Abstract
BRCA2 is a multi-faceted protein critical for the proper regulation of homology-directed repair of DNA double-strand breaks. Elucidating the mechanistic features of BRCA2 is crucial for understanding homologous recombination and how patient-derived mutations impact future cancer risk. Eight centrally located BRC repeats in BRCA2 mediate binding and regulation of RAD51 on resected DNA substrates. Herein, we dissect the biochemical and cellular features of the BRC repeats tethered to the DNA binding domain of BRCA2. To understand how the BRC repeats and isolated domains of BRCA2 contribute to RAD51 binding, we analyzed both the biochemical and cellular properties of these proteins. In contrast to the individual BRC repeat units, we find that the BRC5–8 region potentiates RAD51-mediated DNA strand pairing and provides complementation functions exceeding those of BRC repeats 1–4. Furthermore, BRC5–8 can efficiently repair nuclease-induced DNA double-strand breaks and accelerate the assembly of RAD51 repair complexes upon DNA damage. These findings highlight the importance of the BRC5–8 domain in stabilizing the RAD51 filament and promoting homology-directed repair under conditions of cellular DNA damage.
INTRODUCTION
RAD51 is a key player in homologous recombination, assembling onto single-stranded DNA (ssDNA) as a nucleoprotein filament to catalyze the invasion and search for homology within a donor DNA template, usually the sister chromatid. BRCA2 is proposed to sequester free RAD51 in the nucleus until DNA damage mobilizes this repair complex to DNA double-strand breaks (DSBs) (1,2). Mechanistically, the cellular choice to engage homology-driven repair at a DSB is initiated by DNA resection resulting in ssDNA that is subsequently coated by Replication Protein A (RPA). BRCA2 can then bind to single-strand/double-strand DNA (ss/dsDNA) junctions as well as along ssDNA to stimulate RAD51 filament formation (3–6). Upon delivery of RAD51 to a resected DNA DSB, BRCA2 facilitates the stabilization of the ensuing nucleoprotein filament by maintaining RAD51 in the active ATP-bound form, and thereby, facilitates the exchange of RAD51 for RPA on the ssDNA lattice. Additional functions have been ascribed to BRCA2 including interactions with RAD51 that protect replication forks from MRE11 nucleolytic degradation (7,8). This link to replication fork stability is independent of BRCA2's role in homology-directed repair (HDR) and is dependent upon the S3291 residue located in the carboxy terminus of the protein (7,9–13). In the absence of DNA damage, BRCA2 likely prevents free RAD51 in the nucleus from attempting filament formation on inappropriate substrates such as dsDNA (1,13–16).
Early sequence analysis studies of vertebrate BRCA2 identified eight repeating motifs in exon 11 of ∼35 amino acids (17,18). These centrally located BRC repeat motifs were found to bind RAD51 implicating BRCA2 in the homologous recombination pathway of DNA double-strand break (DSB) repair (19–22). Several residues within each human BRC motif are highly conserved including critical phenylalanine and alanine hydrophobic contacts with RAD51 (17,23). Interestingly, the linear amino acid distances between each of the individual BRC repeats vary considerably and are only moderately conserved between mammalian species (17). Whether these ‘spacer regions’ are important for the regulation and orientation of RAD51 in relation to the polarity of filament formation will likely require high resolution structural studies. The majority of currently available human derived tumor cell lines containing mutant BRCA2 alleles are truncated within the BRC repeats and several tumor-associated missense variants map to this region (24–26). The most frequent germline mutation in BRCA2 that exists in the human population, 6174delT, results in a protein that is truncated at the BRC repeats (27,28). The significance of this region in predicting cancer risk prompted us to further investigate the molecular basis for BRC-mediated regulation of RAD51.
Biochemical analyses of individually purified BRC repeats demonstrated the existence of two separable regions with distinct functions required for binding and stabilizing RAD51 on ssDNA (29). Furthermore, individual BRC repeats were found to possess different affinities for RAD51 implying a potential ‘handoff’ mechanism facilitating deposition and stabilization of the RAD51 nucleoprotein filament (29,30). The BRC1, BRC2, BRC3 and BRC4 repeats were found to bind free RAD51, reduce its ssDNA-dependent ATPase activity, and prevent RAD51 from binding dsDNA. In contrast, BRC5, BRC6, BRC7 and BRC8 exhibited none of these activities but were found to bind and stabilize the RAD51-ssDNA nucleoprotein filament (29).
Previous studies did not directly address the role of the BRC repeats tethered to the DNA binding domain (DBD) of BRCA2 and whether biochemical activities correlated with the ability of these proteins to complement BRCA2 functions in cells (29,31). The BRCA2 DBD contains residues that engage binding to both ssDNA and dsDNA, a residue (S3291) that regulates BRCA2 mediator activity through phosphorylation, the interaction site for regulation by DSS1, and the nuclear localization signals (3,12,13,32,33). We took advantage of our 2XMBP (tandem Maltose Binding Protein) tag strategy to provide a comprehensive biochemical and cellular analysis of BRC repeats fused to the DBD as well as the individual N-terminal and DBD domains (3,34). By systematically testing the ability of these domains to mediate recombination, our results reveal new insights regarding the impact of the BRC repeats upon DNA strand pairing and the cellular response to DNA damage. We not only confirmed that two functional BRC repeat modules exist, BRC1–4 and BRC5–8, we find that BRC5–8 coupled to the DBD provides an unexpected robust complementation phenotype likely driven by a potent stimulation of RAD51-mediated DNA strand exchange activity.
MATERIALS AND METHODS
Constructs
All BRCn-DBD constructs were generated by PCR (AccuPrime Pfx SuperMix, ThermoFisher Scientific) or by restriction enzyme digest sub-cloning. PCR products and digested DNA were purified using the Qiaquick PCR Purification Kit (Qiagen). All constructs were generated in the phCMV1 (Genlantis) mammalian expression vector. The 2XMBP tag has been described previously (3) and was placed in-frame at the N-terminus of all proteins separated by an Asparagine linker and the Precission Protease cleavage sequence. The BRCn-DBD fusions and domain fragments were composed of the following amino acid sequences: N-terminus (1–992), DBD (2153–3418), BRC4 (1496–1589), BRC1–4 (993–1589), BRC5–8 (1596–2152) and BRC1–8 (993–2152). A flexible linker (-G-G-G-S-) was placed in between BRC4 and the DBD in the BRC4-DBD and BRC1–4-DBD fusion constructs. Primer sequences used to generate constructs are available upon request. Constructs were sequence verified by the Keck facility (Yale University).
Protein purification and affinity pull-downs
2XMBP-BRCn-DBD fusions and domain fragments were purified essentially as described for the full-length BRCA2 protein (3). Briefly, 10–20 × 150 mm plates of 70% confluent 293TD cells were transiently transfected using CaPO4 precipitation and cells harvested 36 h post-transfection in 50 mM HEPES (pH 7.5), 250 mM NaCl, 5 mM EDTA, 1% Igepal CA-630, 3 mM MgCl2, 1 mM ATP and protease inhibitor cocktail (Roche). Cell extracts were batch bound overnight to amylose resin (NEB) and 2XMBP tagged proteins eluted with 10 mM maltose. The proteins were further purified on a HiTrap Q (GE Life Sciences) column and eluted in buffer containing: 50 mM HEPES (pH 8.2), 450 mM NaCl, 0.5 mM EDTA, 10% glycerol and 1 mM DTT. Purified proteins were visualized on a coomassie stained 4–15% gradient SDS-PAGE TGX gel (Bio-Rad). A general protocol for purifying 2XMBP tagged proteins from 293TD cells can be found here (34). Contaminant bands in the BRC1–4-DBD and BRC1–8-DBD protein preparations were variable and compromised ∼2% and 8% of the total protein respectively, based on quantification in SYPRO Orange stained gels.
