Abstract
DNA damage tolerance (DDT) is responsible for genomic stability and cell viability by bypassing the replication block. In Saccharomyces cerevisiae DDT employs two parallel branch pathways to bypass the DNA lesion, namely translesion DNA synthesis (TLS) and error-free lesion bypass, which are mediated by sequential modifications of PCNA. Rad5 has been placed in the error-free branch of DDT because it contains an E3 ligase domain required for PCNA polyubiquitination. Rad5 is a multi-functional protein and may also play a role in TLS, since it interacts with the TLS polymerase Rev1. In this study we mapped the Rev1-interaction domain in Rad5 to the amino acid resolution and demonstrated that Rad5 is indeed involved in TLS possibly through recruitment of Rev1. Genetic analyses show that the dual functions of Rad5 can be separated and reconstituted. Crystal structure analysis of the Rad5–Rev1 interaction reveals a consensus RFF motif in the Rad5 N-terminus that binds to a hydrophobic pocket within the C-terminal domain of Rev1 that is highly conserved in eukaryotes. This study indicates that Rad5 plays a critical role in pathway choice between TLS and error-free DDT.
INTRODUCTION
Eukaryotic cells from unicellular yeast to human possess a DNA damage tolerance (DDT) pathway defined as bypassing replication-blocking lesions without removing them. This pathway, which is best understood in the model lower eukaryotic microorganism Saccharomyces cerevisiae, is also known as the post-replication repair (PRR) pathway and the relevant genes belong to the RAD6 epistasis group (1,2).
The budding yeast DDT pathway can be divided into two parallel branches: an error-prone branch, also known as translesion DNA synthesis (TLS), which is mediated by Rev1 and Polζ (3) and an error-free branch, which is thought to rely on newly synthesized sister chromatid as a template to synthesize across the replication block (2,4). In budding yeast, DDT is regulated by sequential ubiquitination of proliferating cell nuclear antigen (PCNA), which is encoded by the POL30 gene and forms a homotrimeric DNA clamp. It is now clear that Rad6 (E2) and Rad18 (E3) catalyze PCNA monoubiquitination at its K164 residue (5), and monoubiquitinated PCNA helps to recruit TLS polymerases for lesion bypass (6). Monoubiquitinated PCNA can be further polyubiquitinated by another E2–E3 complex, Mms2-Ubc13-Rad5 (5). Ubc13 and the Ubc variant Mms2 form a stable heterodimer dedicated to promoting K63-linked polyubiquitination (7–9) while Rad5 serves as a cognate E3 (10,11). It is believed that polyubiquitinated PCNA promotes error-free lesion bypass although the exact mechanism remains elusive (12,13). Interestingly, the same Pol30-K164 residue can also be sumoylated by the Ubc9-Siz1 complex (5,14), and sumoylated PCNA is thought to recruit the anti-Rad51 helicase Srs2 to prevent unwanted homologous recombination (15,16).
Although Rad5 is involved in the error-free branch of the DDT pathway together with Mms2 and Ubc13, its mutant phenotypes are complicated. The rad5 null mutant is less sensitive to DNA-damaging agents than rad18 but much more sensitive than mms2 or ubc13 (17), which are thought to be only involved in error-free DDT, suggesting that Rad5 is a multi-functional protein. Indeed RAD5 was also identified as REV2 because its mutant displayed a compromised mutagenesis phenotype following UV irradiation, although subsequent characterization revealed that, unlike other rev mutants, rev2 only affects reversion of the arg4-17 allele among several mutagenesis assays (18–20). Several attempts have been made to understand the additional function(s) of Rad5. For example, Rad5 has been implicated in the repair of double-strand breaks (21), possibly through avoidance of non-homologous end joining (22) and its helicase activity is thought to promote replication fork regression (23). An Mms2-Ubc13-independent role in TLS was proposed (24), and a physical interaction between Rad5 and Rev1 was subsequently reported (25); however, the physiological significance of this interaction was not explored due to a lack of mutations that selectively affect this physical interaction.
Deduced Rad5 contains seven conserved SWI2/SNF2 chromatin-remodeling motifs, a putative helicase domain as well as a RING finger domain in the C-terminal half of the protein (19,26). The RING finger domain is thought to be required for the physical interaction with Ubc13, as a Rad5-I916A point mutation within this region abolishes Rad5–Ubc13 interaction (10). The N-terminal half of the protein mainly contains a leucine heptad repeat or leucine zipper and a HIRAN domain (27), both of which are thought to play a role in DNA binding or protein–protein interaction, but their actual activities remain unclear. Here we report the fine mapping of the Rev1-binding domain and the creation of allele-specific mutations in Rad5 that only affect its interaction with Rev1. This mutation allowed us to critically test a hypothesis that Rad5 is involved in both error-prone and error-free DDT. Furthermore, we determined the crystal structure of the Rad5-Rev1 interaction motifs, which reveals a consensus motif in Rad5 that binds to the C-terminus of Rev1. The structural analysis also reveals a highly-conserved Rev1 C-terminus in eukaryotes serving as a versatile scaffold and trading place. Together with other reports, we are able to revise the current model of DDT in budding yeast.
MATERIALS AND METHODS
Media and yeast strains
Rich yeast-extract peptone dextrose (YPD) broth medium was used to culture yeast cells and supplemented with 120 mg/l adenine to avoid accumulation of a pink intermediate. Strains requiring selection were cultured in an SD medium containing required amino acids and nucleosides as instructed (28). Plasmids were transformed into yeast cells following a modified lithium acetate method (29).
The yeast strains used in this study are listed in Supplementary Table S1. All the strains used in this study were made from and isogenic to DBY747, HK578 or BY4741 background except PJ69-4a (30). Mutants strains were created by a one-step gene deletion method (31) using disruption cassettes as previously described, and genomic polymerase chain reaction (PCR) and phenotypic analysis were employed to confirm desired genetic markers.
Plasmid construction
The plasmid YCpL-RAD5 was constructed by cloning the RAD5 gene from genomic DNA together with its native promoter and terminator sequences by BamHI and SalI into the single copy vector YCplac111 (32). The SalI site is located 617-bp downstream from the RAD5 stop codon. YCpL-rad5ΔNT30, YCpL-rad5ΔNT60 and point mutations were derived from YCpL-RAD5 by subcloning using primers pRad5ΔNT30Stu1F/pRad5Sal1R, pRad5ΔNT60Stu1F/pRad5Sal1R or site-specific mutagenesis by a modified Quick Change method (33) using primers from Supplementary Table S2.
Yeast two-hybrid (Y2H) analysis
All Y2H plasmids were based on either pGBT9 (Gal4BD) or pGAD424 (Gal4AD) (34). Initial Rad5, Rev1 and Pol30 Y2H clones were used to create deletion and point mutation derivatives. The deletion constructs were made by either available restriction sites or PCR amplification of predetermined fragments flanked by restriction sites in the primers to facilitate cloning. Site-specific mutations were made by the PCR-based method using the mutagenic primers as listed in Supplementary Table S2. Gal4AD and Gal4BD plasmids to be tested were co-transformed into PJ69-4a, individual colonies were picked and then allowed to grow at 30°C on an SD-Leu-Trp plate for 2–3 days, after which transformants were printed on SD-Leu-Trp, SD-Leu-Trp-Ade and SD-Leu-Trp-His selective plates with or without a certain amount of the histidine biosynthesis inhibitor 1,2,4-aminotrizole (3-AT).
Yeast cell survival assays after treatment with DNA-damaging agents
The serial dilution assays and the gradient plate assays of yeast strains were performed following the protocol as previously described (35). Briefly, for the serial dilution assay, overnight cultured yeast cells were used to make a set of tenfold dilutions, and then spotted on freshly-made YPD agar plates with or without MMS. The plates were incubated for 2 days at 30°C before photography. For UV-induced DNA damage, the plate was exposed to 254 nm UV in a UV crosslinker (UVP CL-1000M) and incubated in the dark. For the MMS gradient plate assay, overnight cultures were printed onto a specially-made two-layer YPD agar plate using a glass slide. The bottom layer of the plate contains a predetermined MMS concentration and is poured on a slope. Once solid, the plates are returned to a flat surface, and a top layer without MMS is poured. Plates were incubated at 30°C for 2 days before photography. For assaying sensitivity to acute doses of MMS, overnight yeast cultures were used to inoculate fresh YPD and grown at 30°C until a cell count of ∼2 × 107cells/ml was achieved. MMS was then added to the liquid culture and samples were taken at the indicated times. Cells were pelleted by centrifugation, washed, diluted, and plated on YPD. Colonies were counted after 3 days of incubation at 30°C and scored as a percentage of cell survival against untreated cells. Sensitivity to UV was assessed similarly, with overnight yeast cultures used to inoculate fresh YPD and grown at 30°C until a cell count of ∼2 × 107cells/ml was achieved. Cells were pelleted by centrifugation, washed, diluted, plated on YPD and then exposed to the indicated doses of UV. Plates were incubated in the dark for 3 days at 30°C, and scored as a percentage of cell survival against untreated cells.
