Abstract
DNA double-strand breaks (DSBs) are known to be powerful inducers of homologous recombination (HR), but single-strand breaks (nicks) have also been shown to trigger HR. Both DSB- and nick-induced HR (nickHR) are exploited in advanced genome-engineering approaches based on the bacterial RNA-guided nuclease Cas9. However, the mechanisms of nickHR are largely unexplored. Here, we applied Cas9 nickases to study nickHR in mammalian cells. We find that nickHR is unaffected by inhibition of major damage signaling kinases and that it is not suppressed by nonhomologous end-joining (NHEJ) components, arguing that nick processing does not require a DSB intermediate to trigger HR. Relative to a single nick, nicking both strands enhances HR, consistent with a DSB intermediate, even when nicks are induced up to ∼1kb apart. Accordingly, HR and NHEJ compete for repair of these paired nicks, but, surprisingly, only when 5' overhangs or blunt ends can be generated. Our study advances the understanding of molecular mechanisms driving nick and paired-nick repair in mammalian cells and clarify phenomena associated with Cas9-mediated genome editing.
INTRODUCTION
The reactive byproducts of endogenous metabolic activities as well as exogenous agents, such as ionizing radiation, threaten the integrity of mammalian genome. In response, cells have evolved complex protein networks that can detect and repair DNA damage (1). Although the different types of DNA lesions are typically repaired by dedicated pathways, a functional overlap between these pathways has also been observed. One interesting example of such overlap has been reported for the repair of DNA single-strand breaks (SSBs or nicks). These abundant lesions are typically repaired by specialized SSB repair mechanisms (2), but replication can convert nicks to the more dangerous double-strand breaks (DSBs) (3,4). Multiple mechanisms compete to repair DSBs in mammalian cells, including homologous recombination (HR) and non-homologous end joining (NHEJ) (5,6). Intriguingly, nicks also may trigger HR directly (7–11), even though this pathway is not commonly associated with nick repair. Two recent studies suggested that nick-induced HR (nickHR) utilizes factors involved in classical DSB-induced HR (DSBHR) (12,13).
Recent advances in the field of genome engineering provide a powerful approach to investigate nick and DSB repair mechanisms. The programmed induction of a DSB at a genomic locus has long been known to trigger HR which can precisely edit the genomic sequence when provided with the appropriate repair template (14). Alternatively, the DSB may be processed by error-prone NHEJ to generate random insertions or deletions – a desirable outcome when the targeted gene is to be inactivated. Various tools have been developed to induce DNA damage in a controlled and targeted manner, including homing endonucleases, zinc finger nucleases and TAL effector nucleases (15–17). Recently, the bacterial RNA-guided endonuclease Cas9 has been adapted for application in eukaryotic cells, providing a versatile method to induce SSBs and DSBs at a chosen genomic locus (18,19) and enable efficient HR- or NHEJ-mediated genome manipulation in multiple model organisms (20). Cas9 recognizes a 19 bp target sequence based on its complementarity to a short guide RNA (gRNA) and the presence of a downstream NGG protospacer adjacent motif (PAM). Upon DNA binding, the two catalytic domains of wild-type Cas9 cleave both DNA strands ∼3 bp upstream from the PAM to generate a DSB (18).
Mutation of either catalytic domain can convert Cas9 into a nickase, e.g. Cas9D10A or Cas9H840A (18). Because Cas9 tolerates multiple nucleotide mismatches in the 5′ part of the targeted sequence, off-target NHEJ events are not uncommon (21), such that Cas9-mediated nicking has been developed as an alternative genome engineering approach. Single nicks can stimulate HR without inducing NHEJ events and thus have a much lower mutagenic potential (11,13,22–26). However, it is not well understood how nickHR is regulated, how similar it is to classical HR, and whether NHEJ factors participate in this process. Conversely, paired nicks in both DNA strands can result in a DSB to induce either HR or NHEJ, but with increased specificity since binding of two gRNAs in sufficient proximity is required (27–31).
Here, we applied the RNA-guided Cas9 nickases, derived from the Streptococcus pyogenes CRISPR-Cas9 system, to study the mechanisms of nickHR. Our results show that nickHR is mildly reduced by inhibition of ATM or ATR signaling and that it is stimulated by replication if the lagging template strand is nicked. nickHR is partly overlapping with DSBHR in that it is dependent on the known HR factors BRCA1 and RAD51 (13), but, in contrast to DSBHR, it is not affected by NHEJ deficiency, suggesting that it does not typically proceed through a DSB intermediate. We then focused on the scenario when HR is triggered by nicking of both DNA strands, i.e. paired-nick HR (PNHR). With paired nicks, we detected an increased stimulation of HR as compared to single nicks, which is consistent with a DSB intermediate. Both 3′- and 5′-overhangs resulting from paired nicks were found to stimulate HR, even with nicks separated by up to 940 bp. Surprisingly, however, only HR induced by paired nicks which would result in a 5′ overhang was suppressed by the NHEJ factor Ku. These results advance our understanding of the mechanism behind nickHR and PNHR.
MATERIALS AND METHODS
Plasmids and cloning
The following plasmids were previously described: pDR-GFP (32) hprtDR-GFP (33), pCBASce (I-SceI expression vector) (34), pCD2E (empty vector), PCD2E-XH (XRCC1 vector) (35), EBNA1 expression vector pCEP4 (36), pCAGGS (empty vector) and pCAGGS expression vectors with BARD1-hB202, BRC3 and KU70 (33,37–41).
Cas9WT, Cas9D and empty gRNA expression vectors were purchased from Addgene (ID 41815, 41816 and 41824) (24). The generation of Cas9H nickase and the catalytically dead Cas9DH was described earlier (42). Cas9N863A expression vector was generated by PCR mutagenesis using the Cas9WT vector as template and primers Cas9-N863A-F (5′-GCTAGAGGGAAGAGTGATAACGTCCC-3′) and Cas9-N863A-R (5′-ACTCTTCCCTCTAGCTTTATCGGATCTTGTCAACACTTTATTATC-3′). Subsequently, the PCR product was circularized using In-Fusion kit (Clontech). The cloning of targeting gRNA expression vectors is described in supplementary experimental procedures. The enhanced versions of the gRNAs with adjustments in the stem-loop of the gRNA were described earlier (43) and generated by PCR mutagenesis as detailed in Supplementary experimental procedures. hprtDR-GFP (33) was the basis for generation of the pnDR-GFP reporter plasmids (Supplementary experimental procedures). To generate the DR-Orip-GFP plasmid, the TR-OriP-GFP (36) and DR-OriP-GFP plasmids were digested using AccI and MfeI (New England Biolabs). Subsequently, the restriction fragment containing the OriP cassette from the TR-OriP-GFP was ligated with the fragment of the DR-OriP-GFP containing SceGFP and iGFP using the DNA Dephos & Ligation Kit (Roche).
Cell lines and cell culture
U2OS cells harboring a single, genomically integrated copy of the DR-GFP reporter were described previously (36). In addition, mouse J1 (wild-type), E14 (wild-type), Ku70−/−, Xrcc4−/− and DNA-PK−/− ES cells with a single Hprt-targeted DR-GFP reporter were used, as were mouse J1 (wild-type) ES cells with a single Pim1-integrated DR-GFP reporter (33,37–39). XRCC1−/− CHO cells EM9-V (complemented with an empty vector) and EM9-XH (complemented with the human wild-type XRCC1) as well as empty and XRCC1 expression plasmids were kindly provided by Dr K.W. Caldecott (35). U2OS, HEK293T and EM9 cells were cultured in high glucose DMEM with 10% fetal bovine serum (FBS), the medium for culturing the mouse ES cells was described earlier (44). All cells were incubated at 37°C in an atmosphere containing 5% CO2. The various pnDR-GFP plasmids were stably integrated at the Hprt locus of wild-type (E14) and Ku70−/− ES cells (33). To this end, 2 × 106 cells were transfected with pnDR-GFP plasmids linearized with KpnI/SacI, or PvuII in case of pnDR-GFP-940 bp, (New England Biolabs) using Amaxa nucleofector II (Lonza), program X-005. Selection of clones was performed as described previously (44). Genomic DNA was extracted using Salt-X and digested with PstI (New England Biolabs). Southern blot analysis of the digested DNA, using iGFP fragment from the hprtDR-GFP plasmid as a probe, confirmed correct targeting (Supplementary Figures S3A and S4).