Amylose pull-down assays using purified proteins were performed in binding buffer ‘B’: 50 mM HEPES (pH 7.5), 250 mM NaCl, 0.5 mM EDTA and 1 mM DTT. Purified 2XMBP-BRCn-DBD proteins (2 μg, ∼100–200 nM) were incubated with 1 μg purified RAD51 (540 nM) for 30 min at 37°C and then batch bound to 30 μl of amylose resin for one hour at 4°C. RAD51 was purified as described previously (3). The complexes were washed with buffer B containing 0.5% Igepal CA-630/0.1% Triton X-100, eluted with 30 μl of 10 mM maltose in buffer B, laemmli sample buffer was added, samples were heated at 54°C for 4 min, and loaded onto a 4–15% gradient SDS-PAGE gel (Bio-Rad TGX Stain-Free gel). The gel was run for 2 h at 100 V. The 2XMBP-BRCn-DBD proteins were visualized in the Stain-Free gel on a ChemiDoc MP imaging system (Bio-Rad). RAD51 protein was visualized by staining with SyproOrange (Invitrogen). The Stain-Free protein bands were quantified using Image Lab software (Bio-Rad) and the SyproOrange bands by ImageQuant software on a Storm 860 PhosphorImager (Molecular Dynamics). The amount of RAD51 pulled down with 2XMBP-BRCn-DBD was determined using standard curves generated from known concentrations of RAD51 and 2XMBP-BRCn-DBD run in parallel in the same gel. The input amount in each pull-down for saturation binding analysis was 100–200 nM for BRCn-DBD proteins and 85 nM to 2 μM for RAD51. To determine the ratio of RAD51 bound to 2XMBP-BRCn-DBD, we fit the data to either a segmented linear regression (multiple binding sites) or a hyperbola (single site) using GraphPad Prism 6.0. Cellular amylose pull-down assays were performed by transiently transfecting 1 μg of the indicated constructs into 5 × 105 293TD cells/well seeded in six-well plates using TurboFect (Thermo Fisher Scientific). 36 h post-transfection, the cells were harvested in 500 μl of buffer ‘BB’: (50 mM HEPES [pH 7.5], 250 mM NaCl, 1% Igepal CA-630, 1 mM MgCl2, 1 mM DTT, 250 units/ml Benzonase (EMD Millipore), and 1× EDTA-free protease inhibitor cocktail (Roche). Cell lysates were batch bound to 20 μl of amylose resin for 2 h, washed 3× in buffer B, and eluted in buffer B containing 10 mM maltose. Elutions were processed and run on SDS-PAGE gels as described above for amylose pull-downs with purified proteins. The 2XMBP-BRCn-DBD proteins were visualized by Stain-Free imaging on a ChemiDoc MP imaging system (Bio-Rad). Endogenous RAD51 was visualized by transferring the gel to PVDF (EMD Millipore) membrane overnight at 4°C, blocking for 30 min with 5% milk in 1XTBS-T (50 mM Tris [pH 7.5], 150 mM NaCl, 0.05% Tween20), incubating the membrane with anti-RAD51 antibody (1:1000, 14B4 Novus) in 1XTBS-T for 30 min and then incubating for 45 min with HRP-conjugated anti-mouse secondary (sc-2005, Santa Cruz Biotechnology). Following extensive washing with 1XTBS-T, the membrane was incubated with WesternC (Bio-Rad) ECL reagent and images captured on the ChemiDoc MP system.
Electrophoretic mobility shift assay
Oligonucleotide substrates were obtained PAGE purified from IDT. The following oligonucleotides were utilized: RJ-167-mer (5′-CTG CTT TAT CAA GAT AAT TTT TCG ACT CAT CAG AAA TAT CCG TTT CCT ATA TTT ATT CCT ATT ATG TTT TAT TCA TTT ACT TAT TCT TTA TGT TCA TTT TTT ATA TCC TTT ACT TTA TTT TCT CTG TTT ATT CAT TTA CTT ATT TTG TAT TA TCC TTA TCT TAT TTA-3′) and RJ-PHIX-42-1 (5′-CGG ATA TTT CTG ATG AGT CGA AAA ATT ATC TTG ATA AAG CAG-3′). To generate the 3′ tail DNA substrate, RJ-167-mer was radiolabeled with 32P at the 5′-end using T4 Polynucleotide Kinase (NEB) and then annealed at a 1:1 molar ratio to RJ-PHIX-42-1. The 2XMBP tagged BRCn-DBD proteins, at the indicated concentrations, were incubated with 0.2 nM (molecules) of the radiolabeled DNA substrate for 30 min at 37°C in DNA strand exchange buffer (25 mM TrisOAc [pH 7.5], 1 mM MgCl2, 2 mM CaCl2, 0.1 μg/μL BSA, 2 mM ATP and 1 mM DTT). The reactions were resolved by electrophoresis on a 6% polyacrylamide gel in TAE (40 mM Tris–acetate [pH 7.5], 0.5 mM EDTA) buffer for 90 min at 80 V. The gel was then dried onto DE81 paper and exposed to a PhosphorImager screen overnight. The screen was scanned on a Molecular Dynamics Storm 840 PhosphorImager and bands quantified using ImageQuant software. The percentage of protein-DNA complexes was calculated as the free radiolabeled DNA remaining in a given lane relative to the protein-free lane, which defined the value of 0% complex (100% free DNA).
DNA strand exchange assay
All DNA substrates were obtained PAGE purified from IDT. The 3′ tail DNA substrate was generated by annealing RJ-167-mer to RJ-PHIX-42-1 at a 1:1 molar ratio. The dsDNA donor was generated by first radiolabeling RJ-Oligo1 (5′-TAA TAC AAA ATA AGT AAA TGA ATA AAC AGA GAA AAT AAA G-3′) with 32P (T4 Polynucleotide Kinase) on the 5′-end and annealing it to RJ-Oligo2 (5′-CTT TAT TTT CTC TGT TTA TTC ATT TAC TTA TTT TGT ATT A-3′) at a 1:1 molar ratio. The assay buffer contained: 25 mM TrisOAc (pH 7.5), 1 mM MgCl2, 2 mM CaCl2, 0.1 μg/μl BSA, 2 mM ATP and 1 mM DTT. All pre-incubations and reactions were at 37°C. The DNA substrates and proteins were at the following concentrations unless otherwise indicated in the figure legend: RPA (0.1 μM); RAD51 (0.22 μM); 3′ tail (4 nM molecules); and dsDNA (4 nM molecules). The 3′ tail DNA was incubated first with RPA for 5′, followed by the addition of each BRCn-DBD protein and RAD51, and finally, the radiolabeled donor dsDNA was added for 30 min. Where proteins were omitted, storage buffer was substituted. The reaction was terminated with Proteinase K/0.5% SDS for 10 min. The reactions were loaded on a 6% polyacrylamide gel in TAE buffer and electrophoresis was at 60 V for 80 min. The gel was then dried onto DE81 paper and exposed to a PhosphorImager screen overnight. The percentage of DNA strand exchange product was calculated as labeled product divided by total labeled input DNA in each lane.
Biotinylated DNA pull-down assay
The ssDNA oligonucleotide substrate, RJ-167-mer, was synthesized with a 5′ biotin modification and PAGE purified by IDT (Ultramer). 1 nM (molecules) of the biotinylated ssDNA was incubated with a stoichiometric excess of RAD51 (240 nM) for 5 min at 37°C in Buffer ‘S’: 25 mM TrisOAc (pH 7.5), 1 mM MgCl2, 2 mM CaCl2, 0.1 μg/μl BSA, 2 mM ATP and 1 mM DTT. The BRCn-DBD proteins (20 nM) were then added to the reaction for 1, 5, 15 or 30 min at 37°C. Final volumes were normalized with storage buffer as needed. The DNA-protein complexes were then captured by adding 2.5 μl of pre-washed MagnaLink Streptavidin magnetic beads (SoluLink) in buffer SW (Buffer S supplemented with 0.1% Igepal CA-630 and 0.5% Triton X-100). The bead-DNA-protein complexes were rotated at 25°C for 5 min and then washed 3× with buffer SW (lacking 2 mM ATP), resuspended in 20 μl laemmli sample buffer, heated at 54°C for 4 min, and loaded onto a 4–15% gradient SDS-PAGE gel. The BRCn-DBD proteins bound and eluted from the biotinylated DNA substrate were detected by western blot using a polyclonal antibody (C-18) to MBP (sc-808, Santa Cruz Biotechnology). RAD51 protein bound and eluted from the biotinylated DNA was determined by western blot using a monoclonal antibody specific to human RAD51 (14B4, Novus).