Spontaneous and UV-induced mutagenesis assays
The spontaneous mutagenesis assay was carried out by measuring the reversion rate of the trp1-289 allele in the DBY747 strain. The method used in this study is modified from the Luria and Delbruck fluctuation test as previously described (8). Yeast cells were cultured overnight at 30°C and subcultured into 5 ml of fresh YPD at a starting concentration of 20 cells/ml and incubated at 30°C for 3 days. Each strain was then collected, washed and plated onto both YPD and SD-Trp to score the Trp+ revertants.
To assess UV-induced Trp+ revertants, overnight yeast cultures were used to inoculate fresh YPD and grown at 30°C until a cell count of ∼2 × 107cells/ml was achieved. Cells were collected, washed, diluted and plated on YPD plates to score survival, and plated undiluted or concentrated on SD-Trp plates to determine Trp+ reversion. Both YPD and SD-Trp plates were exposed to UV, and then incubated for 3 days at 30°C. Plates were kept in the dark during incubation to prevent photoreactivation.
In vitro pull-down assay
The Rad5-NT164 coding sequence or Rad5-NT164 containing the FN13,14AA mutation was cloned into pGEX6 (Phamacia) in-frame with GST and the Rev1-CT239 coding sequence and its point mutation derivatives were cloned into pHis-P1 (36) in-frame with His6. Each resulting plasmid was transformed into Escherichia coli BL21 (DE3). The target fusions proteins were induced by 0.2 mM IPTG at 16°C for 22 h and affinity purified to apparent homogeneity. For the GST pull-down assay, 5 μg purified His6-Rev1-CT239 was incubated with immobilized GST-Rad5-NT164 or GST-Rad5-FN13,14AA beads overnight at 4°C. The beads were washed five times with phosphate buffered saline(PBS) and eluted with PBS containing 20 mM reduced glutathione. The eluted fractions were subjected to immunoblotting with anti-GST or anti-His antibody.
Protein engineering, expression and purification
The Rad5 (5–20) and Rev1 (876-985) coding sequences were linked together with the flexible linker sequence GGSSSSLVPRGSGGSGGSP using PCR and subcloned into a pET15b vector, which had been modified to contain a His-tag and a cleavage site for the TEV protease before the fusion gene. The fusion protein was overexpressed in E. coli BL21 (DE3) cells. Cells were grown at 37°C to an OD600 of 0.2–0.5 followed by induction of protein expression by 0.2 mM IPTG for 12–16 h at 18°C. Cells were lysed in 200 mM NaCl, 10 mM HEPES, 5% glycerol (v/v), 1 mM PMSF, pH 7.0 at 4°C using a cell homogenizer (ATS). After centrifugation, the supernatant was loaded onto a Ni2+ affinity column, washed, and eluted with buffer (200 mM NaCl, 10 mM HEPES, 250 mM imidazole, 5% glycerol (v/v), pH 7.0). The fusion tag was removed by treatment with TEV protease at 4°C for 12 h. The protein was further purified using Source 15S chromatography (GE), and then subjected to a Superdex 75 column equilibrated with protein buffer (200 mM NaCl, 10 mM HEPES, 2 mM DTT, pH 7.0), concentrated to ∼20 mg/ml and stored at −80°C.
Crystallization and data collection
Crystals were grown at 18°C by hanging-drop vapor diffusion methods. The native crystals grew from 18–20% PEG 3350, 100 mM potassium citrate, 100 mM HEPES, 5 mM DTT, pH 7.0 after 2–3 weeks. To obtain heavy atom derivative crystals, the native crystals were soaked with ethylmercurithiosalicylic acid (4 mg/ml) for 5–10 min. All crystals were harvested from buffer (100 mM NaCl, 50 mM HEPES, 50 mM potassium citrate, 20% glycerol, 20% PEG 3350, pH 7.0). Diffraction data were collected at −170°C at the beam line BL17u at Shanghai Synchrotron Radiation Facility (SSRF), and processed with the HKL2000.
Data processing and structure solution
The fusion protein crystallized into two different crystal lattices (P21 and P21212). Initial structure of the fusion complex was solved by experimental phasing using single isomorphous replacement using anomalous scattering (SIRAS). The initial phases were obtained with Hg-derivatized crystals obtained in higher symmetry lattice (P21212). Heavy atom sites were found with Shelx C/D, and phases were calculated and improved with Sharp/AutoSharp using both isomorphic and anomalous differences (37). The initial model was built with Phenix (resolve) (38). The partial model was then used as a search model for molecular replacement using the lower symmetry lattice (P21), which diffracted to higher resolution and had better data quality. The rest of the model was built manually using Coot. Refinement was performed with Phenix, using the TLSMD server to generate TLS bodies for refinement (39). The final structure was refined to Rwork/Rfree = 20.1/24.6 with Ramachandran outlier 0%, allowed 1.5% and favored 98.5%.
Calculation of surface conservation
The conservation scores were calculated using the Consurf Sever (40). Increasing conservation (scored from 1 to 9) was color coded in the figure by the spectra of white-to-blue, as described before (41). The sequences used in the calculation are: P12689.2, NP_057400.1, XP_001839880.1, XP_004525720.1, EFN73756.1, CCG21881.1, EGV65736.1, XP_456121.1, XP_003718577.1, XP_964009.1, XP_002902996.1, EMF12452.1, NP_596693.2, ETS04407.1, CCH46613.1, XP_001393536.2, EHL00847.1, KDB16736.1, GAA88822.1, XP_007811671 and CCG83408.1.
RESULTS
The N-terminus of Rad5 interacts with Rev1
There has been convincing evidence that Rad5 participates in error-free DDT by serving as an E3 for K63-linked PCNA polyubiquitination in budding yeast (5), and that this activity is mediated by its physical interaction with the cognate E2 Ubc13 via a RING finger motif (10,11). However, the rad5 null mutant is much more sensitive to DNA-damaging agents than the ubc13 null mutant, suggesting that Rad5 is a multifunctional protein. Prakash et al. first reported that Rad5 is involved in TLS (24) and later found a physical interaction between Rad5 and Rev1 (25). During a systematic yeast two-hybrid (Y2H) analysis of protein-protein interactions within the DDT pathways, we also found the Rad5-Rev1 interaction. Based on the above observations, we hypothesized that Rad5 contains separate domains for interaction with Ubc13 and Rev1 and that these two domains are responsible for the error-free and error-prone DDT, respectively. To critically test this hypothesis, it is necessary to map the Rev1-binding domain within Rad5 and create a mutation that only affects this interaction.