Transfections, inhibitors and repair assays
A total of 2 × 106 cells were transfected using the Amaxa nucleofector II (Lonza) and home-made nucleofection solution (45). The cell lines with corresponding nucleofector programs and expression vector concentrations are described in supplementary experimental procedures. Cas9WT with empty gRNA expression vector at the ratio described in Supplementary experimental procedures were used as a negative control. For paired nicking, the total amount of gRNA expression vectors was kept constant by using an empty gRNA expression construct. After nucleofection, Dimethyl sulfoxide (DMSO) or 1 μM of olaparib (Organic Synthesis Core Facility, MSKCC) were added for 24 or 48 h, while indicated concentrations of KU55933 (46) (EMD Millipore), VE-821 (47) or NU7441 (Selleck Chemical)(48) were added for 48 h. Flow cytometry data were analyzed with FlowJo (Treestar).
Statistics
Unless stated otherwise, the values in the figure graphs are derived from at least three independent experiments and error bars represent the standard deviation; P- values are indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
Cell cycle analysis
Cells were pre-treated with DMSO or 500 nM Nocodazole (Sigma-Aldrich) for 16 h before transfection and for an additional 24 h after transfection. Samples for cell cycle and GFP expression analysis were collected immediately and 24 h after transfection. Cells were fixed in ice-cold 70% ethanol for 30 min, then washed twice with phosphate buffered saline (PBS), resuspended in 100 μg/ml RNAse (Thermo Scientific) with 50 μg/ml propidium iodide (Sigma-Aldrich) and incubated for 30 min at 37°C. After incubation, cells were directly analyzed by flow cytometry.
Western blotting
Cells were collected using a scraper, washed with PBS and resuspended in lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA) containing protease and phosphatase inhibitor cocktail (Roche Molecular Biochemicals). The following antibodies with corresponding concentrations were used: anti-PAR (Trevigen, 1:1000), anti-Chk2 (EMD Millipore, 1:1000), anti-pChk2-T68 (Cell Signaling, 1:1000), anti-Chk1 (Santa Cruz, 1:750), anti-pChk1-S345 (Cell Signaling, 1:1000), anti-pKAP1-S824 (Bethyl Laboratories, 1:2000), anti-Cas9 (Cell Signaling Technology, 1:1000), anti-Clathrin Heavy Chain (BD Transduction Laboratories, 1:10 000) and anti-Tubulin (Sigma, 1:10 000).
RESULTS
Cas9-induced nicks stimulate intrachromosomal HR
To investigate nickHR, we exploited the DR-GFP reporter (32) which is commonly used to study DSBHR in mammalian cells. DR-GFP consists of two tandem, inactive copies of the GFP gene, the first copy (SceGFP) containing the 18 bp I-SceI endonuclease recognition site with an in-frame stop codon and the second copy (iGFP) harboring truncations at both ends (Figure 1A). A DSB at the I-SceI site induces HR using the downstream copy as a donor, restoring the GFP open reading frame and leading to GFP positive (GFP+) cells (Figure 1A).
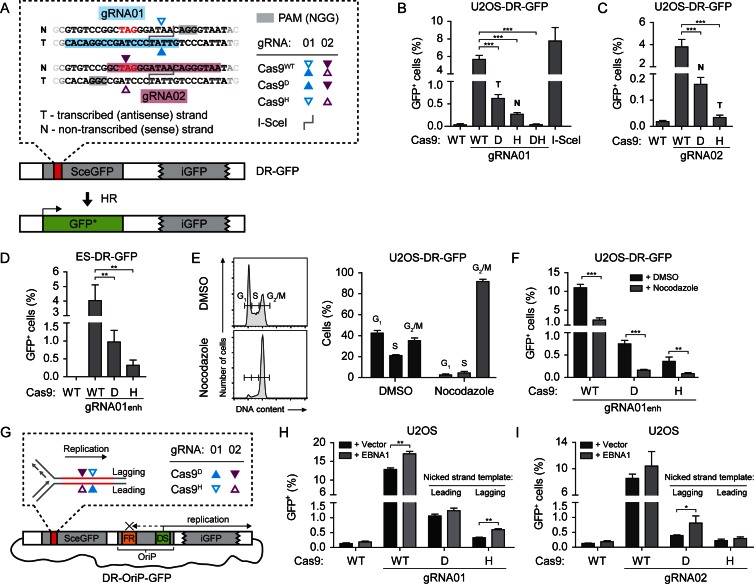
Figure 1.
HR repair of nicks on the lagging-strand template is promoted by replication. (A) The DR-GFP reporter consists of two inactive copies of the GFP gene. Induction of a DSB or nick in SceGFP triggers HR that uses iGFP as a template to restore the GFP open reading frame. The inset shows the SceGFP sequence around the 18 bp I-SceI recognition site, which begins at the in-frame stop codon (red). PAMs required for Cas9 binding are shaded gray and the sequences bound by the indicated gRNAs are shaded in light blue (gRNA01) and light purple (gRNA02). Cas9 variants: wild-type (Cas9WT), D10A (Cas9D) and H840A (Cas9H). The predicted Cas9 cleavage sites are marked by colored triangles. T - transcribed strand, N - non-transcribed strand. (B–D) U2OS-DR-GFP cells (B and C) or ES-DR-GFP cells (D) were transfected with expression vectors for the indicated Cas9 and gRNA variants, or I-SceI. As a negative control (first bar in graphs B–D), Cas9WT was transfected with a gRNA expression vector that lacked the targeting portion of the gRNA sequence. (E) U2OS-DR-GFP cells were arrested at the G2/M phase transition by 16 h incubation in 500 nM nocodazole or mock-treated with DMSO, stained with propidium iodide and analyzed by flow cytometry to confirm the cell cycle arrest. The left panel shows representative histograms of DNA content. The right panel shows the average percentage of cells in the indicated cell cycle phases. (F) U2OS-DR-GFP cells were treated as in (E) and transfected with the indicated Cas9 and gRNA expression vectors, then mock-treated or incubated in the presence of nocodazole for a further 24 h and analyzed by flow cytometry. The cell cycle distribution of cells at the time of flow cytometric analysis is shown in Supplementary Figure S1I. (G) Schematic overview of the DR-Orip-GFP reporter. EBNA1 initiates replication downstream from SceGFP, such that the top strand becomes the lagging-strand template. The inset depicts the predicted sites of cleavage by Cas9D (filled triangles) or Cas9H (empty triangles), in combination with the indicated gRNAs. (H) U2OS cells were transfected with the DR-OriP-GFP reporter, the empty vector (+ Vector) or EBNA1 expression vector (+ EBNA1), gRNA01 and the indicated Cas9 variant. Cas9D nicks the leading-strand template and Cas9H nicks the lagging-strand template, as indicated. (I) Cells were transfected as in (H), except gRNA02 was used, such that Cas9D nicks the lagging-strand template and Cas9H the leading-strand template.
NGG motifs flank the I-SceI cleavage site on both strands such that both strands can be recognized by Cas9 paired with gRNAs that overlap the site (18,19) (Figure 1A). gRNAs were designed to bind either the transcribed (gRNA01) or non-transcribed (gRNA02) strand. We used human codon-optimized versions of wild-type Cas9 (Cas9WT) to induce DSBs, Cas9D10A (Cas9D) and Cas9H840A (Cas9H) nickases to induce SSBs, and catalytically inactive Cas9 (Cas9DH) as a control (18,24,42). Cas9D nicks the strand complementary to the gRNA, while Cas9H nicks the strand identical to the gRNA sequence (18,19) (Figure 1A).
Human osteosarcoma (U2OS-DR-GFP) cells and mouse embryonic stem (ES-DR-GFP) cells harboring a single, genomically integrated copy of the DR-GFP reporter were transiently transfected with Cas9 and gRNA01 expression vectors. To measure the relative efficiency of HR, cells were analyzed by flow cytometry 48 h after transfection. In U2OS-DR-GFP cells, Cas9WT and both nickases stimulated HR; Cas9WT was more efficient than either nickase, and Cas9D was more efficient than Cas9H (Figure 1B). The nuclease-dead Cas9DH failed to induce HR, as did Cas9WT in the absence of a targeting gRNA. Similar results were obtained in U2OS cells transiently transfected with the DR-GFP reporter plasmid (Supplementary Figure S1A) and in mouse ES-DR-GFP cells, although the difference between Cas9D and Cas9WT was less pronounced (Figure 1D).
