Abstract
Accurate variant calling in next generation sequencing (NGS) is critical to understand cancer genomes better. Here we present VarDict, a novel and versatile variant caller for both DNA- and RNA-sequencing data. VarDict simultaneously calls SNV, MNV, InDels, complex and structural variants, expanding the detected genetic driver landscape of tumors. It performs local realignments on the fly for more accurate allele frequency estimation. VarDict performance scales linearly to sequencing depth, enabling ultra-deep sequencing used to explore tumor evolution or detect tumor DNA circulating in blood. In addition, VarDict performs amplicon aware variant calling for polymerase chain reaction (PCR)-based targeted sequencing often used in diagnostic settings, and is able to detect PCR artifacts. Finally, VarDict also detects differences in somatic and loss of heterozygosity variants between paired samples. VarDict reprocessing of The Cancer Genome Atlas (TCGA) Lung Adenocarcinoma dataset called known driver mutations in KRAS, EGFR, BRAF, PIK3CA and MET in 16% more patients than previously published variant calls. We believe VarDict will greatly facilitate application of NGS in clinical cancer research.
INTRODUCTION
Next-generation sequencing (NGS) has revolutionized our understanding of genetic variants in cancer and their role in cancer progression. As a platform for discovery NGS has revealed new genetic drivers of cancer leading to development of targeted cancer therapies (1), and in the clinic NGS provides a tool to detect mutations determining a patient therapy (2). Cancer genomes are known to harbor a wide range of mutations, including single nucleotide variants (SNVs), multiple-nucleotide variants (MNVs), insertions, deletions and complex variants, in addition to even more complex structural variants (SVs) such as duplications (DUPs), inversions (INVs), insertions and translocations. Oncogenes such as KRAS, NRAS, BRAF and EGFR, often contain hotspot missense mutations, which are the focus of most variant callers (3,4). A number of regularly cited variant callers, such as GATK (3), FreeBayes (http://arxiv.org/abs/1207.3907) and VarScan (4) are designed to call SNV and small InDels separately, but not complex combinations of these events. Furthermore tumor suppressors, such as TP53, PTEN, BRCA1/2, RB1, STK11 and NF1, often contain large frameshift insertions and deletions (InDels) or complex mutations and sometimes even SVs (5) and are often missed by those variant callers. To more comprehensively analyze cancer genomes, a variant caller that can identify all these different types of mutations is needed.
In addition, ultra-deep sequencing (>5000×) is increasingly applied in a clinical setting where low allele frequency (AF) mutations are of key interest, for example to discover mutations present in only a small sub-clonal proportion of the tumor cells that might be resistant to targeted therapy (6), or for detection of mutations in the often small proportion of tumor DNA circulating with normal DNA in a patient's blood (7). Most commonly used variant callers do not scale well with increasing depth and typically downsample (randomly remove portions of data) to increase their computational performance. However downsampling can significantly reduce the sensitivity to detect low AF mutations. Coupled with its random nature, downsampling is thus not desired in such situations. Variant callers that can scale computational performance to comprehensively handle ultra-deep sequencing data are urgently required to improve sensitivity.
Here, we present a de novo and versatile variant caller, VarDict, which can simultaneously call SNV, MNV, InDels, complex composite variants, as well as SVs with no size limit. VarDict contains many features that are distinct from other variant callers, including linear performance to depth, intrinsic local realignment, built-in capability of de-duplication, detection of polymerase chain reaction (PCR) artifacts, accepting both DNA- and RNA-Seq, paired analysis to detect variant frequency shifts alongside somatic and loss of heterozygosity (LOH) variant detection and SV calling. We use a number of both simulated and real human tumor sample whole-genome, exome and targeted sequencing data sets to compare VarDict to current gold standard variant callers. VarDict demonstrates consistently improved performance and sensitivity, particularly for InDels calling. We believe VarDict will greatly facilitate application of NGS in cancer research, enabling researchers to use one tool in place of an alternative computationally expensive ensemble of tools.
MATERIALS AND METHODS
Prerequisites
VarDict works on Binary Alignment/Map (BAM) files that contain aligned sequence reads against a reference genome. VarDict is compatible with BAM files generated from common DNA-Seq aligners, such as BWA (8), Novoalign (http://www.novocraft.com), Bowtie (9), and Bowtie2 (10), as well as RNA-Seq aligners, such as Tophat (11) and STAR (12).
Local realignments and InDel calling
VarDict performs two types of local realignments to more accurately estimate allele frequencies for InDels: supervised and unsupervised. InDels that are much shorter than the read length and central to the reads are typically aligned with gaps by most aligners, but result in forced alignment with mismatches, or soft-clipping when mismatches are too many. An example was shown in IGV (13) (Figure 1). This mismatched and soft-clipped sequences are often ignored or mis-treated by most variant callers, but in fact offer important additional evidence of the InDels. When such an InDel is found in an alignment VarDict triggers supervised local realignment to identify mismatched alignment of 3′ and 5′ read ends flanking the InDels and adds them in support of the InDel, resulting in increased allele frequencies. In unsupervised local realignment, VarDict scans the local sequences near soft-clippings to look for larger InDels. VarDict first derives the consensus sequences from soft-clipped reads clipped at the same genomic location. If a consensus sequence can be found, VarDict then uses it to find an un-gapped match within a user definable distance (default to 125 bp), but allowing ≤3 base mismatches. When a match is found in entirety and away from the breakpoint, a deletion is called; and when the end portion of the consensus matches adjacent to the breakpoint, an insertion is called. If no InDels can be called, VarDict identifies sequential well-clipped sequences (typically within 5 bp) with respective 5′ and 3′ soft-clippings, assuming them to flank either side of an insertion, and determines whether they have matched ends. If an ungapped match is found with mismatches (≤3), a large insertion is called. This approach enables calling of insertions that are larger than read length, as well as large complex variants.