Generation of stable cell lines
VC8 BRCA2 mutant hamster cells were cultured in HAMs + 15%FBS. VC8 cells seeded into 60 mm plates were stably transfected with 2 μg of plasmid DNA using GenJet Plus (SignaGen) transfection reagent. After 48 hours, the cells were trypsinized and diluted into 100 mm plates containing 1 mg/ml G418. Single cell colonies were isolated and expanded for use. Human DLD1 BRCA2−/− cells (Horizon Discovery, originally generated by Hucl et al. (35)) were cultured in RPMI + 10%FBS. The DLD1 BRCA2−/− cells were stably transfected with 2 μg of DNA using LipoFectamine3000 (Invitrogen), and single cell clones were derived as described for the VC8 cells. Protein expression was verified in total cell lysates or nuclear extracts by western blot using a rabbit antibody against MBP (sc-808, C18 clone, Santa Cruz Biotechnology) and a mouse monoclonal antibody against BRCA2 (Ab-1, EMD Millipore). Biochemical cell fractionation in the DLD1 BRCA2−/− stable cell lines was performed using the NE-PER Nuclear and Cytoplasmic Extraction Kit (ThermoFisher Scientific) supplemented with protease inhibitor cocktail (Roche). Partitioning efficiency was determined by western blotting against PCNA (nuclear, MCA1558, Bio-Rad) or GAPDH (cytoplasmic, sc-166574, Santa Cruz Biotechnology).
Clonogenic survival
VC8 and DLD1 BRCA2−/− stable cell lines were counted using a TC20 automated cell counter (Bio-Rad), serially diluted, and seeded into six-well plates at 100 and 500 cells/well in triplicate for plating efficiency. A 1.5 mM MMC (Sigma) stock was made by diluting 2 mg powder in 4 mL dH2O, filter sterilized, protected from light, and either used immediately or stored at −80°C in small aliquots. For exposures to MMC, cells were seeded at 100, 1000 and 10 000 cells/well in six-well plates and 100 000 cells in 100 mm plates in triplicate. Sixteen hours after seeding, cells were treated with an acute (1 h) exposure to MMC prepared in 2 mL HAMs (VC8) or RPMI (DLD1) in the absence of serum (in the dark) at the concentrations indicated in the graphs. After 1 h treatment with MMC, media was aspirated off, cells were washed 2× with PBS, cells were re-fed with fresh media containing serum and penicillin (50 I.U./ml)–streptomycin (50 μg/ml), and allowed to form colonies over 12–14 days. Cells were fixed and stained with crystal violet. Plates were dried overnight, colonies containing 50 or more cells were scored, and the surviving fraction was determined.
Immunofluorescence imaging
Human DLD1 BRCA2−/− stable cells were grown on coverslips at 20 000 cells/well in a 24-well plate for 24 h. Cells were washed twice with PBS, fixed in 1% paraformaldehyde (PFA)–2% sucrose in PBS for 15 min at room temperature, washed two times with PBS, permeabilized with methanol for 30 min at −20°C, and washed two times with PBS. Cells were blocked with 5% BSA in PBS for 30 min at room temperature and incubated with primary antibody against MBP (1:200, E8038S, NEB), RAD51 (1:100, H-92 sc8349, Santa Cruz Biotechnology) and/or γ-H2AX (Ser139, 1:200, 05-636, EMD Millipore) in 5%BSA–0.05%Triton at 4°C overnight. After primary antibody incubation, cells were washed three times with PBS, followed by goat anti-rabbit and anti-mouse secondary antibodies conjugated to the fluorophore alexa-488 and alexa-594 (1:1000, Invitrogen). Coverslips were washed three times, incubated with 4′-6-diamidino-2-phenylindole (DAPI, 3.3nM) for 2 min and mounted on slides with FluorSave (345789, Calbiochem). Immunofluorescence images were taken using a Zeiss 710 epifluorescence microscope and confocal microscope (Leica SP5) using an oil 63× objective with fixed optical slice, laser power, and detector/amplifier settings for all samples across each individual experiment to allow comparison. Cells were either left untreated (sham) or irradiated (4 Gy) with an X-Rad 320 Biological Irradiator (Precision X-Ray) and incubated for 8 or 48 h prior to the immunofluorescence protocol. Captured images were analyzed with ImageJ cell counter plugin. We scored the nuclear foci staining as follows: 100 to 200 nuclei were counted on three to five representative areas across each coverslip. Total nuclei were counted with DAPI images. A cell was counted as being RAD51 positive if there were at least three distinct foci per nucleus. A cell was counted as being γ-H2AX positive if there were at least 5 distinct foci per nucleus. These thresholds were selected after examining: untreated, 2, 4 and 8 Gy treatments of vector control versus wild type full-length BRCA2 complemented cells at the following times points: 4, 8, 12, 24 and 48 h. The raw γ-H2AX and RAD51 foci count was taken as the number of γ-H2AX or RAD51 foci positive cells divided by the total number of cells expressed as a percentage per field. Statistical analysis was performed with GraphPad Prism version 6.0.
HDR luciferase assay
The HDR luciferase reporter gene (gWiz.Lux-5′-3′ Luc) was constructed from the parental vector gWiz Luciferase (Genlantis). An I-SceI site was created in the luciferase ORF by ligating the following annealed oligonucleotides: 5′SCEILUC (5′-CGC TAG GGA TAA CAG GGT AAT-3′) and 3′SCEILUC (5′-CGA TTA CCC TGT TAT CCC TAG-3′) into a BstBI site. This seven amino acid insertion at amino acid 56 of the luciferase ORF ablates luciferase activity. A second luciferase ORF was ligated into the XmnI site 700 bp downstream from the first luciferase ORF. The second luciferase ORF lacks a promoter but can be utilized as a donor in homology directed repair of the first luciferase ORF upon generation of a double-strand break by expression of the I-SceI nuclease. The pSce-MJ mammalian I-SceI expression vector was a kind gift from Dr Fen Xia.
To perform the assay, cells were seeded into six-well plates at 2.5 × 105 cells/well. Twenty hours later, cells were transfected with 200 ng of gWiz.Lux-5′-3′Luc vector and 1 μg of the I-SceI expression vector using Lipofectamine 3000. gWiz.Lux-5′-3′Luc or gWiz.Lux vector were transfected alone and used as negative and positive controls, respectively. Cells were harvested 48 hours post-transfection in 200 μl of buffer B prior to luminometer analysis. Luminescence was measured using an integration time of 5 s with 40 μl of the lysate plus 100 μl of luciferin substrate (One-Glo luciferase assay, Promega). Luciferase values were measured as independent duplicates in each experiment. The raw data was normalized to protein levels and the values plotted were calculated by setting the full-length BRCA2 mean value to 100%. The data presented is the average of five independent experiments.
RESULTS
RAD51 binding to BRCA2 domains and BRCn-DBD fusions
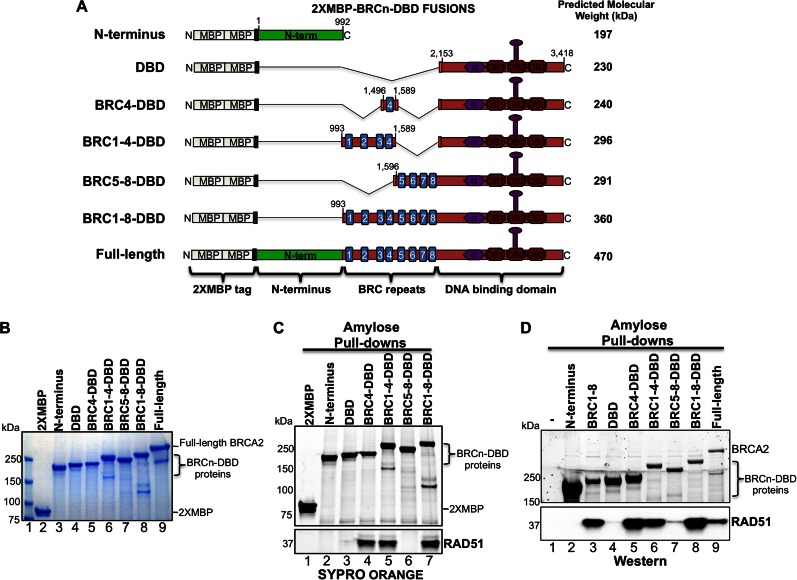
Previous biochemical studies of BRCA2 revealed two distinct modules within the BRC1–8 repeat domain (29). Individually, BRC1, BRC2, BRC3 and BRC4 reduced the ATPase activity of RAD51, prevented RAD51 binding to dsDNA, and increased RAD51 binding activity towards ssDNA. BRC5, BRC6, BRC7 and BRC8, however, exhibited only the latter activity of stabilizing the ssDNA bound form of RAD51. To query how these two modules, BRC1–4 and BRC5–8, contribute to the biochemical functions of BRCA2, we generated fusions tethered to the DBD of BRCA2 (see schematic in Figure 1A). We also generated fusions between BRC4 and the entire BRC1–8 region to the DBD. For simplicity, we will refer to these fusions as BRCn-DBD proteins throughout the text. Our previous work demonstrated that a tandem Maltose Binding Protein (2XMBP) tag located at the N-terminus of BRCA2 did not interfere with in vitro or cellular functions, and therefore, all constructs in this study contained the same N-terminal 2XMBP tag (3). To complete our analyses, we included the N-terminal and DBD domains of BRCA2 (lacking any BRC repeats) to determine whether these protein fragments are capable of RAD51 loading or complementing the cellular response to DNA damage. We expressed the proteins in human 293T cells and purified each for biochemical characterization (Figure 1B).