Our results suggest that, although somewhat variable, the efficiency of nickHR is generally lower than that of DSBHR, although we cannot rule out that SSB formation by Cas9 nickases is less efficient than DSB formation by Cas9WT. Overall, the level of DSBHR induced by Cas9WT was similar to that induced by I-SceI (Figure 1B and Supplementary Figure S1A–C). However, nickHR induced by Cas9D was much more efficient than nickHR induced by I-SceI nickases (unpublished results; (49)).
The higher efficiency of Cas9D relative to Cas9H may be due to an inherently higher nickase activity or may be related to which strand is nicked. To attempt to distinguish between these possibilities, a second gRNA (gRNA02) was used which binds the opposite strand (Figure 1A). Overall, gRNA02 was less efficient at inducing HR than gRNA01, suggesting that this gRNA targets Cas9 less well. As with gRNA01, Cas9D+gRNA02 induced a higher level of nickHR than Cas9H+gRNA02, indicating that Cas9D is a more efficient nickase than Cas9H (Figure 1C). The transcribed strand is nicked by Cas9D+gRNA01 and Cas9H+gRNA02 (T, Figure 1B and C), while the non-transcribed strand is nicked by Cas9D+gRNA02 and Cas9H+gRNA01 (N, Figure 1B and C). The induction of nickHR by Cas9D was higher than Cas9H with gRNA01, but the relative level of induction with Cas9D was even greater with gRNA02. Thus, these results suggest that the transcribed-strand bias recently reported for nick-induced gene targeting events (13) may not be recapitulated with intrachromosomal HR.
nickHR induced by Cas9H in ES cells was very low under the transfection conditions that we used (Supplementary Figure S1C). To improve the efficiency, a redesigned gRNA backbone was used which has been reported to enhance gRNA transcription and stability (43). The enhanced gRNA (gRNA01enh) substantially improved the induction of both DSBHR and nickHR in ES-DR-GFP cells (Figure 1D) as well as in U2OS-DR-GFP cells, especially for the nickases (Supplementary Figure S1B). With the enhanced gRNA design we compared the kinetics of nickHR and DSBHR and found that their overall kinetics were similar (Supplementary Figure S1D).
It has been reported that the Cas9N863A (Cas9N) mutant, like Cas9H, also nicks the strand identical to the gRNA sequence (50). We compared the efficiency of Cas9N with Cas9H at inducing nickHR and found that they were similar (Supplementary Figure S1B). Importantly, differences in HR induction by the different Cas9 variants were not due to varying expression levels (Supplementary Figure S1B).
The generally lower levels of nickHR, as compared to DSBHR, could be due to the activity of dedicated SSB repair mechanisms that may compete with HR for nicked DNA. Inhibition of poly(ADP)ribose polymerase 1 (PARP1), involved in early signaling during SSB repair, had little or no effect on nickHR and DSBHR (Supplementary Figure S1E and F). However, we observed significantly increased nickHR, but not DSBHR, in cells deficient for XRCC1, a molecular scaffold protein essential in later stages of SSB repair (51) (Supplementary Figure S1G and H). These results suggest that Cas9-induced nicks can be processed by SSB repair pathways which thus may indirectly modulate nickHR frequencies.
HR repair of nicks on the lagging-strand template is promoted by replication
DSBHR has been reported to be reduced outside of S phase (52). To determine whether cell cycle phase influences nickHR, U2OS DR-GFP cells were arrested at G2/M by 16 h incubation in the presence of nocodazole, a microtubule depolymerizing agent (53). The arrested cells were transfected with Cas9 and gRNA expression vectors, and nocodazole treatment was continued for another 24 h, at which time cells were collected for flow cytometry. Approximately 85–90% of nocodazole-treated cells were in G2/M, compared with 35% of DMSO-treated control cells (Figure 1E), and the nocodazole-induced arrest persisted until GFP expression was analyzed 24 h later (Supplementary Figure S1I). In agreement with previous results (52), a 4-fold reduction in DSBHR was observed in the arrested cells (Figure 1F). We detected a similar 4-fold reduction in nickHR, regardless of which DNA strand was nicked (Figure 1F), suggesting that nickHR is not active in all phases of the cell cycle. Although we cannot rule out that the absolute levels of break formation were affected, nocodazole treatment had little effect on transfection efficiencies (NZE-GFP, Supplementary Figure S1I).
To explore the effect of DNA replication on nickHR, we used the DR-OriP-GFP reporter (36,54) which has the Epstein–Barr virus origin of replication (OriP) and can replicate in the presence of the EBNA1 protein (36,54) (Figure 1G). Replication from OriP at the dyad symmetry (DS) element is essentially unidirectional, given that EBNA1 binding to the family of repeat (FR) elements acts as a replication fork barrier (55). U2OS cells were transiently transfected with DR-OriP-GFP and an EBNA1 expression vector, together with Cas9 and gRNA01 expression vectors, and the frequency of GFP+ cells was determined 48 h later. In agreement with previous results (36), a modest (∼1.3-fold) increase in the number of GFP+ cells was observed in the presence of the EBNA1 expression vector (Figure 1H), suggesting that DSBHR is weakly stimulated by replication. Alternatively, the modest increase in DSBHR in the presence of EBNA1 could be at least partly explained by enhanced GFP expression due to replication, which is supported by ∼3-fold increase in the mean fluorescence intensity of GFP+ cells (data not shown) and sustained GFP expression at 96 h post-transfection (Supplementary Figure S1J). As with Cas9WT, the fraction of GFP+ cells was only marginally increased with Cas9D with EBNA1 expression (Figure 1H). By contrast, however, the fraction of GFP+ cells was considerably higher with Cas9H when the plasmid could replicate (∼1.8-fold). The much larger increase with Cas9H cannot be explained by enhanced GFP expression due to replication, as the mean fluorescence intensity of GFP+ cells in samples transfected with Cas9D or Cas9H was only ≤30% higher for both nickases (data not shown).
In these experiments performed with gRNA01, Cas9D nicked the leading-strand template, whereas Cas9H nicked the lagging-strand template (Figure 1G). To distinguish whether the apparent difference in the stimulation of nickHR by the two nickases is due to the nicked strand or due to the Cas9 nickase variant, we repeated this experiment using gRNA02, which binds to the opposite strand. In contrast to results with gRNA01, with gRNA02 nickHR was only marginally increased with Cas9H when the plasmid could replicate, but was considerably higher with Cas9D (∼2.1-fold; Figure 1I). These results suggest that replication promotes HR at a nick on the lagging-strand template.
nickHR requires established HR factors but is not suppressed by NHEJ components
Among the early DNA damage responders are ATM and ATR, specialized kinases that typically respond to DSBs and replication stress, respectively (56,57). To test their involvement in nickHR, we applied chemical inhibitors KU55933 (ATM) and VE-821 (ATR) to transfected U2OS-DR-GFP cells. Inhibition of ATM led to a small (up to 25%) decrease in DSBHR induced by either Cas9WT or I-SceI at relatively low inhibitor concentrations (1–5 μM, Figure 2A), analogous to earlier results obtained using I-SceI in mouse cells (58). A similarly moderate (up to 25%) reduction in nickHR was observed when nicks were induced by either Cas9D or Cas9H. Conversely, inhibition of the ATR kinase significantly reduced DSBHR triggered by either Cas9WT or I-SceI; e.g. at concentrations which do not considerably influence the cell cycle (0.5 and 1 μM, (59)), DSBHR was decreased up to ∼3-fold (Figure 2B). In contrast, at these concentrations nickHR induced by either Cas9D or Cas9H was only decreased by ∼40%. The effectiveness and specificity of both inhibitors was confirmed by Western blot analysis of kinase substrates (Supplementary Figure S2A). Thus, neither ATM nor ATR signaling appear to be essential for nickHR, although both kinases may play a role in this pathway.
