = 3, error bars represent SEM). Note that both DUX4 and DUX4ΔC are recruited to the DUX4 site in exon 2, but p300 is not recruited by DUX4ΔC, and acetylation of H3K18 is not increased by DUX4ΔC.
To evaluate epigenetic marks, we first tested enrichment with total histone H3 and found that H3 was relatively depleted from the exon 2 region when DUX4 was present (Figure 4D). This depletion was seen only weakly with DUX4-ΔC. This data suggests that binding of FL DUX4 displaces nucleosomes, at least at this specific site. We then immunoprecipitated with antibodies to H3 marks: K18Ac, K27Ac and K4me3 using H3 occupancy to normalize the H3 modifications. This revealed that H3K18ac and H3K27ac were enhanced at sites where and under conditions when p300 was present (Figure 4E and F). In addition, we found enhancement of H3K4me3 (Figure 4G), a histone mark found in actively transcribed genes.
Global analysis of DUX4-induced changes at target loci
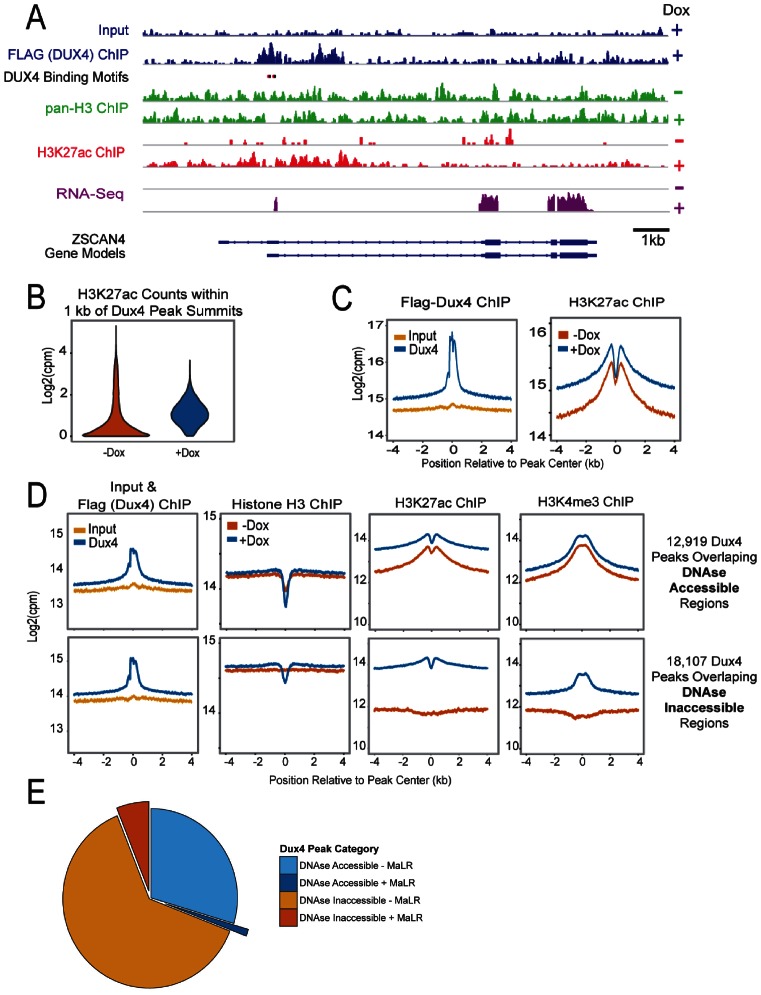
These data prompted us to investigate DUX4 effects on H3K27Ac globally, with the aim of understanding the average or typical response of a generic DUX4 target locus. We therefore performed ChIP-seq on DUX4 (Flag), H3K27Ac, H3K4me3, total H3 and RNA-seq, all at 6 h post-DUX4 induction via dox in the immortalized LHCN-M2 DUX4-inducible myoblasts. Before generalizing, we first reviewed this data over the ZSCAN4 locus. H3K27Ac and H3K4me3 changes at ZSCAN4 were as predicted by Figure 4 above, however the RNA-seq data clearly showed that in these cells, ZSCAN4 transcription was initiated from an alternative promoter to produce a transcripted annotated in ENSEMBL as ENST00000612521. The DUX4 binding site, presumed to be in exon 2, is in fact just upstream of the induced transcript (Figure 5A). A clear peak for DUX4 was observed at this site, and in the presence of this peak, a strong H3K27Ac signal was induced immediately up and downstream. The region under the DUX4 peak itself was depleted for H3K27Ac signal, concordant with the total H3 depletion at this site shown above in Figure 4D. The induced acetylation ranged for several kb both upstream and downstream and encompassed the annotated promoter of ZSCAN4. Globally, we identified 31 042 DUX4 peaks, 17% of which are in common with similar analysis of a previous DUX4 ChIP-seq data set (15).