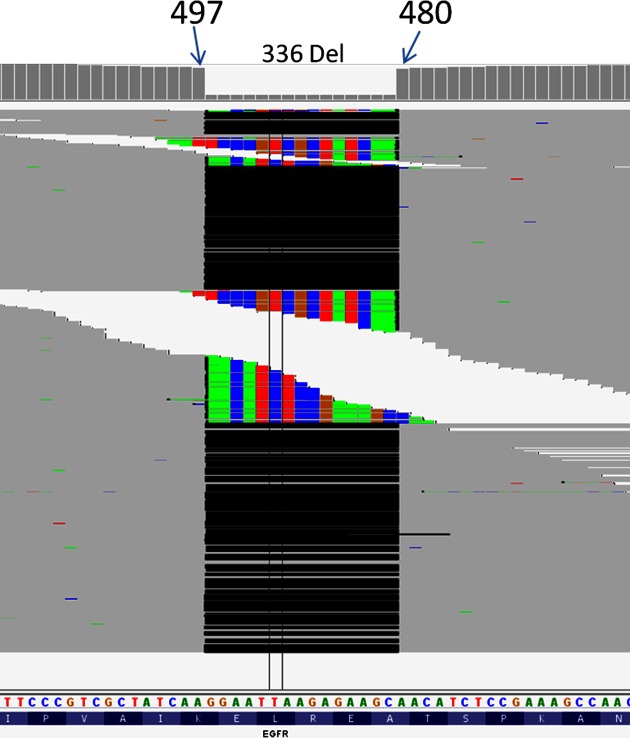
Figure 1.
VarDict uses soft-clipped reads for local realignment to comprehensively estimate allele frequency (AF). This example shows the 15-bp deletion mutation in EGFR exon 19 in the PC-9 lung cancer cell line, as shown in IGV. Top track is the coverage for each base pair. Each thin gray line represents a sequence read. Black lines in the middle indicate gapped alignments due to the 15-bp deletion. The colored portion shows soft-clipped reads that cannot be aligned due to short overhangs. The bottom track shows reference sequence and amino acids for EGFR exon 19.
Detecting complex variants
VarDict calls complex variants that are a combination of insertions and deletions and typically off the limit or miscalled by most currently published variant callers. We observed that composite proximal (<10 bp) InDels and mismatches in the same reads typically work in tandem as one complex variant. VarDict represents complex variant composites as a single variant, rather than as multiple individual variants. Whenever an InDel is detected in a read, VarDict will recursively scan for another InDel (within 10 bp) or mismatches (within 3 bp) in the same read, and if found, combines them as one variant. The same rule also applies to consecutive mismatches, resulting in calling multiple nucleotide variants (MNVs).
Structural variants
VarDict takes a two-step approach to call SVs. First it will use soft-clipped reads as described above to build a consensus sequence from clipped sequences, and then search whether this consensus can be uniquely aligned within 5 kb of the given region. If a match is not found, VarDict searches in the region suggested by discordant mate pair alignment. In order to quickly find a match, VarDict builds a hash table of all 17 and 11 bp seeds from reference sequences in the region, using only unique seeds to avoid false positives. For even larger SVs where the second breakpoint is outside the given region, VarDict will use discordant mates as a guide and search only in the regions suggested by discordant mates to identify the other breakpoint position. When the soft-clipped sequences can be uniquely mapped, VarDict will call the SVs, perform a 5′ shift if necessary and estimate the AF of the detected SV, a distinct feature from other published SV callers. When no soft-clipped reads are found, VarDict will then use only clustered discordant mates to call SVs based on both distance and orientation, and estimate the breakpoints as well as AF based on number of discordant mates. Currently, VarDict implements large deletions (DEL), DUPs and INVs. Insertions larger than two read lengths are not called, and will require future development, including de novo assembly algorithms. Also, inter-chromosome fusion (BND) calling has not yet been implemented.
Paired analysis
In paired sample analysis mode, where two BAM files are given, VarDict will extract the read counts for variant and reference alleles and perform a Fisher's exact test to determine whether a variant has a significant difference in AF between the two samples. Based on the AF difference, a variant is classified as ‘Somatic’ if only present in the first sample, ‘Germline’ if present in both samples, ‘LOH’ if a heterozygous variant in the second sample but becomes homozygous or is lost in the first sample, ‘Deleted’ if present in the second sample but no coverage in the first sample.
Amplicon calling
When given PCR amplicon designs with primer locations, VarDict will trigger ‘amplicon calling mode’. In this mode, VarDict calls variants amplicon by amplicon. It first compares the read mapping positions to the PCR design supplied in a BED file with primer locations and determines whether the read belongs to a particular PCR amplicon. VarDict will then only use those read pairs that have 90% overlap with the amplicon and fall within 10 bp of amplicon's PCR edges, and avoids calling variants overlapping the primers. Variants in regions covered by more than one amplicon that cannot be called in all amplicons are considered amplicon biased and filtered out as PCR artifacts.
De-duplication
PCR duplicates in NGS are a predominant source of false positives in variant calling. A typical NGS workflow involves a separate de-duplication step to mark or remove duplicates. VarDict has a built-in option to perform de-duplication on the fly, removing the necessity for an additional step and so improving efficiency. Any read pairs with the same alignment positions for both reads are deemed duplicates and VarDict will only use the first one encountered for variant calling. VarDict also supports BAM files where duplicates have already been marked and, by default, will exclude those reads marked as duplicates. The de-duplication option is recommended for hybrid capture-based sequencing, whole genome sequencing (WGS), and perhaps RNA-Seq, but not for PCR-based targeted sequencing.
Memory management and run time
VarDict was designed to have efficient memory management and run time for ultra-deep sequencing. VarDict constructs a unique data structure for the regions of interest, such as exons, in memory to represent different types of variants, thus making memory usage only proportional to the region of interest, regardless of sequencing depth. It parses reads mapped to the region sequentially and updates the variant data structure accordingly. By generating a consensus call for the set region to the most degenerative alignment, VarDict's local realignment runs with computational efficiency proportional to the sequencing depth, scaling linearly to depth. These combined features ensure VarDict is uniquely suitable for computationally efficient and sensitive variant calling from ultra-deep targeted sequencing, where low allele frequencies are expected and downsampling is not desired.