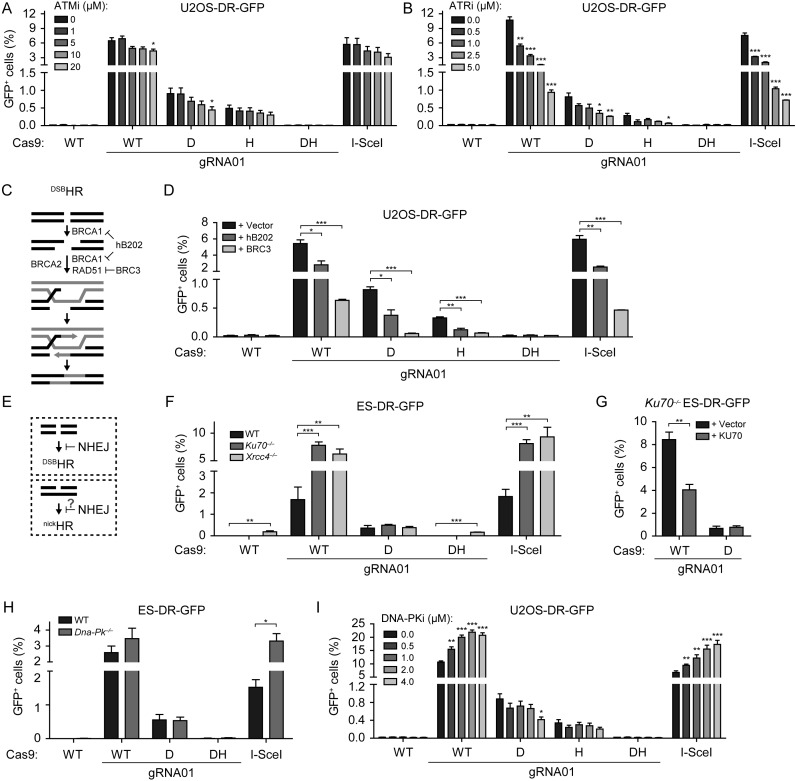
Figure 2.
nickHR requires established HR factors but is not suppressed by NHEJ components. (A and B) U2OS-DR-GFP cells transfected with the indicated Cas9, I-SceI and gRNA expression vectors were incubated for 48 h in the presence of the various concentrations of ATM inhibitor KU55933 (A) or ATR inhibitor VE-821 (B). (C) Schematic representation of HR and its steps that are inhibited by the BRCA1-interacting peptide hB202 (from BARD1) and the RAD51-interacting peptide BRC3 (from BRCA2). (D) U2OS-DR-GFP cells were transfected with the indicated Cas9 and gRNA or I-SceI expression vectors and either with the empty expression vector (pCAGGS) or with the vectors encoding the hB202 and BRC3 peptides. (E) Canonical NHEJ is known to suppress DSBHR, but its effect on nickHR is uncertain. (F) Wild-type (J1), Ku70−/− or Xrcc4−/− ES-DR-GFP cells were transfected with the indicated expression vectors. (G) Ku70−/− ES-DR-GFP cells were transfected with the indicated expression vectors and either the empty (+ Vector) or KU70 (+ KU70) expression vector. (H) Wild-type (E14) or Dna-Pk−/− ES-DR-GFP cells were transfected with the indicated expression vectors. (I) U2OS-DR-GFP cells were transfected and analyzed as in (A and B), but incubated in the presence of various concentrations of a DNA-PK inhibitor (NU7441).
DSBHR involves multiple steps, including end resection, RAD51 filament formation on the resulting 3′ overhang, strand invasion into the homologous template and repair DNA synthesis (60) (Figure 2C). BRCA1, together with its heterodimeric partner BARD1, is implicated in end resection and recruitment of other HR proteins to DNA damage sites. BRCA2 plays a critical downstream role in HR by binding RAD51 and promoting RAD51 filament formation (6). To test the involvement of BRCA1 in nickHR, we expressed a truncated form of BARD1 (hB202), which inhibits BRCA1 function by preventing its interaction with full-length BARD1 (37,40). Similar to the reduction observed in DSBHR, inhibition of BRCA1 function reduced the frequency of nickHR by about 2-fold in both U2OS-DR-GFP and ES-DR-GFP cells (Figure 2D, Supplementary Figure S2B). We then inhibited the interaction between BRCA2 and RAD51 by expressing the BRC3 repeat fragment of BRCA2 which disrupts RAD51 filament formation (37,41). BRC3 expression reduced nickHR even more substantially than hB202 in both U2OS-DR-GFP and ES-DR-GFP cells, as it did DSBHR (Figure 2D, Supplementary Figure S2B). These results suggest that nickHR proceeds, at least in part, via a pathway similar to classical DSBHR.
NHEJ is the other major pathway of DSB repair in mammalian cells (61). The canonical NHEJ pathway involves the rejoining of broken DNA ends through the activity of the KU70/KU80 and LIG4/XRCC4 complexes and the protein kinase DNA-PKcs, as well as a number of other proteins. As NHEJ is active throughout the cell cycle, including S phase when HR is also active, it can ‘compete’ with HR for the repair of broken DNA ends, such that DSBHR is elevated in NHEJ mutants (62) (Figure 2E). We thus asked whether loss of canonical NHEJ components would affect nickHR, as it does DSBHR. As previously observed with I-SceI (33),DSBHR induced by Cas9WT was substantially increased in Ku70–/– and Xrcc4–/– cells relative to the parental J1 ES-DR-GFP cells (∼4 to 5-fold; Figure 2F); re-expression of KU70 in the Ku70–/– cells reduced DSBHR (Figure 2G). However, nickHR induced by Cas9D was not significantly altered in either mutant cell line and was not affected by KU70 expression in the Ku70–/– cells. Similarly, DSBHR was augmented in Dna-Pk–/– ES cells relative to E14 ES-DR-GFP cells, but nickHR was not affected (Figure 2H). We also examined the effect of canonical NHEJ disruption in human cells by using a chemical inhibitor of DNA-PK (NU7441). DSBHR was significantly increased at all inhibitor concentrations tested, but, in contrast, there was a mild inhibition of nickHR induced by either Cas9D or Cas9H (Figure 2I). These results suggest that HR and NHEJ do not compete for repair of nicked DNA. Importantly, they provide evidence that nickHR does not likely proceed via a DSB intermediate.
Paired nicks induce intrachromosomal HR
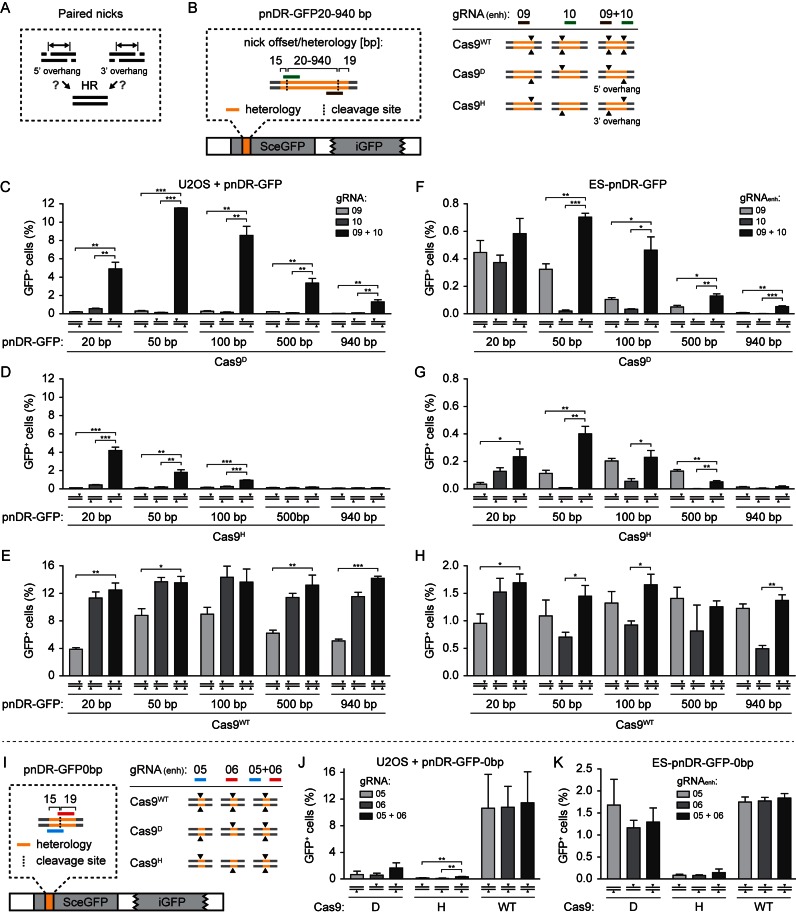
While single nicks are poor inducers of NHEJ, paired nicks induce both NHEJ and HR in genome editing applications, presumably because they are processed to DSBs (27,28) (Figure 3A). Inexplicably, paired nicks that give rise to 3′ overhangs were shown to be poor inducers of both NHEJ and HR, unlike those that give rise to 5′ overhangs. To investigate paired nick-induced HR (PNHR), we designed a series of paired-nick DR-GFP reporters (pnDR-GFP; Figure 3B). Because the distance between nicks has been shown to affect the efficiency of genome editing (27,28), DNA fragments of varying lengths were inserted between the target sites for the two gRNAs, such that paired nicks could be induced at offsets of 20 to 940 bp (pnDR-GFP-20 bp to pnDR-GFP-940 bp with gRNA09 and gRNA10). These inserts were devoid of PAMs such that neither they, nor the donor fragment, could be nicked. Further, the inserts were not homologous to the donor DNA fragment, such that the length of homology was identical in each reporter.