Figure 1.
BRCA2 BRCn-DBD protein purification and binding to RAD51. (A) Schematic of human BRCA2 protein fusions and fragments. All constructs contain the 2XMBP tag located at the N-terminus. Amino acid numbers included in each construct are depicted. (B) Coomassie stained SDS-PAGE gel of purified BRCA2 BRCn-DBD fusion and fragment proteins. Molecular weight markers were run in lane 1. Some protein preparations contained minor species likely due to degradation or truncated expression (see band at ∼160 kDa in BRC1–4-DBD and band at ∼120 kDa in BRC1–8-DBD). (C) Purified 2XMBP tagged BRCA2 BRCn-DBD proteins (lanes 2–7) were incubated with purified RAD51 and bound to amylose. Lane 1 is purified 2XMBP tag alone incubated with RAD51. The protein complexes were washed, eluted, and analyzed by SDS-PAGE. The gel was stained with SYPRO Orange to visualize proteins. (D) Human 293T cells were transiently transfected with the indicated 2XMBP tagged BRCA2 BRCn-DBD constructs (lanes 2–9). Lane 1 is a non-transfected control to visualize non-specific binding of RAD51 to the amylose beads. Thirty six hours post-transfection, cell lysates were harvested and batch bound to amylose beads in the presence of Benzonase, washed, and eluted with maltose. The BRCA2 proteins were detected using StainFree gel imaging (see Materials and Methods section) and the endogenous RAD51 protein was detected by western blotting with an antibody targeted against RAD51.
Our prior work demonstrated that purified full-length BRCA2 protein binds approximately six RAD51 monomers based on biochemical pull-down assays (3). Under the same conditions, we incubated each of our purified BRCn-DBD proteins with purified RAD51. BRC4, BRC1–4 and BRC1–8 fused to the DBD were capable of binding free RAD51 (Figure 1C, lanes 4, 5 and 7) while significant binding to the BRC5–8 region (Figure 1C, lane 6) was notably absent. These findings are in agreement with Carreira et al. (29), such that BRC1, BRC2, BRC3 and BRC4 are competent for binding to free RAD51 whereas BRC5, BRC6, BRC7 and BRC8 display little or no binding. Upon longer exposures, a small amount of RAD51 can be detected bound to both BRC5–8-DBD and the DBD alone (Figure 1C, lanes 3 and 6). Consistent with prior reports of purified RAD51 forming oligomers in solution, these structures may mimic the nucleoprotein filament and account for these binding properties (13,16). The purified N-terminal domain of BRCA2 and the 2XMBP tag alone did not bind free RAD51 (Figure 1C, lanes 1 and 2). To explore the affinities of fusion proteins capable of binding free RAD51, we performed a saturation binding analysis (Supplementary Figure S1) using a fixed concentration of each BRCn-DBD protein with increasing amounts of RAD51 protein. As anticipated, the number of BRC repeats correlated with a higher level of saturation binding to RAD51 (Supplementary Figure S1B). Despite a lack of interaction between BRC5–8 and free RAD51, the inclusion of repeats five to eight into BRC1–8-DBD contributed additional RAD51 binding approaching the levels of full-length BRCA2 protein.
To corroborate our findings with the purified proteins in a cellular assay, we transfected our BRCn-DBD constructs into 293T cells and captured the 2XMBP tagged proteins with amylose beads. The cellular lysates were treated with Benzonase to prevent the detection of DNA-mediated interactions. We then washed and eluted the bound proteins with maltose. We detected negligible binding of endogenous RAD51 to the amylose beads alone in our control samples (Figure 1D, lane 1). Pull-down analysis of BRCA2 separated into three functional units (N-terminus, BRC1–8 and DBD) revealed that only the BRC1-8 region binds endogenous RAD51 (Figure 1D, lanes 2–4) under our conditions (similar to Figure 1C, minor RAD51 binding to DBD alone can be visualized upon longer exposures). Upon further analyses with the BRCn-DBD fusions, we find BRC1–4 imparts the majority of binding to endogenous RAD51 as very little RAD51 is found associated with the BRC5–8-DBD protein (Figure 1D, compare lane 7 to lanes 5, 6, 8 and 9). The residual binding we observe with DBD alone and BRC5–8-DBD (Figure 1D, lanes 4 and 7) could be due in part to RAD51–ssDNA complexes inaccessible to Benzonase treatment. Finally, we note the absence of RAD51 binding to the N-terminal domain of BRCA2 (Figure 1D, lane 2).
BRCn-DBD proteins bind DNA and stimulate RAD51-mediated DNA strand exchange
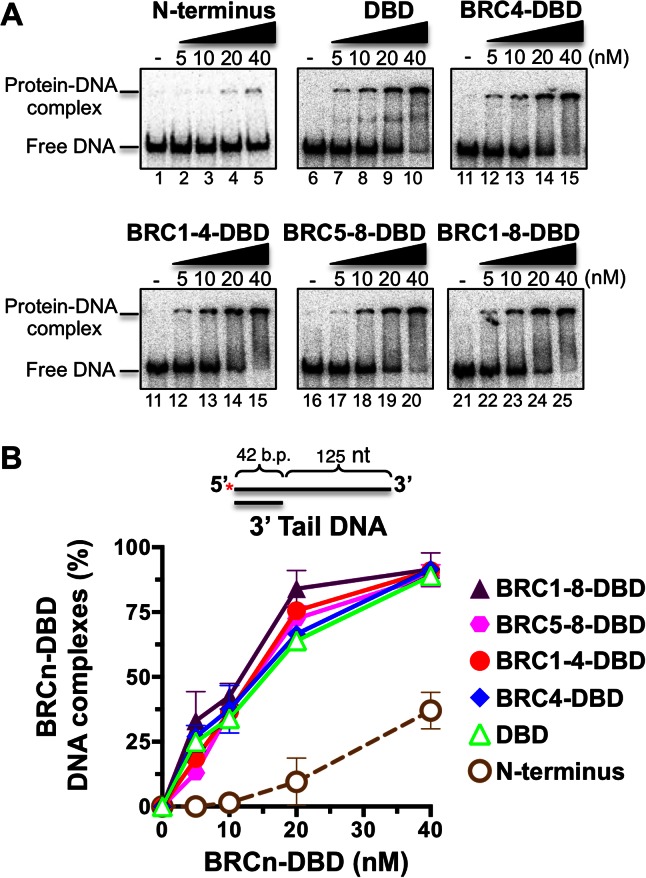
The DBD of BRCA2 binds ssDNA through the oligonucleotide/oligosaccharide binding folds 2 and 3 (OB2 and OB3) domains (36). A three-helix bundle at the tip of the tower domain is predicted to facilitate binding to dsDNA (36). In order to verify that DNA binding activity in our BRCn-DBD proteins remained intact, we performed an electrophoretic mobility shift assay (EMSA) analysis. Each protein was incubated with a radiolabeled substrate containing 42 basepairs of dsDNA and 125 nucleotides of ssDNA referred to as 3′ tail DNA. As anticipated, the DBD alone and all BRC repeats fused to the DBD were capable of binding 3′ tail DNA (Figure 2A and B). The affinities were comparable to our previous DNA binding analysis of the full-length BRCA2 protein (3). The purified 2XMBP tag alone did not confer any DNA binding activity (Supplementary Figure S2A). These results substantiate that the DBD provides the majority of the DNA binding functions for BRCA2 and that inclusion or fusions of various BRC repeats to the DBD does not dramatically alter DNA binding properties. At the highest concentrations tested (20 and 40 nM), we detected modest binding of the N-terminal domain to the 3′ tail DNA substrate. In-silico sequence analysis predicts a potential OB fold at amino acids 134–162 (CloneManagerV.9). To the best of our knowledge, this is the first demonstration of any DNA binding activity located in the N-terminal region of human BRCA2. Interestingly, a second DNA binding domain important for recombination function in the N-terminal region of the Ustilago maydis BRCA2 orthologue, Brh2, has previously been described (37).