Data
Over the course of development, we used several datasets to test VarDict as listed below:
NA12878 and Genome In a Bottle (GIAB) variant calls: NA12878 WGS data was downloaded from Platinum genome project (http://www.illumina.com/platinumgenomes) and aligned using BWA (v0.7.8). Version 17 of published GIAB calls (https://sites.stanford.edu/abms/giab) were used for comparison to VarDict calls.
ICGC-TCGA DREAM Mutation Calling challenge: synthetic challenge 3 and 4. Each challenge was deeply sequenced (60–80× coverage) WGS datasets from a single (e.g. cell line) sample and then randomly sampled into two non-overlapping subsets of equal size. A non-overlapping spectrum of mutations was generated, some randomly selected and some targeting known cancer-associated genes, which were then added to one of the sampled BAM files which becomes the ‘tumor’, with the other being the ‘normal’.
The Cancer Genome Atlas (TCGA) lung dataset (14). We downloaded exome BAM files of TCGA lung adenocarcinoma (LUAD) from CGHub (https://cghub.ucsc.edu/) for a given set of genes. BAM files were used directly without re-alignment. VarDict was run with de-duplication turned on (-t), requiring at least 4 supporting reads (-r 4) with AF ≥ 7.5% and minimum base quality ≥ 23 (-q 23) to call variants, but allowing allele frequencies ≥ 2.5% for known mutations.
RESULTS
VarDict accurately estimates InDel allele frequency
The local realignment capability of VarDict enables accurate estimation of AF for InDels. The local realignment not only recovers unaligned terminal read portions that are otherwise removed (‘soft-clipped’), but also terminal read portions with short overhang that are otherwise forcefully aligned with mismatches and can be miscalled as SNVs. For a known EGFR exon 19 deletion in exome sequencing of cell line PC-9, we compared VarDict's AF estimation to those from several other popular variant callers including GATK (3), FreeBayes and VarScan (4). VarDict consistently returned higher variant AF and depth indicating more comprehensive consideration of all different supporting reads, as illustrated in Figure 1 and Table 1. The AF is 69% when only gap-opening reads are considered, as is the case for GATK's UnifiedGenotyper. VarDict recovers 26% more reads from soft-clipped reads (shown as colors in Figure 1) from both sides of the deletion, as well as forced mismatches when overhangs are too short to be softly clipped by the aligner. The GATK HaplotypeCaller and FreeBayes, on the other hand, seem to only recover soft-clipped reads from one side of the deletion, resulting in lower AF estimations.
Table 1. Comparison of different algorithms on a short 15-bp deletion variant calling.
| Method | Deletion | Depth | Allele Freq (%) |
|---|---|---|---|
| Gapped reads only | 336 | 488 | 69% |
| GATK (UnifiedGenotyper) | 338 | 496 | 68% |
| GATK (HaplotypeCaller) | 404 | 482 | 84% |
| FreeBayes | 339 | 413 | 82% |
| VarDict | 493 | 575 | 86% |
The 15-bp deletion in EGFR exon 19 in lung cancer cell line PC-9 is shown in Figure 1. GATK UnifiedGenotyper does not recover soft-clipped reads, and as expected, produces similar results when only gapped reads are considered. VarDict is able to recover more supporting soft-clipped reads from both sides of deletion, while GATK's HaplotypeCaller and FreeBayes only recover from one side, producing fewer deletion read counts and depth.
VarDict recovers large InDels from soft-clipped reads
Large InDel variants are often underrepresented, particularly in cancer genomes, which have higher prevalence of large InDels. One possible reason is large InDels are often beyond the limits of detection by most variant callers or allele frequencies are under-estimated. The unique local realignment feature of VarDict makes it possible to call large InDels (up to 125 bp by default, but can be user defined) with sensitive AF estimation, even when supported by soft-clipped reads alone. Figure 2 shows an example of a 124 bp deletion (rs67488720) from the human reference DNA NA12878. VarDict not only correctly calls the deletion, as supported by dbSNP and present in both parents (NA12891 at 100%, NA12892 at 44%), but also with estimated 56% AF consistent with expectations for this heterozygous germline variant. GATK-HaplotypeCaller (3), VarScan (4) and FreeBayes all failed to call this deletion variant from the same BAM file using default recommended settings.
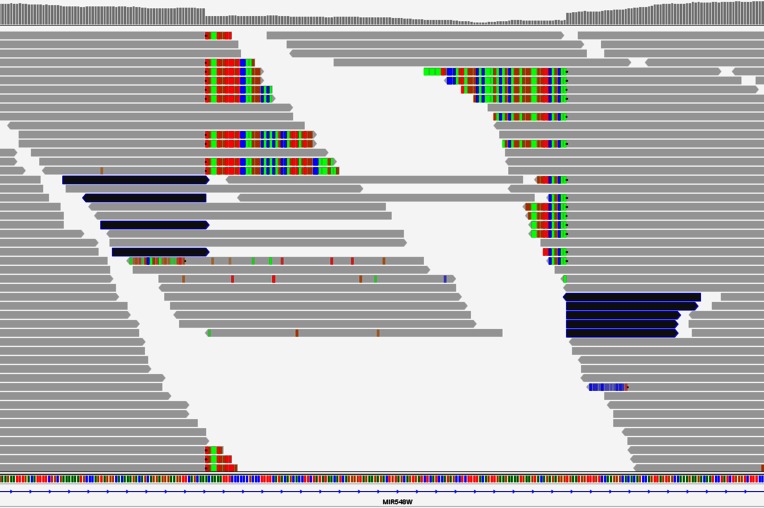
Figure 2.
VarDict calls a large 124-bp deletion in NA12878. The deletion has clear support from both soft-clipped reads (colored reads) at both breakpoints and the apparent drop of the coverage illustrated in the top track. The apparent consensus of the clipped sequences indicates the existence of an relatively large InDel. Dark colored short reads are supplementary alignments from split reads, where individual reads are split into two segments that are aligned at the edges flanking the deletion. The deletion variant was further supported by the existence of an entry in dbSNP (rs67488720). This deletion was detected by VarDict, but not by GATK, VarScan, or FreeBayes.