Figure 3.
Paired nicks induce intrachromosomal HR. (A) Schematic of paired nicks with the potential to generate 5′ or 3′ overhangs to be tested for induction of PNHR. (B) Reporters for measuring PNHR between repeats. The left panel shows a schematic of the pnDR-GFP20-940 bp reporters. The inset shows the relative binding positions of the gRNAs (marked by the green or black horizontal bars) and Cas9 cleavage sites (dotted vertical lines). The lengths of the 5′ or 3′ nick offset as well as the heterology (orange bars) are indicated. The right panel shows expected cleavage positions in the pnDR-GFP reporter with the indicated gRNA/Cas9 combinations. (C-E) U2OS cells were transiently transfected with the indicated pnDR-GFP reporter and Cas9/gRNA expression vectors required to generate single or paired nicks or DSBs (schematically drawn under each bar). (F-H) ES cells with the various pnDR-GFP reporters integrated at the Hprt locus were transfected with the indicated Cas9 and gRNA expression vectors to generate single or paired nicks or DSBs (schematically drawn under each bar). (I) Schematic representation of the pnDR-GFP0 bp reporter, analogous to (B). (J) U2OS cells were transfected with the pnDR-GFP0 bp reporter and the indicated Cas9 and gRNA expression vectors. (K) ES cells with pnDR-GFP0 bp integrated at the Hprt locus were transfected with the indicated Cas9 and gRNA expression vectors.
Transient transfections of the pnDR-GFP reporters, together with the Cas9 and gRNA expression vectors, were performed in U2OS cells and GFP+ cells were quantified by flow cytometry 48 h later. For induction of single nicks, the total amount of gRNA expression vector used was kept equivalent to induction of paired-nicks by adding the non-functional gRNA vector that lacked the targeting sequence. With Cas9D, expression of both gRNAs led to a substantially higher level of HR than either single gRNA at every distance tested (≥10-fold, Figure 3C), demonstrating that paired nicks are better inducers of HR than single nicks. The absolute level of PNHR, as well as the level relative to nickHR, was highest with paired nick offsets of 50 and 100 bp, although PNHR events were also observed with larger offsets of 500 and 940 bp. These results suggest that in this plasmid setting, paired nicks that can be processed to 5′ overhangs are potent inducers of PNHR. With Cas9H, expression of both gRNAs led to a higher level of HR than expression of either gRNA at offsets of 20 to 100 bp (Figure 3D), suggesting that PNHR can also be induced by DNA ends with 3′ overhangs. Overall, the absolute levels with Cas9H were lower than with Cas9D, similar to what was observed with single nicks (Supplementary Figure S1), such that PNHR with the largest nick offsets approached background levels (i.e. without targeting gRNA).
To determine if PNHR can occur between chromosomal sequences, we targeted the pnDR-GFP reporters to the Hprt locus in ES cells (33) and confirmed correct, single-copy integration by Southern blotting (Supplementary Figure S3A). With Cas9D, HR was significantly higher when two gRNAs were expressed in reporters with nick offsets ≥50 bp (Figure 3F). The fold increase, relative to HR induced by a single nick, was not as high as observed with the plasmid reporters in U2OS cells, indicating either that paired nicks are more efficiently converted to DSBs in plasmid substrates or that single nicks can be inducers of HR in the chromosomal setting, even when heterology is present adjacent to the nicks (see Supplementary Figure S3B and legend). With Cas9H, higher levels of HR were also typically observed with expression of both gRNAs compared with either single gRNA, although it was significantly higher only at nick offset of 50 bp (Figure 3G), indicating that PNHR can also occur at DNA ends with 3′ overhangs in the chromosomal context.
Efficiency of PNHR was considerably decreased at larger nick offsets, suggesting that the additional heterology may inhibit HR. Removal of the long heterology should, therefore, stimulate HR. To test this, we overexpressed exonuclease I (EXO1 (63)), which can cleave 5′ flaps and resect a nicked strand in a 5′ to 3′ direction (64), potentially generating a blunt-ended DSB. Surprisingly, however, overexpression of EXO1 in mES-pnDR-GFP-940 bp cells reduced both DSBHR and nickHR (Supplementary Figure S3C).
With Cas9WT, paired DSBs could be introduced into the pnDR-GFP reporters when both gRNAs were expressed (Figure 3B). Unlike in many of the paired nick configurations, however, paired DSBs did not substantially increase HR compared with a single DSB in either the plasmid or chromosome setting (Figure 3E and H).
An additional reporter was tested in which paired nicks could give rise to blunt ends, should both gRNA/Cas9 complexes bind (pnDR-GFP-0 bp, gRNA05 and gRNA06; Figure 3I). For the plasmid reporter in U2OS cells, HR induced by Cas9D or Cas9H and both gRNAs was only ∼3-fold higher than with either single gRNA and the absolute levels were much lower than with Cas9WT (Figure 3J), suggesting that paired nicks were not efficiently induced, possibly as a result of occlusion of the target site by the binding of a single gRNA/Cas9 complex. For the chromosome reporter in ES cells, HR induced by either nickase was similar regardless of whether one or both gRNAs were expressed, also consistent with cleavage site occlusion preventing DSB formation. However, in this case, interestingly, Cas9D induced HR to nearly the same extent as Cas9WT (Figure 3K). While single nicks are generally less efficient at inducing HR than DSBs, at least one study has suggested an equivalence between nicks and DSBs (24), although the underlying mechanisms remain unclear.
NHEJ components suppress paired nick HR, depending on overhang type and length
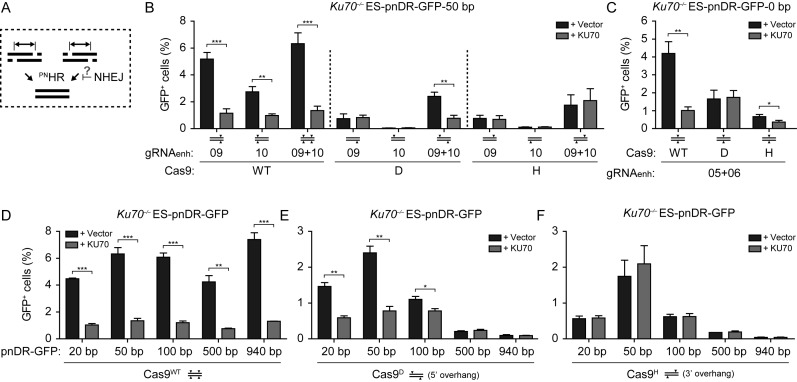
Our results demonstrating that paired nicks are more potent inducers of HR than single nicks are consistent with the notion that paired nicks are converted to DSBs. This interpretation is supported by previous work which showed that paired nicks induce mutagenic NHEJ events, while NHEJ is rarely detected at single nicks (27,28). Given that HR and NHEJ can compete for the repair of a DSB induced by Cas9WT or I-SceI (Figure 2F–I) (33), we asked whether the two pathways compete for the repair a DSB induced by paired nicks and whether the overhang type influences this process (Figure 4A). To test this, we first focused on the pnDR-GFP-50 bp reporter, since PNHR was significantly higher than nickHR for both overhang types in ES-pnDR-GFP-50 bp cells (Figure 3F and G). The reporter was targeted to the Hprt locus of Ku70–/– ES cells (Supplementary Figure S4) and HR frequencies were compared in the absence or presence of exogenous KU70 expression 48 h after transfection.