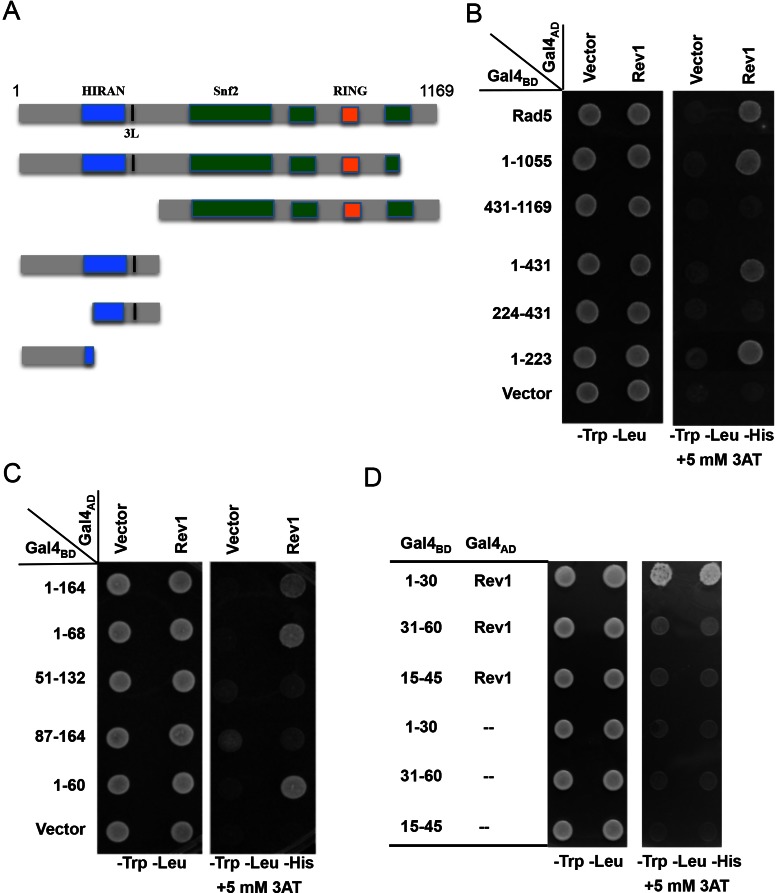
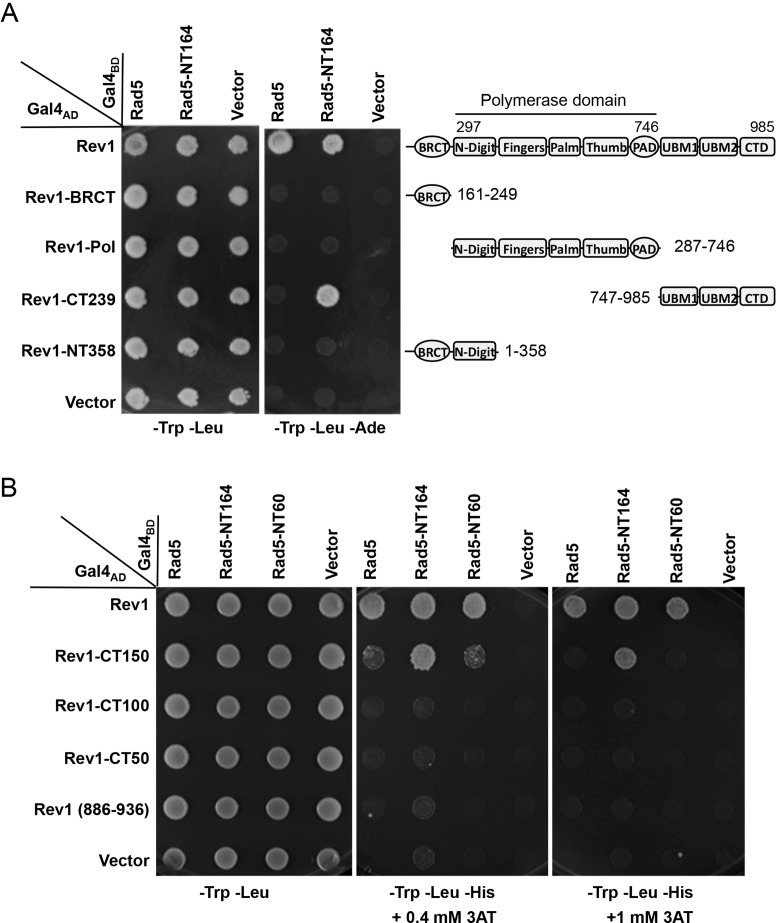
A Y2H assay was first established with full-length RAD5 (19) and REV1 (42) ORFs cloned into both pGBT9 (Gal4BD) and pGAD424 (Gal4AD) vectors. The pGBT-Rad5 and pGAD-Rev1 co-transformants displayed the gene-specific interaction (Supplementary Figure S1A), whereas pGBT-Rev1 co-transformed with the pGAD424 vector displayed false-positive results (data not shown). Hence a set of deletion constructs were made in the pGBT-Rad5 plasmids based on available restriction sites (Figure 1A) and tested against pGAD-Rev1. As seen in Figure 1B, a Rad5 truncation construct containing the N-terminal 223 amino acids is sufficient to interact with the full-length Rev1, whereas a Rad5 deletion lacking the N-terminal region fails to interact with Rev1. Since both predicted HIRAN and the leucine zipper (3L) domains are located at the N-terminus, we made two constructs with one containing both domains (aa. 161–295) and the other containing neither (aa. 1–164), and surprisingly found that the Rev1-binding domain is located within the N-terminus distinct from putative HIRAN and leucine zipper motifs (Supplementary Figure S1B). The Rad5 N-terminal 164 amino acid residues were further divided into overlapping fragments and the Rev1-binding region was mapped to the N-terminal 60 amino acids (Figure 1C). Further truncation of this 60-residue region narrowed the Rev1-binding domain within the N-terminal 30 amino acids (Figure 1D). Since no truncation without the very N-terminus displays positive results, we cautiously conclude that the N-terminal 30 amino acids of Rad5 is necessary and sufficient to mediate physical interaction with Rev1. It is noted that this region does not contain a previously known or predicted functional motif (27,43).
Figure 1.
Mapping the Rev1-binding region in Rad5 by a yeast two-hybrid assay. (A) A diagram indicating Rad5 putative functional domains and the sites of truncation. (B) The Rev1-binding region is mapped to the N-terminal 223 amino acids of Rad5. (C) The Rev1-interacting domain is restricted to the N-terminal 60 amino acids of Rad5. (D) The N-terminal 30 amino acids of Rad5 are sufficient to interact with Rev1. Various combinations of Rad5 truncations and Rev1 as indicated were co-transformed into PJ69-4a. The transformants were spotted on control plates (SD-Leu-Trp) and selective plates (SD-Leu-Trp-His+3AT), which were incubated at 30°C for 4 days before photography. Numbers on the left panel indicate Rad5 amino acid sequences encoded by the pGBT plasmids. Only images from the control plates and selective plates containing 5 mM 3AT are presented.
Point mutations in Rad5 that affect Rad5–Rev1 interaction
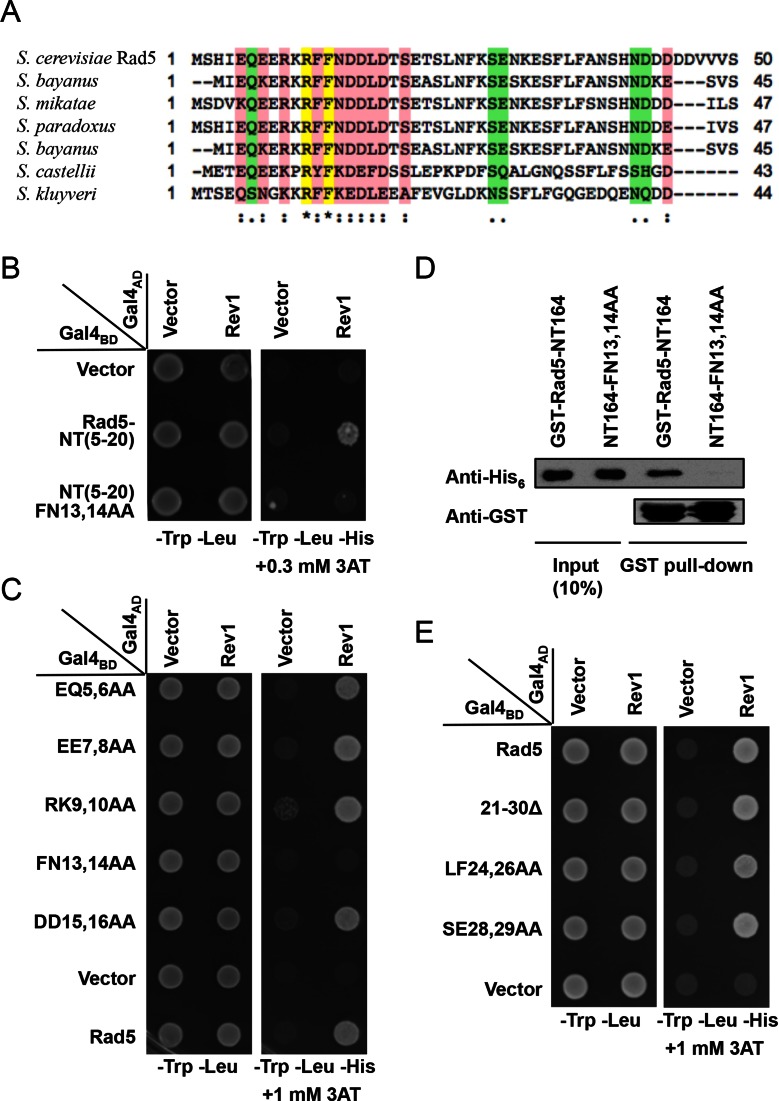
To further identify residue(s) in the Rad5 N-terminus responsible for Rev1 interaction, we searched for highly conserved Rad5 homologs from the genome database and made a multi-sequence alignment. It is noticed that the Rad5 N-terminal sequences are highly conserved within the Saccharomyces family members and that the conserved amino acid sequences are enriched between residues 5 and 20 relative to S. cerevisiae Rad5 (Figure 2A). Indeed, a polypeptide containing these 16 amino acids from the Rad5-N-terminus is sufficient to interact with Rev1, although with a reduced affinity (Figure 2B). To further map amino acid residues required for the interaction with Rev1, we made systematic ‘double alanine scan’ mutations in pGBT-Rad5 based on this alignment and obtained five such mutant clones, namely EQ5,6AA, EE7,8AA, RK9,10AA, FN13,14AA and DD15,16AA. Among the five mutant clones, only Rad5-FN13,14AA abolishes interaction with Rev1, while other mutations have no obvious effect on the interaction compared with wild-type Rad5 (Figure 2C). To independently confirm the above observations, we expressed and purified the Rad5-NT164 fragment as a GST fusion protein and Rev1-CT239 (see below for the mapping of Rad5 binding domain within Rev1) as a His6 fusion protein in bacterial expression systems and performed an in vitro pull-down assay. As seen in Figure 2D, The FN13,14AA mutation severely affected the ability of Rad5-NT164 to bind the Rev1-CT239 fragment, confirming the direct interaction between Rad5-NT and Rev1-CT and the critical role of Rad5-FN13,14 in this interaction.