VarDict calls a new class of complex variants
Complex composite variants, where multiple InDels and/or substitutions occur in close proximity, are also underrepresented. Most variant callers we tested either call multiple variants or give no call. Currently only FreeBayes has some limited ability to call small complex variants with lengths under 5 base pairs. VarDict, on the other hand, is able to call much larger complex variants, involving insertion and/or deletion of dozens of base pairs. Figure 3 illustrates a complex composite variant, where an allele shows deletion of 29 bp followed by insertion of 13 bp (rs386762976, a recent addition to dbSNP 138). This allele is in fact very common in the population. Analysis of hundreds of CCLE exomes and TCGA germline exomes indicated it has prevalence over 40% in the population. The presence of a cluster of dbSNP entries proximal to this variant is likely the result of erroneous calls from variant callers that were unable to handle such complex variants, as evidenced by their absence in hundreds of germline exomes from TCGA that we analyzed (data not shown).
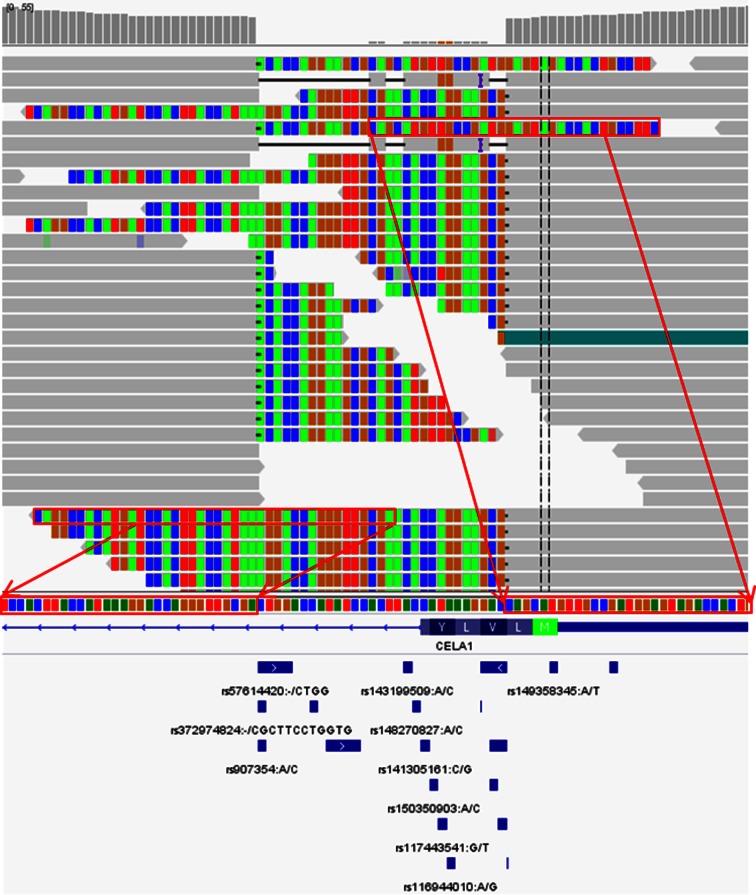
Figure 3.
VarDict calls a new class of complex variants. The clear drop out in coverage (top track) indicates a deletion from a complex variant example called in NA12878 at chr12:51740388. Only the end portion of soft-clipped sequences, indicated by red arrows, can be aligned to the other side of the deletion breakpoint, suggesting an additional proximal InDel may be present contributing to a complex composite variant. VarDict calls one single homozygous complex variant, comprising of a 29-bp deletion followed by a 13-bp insertion (CTGGACCATATCCACTTACCATAAAGGAC>ACACCAGGAAGCG). This is further supported by a recent entry in dbSNP (rs386762976). Many clustered dbSNP entries within the gap from dbSNP138 (bottom track), are likely from mis-interpretation of mis-alignments.
Complex variant calling can have profound impact on downstream clinical interpretation. It is known that in-frame deletions in exon 19 in lung cancer patients activate EGFR, and these patients would benefit from EGFR inhibiting drugs, such as gefitinib, erlotinib or AZD9291 (15–17). Figure 4 shows such an example of EGFR exon 19 from a lung cancer patient (15). Many variant callers return a series of individual out-of-frame EGFR InDel calls for these events, suggesting the patient was ineligible for EGFR therapy. VarDict is the only algorithm that correctly calls this composite of variants as a single mutation, resulting in an in-frame deletion event, likely to activate EGFR, rendering eligibility of EGFR inhibitor treatment. Furthermore, VarDict rescues all aligning reads (including soft-clipped) to sensitively quantify the variant AF.
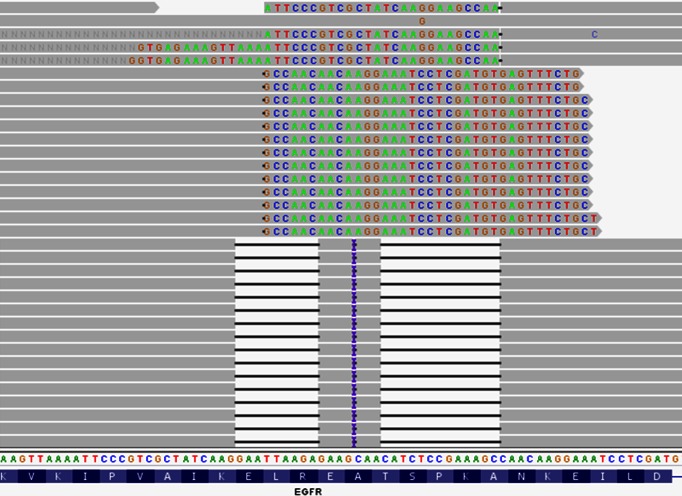
Figure 4.
Complex variants can impact clinical interpretation. For this example of EGFR exon 19 deletion from a lung cancer patient (15), BWA produced different alignments for reads with different lengths, some with two deletions and on insertion, while others are soft-clipped. The misalignment can be incorrectly interpreted as insertion of a single base of C followed by an out of frame deletion. VarDict correctly calls a single complex mutation comprising of a 26-bp deletion and a 5-bp insertion (TTAAGAGAAGCAACATCTCCGAAAGC>GCCAA), which explains all alignments, including soft-clipped reads. The mutation will be thus annotated as an in-frame deletion of exon 19, which would be clinically actionable for EGFR inhibitor therapy.