Figure 2.
The BRCn-DBD proteins bind ssDNA tails with similar affinities. (A) Electrophoretic mobility shift assay (EMSA) gels depicting increasing concentrations of BRCn-DBD proteins incubated with a 3′ tail DNA substrate. (B) Quantification of EMSA results from three independent experiments. Errors bars are SD.
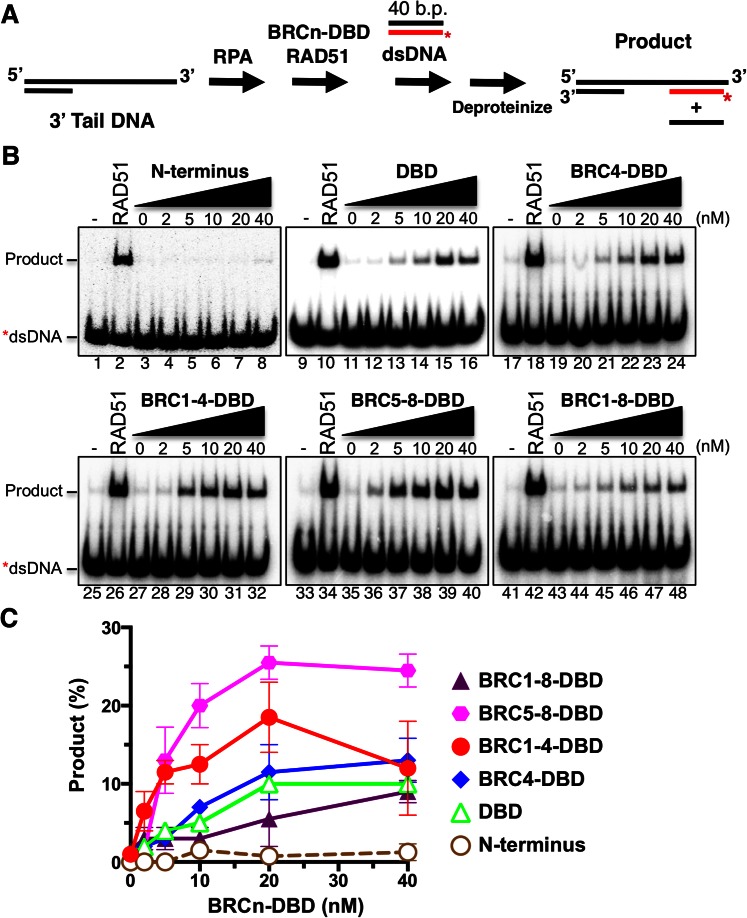
Cells that encounter DNA DSBs during or after S phase are poised to repair these breaks by homologous recombination using the sister chromatid as a template. A DSB that is destined for repair by recombination must first be resected by nucleases resulting in 3′ tailed ssDNA. The ssDNA that is generated following resection is immediately coated with the ssDNA binding protein, Replication Protein A (RPA). While RPA can stabilize and remove secondary structure in ssDNA, its high affinity for ssDNA can block RAD51 binding and filament formation. One of the mediator functions of BRCA2 is to overcome this RPA blockade and replace RPA-bound ssDNA with RAD51. We previously devised a DNA strand exchange assay utilizing a 3′ tail DNA oligonucleotide substrate to mimic the cellular process of RAD51 loading and strand invasion (see schematic in Figure 3A) (3). We utilized this same DNA strand exchange assay with the BRCn-DBD proteins to assess the contribution of the BRC repeats to mediate stimulation of RAD51 loading in the presence of RPA first. The 3′ tail DNA was the same substrate used in the EMSA analysis (Figure 2) confirming that each of the BRCn-DBD proteins and the DBD alone possessed comparable affinities for this DNA.
Figure 3.
RAD51-mediated DNA strand exchange activity is stimulated by increasing amounts of BRCn-DBD proteins. (A) Diagram of DNA strand exchange assay. RPA was pre-bound to the 3′ tail DNA substrate for 5 min. Increasing amounts of each BRCn-DBD protein was then added in combination with RAD51 for 5 min, followed by the addition of radiolabeled donor dsDNA for 30 min. All reactions were incubated at 37°C. The reaction was then deproteinized and run on a 6% TAE polyacrylamide gel. (B) Autoradiograms of PAGE gels used to analyze the products of DNA strand exchange. The first lane in each gel is a no protein control. The second lane in each gel contains RAD51 in the absence of RPA or BRCn-DBD. (C) Quantification of the product formation in (B). Mean values from three independent experiments were plotted. Error bars are SD.
To investigate the stimulation of RAD51-mediated DNA strand exchange, we first incubated the 3′ tail DNA with RPA, followed by the addition of a BRCn-DBD protein simultaneously with RAD51, and finally, a radiolabeled donor dsDNA was added to initiate the strand pairing reaction. While the BRCn-DBD and DBD proteins were able to overcome RPA inhibition and mediate stand exchange to various levels, the BRC5–8-DBD fusion promoted the highest level of activity (Figure 3B). The aggregate results of our DNA strand exchange analysis placed the purified proteins in the following order from highest to lowest activity: BRC5–8-DBD > BRC1–4-DBD > BRC4–DBD > DBD > BRC1–8-DBD (Figure 3C). Under these conditions, the N-terminal fragment of BRCA2 and the 2XMBP tag alone were not capable of stimulating DNA strand exchange (Figure 3B and C, Supplementary Figure S2B). A BRCn-DBD protein concentration range of 2–40 nM was utilized for direct comparison to our previously characterized full-length BRCA2 protein (3). BRC5–8-DBD activity plateaued at 20 nM while BRC1–4-DBD peaked at a concentration of 20 nM and consistently dropped at the higher 40 nM concentration (Figure 3C). The strand pairing activity of BRC4-DBD and DBD alone tracked closely with one another with the addition of the BRC4 repeat fused to the DBD providing a modest additional stimulation as expected. While the DBD of BRCA2 has previously been shown to stimulate DNA strand exchange (36), we were surprised that BRC1–8-DBD exhibited very little activity reaching comparable levels to DBD, BRC4-DBD and BRC1–4-DBD only at the highest concentrations utilized (40 nM). This result was unexpected as BRC1–8-DBD comprises the largest fragment of BRCA2 in its native state and is competent for binding RAD51 (Figure 1 and Supplementary Figure 1). Nonetheless, the results of our DNA strand exchange analysis suggest that BRC5–8 fused to the DBD accelerates formation and/or stabilization of the RAD51 presynaptic filament, and thereby, alleviates the blockade posed by RPA.