Figure 5.
Genome wide changes in histone H3 and H3K27ac occupancy by DUX4 in LHCN-M2-iDUX4 cells. (A) Pan-histone H3 and H3K27Ac changes at ZSCAN4 after DUX4 expression. Red and black ovals represent the TAATTCAATCA and TAAATCAATCA DUX4 homeodomain binding motifs respectively. A strong DUX4 peak, shown in blue, is observed at these motifs proximal to an alternate promoter for ZSCAN4 (Ensembl Transcript ENST00000612521); pan-Histone H3 signal before (–) and after (+) doxycycline treatment are shown in green; H3K27Ac signals before and after doxycycline are shown in red. Note the loss of histone H3 signal and the appearance of H3K27Ac peaks that form adjacent to the site of DUX4 binding. (B) Genome wide changes in chromatin upon DUX4 induction. The distribution in the number of sequence reads in the H3K27ac datasets that map to within 1 kb of 31 042 Dux4 peak summits is shown as a violin plot in absence or presence of doxycycline. (C) The ChIP signals 4 kb upstream and downstream of the peak summits were summed across all Dux4 binding sites, normalized to the mapped sequencing depth and log2 transformed. The upper panel contains reads for the input (yellow) and Flag-DUX4 ChIP (blue) samples. The lower panel contains the H3K27Ac ChIP signals that were observed before (orange) and after (blue) doxycycline treatment. (D) The regions bound by DUX4 used in panels (B) and (C) were separated into 12 912 peaks that overlap DNase hypersensitivity sites (left panels) and 18 130 peaks that do not (right panels). ChIP signals are shown for input, Flag-DUX4 ChIP, histone H3 ChIP, H3K27Ac ChIP and H3K4me3 ChIP. Data are colored as in panel C: input = yellow, absence of doxycycline = orange, presence of doxycycline = blue. Note that a decrease is observed in the histone H3 signal upon DUX4 induction in the DNase in-accessible regions and that a similar decrease preexisted in the DNase accessible DUX4 binding regions. A saddle-shaped distribution of H3K27Ac ChIP signal is observed at DUX4 peaks at DNase accessible regions and upon DUX4 induction in DNase in-accessible regions.
Globally, sites recognized by DUX4 showed much greater H3K27Ac in the presence of DUX4 than in the absence (Figure 5B), and although most sites showed low acetylation prior to binding, there was clearly a population of sites that were already acetylated, prior to DUX4 binding. The average pattern of acetylation across the region bound by DUX4 showed a volcano shape, with the dip in H3K27Ac in center of the DUX4 peak consistent with nucleosome depletion at the site of DUX4 binding, however, this dip was also present prior to binding, suggesting that at a large subset of sites, nucleosomes are already depleted (Figure 5C).