To comprehensively evaluate VarDict's complex variant calling capability, we synthesized a dataset with 1,122 complex variants within or close to every coding exon of common cancer genes (highlighted in bold in Supplementary Table S2). Each complex variant combines a random deletion of 1–50 bp with a random insertion of 1–50 bp of different sequences. Illumina HiSeq 2500 2 × 100 pair end reads were simulated using ART (18) with 50× targeted depth and aligned to hg19 using BWA MEM. VarDict, Pindel (19) and Scalpel (20) were run against this synthetic dataset and the results are shown in Supplementary Table S1. VarDict was able to call 1113 (99%) of these events, 1073 of which matched exactly with at nucleotide level, significantly more than Pindel and Scalpel. Those that did not match genotypes are due to microhomology with alternative alignments. This suggests VarDict is able to call a new class of complex variants that would be otherwise missed.
VarDict is a structural variant caller
VarDict currently implements detection of three types of SVs with no size limit: DEL; DUPs; and INVs. In the TCGA-ICGC DREAM Mutation Calling challenge dataset 4 (https://www.synapse.org/#!Synapse:syn312572/), we demonstrated that VarDict has improved sensitivity over Manta (21) and equivalent sensitivity to Lumpy (22) (Table 2). Of all methods tested, VarDict had the highest proportion of exact matches for the breakpoints.
Table 2. VarDict calls structural variants on the DREAM Mutation Calling challenge set #4 for DEL, DUP and INV.
| Caller | SV type | Calls | Exact hits | Approximate hits | True events |
|---|---|---|---|---|---|
| DEL | 3536 | 426 | 898 | 1165 | |
| Lumpy | DUP | 1330 | 476 | 972 | 1142 |
| INV | 877 | 470 | 837 | 1029 | |
| DEL | 695 | 479 | 686 | 1165 | |
| Manta | DUP | 875 | 539 | 840 | 1142 |
| INV | 1409 | 533 | 733 | 1029 | |
| DEL | 1144 | 823a | 877 | 1165 | |
| VarDict | DUP | 1139 | 883a | 893 | 1142 |
| INV | 1277 | 616a | 842 | 1029 |
We used keys provided by the DREAM challenge for exact hit (precise breakpoint) comparison. VarDict has similar sensitivity to Lumpy and higher than Manta, although Manta has higher specificity for DEL and DUP. VarDict has much higher exact hit matches, demonstrating the preciseness of VarDict's algorithm.
aAll structural variants reported by VarDict have precise breakpoints, taking into account the microhomology around the breakpoint and perform 5′ adjustments according to HGVS recommendations.
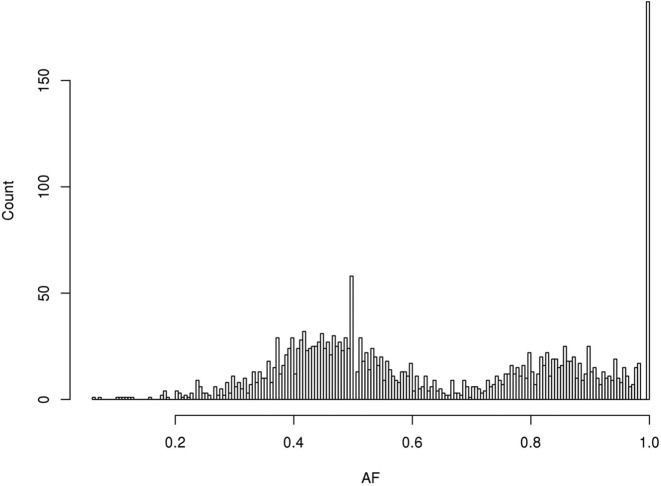
We also applied VarDict on NA12878 for which 2676 high quality DEL variants were recently reported (http://biorxiv.org/content/early/2015/05/22/019372), with sizes ranging from 50 bp to 139 kb. VarDict was able to call 2407 (90%) of these deletions. We also evaluated VarDict's AF estimation for SVs. As NA12878 is a non-diseased sample assumed to be diploid, we expected the AF of any event to peak at 50 and 100%. As expected, the allele frequencies of SVs called by VarDict do indeed show peaks at 50% and 100% (Figure 5). A seemingly smaller peak at 85%, however, suggests room for further improvement in AF estimation.
Figure 5.
The distribution of allele frequencies for structural variants estimated by VarDict. About 2407 high confidence large deletions from NA12878 are called by VarDict. The x-axis shows the AF estimated by VarDict while the y-axis shows the number of variants for a given AF. Two expected peaks at 50 and 100% are visible, consistent with the germline origin of the reference NA12878 sample.
VarDict performs paired analysis for both somatic and LOH variants
VarDict paired analysis mode detects the emergence of new mutations (typical of somatic mutation callers), and extends to other variant classes such as LOH variants (where allele specific loss results in a heterozygous variant becoming either homozygous or lost completely), deleted variants, as well as variants with significant shift in AF. This makes VarDict more broadly applicable in cancer studies where it is often necessary to go beyond typical tumor-normal somatic variant calling to the comparison of patient longitudinal samples to monitor tumor evolution or the impact of therapeutic treatments. Using ICGC-TCGA DREAM Mutation Calling challenge sets 3 and 4 we showed VarDict to be a highly competitive variant caller with superior sensitivity and specificity for InDel calling over popular variant callers, MuTect, FreeBayes and VarScan (23), Figure 6. Notably MuTect only calls somatic SNVs.
Figure 6.
Receiver operating characteristic (ROC) curve for comparison of variant callers on DREAM synthetic dataset #4. The ROC curve is drawn using quality scores for calls of somatic SNV and InDels provided by FreeBayes, VarScan, MuTect and VarDict. MuTect does not report quality and depth was used in its place. There are total 21 913 of synthetic somatic SNV and InDel mutations evaluated. VarDict outperforms other callers with higher sensitivity and specificity. Variants were called and filtered using the default setting of each caller. It is worth noting that MuTect does not call InDels.