Figure 2.
Mapping amino acid residues in Rad5 required for the Rev1 interaction. (A) Multi-sequence alignment of budding yeast Rad5 N-terminus with Rad5 orthologs utilizing the ClustalW alignment tool on the Saccharomyces genome database. Residues with yellow highlight indicate complete conservation; those with pink highlight indicate high conservation; while those with green highlight indicate consensus. (B) The interaction region is limited to the conserved 16 amino acids. (C) Conserved residues were selected to make dual Ala point mutations in Rad5 and tested for their ability to interact with Rev1 in a Y2H assay. (D) A GST pull-down assay to examine physical interaction of purified Rad5-NT164 or its mutant form with His6-tagged Rev1-CT239. (E) Effects of deletion or point mutations within Rad5-(21-30) residues. The Y2H experimental conditions were as described in Materials and Methods and Figure 1.
As our work was in progress, a report was published stating that Rad5-(21-360) is responsible for the Rev1 interaction (44), which overlaps 10 residues with Rad5-(1-30) but contradicts the suspected role of Rad5-NT(5-20) and Rad5-FN13,14 in such an interaction. To address the possibility that certain residue(s) within this overlapping region are also required for the Rad5–Rev1 interaction, we made ‘double alanine scan’ of conserved residues within this region and even deleted residues 21–30 from the full-length Rad5. Neither the Rad5-(Δ21-30) deletion nor selected point mutations within this region interfered with Rev1 interaction (Figure 2E), which, together with previous observations (Figure 1 and Supplementary Figure S1), effectively rules out the involvement of residues 21–360 of Rad5 in the Rev1 binding.
The Rad5-FN13,14AA mutation compromises TLS activity
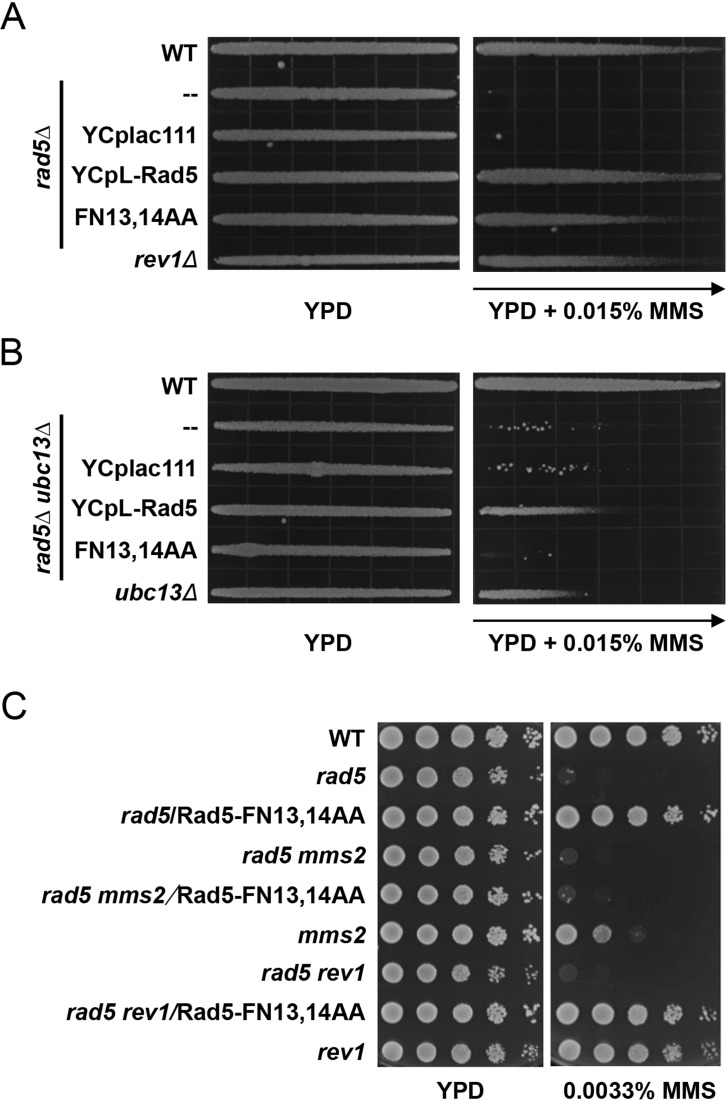
Rad5 is a multi-functional protein and its null mutant displays a pronounced level of sensitivity to DNA-damaging agents (27). To critically examine whether the Rad5-FN13,14AA point mutation affects and only affects TLS activity, we cloned the RAD5 ORF along with its native promoter and terminator sequences into a centromere-based single-copy vector and then introduced the FN13,14AA point mutations into the resulting plasmid. The wild-type and mutant plasmids were used to transform the rad5 null mutant and the sensitivity of transformants to MMS was measured by a semi-quantitative gradient plate assay. As seen in Figure 3A, expression of the wild-type RAD5 gene complemented the severe MMS sensitivity to a level indistinguishable from that of wild-type cells, while expression of the rad5-FN13,14AA mutant allele complements the rad5 null cells to a level that appears to be slightly less than the wild-type but comparable to that of the rev1 mutant.
Figure 3.
Genetic interactions of the rad5-FN13,14AA point mutation with error-free DDT and TLS pathway mutations. (A) The rad5-FN13,14AA point mutation causes moderate sensitivity to MMS, as shown by a gradient plate assay. (B) Complementation of the rad5 ubc13 double mutant by single-copy plasmid carrying RAD5 or its rad5-FN13,14AA mutant form. (A and B) Overnight cultured yeast cells were imprinted onto the premade YPD or YPD + 0.015% MMS gradient plates and the plates were incubated at 30°C for 2 days before photography. Arrows indicate increasing MMS concentration. (C) Genetic interactions between rad5-FN13,14AA and mms2 or rev1 by a serial dilution assay. Overnight-cultured yeast cells were used to make a series of tenfold dilutions and then spotted to YPD or YPD plus various concentrations of MMS. The plates were incubated at 30°C for 2 days before photography. Only one representative MMS plate is shown. All strains are isogenic derivatives of HK578-10D.
The tls single mutants only display moderate sensitivity to MMS. To enhance the tls phenotype, we conducted the complementation experiment in a ubc13 background. One of the characteristic phenotypes of tls mutations is their synergistic interaction with error-free DDT pathway mutations like mms2 (8,45) or ubc13 (9). Expression of wild-type RAD5 rescues the rad5 ubc13 double mutant cells to a level comparable to that of the ubc13 single mutant whereas in contrast, expression of rad5-FN13,14AA fails to rescue the double mutant (Figure 3B), which is a characteristic ubc13 tls double mutant phenotype (9).