DUX4 binds two classes of target site: accessible and inaccessible
The presence of prior acetylation and a central dip in H3K27Ac prior to DUX4 binding were unexpected and prompted us to investigate the prior accessibility of DUX4 target loci. ENCODE data on global DNaseI hypersensitivity in the same LHCN-M2 immortalized myoblast line revealed that 12 919 DUX4 target loci are in DNase accessible sites, while 18 107 are inaccessible prior to DUX4 binding. We therefore analyzed ChIP-seq data independently for these two classes of site. The accessible sites were histone-depleted before DUX4 binding and remained so after, while the inaccessible sites showed histone depletion only after DUX4 binding (Figure 5D, second row and Supplemental Figure S2A). The inaccessible sites showed very low prior acetylation that was greatly elevated by DUX4 binding, while acetylation changes at accessible sites were less dramatic (Figure 5D, third row, and Supplemental Figure S2A). The H3K4me3 mark behaved broadly similarly, present prior to binding at accessible sites and increased greatly after binding at inaccessible sites, which is consistent with the doxyclycline dependent enrichment at the E2 site in ZSCAN4 (Figure 4G) and may reflect the activation of nearby genes via the recruitment of histone methyltransferases. In the DNase-inaccessible subset, there was actually a depression in H3K27Ac and H3K4me3 at the center of the region. This is due to the fact that although the DUX4 binding site at the core of each of these regions is DNase inaccessible, in some regions there are flanking DNase accessible sites, which are associated with local increased H3K27Ac and H3K4me3. If such regions are eliminated from analysis, the profile is flat (Supplemental Figure S2B). An further interesting feature of the DNase inaccessible set is that this is where the great majority of MaLR (mammalian apparent long terminal repeat)-containing DUX4 peaks are contained (Figure 5E). These data suggest that DUX4 can function as a pioneer factor, able to bind at sites within conformationally inaccessible chromatin that are replete with histones lacking markers of activation, and thereafter recruits histone acetyl- and methyl-transferases to enable gene expression.
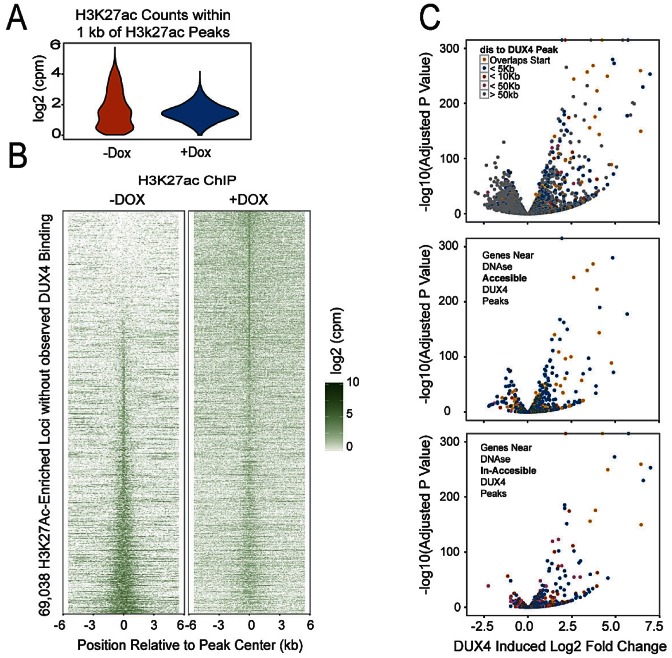
We wondered what effect DUX4 recruitment of H3K27-acetyltransferase activity would have at sites not associated with DUX4 but previously acetylated. We therefore identified all H3K27Ac peaks not associated with DUX4 (both + and – dox) and evaluated levels of enrichment before and after DUX4 expression. This showed a loss of regions of very high acetylation and a general narrowing of the H3K27Ac profile after DUX4 expression (Figure 6A). Stacking of all non-DUX4 associated H3K27Ac peaks revealed that although most prior-acetylated peaks retain a signal of acetylation, there is great reduction in intensity, and in addition, a significant number of new peaks form (Figure 6B). New H3K27Ac peaks not associated with DUX4 may be indirect, i.e. secondary, DUX4 targets, or may be DUX4 targets whose peaks were weak and missed by our algorithm. These data demonstrate a large scale reallocation of H3K27Ac across the genome after DUX4 expression, with a particular reduction in the strongest peaks.
Figure 6.