VarDict calls more actionable mutations in lung cancer
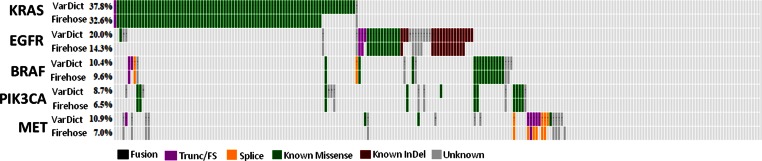
TCGA analysis of cancer samples is considered the gold standard in the field. To evaluate VarDict's performance, we downloaded exome and some whole genome data from TCGA for 208 cancer genes of interests in 230 LUAD patient samples (Supplementary Table S2), processed using VarDict and compared VarDict calls to the equivalent published data (14). We demonstrated that, through sensitive AF estimation and identification of missed complex variants and indels, VarDict calls known driver mutations in the classical lung cancer oncogenes KRAS, EGFR, BRAF, PIK3CA and MET in 16% more patients with mutations (Figure 7, Supplementary Table S3). In addition, VarDict reduces false negative calls (Figure 7) by filtering out variants of unknown significance with AF below 7.5%.
Figure 7.
The comparison of VarDict and Firehose calls for KRAS, EGFR, BRAF, PIK3CA and MET in 230 TCGA LUAD patients. Each column represents a patient. Each gene has two rows, with the top showing calls from VarDict and the bottom showing calls from Firehose. Different colors indicate different mutation types. Patients without matches in Firehose tracks indicate the mutations are only called by VarDict. As expected, all but one KRAS mutations are missense, while EGFR has known in-frame InDels. The patient with a truncating mutation of KRAS also contains an activating G12V mutation, suggesting the heterogeneous nature of the sample. Dark gray indicates mutations that are deemed as VUS (variant of unknown significance). Trunc: Truncation; FS: Frameshift.
VarDict called known KRAS activating mutations (two G12D, four G12V, three G12C, two G13D and one G12F) in twelve additional patients, with AF ranging from 9–43%. As expected, VarDict called more activating InDels in EGFR, such as three additional exon 19 mutations (one deletion and two complex InDel composites) and two additional exon 20 insertions (Supplementary Figure S1). The supporting TCGA manuscript (14) highlighted ten samples showing MET exon 14 skipping as one mechanism for MET activation. Interestingly, in cBio (24) three samples (TCGA-44-6775, TCGA-55-6978 and TCGA-55-6986) are listed as MET ‘exon 14 skip’ from analysis of RNA-Seq. Published DNA-seq calls did not reveal any MET mutations causative for the exon 14 skip for these three samples. However, VarDict called InDel and splice site mutations causative of MET exon 14 skipping in two of the three samples (TCGA-44-6775 and TCGA-55-6978). Furthermore, VarDict indicated RNA-seq calls for the third sample (TCGA-55-6986) may be false positive since no difference in exon 14 coverage is evident (Supplementary Figure S2) and no mutations were detected. More differences between VarDict and Firehose/cBio variant calls were observed in tumor suppressors, where InDel mutations are frequent. For example, VarDict called an additional 11% (57 versus 46%) and 10% (27 versus 17%) mutations in two common tumor suppressors, TP53 and STK11 respectively (Supplementary Figure S3 and Table S3), many of which are truncating InDels likely causing loss of function of the tumor suppressor. It is notable that in all cases the newly detected mutations maintain mutually exclusivity across samples.
Collectively these data demonstrate the potential of VarDict to improve sensitivity of variant calling over popular variant callers as used in the TCGA's analysis pipeline. Through VarDict re-calling of the TCGA data set we have significantly altered the previously reported mutation landscape for LUAD, with potentially significant clinical implications where variant calls in these genes determine current and emerging therapeutic choices.
VarDict performance is linear to depth
When sequencing plasma circulating free DNA (cfDNA) where tumor content can be low, it is desirable to detect somatic mutation allele frequencies as low as 0.1%. As a result, targeted gene panels enabling ultra-high depth (>5000×) sequencing are increasingly being used in the clinical setting. Computational run-time of most popular variant callers scales exponentially with read depth, meaning these algorithms are unable to handle ultra-high depth sequencing data on normal high-performance-computes without downsampling (randomly removing reads) and compromising sensitivity of detection. VarDict's unique algorithm design, which sequentially processes reads and performs local realignment using aggregates, ensures performance linear with depth (Supplementary Figure S4). As a result, VarDict has been successfully applied to longitudinally monitor tumor variants in cfDNA in a recent non-small-cell lung cancer clinical study of treatment with the EGFR inhibitor Tagrisso AZD9291 (15). The study revealed a novel somatic variant EGFR C797S responsible for emergence of resistance to the drug. In this study, the median depth of EGFR coverage was ∼30k, with peak depth >2M, otherwise impractical or not possible to process with other callers.
VarDict detects artifacts in PCR-based enrichment
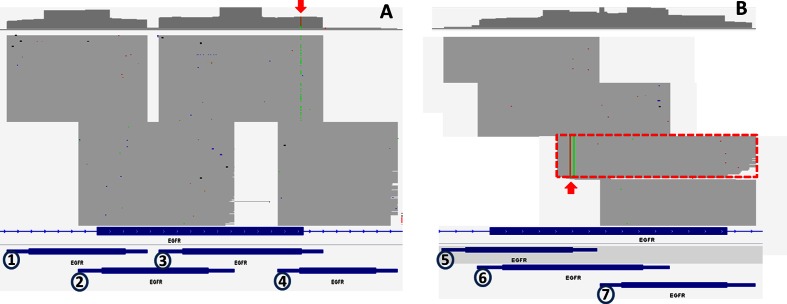
PCR is often used to enrich targeted regions before library construction, and is a known source of false positives in NGS. Figure 8A demonstrates how VarDict is able to detect variants that show amplicon bias when variants are covered by more than one PCR amplicon. In addition, VarDict avoids false positives from primers that are mis-paired due to high similarity (Figure 8B), deeming those reads to not belong to any amplicon and thus not be used for variant calling. Furthermore, when given the amplicon design with PCR primer locations, VarDict will mask them in silico for both depth calculation and variant calling, further improving sensitivity and specificity.