We also made Rad5 clones missing either 30 or 60 N-terminal amino acid sequences and found that rad5-ΔNT30 and rad5-ΔNT60 behave like rad5-FN13,14AA (Supplementary Figure S2), suggesting that the N-terminus of Rad5 is dedicated to Rev1 interaction.
It was consistently observed that MMS-resistant colonies appear much more frequently in the rad5 ubc13 double mutant along the MMS gradient than in the rad5 single mutant (cf. Figure 3A and B). Interestingly, although expression of rad5-FN13,14AA does not rescue the rad5 ubc13 double mutant from MMS sensitivity, it effectively reduces the appearance of MMS-resistant colonies (Figure 3B). We do not know the exact mechanism underlying this phenotype, but suspect that it is due to an uncharacterized activity of Rad5 still functional in the absence of the Rad5 N-terminus.
We also compared the genetic interactions of rad5-FN13,14AA with TLS and error-free DDT. In a serial dilution assay the rad5-FN13,14AA single mutant did not display apparent sensitivity to 0.0033% MMS and the mms2 single mutant was moderately sensitive; however, the MMS sensitivity of the corresponding double mutant is at least 100-fold more sensitive than wild-type or either single mutant. In sharp contrast, the rev1 rad5-FN13,14AA double mutant is as sensitive to MMS as the corresponding single mutants (Figure 3C), confirming that rad5-FN13,14AA is epistatic to rev1 while synergistic with mms2. Hence, rad5-FN13,14AA is a characteristic tls mutant.
Intragenic interaction between error-free and error-prone DDT within the RAD5 gene
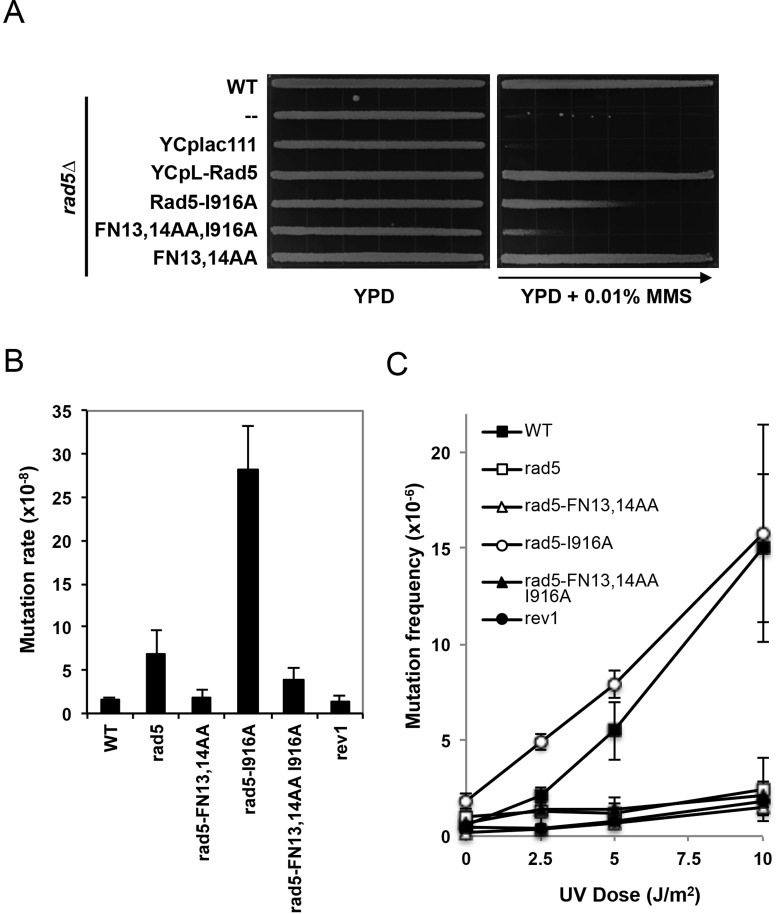
Validation of the rad5-FN13,14AA mutation as a tls allele-specific mutation allowed us to test whether the initial hypothesis holds true. To create an error-free DDT specific mutation within Rad5, we made a rad5-C914A mutation but found that this mutant is much more sensitive than the mms2 or ubc13 single mutant (data not shown), indicating that it either affects other Rad5 activities or interferes with Rad5 folding/stability. A Rad5-I916A substitution has been shown to affect physical interaction with Ubc13 and the mutant phenotype is comparable to that of ubc13 (10). In our hands, the Rad5-I916A substitution reduces its interaction with Ubc13 below a detectable level by a Y2H assay, and the rad5-I916A mutant is less sensitive to MMS and UV than a ubc13 or mms2 single mutant, suggesting that it is partially defective in the recruitment of Ubc13-Mms2. Nevertheless, mms2 and ubc13 are epistatic to rad5-I916A, indicating that rad5-I916A is only defective in error-free DDT (46). In this study, we created a YCp-Rad5 plasmid containing both rad5-FN13,14AA and I916A mutations and compared it to the corresponding single mutant for the rescuing of rad5Δ mutant phenotypes. Indeed, the rad5 double mutant is much more sensitive to MMS-induced killing than either of the corresponding single mutants (Figure 4A), confirming the intragenic complementation between TLS and error-free DDT within RAD5.
Figure 4.
Intragenic genetic interactions of RAD5 point mutations. (A) rad5-FN13,14AA and rad5-I916A mutations are synergistic with respect to MMS sensitivity. Experimental conditions were as described in Figure 3A and B. (B) Effects of rad5-FN13,14AA and rad5-I916A on spontaneous mutagenesis. (C) Effects of rad5-FN13,14AA and rad5-I916A on UV-induced mutagenesis. Strains in (A) are isogenic derivatives of HK578-10D and in (B and C) are isogenic derivatives of DBY747. Data in (B and C) are the average of at least three independent experiments with standard deviation.
Another hallmark of TLS defect is compromised spontaneous and DNA-damage induced mutagenesis. To examine whether the rad5-FN13,14AA mutation interferes with mutagenesis like rev1, we created yeast strains carrying rad5 point mutations at the RAD5 chromosomal locus and compared these mutants with rad5 and rev1 null mutants. As shown in Figure 4B, the rad5-FN13,14AA mutation reduces rad5 spontaneous mutagenesis to a level comparable to that of wild-type and rev1, while the rad5-I916A mutation increases spontaneous mutagenesis by >17-fold. Interestingly, the rad5-FN13,14AA I916A double mutant displays a spontaneous mutation rate between the two corresponding single mutant levels, indicating that the error-free DDT and TLS dual functions of Rad5 balance spontaneous mutagenesis. In contrast, it appears that the rad5-I916A mutation further enhances UV-induced mutagenesis at low doses, while rad5-FN13,14AA completely abolishes induced mutagenesis like rev1 and rad5 cells (Figure 4C). Interestingly, the double point mutations also abolish UV-induced mutagenesis (Figure 4C). Since the rad5-FN13,14AA mutation is epistatic to the rad5-I916A mutation with respect to both spontaneous and UV-induced mutagenesis, we conclude that the Rad5 TLS activity is responsible for mutagenesis.
Mapping the Rad5-binding domain in Rev1
A series of truncations were also made in Rev1 in an attempt to map the Rad5-binding domain. This domain is located in the C-terminal 239 amino acids (Figure 5A) and further restricted to the C-terminal 150 amino acids (Figure 5B) under low stringency conditions (0.4 mM 3AT). It is interesting to note that Rev1-CT239, which interacts with the full-length Rad5 on a -His plate (data not shown) but not under high stringency conditions (-Ade), is capable of interacting with Rad5-NT164 (Figure 5A). Similarly, Rev1-CT150 can also interact with Rad5-NT164 under higher stringency (1 mM 3AT) than with full-length Rad5 (Figure 5B). However, we were unable to further narrow the Rad5-binding domain to less than 150 amino acids within the Rev1 C-terminus (Figure 5B). Hence, our results are consistent with a recent report (44) that the C-terminus of Rev1 is required for the interaction with Rad5, but differ in that we have narrowed the binding domain to the C-terminal 150 amino acids instead of 2/3 of the Rev1 protein as reported.