Global changes in H3K27Ac distribution lead to direct and indirect gene expression changes in LHCN-M2-iDUX4 cells. (A) Redistribution in the number of sequence reads from the H3K27Ac ChIP signals at 68 506 equal width genomic loci that are enriched for H3K27Ac signal in either the absence or presence of doxycycline but are not bound by detectable levels of DUX4 is shown as a violin plot for both conditions. (B) A heatmap view of the regions in panel A sorted by the ratio of signal observed in the absence or presence of doxycycline. The top and bottom of the heatmap contains regions that have a marked increase or decrease in H3K27Ac upon Dux4 expression respectively. (C) Differential gene expression upon DUX4 induction. Minus log10p-values are plotted against the log2 fold change between the control and doxycycline treated samples. The points are colored according the nearest distance between the transcription start site to a DUX4 peak: overlapping (yellow), <5 kb (blue), <10 kb (red), <50 kb (pink) and > 50 kb (gray). The lower panels indicate whether the DUX4 peak associated with each gene (<50 kb) is in a DNase Accessible (middle panel) or DNase In-Accessible region (lower panel). Note that genes with very high expression changes are associated with DUX4 peaks in DNase In-Accessible Regions.
RNA-seq identified a large set of gene expression changes in response to DUX4, both up- and down-regulation, however the majority of strongly upregulated genes were in the vicinity of a DUX4 peak (colored dots, Figure 6C, top). When those changes associated with a DUX4 peak were subdivided by prior accessibility of the DUX4 target locus, upregulation was seen at both accessible and inaccessible loci, but the most strongly upregulated genes were in the prior inaccessible set (Figure 6C bottom panel).
Dominant negative activity of the DUX4 C-terminus
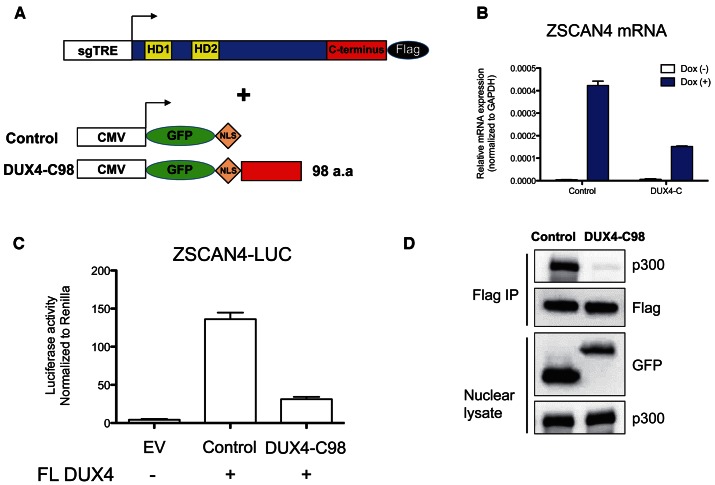
Because of the clear induction of H3K27 acetylation at sites of DUX4 binding, and the interaction of the DUX4 C-terminus with p300/CBP, we wondered whether competing away this interaction from full length DUX4 by overexpressing the DUX4 C-terminus would affect the ability of DUX4 to induce transcription. To test this, we generated a 293T derivative with inducible full length DUX4, using the same lentiviral strategy described above, and transfected these DUX4-inducible 293T cells with nuclear-targeted GFP alone, or fused to the C-terminus of DUX4 (Figure 7A). Eight hours after transfection, we then induced full length DUX4 expression with doxycycline and assayed the endogenous DUX4 target gene, ZSCAN4. Transfection of the nuclear-targeted DUX4 C-terminus inhibited the DUX4-induced expression of endogenous ZSCAN4 (Figure 7B). It also inhibited expression of a cotransfected ZSCAN4-luciferase reporter (Figure 7C), demonstrating that the C-terminus does indeed have dominant negative activity. Finally, to test whether this dominant negative activity involves the interfering with the p300 interaction, we performed co-IPs of FL DUX4 using the Flag antibody from cells overexpressing the GFP-NLS-C-terminus. Indeed, in the presence of the overexpressed C-terminus, the interaction of full length DUX4 with p300 was substantially diminished (Figure 7D). Together, these data suggest that the DUX4 C-terminus exhibits dominant negative activity by competing with full length DUX4 for p300/CBP.