Figure 8.
Common artifacts from PCR based target enrichment. (A) Amplicon biased variants. Four overlapping PCR amplicons were designed against EGFR exon 12. The red arrow highlights a variant with AF of 24% and predicted to be C499Y, which has an entry is COSMIC. However, it is detectable only in amplicon 3 and will be flagged by VarDict and filtered. (B) Mis-paired primers amplified a region with EGFR exon 20. The read pairs highlighted by a red rectangle can not be mapped to any of the amplicons (5-7) below. The two mismatches at the left are actually a primer from ERBB2, which has high sequence similarity to EGFR resulting in primer mis-pairing. VarDict will filter out all those reads and thus no variant will be called from those mismatches in the mis-paired primer. Numbers (1-7) at the bottom indicate PCR amplicons, with thick middle portion for inserts and thin edges for PCR primers.
DISCUSSION
NGS is increasingly being applied in the clinic (25). As there are many types of NGS, ranging from targeted PCR-based sequencing to WGS, an accurate and versatile variant caller is needed that can easily adapt to different situations. This is especially important in cancer research where the complex nature of the cancer genome coupled with variable tumor content and sample quality (e.g. formalin fixation) demands a variant caller that can meet these challenges (2). We have demonstrated the versatility of VarDict over other commonly used variant callers to better handle many of these characteristics of cancer NGS data, proving it a valuable tool to facilitate cancer research.
Amongst the distinctive features of VarDict is its ability to call complex variants, ranging from consecutive di-/tri-nucleotide variants, to much more complex composites of insertions and deletions. In breast and ovarian cancer patients di-nucleotide mutations have been found to cause a stop codon and truncation of the tumor suppressor gene BRCA1 actionable for PARP inhibitor therapy (26) but often misclassified as two inactionable missense mutations. In >1% of all LUAD patients two neighboring frameshift InDels are found on the same allele of EGFR exon 19 that are in fact a single complex composite variant producing an in-frame deletion that activates the EGFR protein and renders patients eligible for EGFR inhibitor therapy. In fact, in TCGA LUAD, 8 out 32 (25%) patients with exon 19 deletion mutations are complex variants (Supplementary Table S3). Alongside this significant clinical relevance, an inability to call complex variants also has a considerable impact on annotation presented in current reference databases including dbSNP and COSMIC (5). We often observe proximal clusters of entries in dbSNP originating from the same sample, all of which in fact contribute to a single complex variant call at the same position by VarDict (Figure 3). Analysis of large sample cohorts often failed to find the existence of most of these entries, further supporting them to be erroneous calls created either due to the inability of Sanger sequencing for phasing, or the inability of other variant callers to call complex variants.
NGS is now being adopted to address more complex problems such as disseminating the genetic sub-clonality of heterogeneous tumor samples. Furthermore these questions are being asked in increasingly difficult samples such as FFPE and cfDNA from cancer patients. Researchers and clinicians are therefore increasingly interested in sensitive detection of low AF mutations. To achieve this, it is necessary to sequence at coverage depth >1000×, however, many popular variant callers do not scale well at such coverage depth. VarDict's ability to scale linearly to depth makes it suitable for ultra-deep targeted sequencing, as demonstrated in the discovery of the EGFR C797S mutation as a resistant mechanism to AZD9291 in NSCLC harboring EGFR T790M mutation (15). In fact, in that study, VarDict was the only variant caller that successfully completed in time (mostly overnight) with the desired sensitivity, while other callers attempted either failed to run or were not sensitive enough to detect low AF EGFR mutations resistant to AZD9291 (15).
As many targeted cancer drugs approved or in development use mutations in a handful of genes as patient selection biomarkers, PCR is often the technology of choice to amplify those genes before submitting them to sequencing. However, it is known that PCR can produce artifacts and if not dealt with carefully can lead to false positive/negative variant calls in downstream analysis, impacting clinical decisions. VarDict's abilities to detect PCR artifacts, such as amplicon bias and mis-paired primers, together with the linear scalability to depth, make it desirable in such studies to reduce both false positives and false negatives.
Somatic mutations are of high interest in cancer, as well as mutations that change allele frequencies significantly after treatment or during tumor evolution over time. VarDict is also able to perform paired analyses to identify such variant changes. VarDict was demonstrated to be a very effective somatic mutation calling for both SNV and InDels (27). In addition to somatic mutations, VarDict will identify germline, LOH and/or deleted variants, making it more suitable for comparison of paired longitudinal samples, for example pre- and post-treatment comparisons routinely investigated to understand drug resistance mechanisms in cancer research.
We further demonstrated the accuracy of VarDict calls through re-analysis of exome sequencing of 230 TCGA LUAD patient samples and comparison to data in Firehose and cBio, a commonly accepted gold standard in the field. To our surprise, VarDict calls 16% more patients with known activating mutations in well known lung cancer driver oncogenes, KRAS, EGFR, BRAF, PIK3CA and MET. In addition a number of somatic mutations with unknown functional significance in these genes were also called by VarDict, potentially revealing actionable mutations in even more patients missed by the gold standard analyses. This clearly has big impact in clinics where NGS is used to identify mutations for patient enrolment. For example, failure to detect a complex EGFR exon 19 deletion might prevent a lung cancer patient receiving clinically proven beneficial therapy, such as gefitinib or erlotinib (16,17). On the other hand, failure to detect a KRAS mutation might direct a colon cancer patient into EGFR therapies, such as cetuximab or panitumumab, which has been proven to not be beneficial (28,29). The effect might be even larger for tumor suppressors, such as BRCA1, BRCA2, TP53, RB1 and STK11, where InDel mutations are a common mechanism of loss of function but are poorly detected by other callers.