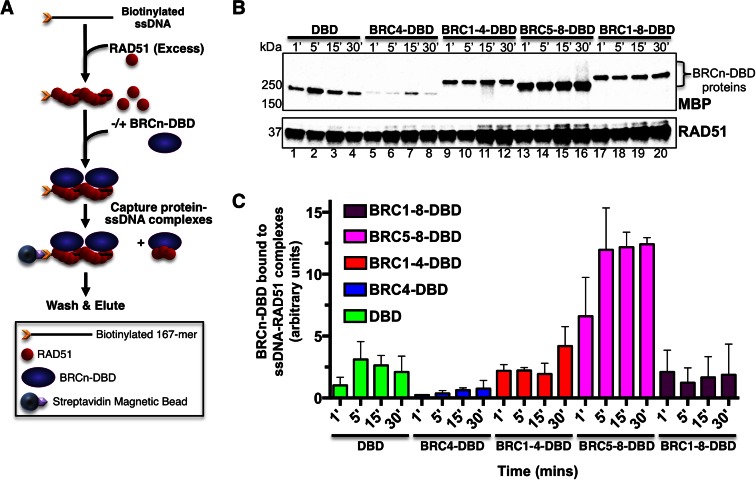
BRC5–8-DBD binds and stabilizes the RAD51 nucleoprotein filament
Our protein interaction studies indicated that BRC5–8-DBD was incapable of binding significant levels of free RAD51 using either purified proteins or cellular lysates (Figure 1C and D). To further understand the mechanism underlying BRC5–8-DBD mediated promotion of DNA strand pairing, we designed a biotinylated DNA pull-down assay (Figure 4A) to interrogate binding to the RAD51 filament on ssDNA, rather than free RAD51 protein. Following filament formation on ssDNA, we performed a time course with the addition of each BRCn-DBD protein to the RAD51–ssDNA complex. We utilized an excess concentration of RAD51 such that the ssDNA lattice would be fully saturated and free RAD51 would be available for binding to the BRCn-DBD proteins. Based on the results of Carriera et al. (29), we reasoned that BRC5–8-DBD would bind preferentially to the RAD51 filament while excess free RAD51 could effectively compete for binding to fusions containing BRC repeats 1–4. Binding events to free RAD51 would not be detected as these complexes are washed away before the elution step. We confirmed that the DBD and BRCn-DBD proteins were competent for binding to the biotinylated ssDNA in the absence of RAD51 (Supplementary Figure S3). As shown in Figure 4B and C, all fusion proteins and the isolated DBD domain were capable of binding to the RAD51–ssDNA complex, however, the level of BRC5–8-DBD binding (Figure 4B, lanes 13–16) was substantially higher. These results suggest that BRC5–8-DBD promotes the enhanced DNA strand exchange activity of RAD51 by both binding and stabilizing the RAD51 nucleoprotein filament. Consistent with prior results, the BRC1–8-DBD fusion performed no better than BRC1–4-DBD in this binding assay (Figure 4B, compare lanes 17–20 to 9–12). We do note an increase in RAD51 pulled down with biotinylated ssDNA in reactions containing BRC1–4-DBD, BRC5–8-DBD and BRC1–8-DBD (Figure 4B, compares lanes 1–8 to 9–20) indicating stabilization of the nucleoprotein filament, however, the increase was modest at ∼1.5-fold.
Figure 4.
BRC5–8-DBD binds preferentially to the RAD51 nucleoprotein filament. (A) Schematic of biotinylated DNA pull-down assay. Biotinylated ssDNA was pre-incubated with an excess of purified RAD51 for 5 minutes. Purified BRCn-DBD or DBD proteins were then incubated with the RAD51–ssDNA complexes for the indicated time points in (B) and (C) and captured on magnetic streptavidin beads. The beads were then washed, eluted in sample buffer, and analyzed by SDS-PAGE western blotting. (B) Top panel is a western blot against MBP to detect BRCn-DBD and DBD proteins eluted from the biotinylated ssDNA–RAD51 complexes. Lower panel is the same western blot probed with anti-RAD51. (C) Quantification of time course results in (B) by band densitometry (ImageLab). Densitometry analyses of proteins bound to RAD51–ssDNA compexes were analyzed in the linear range of detection (ChemiDocMP) and arbitrary units used to compare values from three independent experiments. Errors bars are SD.
BRCn-DBD proteins complement mammalian cells deficient for BRCA2
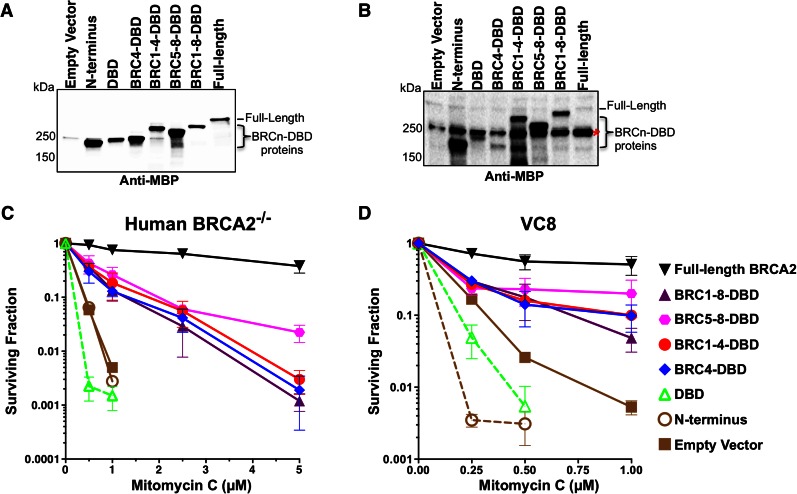
To corroborate our biochemical findings with genetic analyses, we generated stable complemented cell lines expressing the same constructs that were used for purification. To rule out cell line specific effects, we utilized both human BRCA2 knockout cells and the hamster VC8 BRCA2 mutant cell line (35,38). We confirmed expression of each construct taking advantage of the MBP tag as an epitope for detection by western blot (Figure 5A and B). A non-specific band in the VC8 cells at 250 kDa cross-reacted with the MBP antibody (red asterisk) migrating at a similar position to the BRC4-DBD protein (Figure 5B). We analyzed several single-cell derived stable clones for each construct in both cell lines and found that while expression varied somewhat between clones, the level of complementation remained equivalent (Supplementary Figure S4). As previously reported, larger constructs containing more of the BRCA2 sequence tended to express at lower levels (31). To verify our fusion constructs encompassing the DBD localized to the nucleus, we examined the human BRCA2−/− stables by biochemical fractionation and confocal immunofluorescence microscopy (Supplementary Figure S5A and B). Three putative nuclear localization signals reside in the DBD (KKRR a.a. 3266–3269, PIKKK a.a 3311–3315 and RLKRR a.a 3347–3351) and a previous study demonstrated BRCA2 peptides truncated upstream of these residues localize to the cytoplasm (39). As expected, all constructs containing the DBD domain localized to the nucleus (Supplementary Figure S5B). Only the N-terminal fragment of BRCA2 was excluded from the nucleus illustrating the importance of the NLS signals within the DBD. Despite an intact interaction domain, PALB2 (Partner and Localizer of BRCA2) is not sufficient to deliver the N-terminal fragment of BRCA2 into the nucleus (see discussion below). We further confirmed the BRCn-DBD proteins stably expressed in human BRCA2−/− cells were capable of interacting with endogenous RAD51 (Supplementary Figure S6).
Figure 5.
Mitomycin C clonogenic survival analyses of BRCA2 deficient cells complemented with BRCn-DBD proteins. (A) Western blot of total cell lysates from human BRCA2−/− cells stably expressing BRCA2 and BRCn-DBD constructs. (B) Western blot of total cell lysates from hamster VC8 brca2 mutant cells stably expressing BRCA2 and BRCn-DBD constructs (red asterisk denotes a non-specific band that cross-reacts with the MBP antibody in VC8 protein extracts). (C) Clonogenic survival graph of human BRCA2−/− cells stably expressing constructs in (A) treated with increasing concentrations of the DNA cross-linking agent Mitomycin C (MMC). (D) Clonogenic survival graph of VC8 cells with stable expression of constructs in (B) treated with increasing concentrations of MMC. Error bars are SD (n = 3).
Mammalian cells deficient in BRCA2 function are exquisitely sensitive to the crosslinking agent Mitomycin C (MMC). In agreement with our biochemical analysis, the BRC5–8-DBD construct provided the highest level of complementation in both human and hamster cells upon DNA damage with MMC (Figure 5C and D). The order of complementation from highest to lowest was: BRC5–8-DBD > BRC1–4-DBD > BRC4-DBD > BRC1–8-DBD > N-terminus > DBD. The cellular complementation results track well with our biochemical DNA strand exchange analysis implying that the recombination mediator activities of the BRCn-DBD proteins play an important role in repairing MMC cross-link damage. As expected, the full-length BRCA2 protein provided the highest level of complementation in both cell lines, despite the lower level of expression compared to the BRCn-DBD constructs. We were again surprised that the BRC1–8-DBD protein performed poorly in these MMC clonogenic survival assays compared to other BRCn-DBD proteins. Interestingly, expression of the N-terminus or DBD alone could not rescue any MMC sensitivity, and in fact, displayed a reproducible modest reduction in survival compared to the empty vector control cells (Figure 5C and D). The N-terminal and DBD fragments lack sufficient activity to promote HDR and may further antagonize cellular pathways responsible for MMC-induced DNA cross-link repair.