Figure 7.
Dominant negative activity of the p300 interacting C-terminal domain of DUX4. (A) Schematic of DUX4 expression constructs. 293T cells modified to express full-length DUX4 driven from the dox-inducible sgTRE promoter, shown above, were transfected with transient expression constructs, shown below. The number of C-terminal DUX4 amino acids is indicated. sgTRE: second generation tet-response element for dox-induced expression; CMV: CMV promoter for high level constitutive expression; NLS: SV40-nuclear localization signal; HD: homeodomain. GFP was added to stabilize DUX4 fragments. NLS was added to deliver DUX4 fragment in the nucleus. (B) DUX4 mediated ZSCAN4 mRNA expression. Cells were transfected, 24 h later 250 ng/ml dox was applied, and mRNA extracted 6 hours after that (n = 3, error bars represent SEM). (C) Activity of a cotransfected DUX4-luciferase reporter. 293T cells were co-transfected with both full length DUX4 and empty vector (EV) or the tested fragments, together with DUX4 luciferase and Renilla control reporters (n = 3, error bars represent SEM). (D) Western blots of DUX4 immunoprecipitations performed in the presence of GFP-NLS control or GFP-NLS-DUX4-C-terminal 98 amino acids. Interaction of full length DUX4 with p300 is inhibited by overexpression of the C-terminus of DUX4.
DISCUSSION
Although target genes of DUX4 have been identified in a number of studies (14,15,32), the mechanisms used by DUX4 to drive these gene expression changes has not been investigated. Our data show that p300 and CBP are DUX4 interacting proteins, that they interact with the DUX4 C-terminus, and that they are recruited to upregulated target genes such as ZSCAN4, when DUX4 is bound. Both at ZSCAN4 specifically, and across the genome at all DUX4-bound regions, histone H3 is depleted from the central region of the DUX4 peak, suggesting that DUX4 binding displaces nucleosomes. Consistent with this, approximately 60% of DUX4 bound regions were previously inaccessible to DNaseI, but are bound by DUX4 nonetheless, and are then centrally depleted of H3, suggesting that DUX4 can act as a pioneer factor. Interestingly, in the 40% of sites that were previously accessible to DNase, the central depletion of H3 is present before binding, suggesting the presence of other binding factors at these sites prior DUX4 binding. The possibility that DUX4 is tethered to these sites by the resident factors rather than by binding to DNA is unlikely: the consensus DNA motif recognized by DUX4 (15) is equally present in both classes of target site. H3K27Ac is increased in the flanking regions of both classes of DUX4 target locus, but the increase is most pronounced at sites in previously inaccessible chromatin, as they showed prior depletion of H3K27Ac. In addition to enrichment of H3K27Ac, H3K4me3 is also locally enriched. This is consistent with the idea that the acetylation activity of p300/CBP effects epigenetic changes promoting gene activation by recruiting histone methyltransferases such as the SET1 complex (33).
While the increase in acetylation in the vicinity of DUX4 peaks was pronounced, the amount of depletion of H3K27Ac at non-DUX4 associated sites of peak enrichment prior to DUX4 expression was equally striking. Transcriptional profiling showed downregulation of many transcripts, almost exclusively within the set that were not associated with DUX4 binding, but this downregulation was modest compared to the upregulation of DUX4 targets. It is likely that the very short time between induction of DUX4 and assay (6 h) combined with perdurance of these transcripts explains their modest downregulation. Although there has been much thought given to the potential role particular DUX4-induced target genes in the cytopathology of DUX4 (9,14,15,34), it may well be the disrupted expression of the large number of non-targets that is the more lethal. However, it is important to keep in mind that the level of expression of DUX4 in those FSHD cells that do express it is unknown, and the global reductions in the major H3K27Ac peaks described here may depend on relatively high levels of DUX4.