Many known cancer driver fusions, such as EML4-ALK (30) in lung cancer, are the result of SVs due to large rearrangements and are clinically actionable. Oncogenes involved in fusions, such as ROS1 in lung cancer, often have different fusion partners in different patient subsets (31), which prevents design of a universal assay to detect all potential fusions. Since NGS is unbiased to any particular fusion partnership it is increasingly being used to detect fusions. Internal gene rearrangements involving one or more exons are also a common mechanism by which tumor suppressors such as BRCA1 and BRCA2 to lose functions (32), and are eligible for targeted therapies (26). Identification of fusions and rearrangements typically involves a custom step of analysis requiring different software. This will be simplified by the ability to accurately call multiple types of SVs in NGS in a single step with VarDict. VarDict can also estimate allele frequencies for SVs, further facilitating the interpretation of their clinical relevance. However, in order to increase the specificity, VarDict currently heavily relies on split reads, coupled with discordant mate pairs to detect SVs at precise breakpoints. This does limit its sensitivity in lower coverage WGS where split reads are less likely to be sequenced and aligned. Further work is needed to improve its sensitivity in such scenario while maintaining specificity.
These properties, and the value to detect otherwise missed variants in cancer samples demonstrated in this manuscript, highlight VarDict as a unique variant caller of high value in cancer translational research. The algorithm is open source and freely available for public use. As the only published variant caller scalable to ultra-deep sequencing and capable of calling all variant types, including complex variants and SVs, we believe VarDict fills a gap in NGS analysis critical to interpretation of tumor genome complexity to advance our understanding and treatment of cancer.
AVAILABILITY
VarDict is implemented in Perl and is publicly available in GitHub (https://github.com/AstraZeneca-NGS/VarDict). A Java re-implementation of VarDict with improved performance is also publicly available in GitHub (https://github.com/AstraZeneca-NGS/VarDictJava), but currently without the SV calling capability.
Supplementary Material
Acknowledgments
We would like to thank Rory Kirchner, Harvard University Department of Biostatistics, for his contributions to the incorporation of VarDict into their BCBio framework that made VarDict more broadly available to the public; David Jenkins, a graduate student at Boston University, who helped evaluate VarDict's SV calling and compare to other SV callers; Hugo Lam and Li Tai Fang at Bina who tested and incorporated VarDict into their RAVE platform; and Roman Valls at the WWCRC for his help visualizing the computational resource utilization. Finally, Zaal Lyanov, Viktor Kirst, Mikhail Alperovich at EPAM and Vitaly Rozenman helped to re-implement VarDict in Java to optimize performance, which showed significant improvements on runtime.
FUNDING
AstraZeneca. Funding for open access charge: AstraZeneca.
Conflict of interest statement. None declared.
REFERENCES
- 1.TCGA. Integrated genomic analysis of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J., Schnall-Levin M., White J., Sanford E.M., An P., et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat. Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koboldt D.C., Zhang Q., Larson D.E., Shen D., Mclellan M.D., Lin L., Miller C.A, Mardis E.R, Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harismendy O., Schwab R.B., Alakus H., Yost S.E., Matsui H., Hasteh F., Wallace A.M., Park H.L., Madlensky L., Parker B., et al. Evaluation of ultra-deep targeted sequencing for personalized breast cancer care. Breast Cancer Res. 2013;15:R115. doi: 10.1186/bcr3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchetti A., Palma J.F., Felicioni L., De Pas T.M, Chiari R., Del Grammastro M., Filice G., Ludovini V., Brandes A.A., Chella A., et al. Early prediction of response to tyrosine kinase inhibitors by quantification of EGFR mutations in plasma of NSCLC patients. J. Thorac Oncol. 2015;10:1437–1443. doi: 10.1097/JTO.0000000000000643. [DOI] [PubMed] [Google Scholar]
- 8.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Gen Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Meth. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative Genomics Viewer. Nat. Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.TCGA. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thress K.S, Paweletz C.P., Felip E., Cho B.C., Stetson D., Dougherty B., Lai Z., Markovets A., Vivancos A., Kuang Y., et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harbouring EGFR T790M. Nat. Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 17.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Rulton L., et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W., Li L., Myers J.R., Marth G.T. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect breakpoints of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narzisi G., O'Rawe J.A., Iossifov I., Fang H., Lee Y., Wang Z., Wu Y., Lyon G.J., Wigler M., Schatz M.C. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods. 2014;11:1003–1036. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Källberg M., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2015;2015:btv710. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 22.Layer R.M, Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ewing A.D., Houlahan K.E., Hu Y., Ellrott K., Caloian C., Yamaguchi T.N., Bare J.C., P'ng C., Waggott D., Sabelnykova V.Y., et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods. 2015;12:623–630. doi: 10.1038/nmeth.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park J.Y., Kricka L.J., Fortina P. Next-generation sequencing in the clinic. Nat. Biotechnol. 2013;31:990–992. doi: 10.1038/nbt.2743. [DOI] [PubMed] [Google Scholar]
- 26.Ledermann J., Harter P., Gourley C., Friedlander M., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T., et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 27.Fang L.T., Afshar P.T., Chhibber A., Mohiyuddin M., Fan Y., Mu J.C., Gibeling G., Barr S., Asadi N.B., Gerstein M.B., et al. An ensemble approach to accurately detect somatic mutations using SomaticSeq. Genome Biol. 2015;16:197. doi: 10.1186/s13059-015-0758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karapetis C.S., Khambata-Ford S., Jonker D.J., O'Callaghan C.J., Tu D., Tebbutt N.C., Simes R.J., Chalchal H., Shapiro J.D., Robitaille S., et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N. Engl. J. Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 29.Bardelli A., Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J. Clin. Oncol. 2010;28:1254–1261. doi: 10.1200/JCO.2009.24.6116. [DOI] [PubMed] [Google Scholar]
- 30.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 31.Davies K.D., Le A.T., Theodoro M.F., Skokan M.C., Aisner D.L., Berge E.M., Terracciano L.M., Cappuzzo F., Incarbone M., Roncalli M., et al. Identifying and targeting ROS1 gene fusion in non-small cell lung cancer. Clin. Cancer Res. 2012;18:4570–4579. doi: 10.1158/1078-0432.CCR-12-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judkins T., Rosenthal E., Amell C., Burbidge L.A., Geary W., Barrus T., Schoenberger J., Trost J., Wenstrup R.J., Roa B.B. Clinical significance of large rearrangements in BRCA1 and BRCA2. Cancer. 2012;118:5210–5216. doi: 10.1002/cncr.27556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.