Figure 5.
The Rad5 interaction region is mapped to the C-terminus of Rev1 by yeast two-hybrid assays. (A) Physical interaction of Rev1 and its truncations with the full-length Rad5 or Rad5-NT164. Fragments remaining after Rev1 deletion are shown in the right panel relative to known functional domains. (B) Rev1-CT150 is the minimum region capable of interaction with Rad5. The Y2H conditions are as described in Figure 1.
Structure of the Rev1–Rad5 fusion complex
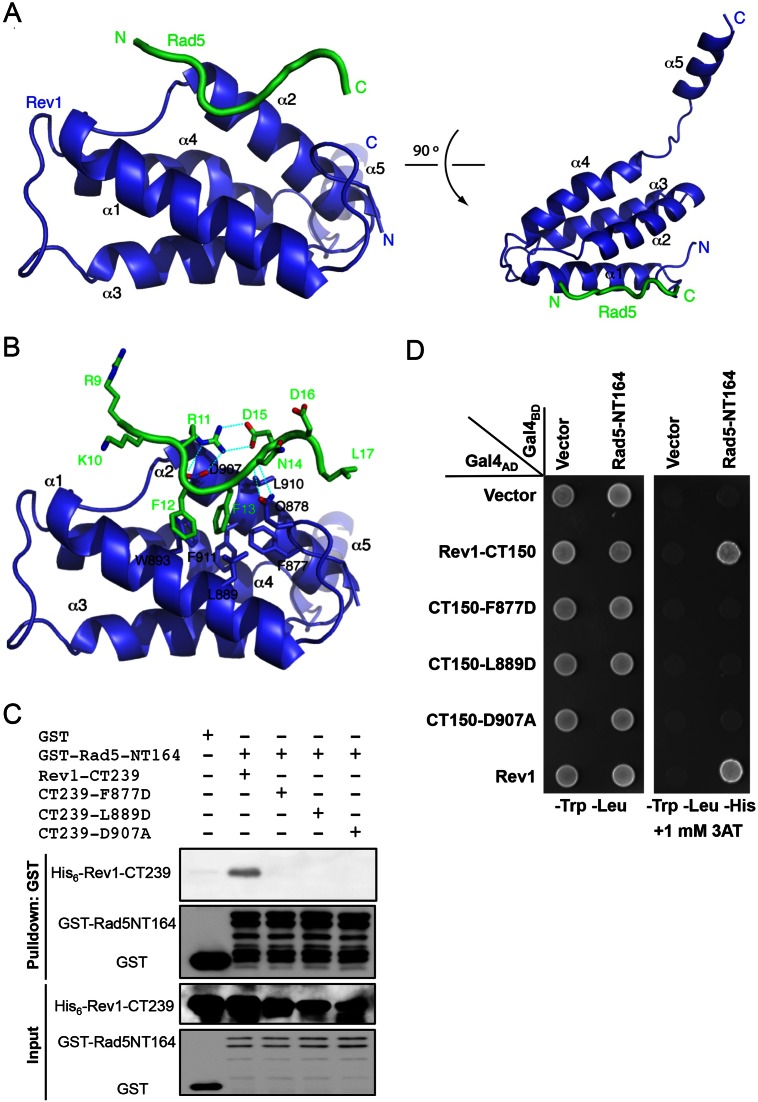
Based on the above fine mapping of the Rad5-Rev1 interaction regions, we attempted to determine the complex structure. The C-terminal domain (CTD) of Rev1 (876–985) alone could not be expressed in soluble form. Fusion of the Rad5 peptide (5–20) at its N-terminus greatly enhanced its solubility, and this fusion strategy was employed to determine the complex structure as previously reported (47). Crystals were obtained using this fusion protein, which diffracted to high resolution of 2.0 Å (Supplementary Table S3). There are four copies of the complex in one asymmetric unit (AU), in which all show similar Rev1–Rad5 interactions (Supplementary Figure S3A). There is no traceable electron density for the linker sequence, suggesting that this sequence is highly mobile in space and should not interfere with the Rev1–Rad5 interactions.
Consistent with previous structural studies on the vertebrate Rev1 (48), the CTD of yeast Rev1 folds into a four-helix bundle structure, with an extra helix (α5) tethering at the C-terminal end through a flexible sequence (Figure 6A, residues 968–974). The four copies of Rev1 in one AU are essentially identical except the tethering helix α5, which orientates toward different directions due to crystal packing interactions (Supplementary Figure S3A). It appears that α1, α2 and an N-terminal hairpin structure of Rev1-CTD forms a hydrophobic pocket that interacts with key Rad5 residues (Figure 6B). To validate the above structure, we made amino acid substitutions to the critical hydrophobic residues and examined their effects on the Rad5–Rev1 interaction. While His6-tagged wild-type Rev1-CT239 is able to bind GST-tagged Rad5-NT164 in an in vitro pull down assay, each of the F877D, L889D and D907A substitutions abolishes this interaction (Figure 6C). A Y2H assay using truncated Rad5-NTD and Rev1-CTD independently confirms the above pull down result (Figure 6D).
Figure 6.
Characterization of Rev1-CTD residues critical for the interaction with Rad5-NTD. (A) Two orthogonal views of the structure of Rev1-CTD bound with Rad5 fusion-peptide. Rev1 and Rad5 are colored blue and green, respectively. The N- and C-termini of the proteins are labeled. (B) Detailed interactions between Rev1 and Rad5 with labeled residues. (C) A GST pull-down assay to examine physical interaction between purified His6-tagged Rev1-CT239 or its mutant derivatives and GST-tagged Rad5-NT164. (D) A Y2H assay to examine physical interaction between Rev1-CT150 or its mutant derivatives and Rad5-NT164. The Y2H experimental conditions were as described in Figure 1.
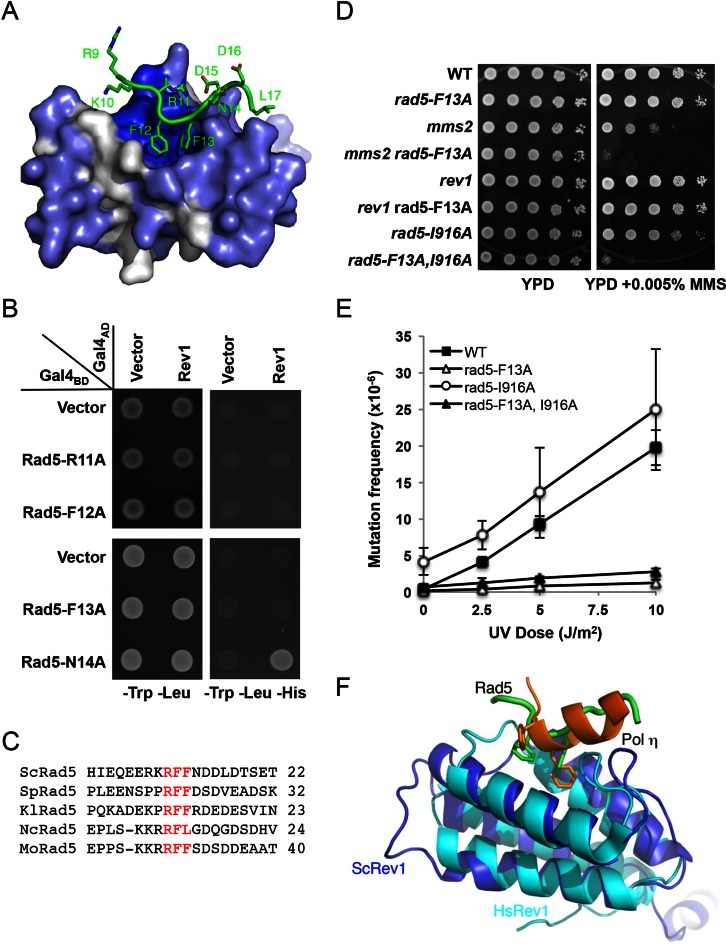
Identification and characterization of an RFF motif in Rad5-NTD
The Rad5-NTD amino acids inserting into the Rev1-CTD hydrophobic pocket are two Phe residues (F12 and F13) (Figure 7A and Supplementary Figure S3B), while R11 makes two hydrogen bonds to the D907 of Rev1 (Figure 6B), which further stabilizes the binding of the peptide. To ask whether all three residues of the RFF motif in Rad5 are critical for Rev1 binding, we made single amino acid substitutions and found in the Y2H assay that each of the mutations R11A, F12A and F13A disrupts Rev1 binding, whereas an adjacent N14A mutation does not affect Rev1 binding (Figure 7B). The lack of binding to Rev1 by the above RFF motif mutations was not due to altered expression or mis-folding of the altered proteins, as these fusion proteins are capable of interacting with Ubc13 indistinguishably from their wild-type counterpart (Supplementary Figure S4A), and these mutant alleles were able to complement the rad5 null mutant to the level characteristic of the tls mutant (Supplementary Figure S4B). The RFF motifs in Rad5 are highly conserved in different eukaryotic microorganisms (Figure 7C), suggesting that this interaction is a general phenomenon in fungi.