We show that the C-terminus of DUX4 is essential for the interaction with p300. It was previously reported that a fusion protein combining the DNA binding domain of CIC and the C-terminus of DUX4 acts as a transactivator to promote tumorigenesis (19). By deleting the middle half of the DUX4 protein sequence, we show that the only domains necessary for cytotoxicity and transcriptional activation by DUX4 are the N-terminal DNA-binding homeodomains and the C-terminal 98 amino acid transcriptional activation domain.
The importance of the C-terminal domain is underscored by the fact that it can act as a dominant negative. This dominant negative activity is presumably due to the formation of non-productive complexes of the C-terminus with coactivators, depriving FL DUX4 of opportunities for productive interactions. Indeed, we find that the interaction of FL DUX4 with p300 is greatly diminished in the presence of the overexpressed C-terminus. This approach to inhibiting DUX4 activity may open up avenues for therapy development, particularly if the dominant negative activity can be reduced to small peptides from the DUX4 C-terminus or from p300 that would compete for this interaction, a strategy that has been successful in other systems (35).
Supplementary Material
Acknowledgments
We thank the Dr Bob and Jean Smith Foundation for their generous support.
Footnotes
Present address: Darko Bosnakovski, University Goce Delčev – Štip, Faculty of Medical Sciences, Krste Misirkov b.b., 2000 Štip, R. Macedonia.
FUNDING
National Institutes of Health [R01 AR055685 to M.K.]; Muscular Dystrophy Association [MDA 4361 to D.B.]; FSH Society [Marjorie Bronfman Fellowship FSHS-MGBF-016 and FSHS-82010–01, to D.B.]. Funding for open access charge: NIH/NIAMS [R01 AR055685]; Muscular Dystrophy Association [MDA 4361 to D.B.]; FSH Society [FSHS-MGBF-016 to D.B.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gabellini D., Green M.R., Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- 2.Zeng W., de Greef J.C., Chen Y.Y., Chien R., Kong X., Gregson H.C., Winokur S.T., Pyle A., Robertson K.D., Schmiesing J.A., et al. Specific loss of histone H3 lysine 9 trimethylation and HP1gamma/cohesin binding at D4Z4 repeats is associated with facioscapulohumeral dystrophy (FSHD) PLoS Genet. 2009;5:e1000559. doi: 10.1371/journal.pgen.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Overveld P.G., Lemmers R.J., Sandkuijl L.A., Enthoven L., Winokur S.T., Bakels F., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Hypomethylation of D4Z4 in 4q-linked and non-4q-linked facioscapulohumeral muscular dystrophy. Nat. Genet. 2003;35:315–317. doi: 10.1038/ng1262. [DOI] [PubMed] [Google Scholar]
- 4.de Greef J.C., Lemmers R.J., van Engelen B.G., Sacconi S., Venance S.L., Frants R.R., Tawil R., van der Maarel S.M. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum. Mutat. 2009;30:1449–1459. doi: 10.1002/humu.21091. [DOI] [PubMed] [Google Scholar]
- 5.Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- 6.van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- 7.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R., et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabriels J., Beckers M.C., Ding H., De Vriese A., Plaisance S., van der Maarel S.M., Padberg G.W., Frants R.R., Hewitt J.E., Collen D., et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. doi: 10.1016/s0378-1119(99)00267-x. [DOI] [PubMed] [Google Scholar]
- 9.Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Matteotti C., van Acker A.M., Leo O., et al. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18157–18162. doi: 10.1073/pnas.0708659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P., Jr Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 2012;21:4419–4430. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rickard A.M., Petek L.M., Miller D.G. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum. Mol. Genet. 2015;24:5901–5914. doi: 10.1093/hmg/ddv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosnakovski D., Xu Z., Gang E.J., Galindo C.L., Liu M., Simsek T., Garner H.R., Agha-Mohammadi S., Tassin A., Coppee F., et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–2779. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Lee J.K., Toso E.A., Lee J.S., Choi S.H., Slattery M., Aihara H., Kyba M. DNA-binding sequence specificity of DUX4. Skeletal Muscle. 