Figure 4.
Ku suppression of PNHR depends on paired nick offset type and distance. (A) Schematic representation of the distinct genetic control of HR triggered by different DNA lesions. The NHEJ component Ku suppresses DSB and PNHR resulting from 5′ overhangs but does not affect nickHR or PNHR resulting from 3′ overhangs. (B and C) Ku70−/− ES-pnDR-GFP-50 bp (B) or Ku70−/− ES-pnDR-GFP-0 bp (C) cells were transfected with the indicated Cas9 and gRNA expression vectors required to generate single or paired lesions (drawn schematically under each bar) and either the empty (+ Vector) or KU70 (+ KU70) expression vector. (D–F) Ku70−/−ES cells with the various pnDR-GFP reporter variants were transfected with the expression vectors for Cas9WT (D), Cas9D (E) or Cas9H (F), gRNA09enh, gRNA10enh and either the empty (+ Vector) or KU70 (+ KU70) expression vector.
As expected, DSBHR was significantly reduced with KU70 expression when either single or paired DSBs were induced by Cas9WT (Figure 4B). A significant reduction in Cas9D-induced PNHR was also observed with KU70 expression, indicating that NHEJ components can suppress HR involving DSBs with 5′ overhangs (Figure 4B). Surprisingly, however, Cas9H-induced PNHR was not suppressed, suggesting that NHEJ cannot compete with HR at 3′ overhangs. As observed with DR-GFP (Figure 2G), levels of nickHR were not influenced by KU70 expression, regardless of the Cas9/gRNA combination used to induce the single nicks.
To examine whether the effect of NHEJ deficiency on PNHR is influenced by nick offset, we generated a panel of Ku70–/– ES cell lines harboring the remaining pnDR-GFP reporters at the Hprt locus (Supplementary Figure S4). Cells were transfected with a Cas9 expression vector and the appropriate gRNA pairs. In each case, DSBHR induced by paired DSBs was considerably reduced (∼>4-fold) by KU70 expression (Figure 4D). Similarly, KU70 expression significantly reduced PNHR at Cas9D-generated nicks offset by 20 to 100 bp (Figure 4E), suggesting that paired nicks at these distances are converted to DSBs. However, KU70 expression did not affect PNHR at nick offsets of 500 or 940 bp, raising the possibility that paired nicks are not converted to DSBs at these distances or that DSBs with long 5′ overhangs cannot be processed by NHEJ. In contrast, HR of paired nicks generated by Cas9H was not affected by KU70 expression, regardless of the distance between nicks (Figure 4F). However, given that PNHR induced by Cas9H was significantly higher than nickHR only at a nick offset of 50 bp in wild-type ES cells (Figure 3G), we cannot exclude that the lack of an effect with KU70 expression at other distances is due to the lack of robust PNHR.
Interestingly, at nick offset of 0 bp, KU70 overexpression reduced frequency of HR when we used Cas9H but not Cas9D (Figure 4C). In the case of Cas9D, HR was equivalently high in wild-type ES cells with a single nick as with two nicks (Figure 3K), such that the bulk of the events may be attributable to nickHR. However, in the case of Cas9H, HR was somewhat higher with two nicks than with a single nick (Figure 3K), such that a low level of HR may be attributable to DSB formation which is suppressed by KU70.
These results suggest that although short 5′ overhangs of 20–100 bp generated by paired nicking can be processed by NHEJ as well as HR, neither longer 5′ overhangs nor 3′ overhangs of any length can engage the NHEJ machinery.
DISCUSSION
Despite the recent development of genome engineering techniques relying on the CRISPR-Cas9 system for the induction of single or paired nicks, the mechanisms of nickHR and PNHR and their importance in vivo remain largely unexplored. Here, we utilize the HR reporter DR-GFP and Cas9 nickases to study HR induced by either single or paired nicks in comparison to Cas9-induced DSBs in mammalian cells and determine that there is distinct genetic control of HR triggered by these different lesions.
nickHR requires core HR proteins and is not suppressed by NHEJ factors
BRCA1, BRCA2 and RAD51 are established core HR factors for the repair of DSBs in mammalian cells (65–67). The importance of RAD51 and BRCA2 in nickHR has been reported recently (13) and our results confirm that nickHR is reduced upon disruption of RAD51 function and extend these observations to BRCA1. Thus, nickHR requires the same core factors as DSBHR and proceeds via RAD51/BRCA2-mediated strand invasion (6). BRCA1 may be required only at this invasion step, possibly to promote the recruitment of PALB2-BRCA2, although we cannot rule out its involvement in an earlier step (6).
NHEJ is well established to compete with HR for the repair of DSBs (62). Our results with Cas9-induced DSBs concur with these earlier findings. Notably, nickHR is not increased in the absence of canonical NHEJ factors, suggesting that it does not necessarily rely on DSB intermediates. In agreement, single Cas9-generated nicks fail to induce oncogenic chromosomal translocations, which require DSB formation (68). However, nicks may reportedly become NHEJ substrates in some contexts, e.g. when nickHR is impaired by reduction of BRCA2 activity (12,13).
Two master kinases, ATM and ATR, control the cellular response to DNA damage (57). Inhibition of ATM results in a limited reduction in DSBHR (58), possibly by impairing Mre11-mediated end resection (69). Our results show a similarly limited reduction in nickHR upon ATM inhibition. Since ATM is not likely activated by DNA nicks or gaps (70), the small reduction in nickHR might be attributable to an involvement in later steps of HR (71) or to its role in activation of ATR (69,70). In cells treated with concentrations of an ATR inhibitor that are not likely to affect the cell cycle, we observe up to a ∼3-fold reduction of DSBHR but only a small and non-significant reduction of nickHR, more comparable to that observed after ATM inhibition. This suggests that, in contrast to DSBHR, involvement of ATR in nickHR is also limited. Since a robust activation of ATR by gapped DNA has been reported (72), either nicks are not converted to gaps or activation of ATR is not critical for nick repair by HR. As with ATM and ATR, a similarly mild reduction of nickHR is observed upon inhibition of DNA-PK. Again, this contrasts with DSBHR which was considerably increased. Thus, while inhibitors of ATM, ATR and DNA-PK have distinct effects on DSBHR (mildly inhibitory, strongly inhibitory, and promoting, respectively), each of the inhibitors exerts relatively mild inhibitory effects on nickHR. All three kinases have been shown to interact at replication-related lesions in vitro (72) and it is feasible that a fraction of Cas9-induced nicks may be encountered by incoming replication forks and require their concerted action for HR repair.
Ku suppresses PNHR induced by 5′ but not 3′ overhangs
A recently developed genome engineering approach involves nicks on opposing DNA strands—paired nicks—to stimulate gene targeting and NHEJ (27,28). Our experimental design builds on this approach. We generated paired nicks with the potential to form 5′ or 3′ overhangs and found that either configuration stimulates HR in both plasmid and chromosomal substrates, especially when nicks are generated in proximity (≤100 bp). HR is higher with paired nicks that could give rise to 5′ overhangs, although we cannot rule out that this is due to the overall higher efficiency of Cas9D relative to Cas9H. In contrast to our results, previous studies reported that only 5′ overhangs efficiently stimulate PNHR. While the previous studies used Cas9D with different gRNA pairs to create both types of overhangs (27,28), our study uses Cas9D and Cas9H with a single set of gRNAs to generate 5′ and 3′ overhangs, respectively. In principle, the PNHR we observe with 3′ overhangs could be due to how Cas9H cleaves DNA: Cas9D and Cas9H use distinct catalytic domains, and cleavage by Cas9H has been reported to be accompanied by exonuclease activity at the 3′ ends (18). However, this minimal exonuclease activity is unlikely to promote HR as it would be directed at the long overhangs. Thus, it seems more likely that contrasting results with 3′ overhangs are due to other differences in experimental design, e.g. nicking of the donor homology region (28), which has been shown to affect HR (13).