Figure 7.
Characterization of the Rad5-NTD RFF motif. (A) Rad5-NTD residues involved in binding to a conserved pocket of Rev1. Rev1 is shown as gray surface with conserved residues in blue and the Rad5 polypeptide is shown in green. (B) The ‘RFF’ motif of Rad5 is essential for the binding of Rad5 to Rev1, as judged by a Y2H assay. The experimental conditions were as described in Figure 1. (C) Sequence alignments around the ‘RFF’ motif among several fungal Rad5 homologs. The RFF motif is in red. Source of sequences: ScRad5, Saccharomyces cerevisiae, P32849.1; SpRad5, Schizosaccharomyces pombe, XP_001713034.1; KlRad5, Kluyveromyces lactis, XP_455865.1; NcRad5, Neurospora crassa, XP_958511.1; MoRad5, Magnaporthe oryzae, XP_003712540.1. (D) Inter- and intragenic interactions between rad5-F13A and DDT pathways mutations with respect to MMS-induced killing. (E) Effects of rad5-F13A and rad5-I916A on UV-induced mutagenesis. Strains are isogenic derivatives of DBY747. Data are the average of at least three independent experiments with standard deviation. (F) Structural alignment of Rev1 between budding yeast (blue) and human (cyan, PDB code 2LSK). hPolη (orange) binds to the same pocket of hRev1 as yRad5 (green) does with yRev1.
To further address whether a single amino acid substitution within the RFF motif is sufficient and specific to disrupt the TLS activity in Rad5, we created a rad5-F13A single mutation at the chromosomal RAD5 locus and examined its biological effects. Analogous to our previous analysis of the rad5-FN13,14AA mutation, this single amino acid substitution is synergistic with mms2 or rad5-I916A and epistatic to rev1 with respect to MMS-induced killing (Figure 7D), and abolishes UV-induced mutagenesis in both wild-type and rad5-I916A mutant cells (Figure 7E). Rad5 is also known to interact with Pol30 (11) and it was recently reported that the N-terminal 393 amino acids of Rad5 is able to interact with both Rev1 and Pol30 (49), raising a possibility that the two interactions are related. In a Y2H assay, we found that Rad5-F13A is unable to interact with Rev1, but its interaction with Pol30 (Supplementary Figure S5A) and the level of expression (Supplementary Figure S5B) are not altered. Hence, Rad5 appears to use distinct motifs to bind Rev1 and Pol30.
The CTD of yeast Rev1 superimposes well with that of human Rev1, with root-mean-square-deviation (r.m.s.d.) of 1.9 Å over 77 Ca (Figure 7F). Interestingly, the Rad5-binding pocket of Rev1 in budding yeast maps to the same binding site for Polη and Polκ in human Rev1 CTD (Figure 7F), which also binds two Phe residues of the TLS polymerases (48,50). As Polη is also conserved from yeast to humans, we speculate that yeast Polη binds to Rev1 in a manner analogous to its counterpart in human.
DISCUSSION
Despite the fact that Rad5 has been well characterized as a RING finger domain-containing E3 that physically interacts with Ubc13 and is a member of error-free DDT (11,45), Rad5 must possess additional functions in DNA damage response since the rad5 null mutant is much more sensitive to DNA-damaging agents than the mms2 or ubc13 mutant (51). Prior to this study, there have been reports indicating its possible involvement in TLS, as rad5 is found to be allelic to rev2 (22), and some recent reports suggest that Rad5 promotes TLS through a physical interaction with Rev1 (24,25). In this report, we independently confirmed this interaction and went further to define the region of interaction to the Rad5 N-terminal 30 amino acid residues. Coincidentally, a recent report narrowed the Rev1-interaction to the N-terminus 21–360 amino acids of Rad5 (44). To clarify the discrepancy, we went further to show that deletion of residues 21–30 from Rad5 did not compromise Rev1 interaction and that certain amino acid substitutions within the Rad5 N-terminal 20 residues completely abolished the Rev1 interaction. With the crystal structure of the Rev1-CTD in complex with the Rad5 peptide and additional Y2H assays, we are able to conclude that as short as a 16-amino-acid peptide (aa. 5–20) of Rad5 is necessary and sufficient to bind Rev1.
The fine mapping and identification of point mutations within the Rad5 N-terminus allowed us to critically test a hypothesis that Rad5 possesses both error-free and error-prone DDT functions via separate domains. A series of analyses allows us to conclude that (i) the Rad5 N-terminus is required for and only involved in TLS; (ii) inactivation of the Rad5 N-terminal activity is epistatic to the inactivation of Rev1 function; (iii) the Rad5 N-terminal mutation is synergistic to both mms2/ubc13 and the rad5-I916A point mutations with respect to DNA damage sensitivity; (iv) lack of the Rad5 N-terminal activity severely compromises spontaneous and DNA-damage-induced mutagenesis; and (v) the Rad5 N-terminal mutation is epistatic to its E3 activity with respect to mutagenesis. These observations collectively confirm our hypothesis that Rad5 has dual functions in DDT.
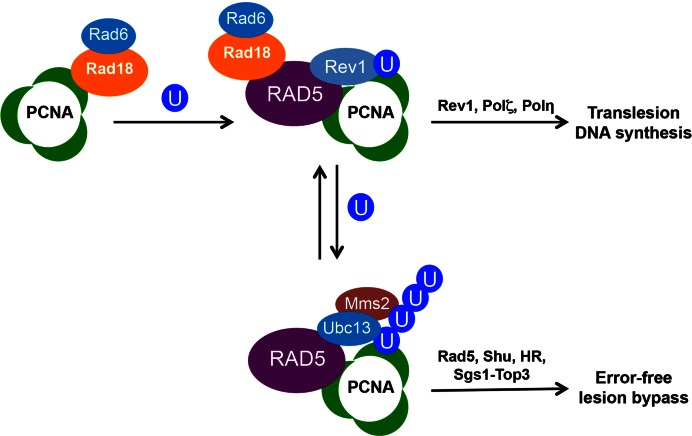
Based on available data, we propose a modified working model of DDT (Figure 8), in which Rad5 is required for both branches of DDT reminiscent of Rad18. However, unlike Rad18 which is required for PCNA monoubiquitination and hence the subsequent polyubiquitination at the same K164 residue (5), Rad5 is not required for PCNA monoubiquitination but it binds to Rad18 (11) and is required for the recruitment of Rev1 after PCNA monoubiquitination. Since the Rad5 N-terminal deletion or point mutants behave almost like the rev1 null mutant under several assay conditions, Rad5 must play a critical role in the recruitment of Rev1 and initiation of TLS. The dual requirements of PCNA monoubiquitination and availability of Rad5 for the initiation of TLS in budding yeast is reminiscent of a ‘matchmaker’ mechanism and implies that it may play a role in the pathway choice and balance within DDT, although exactly how the two DDT pathways are regulated by Rad5 remains to be investigated.
Figure 8.
A revised working model of DNA-damage tolerance in budding yeast. Under DNA-damage conditions, the Rad6-Rad18 complex monoubiquitinates PCNA at the K164 residue. Rad5 interacts with both Rad18 (11) and PCNA (5,49), which is recruited to the damage site. Rad5-Rev1 physical interaction facilitates recruitment of Rev1 and other TLS polymerases for TLS, while Rad5-Ubc13-Mms2 promotes PCNA polyubiquitination and subsequent error-free lesion bypass, in which Rad5 serves as an E3 ligase and possibly a DNA helicase.