BRCn-DBD proteins promote homology-directed repair of a nuclease-induced DSB
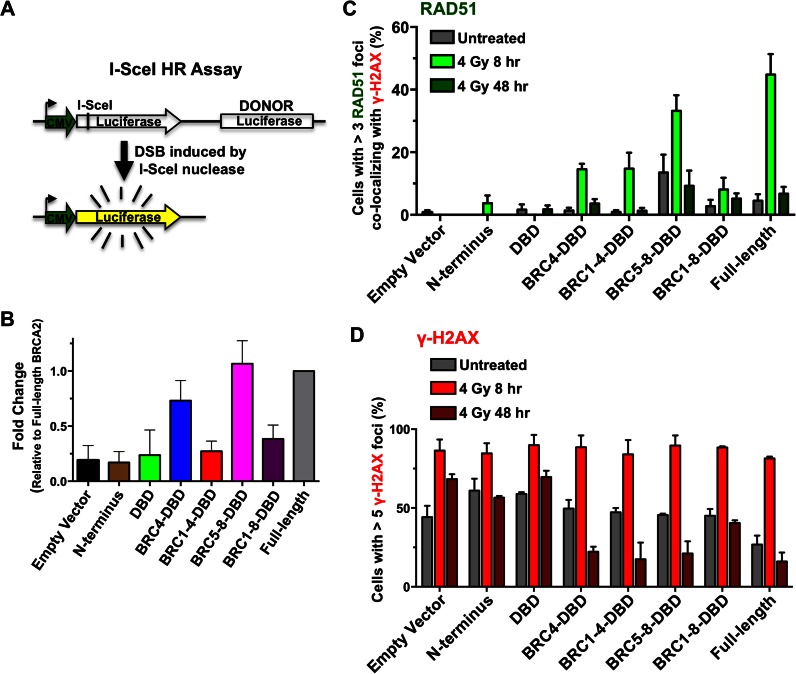
To delineate further the role of BRCn-DBD proteins in HDR, we examined the ability of cells to repair a site-specific DNA DSB. We generated a luciferase based reporter construct based on the I-SceI DR-GFP system pioneered by the lab of Maria Jasin (40). We integrated the I-SceI recognition site into the open reading frame of a luciferase reporter such that luciferase activity was abolished. We then placed a promoterless luciferase open reading frame 700 base pairs downstream to provide a donor template for HDR (Figure 6A). This luciferase HDR reporter was transiently transfected into each of the stably complemented human BRCA2 knockout cell lines with or without expression of the I-SceI nuclease. For relative comparison (as a benchmark), we set the re-activation of luciferase activity in the full-length BRCA2 complemented cells to 100%. Consistent with our MMC survival analyses, the BRC5–8-DBD complemented cells demonstrated the highest level of HDR of the I-SceI nuclease-induced DSB in the luciferase reporter (Figure 6B). The isolated N-terminal and DBD domains of BRCA2 restored minimal luciferase activity, similar to empty vector control cells. In contrast to the MMC survival analyses, the BRC1–4-DBD complemented cells did not perform as well as BRC1–8-DBD or BRC4-DBD. This discrepancy may reflect intrinsic differences in the assays or the specific repair pathways required to restore a nuclease-induced DSB versus MMC cross-link repair.
Figure 6.
Repair of I-SceI nuclease induced DSBs and ionizing radiation induced DNA damage. (A) Schematic of the I-SceI luciferase reporter assay. The I-SceI recognition site is located within the 5′ end of the firefly luciferase ORF and disrupts luciferase activity. A promoter-less luciferase donor DNA is located downstream as a template for HDR. The DSB is generated by co-expression of the I-SceI nuclease. (B) The I-SceI luciferase reporter and I-SceI nuclease were co-transfected transiently into human BRCA2−/− cells stably expressing the indicated BRCn-DBD constructs. Cell lysates were harvested 48 h post-transfection and assayed for luciferase activity. For comparison, the normalized luciferase activity of human BRCA2−/− cells complemented with full-length BRCA2 was set to 100%. Error bars are SD (n = 5). (C) Quantification of RAD51 foci co-localized with γ-H2AX foci in human BRCA2−/− cells stably expressing the indicated BRCn-DBD constructs. Cells were fixed and analyzed at 8 and 48 h following exposure to 4 Gy of ionizing radiation. The untreated samples were sham irradiated. (D) Quantification of γ-H2AX foci in human BRCA2−/− cells stably expressing the indicated BRCn-DBD constructs. Cells were treated exactly as in (C). Nuclei containing >5 (γ-H2AX) or 3 (RAD51) foci were scored as positive. A representative experiment was plotted. Errors bars are SD (n = 200).
Kinetics of IR-induced damage repair in BRCn-DBD fusion cells
Ionizing radiation (IR) generates DNA DSBs to which BRCA2 is recruited (visualized as foci) specifically in the S and G2 phases of the cell cycle (41,42). To follow the kinetics of repair activity by human cells expressing the BRCn-DBD proteins, we scored RAD51 and γ-H2AX foci by confocal microscopy following exposure to IR. Phosphorylation of Serine 139 on H2AX (γ-H2AX) is a commonly used surrogate marker for DNA DSBs (43–45). RAD51 foci are thought to reflect high local concentrations of the RAD51 protein that correlate with HDR-mediated repair of DNA DSBs (46,47). To ensure the RAD51 foci we scored coincided with DNA damage, we scored only those RAD51 foci that co-localized with γ-H2AX foci (Supplementary Figure S7). In the complemented BRCA2 knockout cell lines, RAD51 foci levels peaked at 8 hours following treatment with IR and were mostly resolved by 48 h. In the absence of BRCA2, cells are substantially compromised in their ability to form nuclear RAD51 foci in response to DNA damage (see Empty Vector panel in Supplementary Figure S8). As expected, complementation with full-length BRCA2 restored RAD51 foci after IR damage (Figure 6C and Supplementary Figures S7 and S8). Cells expressing each of the BRCn-DBD proteins induced RAD51 foci 8 h post-IR although levels did not reach that of full-length BRCA2 (Figure 6C). γ-H2AX foci represent sites of DNA damage and as expected, the percentage of cells and number of γ-H2AX foci per nuclei increased dramatically following IR damage (Figure 6D) (44,45). To infer completion of repair, we tracked the disappearance of γ-H2AX foci. Employing full-length BRCA2 as a benchmark, the γ-H2AX foci peaked at 8 h after IR and returned to baseline by 48 hours (similar to RAD51). Those cells expressing the N-terminus, DBD, or empty vector retained significant γ-H2AX foci at 48 hours after IR indicating incomplete repair (Figure 6D and Supplementary Figures S7 and S8). Similar to our results with RAD51 foci, expression of BRCn-DBD proteins resolved most γ-H2AX foci by 48 h (Figure 6D). In agreement with our biochemical and survival data, BRC5–8-DBD expressing cells trended higher in terms of RAD51 foci compared to other BRCn-DBD proteins indicating a higher capacity to resolve IR damage. In contrast, BRC1-8-DBD cells exhibited residual γ-H2AX foci at 48 h following IR (Figure 6D and Supplementary Figure S8). Interestingly, cells expressing only the N-terminus and DBD fragments restored low levels of RAD51 foci, although based on the persistent levels of γ-H2AX foci and failure to complement damage induced survival, these foci might not represent productive repair events. Enigmatic qualitative characteristics we observed in some BRC5–8-DBD nuclei were very bright, focal RAD51 foci. Additional curious features of BRCn-DBD cells were RAD51 foci visible on a background of diffuse or pan-nuclear staining (Supplementary Figure S8). Strikingly, full-length BRCA2 complemented cells formed discrete nuclear RAD51 foci with very little pan-nuclear staining (Supplementary Figure S8). These results suggest that intact full-length BRCA2 protein can sequester and mobilize the majority of RAD51 in the nucleus to sites of DNA damage.