Even though it seems surprising that directional DNA unwinding or resection can occur from nicks separated by tens or hundreds of nucleotides, paired nicks with the potential to form 5′ overhangs appear to be processed to DSBs because they can give rise to NHEJ-mediated insertions and deletions (27,28,68). Supporting the generation of a DSB intermediate also for HR, the level of HR induced by paired nicks is higher than by single nicks. Our results in Ku-deficient cells confirm a DSB intermediate with 5′ overhangs: HR is significantly increased in the absence of Ku with paired nicks, but not with single nicks, for offsets up to 100 bp. In agreement, Ku has been shown to efficiently bind double-stranded DNA with long 5′ overhangs in vivo (200 nt; (73)). Thus, HR is induced by DSBs with long 5′ non-homologous tails and this can be antagonized by Ku.
Remarkably, paired nicks with the potential to generate 5′ overhangs of 500 and 940 bp were still able to induce HR at significantly higher levels than single nicks, although the overall levels were reduced compared to more closely-spaced nicks. PNHR at these offsets was not, however, suppressed by Ku. Even at 100 bp, the suppression by Ku was much weaker than at shorter offsets, raising the possibility that not all paired nicks are converted to DSBs. Alternatively, the long overhangs may not be recognized by Ku as DSB ends, but rather as ssDNA, which is a poor substrate for Ku binding (74). Since nicks are the primary DNA lesion induced by ionizing radiation (75), the ability of distant paired nicks to generate a DSB advances our understanding of DSB induction by ionizing radiation and other agents or processes that result in DNA nicks.
In contrast to 5′ overhangs, HR was not affected by loss of Ku when we used Cas9H which should generate 3′ overhangs. For example, at the 50 bp nick offset which gave the highest level of PNHR, no significant difference was observed in the presence or absence of Ku. One possibility is that DNA unwinding (or resection) required to generate DSBs at paired nicks can only occur efficiently in a single direction (i.e. 5′ > 3′). An alternative explanation is that DSBs with long 3′ overhangs are formed but they are not processed by NHEJ, such that the HR and NHEJ machineries are not competing for this type of DNA end. The poor recovery of NHEJ events from DSBs with 3′ overhangs in human cells (generated in vivo by Cas9D; (27,28)) and in yeast cells (generated by an exonuclease in vitro; (76)) supports this interpretation. Mechanistically, weaker binding of Ku to 3′ overhangs in vitro as compared with 5′ overhangs (∼20 nt; (77)) would support a greater role for Ku at 5′ overhangs; further, one or more polymerases could fill in 5′ overhangs (68,78) to make them more readily bound by Ku. Because 3′ overhangs are generated during HR-associated DSB end resection, inefficient Ku binding could be important for channeling these intermediates into HR.
Paired nicks induced directly opposite each other should, in principle, result in a blunt-ended DSB. However, Cas9-gRNA complexes may remain tightly bound to DNA after the induction of the initial nick (79) which would hamper the nicking of the second strand. Consistent with this, HR is only slightly increased by gRNA pairs that would have the potential to produce blunt ends, suggesting a low level of DSB formation. Accordingly, Ku suppresses PNHR when blunt ends are generated by Cas9H, which also indicates that the modulation of PNHR by Ku is not limited to lesions generated by Cas9D.
Influence of replication on nickHR
Our experiments confirm previous reports that nicks trigger HR (8,9,11–13,27,49,80). nickHR is generally lower than DSBHR; nicks may thus be inherently less recombinogenic, or, alternatively, they may be preferentially repaired by dedicated SSB repair pathways, which could limit the availability of nicked DNA for nickHR. Indeed, it has been reported that inhibition of PARPs, some of which are involved in early steps of SSB repair (81), increases nickHR repair of I-AniI-induced nicks (12). Although in our system nickHR was not clearly affected by PARP inhibition, we did detect increased nickHR in cells lacking the key SSB repair factor XRCC1 (51). Our results suggest, therefore, that Cas9-induced nicks are legitimate substrates for SSB repair pathways and that SSB repair and HR can compete for repair of nicked DNA, analogous to the competition between NHEJ and HR for a DSB (62).
Lesions that arise during DNA replication have been proposed to trigger HR (82). We indeed observe that replication stimulates nickHR, but only when the lagging-strand template was nicked. While further experiments will need to be performed to determine the generality of this result, it is interesting to speculate on its potential implications. Since DSBHR is generally more efficient than nickHR, conversion to a DSB could explain the increase in nickHR on a replicating plasmid, as proposed in other studies (4,83–85). Indeed, physical evidence has been provided for conversion of lagging-strand and leading-strand nicks induced by the HO endonuclease to a DSB at the yeast mat1 locus (84). But the question then arises as to why repair of the leading-strand template nick is unaffected by replication in our system. The authors of this previous study suggested that a nick on the lagging-strand template would be converted to a two-ended DSB by replication upstream of the nick, however, a similar conversion of the nick on the leading-strand template would require a second replication fork arriving from the opposite direction (84). In the absence of the second fork, the leading-strand template nick could only be converted to a one-ended DSB. Since in our experimental system forks should arrive from a single direction dictated by the orientation of the OriP cassette (Figure 1G), only the lagging-strand template nick would produce a two-ended DSB. Such a DSB might lead to a stronger stimulation of HR than a one-ended DSB generated by a leading-strand template nick. An interesting alternative explanation is that incomplete replication of the lagging-strand template results in an inherently recombinogenic 3′ overhang, while replication to the nick on the leading-strand template generates a blunt-ended DSB that would require additional processing prior to engaging the HR machinery. In either case, these results indicate the power of the Cas9 nickase system for probing the relationship between replication and HR. It should be noted, however, that gRNA binding causes DNA unwinding which could potentially influence repair of single or paired nicks. While other nuclease platforms (e.g. zinc-finger nucleases, (22,23) or I-AniI (9,13)) lack the flexibility of Cas9, they could thus further our understanding of various aspects of nickHR.
The data presented in this study begin to unravel the complex relationships between nick and DSB repair pathways during processing of nicked DNA in mammalian cells and may advance the genome engineering capabilities that are essential in developing effective and safe gene therapies.
Supplementary Material
Acknowledgments
This work was performed at MSK. We thank Dr Keith Caldecott (University of Sussex) for providing the XRCC1 expression plasmids, Dr Winfried Edelmann (Albert Einstein College of Medicine) and Dr Jan LaRocque (Georgetown University) for providing the EXO1 expression plasmid.