The detailed characterization of the Rev1-binding region within Rad5 identified a RFF motif critical for the Rev1 interaction, indicating a hydrophobic interaction in which the side chains of two Phe residues play critical roles. It was previously reported that the Rev1 C-terminal 21 amino acids are required for its interaction with Rad5, and the minimum sufficient interaction was only mapped to the C-terminal 2/3 of the protein including the entire Rev1 polymerase domain (44). In this study, we narrowed the Rad5-binding domain to the C-terminal 150 amino acid region including UBM but not the polymerase domain and further demonstrated that the C-terminal 110 amino acid region is sufficient to bind the Rad5 N-terminus. As a matter of fact, our structure and site-specific mutagenesis analyses reveal that Rev1 residues making contact to the Rad5 NTD are clustered around aa. 876–911, indicating that the very C-terminus, if required, may only serve to stabilize the Rev1 overall structure. Of great interest is the fact that the Rad5-binding domain of yRev1 superimposes well with the hRev1 C-terminal motif interacting with Y-family polymerases containing the FF motif, raising a possibility that the yRev1 CTD can also bind yPolη through one of two conserved FF/MF motifs. If this is indeed the case, Polη may compete with Rad5 in mediating DDT pathway choice particularly under UV-induced DNA damage conditions.
Supplementary Material
FUNDING
Chinese National 973 Project [2013CB911003]; Capital Normal University start-up fund [13530530209]; Natural Sciences and Engineering Research Council of Canada Discovery Grant [RGPIN-2014-04580 to W.X.]; ‘Junior One Thousand Talents’ program (to Z.C.). Funding for open access charge: Capital Normal University start-up fund [13530530209].
Conflict of interest statement. None declared.
REFERENCES
- 1.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 2.Broomfield S., Hryciw T., Xiao W. DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat. Res. 2001;486:167–184. doi: 10.1016/s0921-8777(01)00091-x. [DOI] [PubMed] [Google Scholar]
- 3.Lawrence C.W. Cellular functions of DNA polymerase zeta and Rev1 protein. Adv. Protein Chem. 2004;69:167–203. doi: 10.1016/S0065-3233(04)69006-1. [DOI] [PubMed] [Google Scholar]
- 4.Prakash L. The RAD6 gene and protein of Saccharomyces cerevisiae. Ann. N.Y. Acad. Sci. 1994;726:267–273. doi: 10.1111/j.1749-6632.1994.tb52825.x. [DOI] [PubMed] [Google Scholar]
- 5.Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 6.Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–1824. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann R.M., Pickart C.M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 8.Broomfield S., Chow B.L., Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brusky J., Zhu Y., Xiao W. UBC13, a DNA-damage-inducible gene, is a member of the error-free postreplication repair pathway in Saccharomyces cerevisiae. Curr. Genet. 2000;37:168–174. doi: 10.1007/s002940050515. [DOI] [PubMed] [Google Scholar]
- 10.Ulrich H.D. Protein-protein interactions within an E2-RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J. Biol. Chem. 2003;278:7051–7058. doi: 10.1074/jbc.M212195200. [DOI] [PubMed] [Google Scholar]
- 11.Ulrich H.D., Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbour L., Xiao W. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model. Mutat. Res. 2003;532:137–155. doi: 10.1016/j.mrfmmm.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 13.Pastushok L., Xiao W. DNA postreplication repair modulated by ubiquitination and sumoylation. Adv. Protein Chem. 2004;69:279–306. doi: 10.1016/S0065-3233(04)69010-3. [DOI] [PubMed] [Google Scholar]
- 14.Stelter P., Ulrich H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 15.Pfander B., Moldovan G.L., Sacher M., Hoege C., Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 16.Papouli E., Chen S., Davies A.A., Huttner D., Krejci L., Sung P., Ulrich H.D. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Xiao W., Chow B.L., Broomfield S., Hanna M. The Saccharomyces cerevisiae RAD6 group is composed of an error-prone and two error-free postreplication repair pathways. Genetics. 2000;155:1633–1641. doi: 10.1093/genetics/155.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemontt J.F. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971;68:21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson R.E., Henderson S.T., Petes T.D., Prakash S., Bankmann M., Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol. Cell. Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson R.E., Prakash S., Prakash L. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem. 1994;269:28259–28262. [PubMed] [Google Scholar]
- 21.Chen S., Davies A.A., Sagan D., Ulrich H.D. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 2005;33:5878–5886. doi: 10.1093/nar/gki902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahne F., Jha B., Eckardt-Schupp F. The RAD5 gene product is involved in the avoidance of non-homologous end-joining of DNA double strand breaks in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:743–749. doi: 10.1093/nar/25.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gangavarapu V., Haracska L., Unk I., Johnson R.E., Prakash S., Prakash L. Mms2-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:7783–7790. doi: 10.1128/MCB.01260-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pages V., Bresson A., Acharya N., Prakash S., Fuchs R.P., Prakash L. Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics. 2008;180:73–82. doi: 10.1534/genetics.108.091066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lorick K.L., Jensen J.P., Fang S., Ong A.M., Hatakeyama S., Weissman A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unk I., Hajdu I., Blastyak A., Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Sherman F., Fink G.R., Hicks J. Methods in Yeast Genetics. NY: Cold Spring Harbor Laboratory Press; 1983. [Google Scholar]
- 29.Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.James P., Halladay J., Craig E.A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothstein R.J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 32.Gietz R.D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 33.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 35.Barbour L., Ball L.G., Zhang K., Xiao W. DNA damage checkpoints are involved in postreplication repair. Genetics. 2006;174:1789–1800. doi: 10.1534/genetics.106.056283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheffield P., Garrard S., Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of ‘parallel’ expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 37.Vonrhein C., Blanc E., Roversi P., Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 38.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Painter J., Merritt E.A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 40.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z., Borek D., Padrick S.B., Gomez T.S., Metlagel Z., Ismail A.M., Umetani J., Billadeau D.D., Otwinowski Z., Rosen M.K. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larimer F.W., Perry J.R., Hardigree A.A. The REV1 gene of Saccharomyces cerevisiae: isolation, sequence, and functional analysis. J. Bacteriol. 1989;171:230–237. doi: 10.1128/jb.171.1.230-237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer L.M., Babu M.M., Aravind L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle. 2006;5:775–782. doi: 10.4161/cc.5.7.2629. [DOI] [PubMed] [Google Scholar]
- 44.Kuang L., Kou H., Xie Z., Zhou Y., Feng X., Wang L., Wang Z. A non-catalytic function of Rev1 in translesion DNA synthesis and mutagenesis is mediated by its stable interaction with Rad5. DNA Repair (Amst) 2013;12:27–37. doi: 10.1016/j.dnarep.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Xiao W., Chow B.L., Fontanie T., Ma L., Bacchetti S., Hryciw T., Broomfield S. Genetic interactions between error-prone and error-free postreplication repair pathways in Saccharomyces cerevisiae. Mutat. Res. 1999;435:1–11. doi: 10.1016/s0921-8777(99)00034-8. [DOI] [PubMed] [Google Scholar]
- 46.Ball L.G., Xu X., Blackwell S., Hanna M.D., Lambrecht A.D., Xiao W. The Rad5 helicase activity is dispensable for error-free DNA post-replication repair. DNA Repair (Amst) 2014;16:74–83. doi: 10.1016/j.dnarep.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 47.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 48.Wojtaszek J., Liu J., D'Souza S., Wang S., Xue Y., Walker G.C., Zhou P. Multifaceted recognition of vertebrate Rev1 by translesion polymerases zeta and kappa. J. Biol. Chem. 2012;287:26400–26408. doi: 10.1074/jbc.M112.380998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choi K., Batke S., Szakal B., Lowther J., Hao F., Sarangi P., Branzei D., Ulrich H.D., Zhao X. Concerted and differential actions of two enzymatic domains underlie Rad5 contributions to DNA damage tolerance. Nucleic Acids Res. 2015;43:2666–2677. doi: 10.1093/nar/gkv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pozhidaeva A., Pustovalova Y., D'Souza S., Bezsonova I., Walker G.C., Korzhnev D.M. NMR structure and dynamics of the C-terminal domain from human Rev1 and its complex with Rev1 interacting region of DNA polymerase eta. Biochemistry. 2012;51:5506–5520. doi: 10.1021/bi300566z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X., Blackwell S., Lin A., Li F., Qin Z., Xiao W. Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutat. Res. Rev. Mutat. Res. 2015;764:43–50. doi: 10.1016/j.mrrev.2015.02.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.