2016;6:8. doi: 10.1186/s13395-016-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clapp J., Mitchell L.M., Bolland D.J., Fantes J., Corcoran A.E., Scotting P.J., Armour J.A., Hewitt J.E. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:264–279. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawamura-Saito M., Yamazaki Y., Kaneko K., Kawaguchi N., Kanda H., Mukai H., Gotoh T., Motoi T., Fukayama M., Aburatani H., et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum. Mol. Genet. 2006;15:2125–2137. doi: 10.1093/hmg/ddl136. [DOI] [PubMed] [Google Scholar]
- 19.Bosnakovski D., Lamb S., Simsek T., Xu Z., Belayew A., Perlingeiro R., Kyba M. DUX4c, an FSHD candidate gene, interferes with myogenic regulators and abolishes myoblast differentiation. Exp. Neurol. 2008;214:87–96. doi: 10.1016/j.expneurol.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 20.Roeder R.G. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Ogryzko V.V., Schiltz R.L., Russanova V., Howard B.H., Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 22.Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- 23.Iacovino M., Bosnakovski D., Fey H., Rux D., Bajwa G., Mahen E., Mitanoska A., Xu Z., Kyba M. Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells. 2011;29:1580–1588. doi: 10.1002/stem.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L., Magli A., Catanese J., Xu Z., Kyba M., Perlingeiro R.C. Modulation of TGF-beta signaling by endoglin in murine hemangioblast development and primitive hematopoiesis. Blood. 2011;118:88–97. doi: 10.1182/blood-2010-12-325019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stols L., Zhou M., Eschenfeldt W.H., Millard C.S., Abdullah J., Collart F.R., Kim Y., Donnelly M.I. New vectors for co-expression of proteins: structure of Bacillus subtilis ScoAB obtained by high-throughput protocols. Protein Expr. Purif. 2007;53:396–403. doi: 10.1016/j.pep.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Zhu C.H., Mouly V., Cooper R.N., Mamchaoui K., Bigot A., Shay J.W., Di Santo J.P., Butler-Browne G.S., Wright W.E. Cellular senescence in human myoblasts is overcome by human telomerase reverse transcriptase and cyclin-dependent kinase 4: consequences in aging muscle and therapeutic strategies for muscular dystrophies. Aging Cell. 2007;6:515–523. doi: 10.1111/j.1474-9726.2007.00306.x. [DOI] [PubMed] [Google Scholar]
- 27.Agha-Mohammadi S., O'Malley M., Etemad A., Wang Z., Xiao X., Lotze M.T. Second-generation tetracycline-regulatable promoter: repositioned tet operator elements optimize transactivator synergy while shorter minimal promoter offers tight basal leakiness. J .Gene Med. 2004;6:817–828. doi: 10.1002/jgm.566. [DOI] [PubMed] [Google Scholar]
- 28.Huber W., Carey V.J., Gentleman R., Anders S., Carlson M., Carvalho B.S., Bravo H.C., Davis S., Gatto L., Girke T., et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dandapat A., Bosnakovski D., Hartweck L.M., Arpke R.W., Baltgalvis K.A., Vang D., Baik J., Darabi R., Perlingeiro R.C., Hamra F.K., et al. Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep. 2014;8:1484–1496. doi: 10.1016/j.celrep.2014.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin Q., Yu L.R., Wang L., Zhang Z., Kasper L.H., Lee J.E., Wang C., Brindle P.K., Dent S.Y., Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krom Y.D., Thijssen P.E., Young J.M., den Hamer B., Balog J., Yao Z., Maves L., Snider L., Knopp P., Zammit P.S., et al. Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet. 2013;9:e1003415. doi: 10.1371/journal.pgen.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z., Chen W.Y., Shimada M., Nguyen U.T., Kim J., Sun X.J., Sengoku T., McGinty R.K., Fernandez J.P., Muir T.W., et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297–310. doi: 10.1016/j.cell.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tawil R., van der Maarel S.M., Tapscott S.J. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skeletal Muscle. 2014;4:12. doi: 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moellering R.E., Cornejo M., Davis T.N., Del Bianco C., Aster J.C., Blacklow S.C., Kung A.L., Gilliland D.G., Verdine G.L., Bradner J.E. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.