FUNDING
NIH/NCI Cancer Center Support Grant P30 CA008748 to MSK; R01 GM054668 to M.J.; Dutch Cancer Society fellowship [4962 to P.M.K.]. Funding for open access charge: Dutch Cancer Society fellowship [4962 to P.M.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caldecott K.W. DNA single-strand break repair. Exp. Cell Res. 2014;329:2–8. doi: 10.1016/j.yexcr.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 3.Haber J.E. DNA recombination: the replication connection. Trends Biochem. Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 4.Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 5.Chapman J.R., Taylor M.R.G., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- 6.Prakash R., Zhang Y., Feng W., Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galli A., Schiestl R.H. Effects of DNA double-strand and single-strand breaks on intrachromosomal recombination events in cell-cycle-arrested yeast cells. Genetics. 1998;149:1235–1250. doi: 10.1093/genetics/149.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee G.S., Neiditch M.B., Salus S.S., Roth D.B. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 9.McConnell Smith A., Takeuchi R., Pellenz S., Davis L., Maizels N., Monnat R.J., Jr, Stoddard B.L. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakahara M., Sonoda E., Nojima K., Sale J.E., Takenaka K., Kikuchi K., Taniguchi Y., Nakamura K., Sumitomo Y., Bree R.T., et al. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 2009;5:e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metzger M.J., McConnell-Smith A., Stoddard B.L., Miller A.D. Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res. 2011;39:926–935. doi: 10.1093/nar/gkq826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metzger M.J., Stoddard B.L., Monnat R.J., Jr PARP-mediated repair, homologous recombination, and back-up non-homologous end joining-like repair of single-strand nicks. DNA Repair. 2013;12:529–534. doi: 10.1016/j.dnarep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis L., Maizels N. Homology-directed repair of DNA nicks via pathways distinct from canonical double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E924–E932. doi: 10.1073/pnas.1400236111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouet P., Smih F., Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keith Joung J., Sander J.D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2012;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 17.Stoddard B.L. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Friedman G., Doyon Y., Wang N.S., Li C.J., Miller J.C., Hua K.L., Yan J.J., Babiarz J.E., Gregory P.D., et al. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22:1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez C.L., Certo M.T., Mussolino C., Goodwin M.J., Cradick T.J., McCaffrey A.P., Cathomen T., Scharenberg A.M., Joung J.K. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muñoz M.C., Yanez D.A., Stark J.M. An RNF168 fragment defective for focal accumulation at DNA damage is proficient for inhibition of homologous recombination in BRCA1 deficient cells. Nucleic Acids Res. 2014;42:7720–7733. doi: 10.1093/nar/gku421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran F.A., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujii W., Onuma A., Sugiura K., Naito K. Efficient generation of genome-modified mice via offset-nicking by CRISPR/Cas system. Biochem. Biophys. Res. Commun. 2014;445:791–794. doi: 10.1016/j.bbrc.2014.01.141. [DOI] [PubMed] [Google Scholar]
- 30.Rong Z., Zhu S., Xu Y., Fu X. Homologous recombination in human embryonic stem cells using CRISPR/Cas9 nickase and a long DNA donor template. Protein Cell. 2014;5:258–260. doi: 10.1007/s13238-014-0032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duda K., Lonowski L.A., Kofoed-Nielsen M., Ibarra A., Delay C.M., Kang Q., Yang Z., Pruett-Miller S.M., Bennett E.P., Wandall H.H., et al. High-efficiency genome editing via 2A-coupled co-expression of fluorescent proteins and zinc finger nucleases or CRISPR/Cas9 nickase pairs. Nucleic Acids Res. 2014;42:e84. doi: 10.1093/nar/gku251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce A.J., Hu P., Han M., Ellis N., Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson C., Moynahan M.E., Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocations. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor R.M., Moore D.J., Whitehouse J., Johnson P., Caldecott K.W. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol. Cell. Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakanishi K., Cavallo F., Perrouault L., Giovannangeli C., Moynahan M.E., Barchi M., Brunet E., Jasin M. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nat. Struct. Mol. Biol. 2011;18:500–503. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stark J.M., Pierce A.J., Oh J., Pastink A., Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol. Cell. Biol. 2004;24:9305–9316. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinstock D.M., Richardson C.A., Elliott B., Jasin M. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair. 2006;5:1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 39.Elliott B., Jasin M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 2001;21:2671–2682. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Westermark U.K., Reyngold M., Olshen A.B., Baer R., Jasin M., Moynahan M.E. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol. Cell. Biol. 2003;23:7926–7936. doi: 10.1128/MCB.23.21.7926-7936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies A.A., Masson J.Y., McIlwraith M.J., Stasiak A.Z., Stasiak A., Venkitaraman A.R., West S.C. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 42.Vriend L.E.M., Jasin M., Krawczyk P.M. Assaying break and nick-induced homologous recombination in mammalian cells using the DR-GFP reporter and Cas9 nucleases. In: Doudna J, editor. The Use of CRISPR/Cas9, ZFNs,and TALENs in Generating Site-Specific Genome Alterations. Vol. 546. Amsterdam: Elsevier; 2014. pp. 145–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.-W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pierce A.J., Jasin M. Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol. Biol. 2005;291:373–384. doi: 10.1385/1-59259-840-4:373. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Vanoli F., LaRocque J.R., Krawczyk P.M., Jasin M. Biallelic targeting of expressed genes in mouse embryonic stem cells using the Cas9 system. Methods. 2014;69:171–178. doi: 10.1016/j.ymeth.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M.B., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.M. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 47.Reaper P.M., Griffiths M.R., Long J.M., Charrier J.-D., Maccormick S., Charlton P.A., Golec J.M.C., Pollard J.R. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 48.Leahy J.J.J., Golding B.T., Griffin R.J., Hardcastle I.R., Richardson C., Rigoreau L., Smith G.C.M. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 49.Katz S.S., Gimble F.S., Storici F. To nick or not to nick: comparison of I-SceI single- and double-strand break-induced recombination in yeast and human cells. PLoS One. 2014;9:e88840. doi: 10.1371/journal.pone.0088840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caldecott K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 52.Saleh-Gohari N., Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zieve G.W., Turnbull D., Mullins J.M., McIntosh J.R. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980;126:397–405. doi: 10.1016/0014-4827(80)90279-7. [DOI] [PubMed] [Google Scholar]
- 54.Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gahn T.A., Schildkraut C.L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989;58:527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- 56.Paulsen R.D., Cimprich K.A. The ATR pathway: fine-tuning the fork. DNA Repair. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 57.Shiloh Y., Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 58.Kass E.M., Helgadottir H.R., Chen C.-C., Barbera M., Wang R., Westermark U.K., Ludwig T., Moynahan M.E., Jasin M. Double-strand break repair by homologous recombination in primary mouse somatic cells requires BRCA1 but not the ATM kinase. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5564–5569. doi: 10.1073/pnas.1216824110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujisawa H., Nakajima N.I., Sunada S., Lee Y., Hirakawa H., Yajima H., Fujimori A., Uesaka M., Okayasu R. VE-821, an ATR inhibitor, causes radiosensitization in human tumor cells irradiated with high LET radiation. Radiat. Oncol. 2015;10:175. doi: 10.1186/s13014-015-0464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jasin M., Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kass E.M., Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolderson E., Richard D.J., Edelmann W., Khanna K.K. Involvement of Exo1b in DNA damage-induced apoptosis. Nucleic Acids Res. 2009;37:3452–3463. doi: 10.1093/nar/gkp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Potenski C.J., Niu H., Sung P., Klein H.L. Avoidance of ribonucleotide-induced mutations by RNase H2 and Srs2-Exo1 mechanisms. Nature. 2014;511:251–254. doi: 10.1038/nature13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moynahan M.E., Chiu J.W., Koller B.H., Jasin M. Brca1 controls homology-directed DNA repair. Mol. Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 66.Moynahan M.E., Pierce A.J., Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 67.Stark J.M., Hu P., Pierce A.J., Moynahan M.E., Ellis N., Jasin M. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J. Biol. Chem. 2002;277:20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 68.Ghezraoui H., Piganeau M., Renouf B., Renaud J.-B., Sallmyr A., Ruis B., Oh S., Tomkinson A.E., Hendrickson E.A., Giovannangeli C., et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C.M., Lukas J., Jackson S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 70.Shiotani B., Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bakr A., Oing C., Köcher S., Borgmann K., Dornreiter I., Petersen C., Dikomey E., Mansour W.Y. Involvement of ATM in homologous recombination after end resection and RAD51 nucleofilament formation. Nucleic Acids Res. 2015;43:3154–3166. doi: 10.1093/nar/gkv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vidal-Eychenié S., Décaillet C., Basbous J., Constantinou A. DNA structure-specific priming of ATR activation by DNA-PKcs. J. Cell Biol. 2013;202:421–429. doi: 10.1083/jcb.201304139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ristic D., Modesti M., Kanaar R., Wyman C. Rad52 and Ku bind to different DNA structures produced early in double‐strand break repair. Nucleic Acids Res. 2003;31:5229–5237. doi: 10.1093/nar/gkg729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mimori T., Hardin J.A. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 75.Goodhead D.T. The initial physical damage produced by ionizing radiations. Int. J. Radiat. Biol. 1989;56:623–634. doi: 10.1080/09553008914551841. [DOI] [PubMed] [Google Scholar]
- 76.Frank-Vaillant M., Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol. Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 77.Falzon M., Fewell J.W., Kuff E.L. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J. Biol. Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 78.Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.-S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arcangioli B. A site- and strand-specific DNA break confers asymmetric switching potential in fission yeast. EMBO J. 1998;17:4503–4510. doi: 10.1093/emboj/17.15.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fisher A.E.O., Hochegger H., Takeda S., Caldecott K.W. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 83.Saleh-Gohari N., Bryant H.E., Schultz N., Parker K.M., Cassel T.N., Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cortés-Ledesma F., Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roseaulin L., Yamada Y., Tsutsui Y., Russell P., Iwasaki H., Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.