DISCUSSION
An essential feature of BRCA2 as a mediator of homologous recombination is the ability to deliver RAD51 to a resected DNA DSB and stimulate RAD51 nucleoprotein filament formation. All of the available evidence to date across multiple species and using various fusion strategies indicate that this activity requires the presence of at least one BRC repeat (17,31,32,48,49). Recent studies focused on the individual human BRC repeats provide evidence that BRC1–4 and BRC5–8 can be separated into two modules with disparate functions (29,50). Individually, BRC1, BRC2, BRC3 and BRC4 bind free RAD51, inhibit RAD51-ssDNA dependent ATPase activity, and effectively prevent RAD51 binding to dsDNA (29). These functions were previously described in an isolated BRC4 repeat as well as the full-length BRCA2 protein (3,30). In contrast, BRC5, BRC6, BRC7 and BRC8 bound a negligible amount of free RAD51, failed to inhibit RAD51 ATPase activity or suppress binding to dsDNA, and did not promote DNA strand exchange activity in the presence of excess RAD51. However, BRC5, BRC6, BRC7 and BRC8 augmented RAD51–ssDNA complex formation more efficiently than BRC1, BRC2, BRC3 or BRC4 by EMSA analysis (29). The increased binding affinity of the BRC5–8 repeats to the RAD51–ssDNA filament is reminiscent of studies concerning the carboxy terminus (referred to as the TR2 domain) of BRCA2 (13,16). Taken together, these data imply that both BRC5–8 and TR2 engage RAD51 that is pre-bound to ssDNA and stabilize the nucleoprotein filament.
Our biochemical and cellular analyses imply basal interactions between BRC repeats 1–4 and free RAD51 in the nucleus (Figure 1C and D). The BRC5–8 module appears to play a minimal role in binding this free pool of RAD51. Considering that many of the stimulatory activities of BRCA2 reside in the BRC1–4 module, we were surprised to find that BRC5–8-DBD mediated a higher level of RAD51-dependent DNA strand exchange activity (Figure 3C) and complementation functions (Figure 5C and D) than all other BRCn-DBD proteins tested. In our study, inclusion of the DBD engendered BRCA2 with two important properties: the ability to bind directly to DNA and the NLS signals necessary for BRCA2 nuclear localization. An unanticipated finding was the relative poor activity of the BRC1–8-DBD fusion. Our results demonstrate that reconstitution of BRC repeats 1–8 in conjunction with the DBD, but lacking the N-terminus, cripples stimulation of strand pairing and complementation functions (Figures 3 and 5). While the RAD51 binding functions of BRC1–8-DBD protein remained intact (Figure 1 and Supplementary Figure S1), it was unable to deliver or stabilize RAD51 on ssDNA to stimulate subsequent DNA pairing activity (Figures 3 and 4). The full-length BRCA2 protein outperformed all BRCn-DBD constructs in both biochemical and cellular assays implying that the N-terminus rescues the functional deficits of BRC1–8-DBD. How the N-terminus of BRCA2 regulates repair function remains elusive. Perhaps the N-terminus exerts conformational regulation of the two BRC modules impacting the binding, delivery, or stabilization of RAD51 on DNA. An alternative hypothesis is that interactions between the N-terminal domain of BRCA2 and protein partners such as PALB2 or EMSY facilitate RAD51 deposition at sites of DNA damage (51,52). The BRCn-DBD constructs utilized in this study lack the N-terminal domain of BRCA2, and thus, have presumably lost the ability to interact with PALB2, EMSY, and other proteins that assist BRCA2 in localization and mediator activities at DNA breaks. Future studies employing the full-length BRCA2 protein with selected mutations in one or multiple BRC repeats will illuminate further how these two BRC modules synergize to promote HDR.
As noted above, previous reports using purified fragments of the carboxy terminus of BRCA2 (TR2) demonstrate specific binding to the RAD51 nucleoprotein filament regulated by the phosphorylation status of S3291 (13,16). Utilizing a biotin-DNA pull-down assay to pre-assemble the filament, we found BRC5–8-DBD preferentially bound to RAD51 only when complexed with ssDNA (Figure 4B, lanes 13–16). Fusions containing BRC4 or BRC1–4 were competed away with excess free RAD51 (Figure 4B, lanes 5–12). These results support the conclusion that BRC5–8 provides additional binding affinity to the RAD51 filament compared to DBD alone. The DBD in isolation (including the TR2 region) failed to complement BRCA2 deficient cells consistent with a negligible stimulation of strand pairing activity (Figures 3 and 5). Our results suggest that while both the TR2 region and BRC5–8 bind and stabilize the RAD51 nucleoprotein filament, the complete BRC5–8-DBD module is necessary to regain significant complementation functions. Phosphorylation of S3291, contained within the TR2 domain, prevents binding to the RAD51 filament (13,16). While our biochemical studies do not address the physiological consequences of S3291 phosphorylation, this post-translational modification plays an important role in the cell cycle regulation of homologous recombination (12).
Breast and ovarian tumors in patients with germline or somatic mutations in BRCA2 respond favorably to therapeutic treatment with DNA cross-linking agents including MMC and cisplatin (53,54). Accordingly, response to MMC is a useful hallmark of BRCA2 deficiency as patient-derived BRCA2 mutant tumor cell lines and cells experimentally depleted of BRCA2 by RNA interference are highly sensitive to MMC-induced DNA damage. Utilizing clonogenic survival as our endpoint, we ranked the ability of BRCn-DBD proteins to complement two independent mammalian cell models of BRCA2 deficiency as follows: BRC5–8-DBD > BRC1–4-DBD > BRC4-DBD > BRC1–8-DBD. Not surprisingly, the full-length BRCA2 protein restored complementation functions exceeding all BRCn-DBD proteins (Figure 5). Expression of the N-terminus or DBD alone provided no complementation activity, and in fact, displayed a consistent modest decrease in survival compared to the empty vector control suggesting a dominant negative function. Despite a modest differential in stimulation of RAD51-mediated strand pairing activity (Figure 3C), there was a clear separation in complementation ability when BRC1–8-DBD was compared to DBD alone in the cellular response to MMC-induced DNA damage (Figure 5C and D). These data underscore the importance of correctly interpreting mechanistic biochemical analysis in the context of genetic complementation studies to justify specific conclusions. In this regard, BRC1-8-DBD may provide cellular functions in response to MMC damage, such as protein recruitment, independent of its role in mediating RAD51 loading and/or stabilization. Conversely, expression of the N-terminus or DBD in the absence of other functional domains may interfere with alternative repair pathways that engage DNA damage in the absence of normal BRCA2 function. Amongst the disparate activities of each protein that were assay dependent, we noted an apparent robust ability of the BRC4-DBD protein to repair I-SceI-induced DSBs (Figure 6B). While we do not fully understand the increased activity of this particular fusion protein in this assay, this experimental discrepancy may relate to the specific recognition and enzymatic processing of the DSB created by the I-SceI nuclease. Our results imply that stimulation of RAD51-mediated strand pairing by BRC5–8-DBD is the probable mechanism that underlies the ability of this protein to complement the cellular response to DNA damage. Given the aforementioned caveats, we still cannot rule out other mechanisms for BRC5–8-DBD function including the recruitment and stabilization of RAD51 at stalled replication forks (7,9).
Inside cells, the majority of nuclear RAD51 is likely constrained by multiple binding partners to prevent aggregation or inappropriate engagement of DNA substrates. RAD51 may be held in a non-oligomeric state by the BRC1–4 module or even other HR proteins such as PALB2, RAD51AP1 or the RAD51 paralogs (55–58). However, our pull-down experiments with exogenously expressed fusions and fragments containing BRC repeats 1–4 demonstrate significant binding to endogenous RAD51 suggesting that a free pool of RAD51 proteins exist within cells (Figure 1D and Supplementary Figure S6). The inability of BRC5–8-DBD to interact with endogenous RAD51 further supports the hypothesis that this pool of RAD51 is non-oligomeric. Upon cellular DNA damage leading to a resected DSB, RAD51 is mobilized to RPA-coated ssDNA and only then oligomerizes into the nucleoprotein filament with the aid of BRCA2 and other repair assembly complexes. It remains unclear how BRC5–8-DBD obviates the need for BRC repeats 1–4 to bind and load RAD51 onto resected DNA. Given the intrinsic dynamic nature of RAD51, we speculate that short stretches of nucleation events along ssDNA are bound and stabilized by the BRC5–8-DBD protein. The modest stimulation supplied by the DBD alone in strand pairing reactions may also reflect limited RAD51 filament growth and stabilization. In cellular assays, the availability of accessory repair factors may further assist BRC5–8-DBD in the stabilization and acceleration of filament assembly on ssDNA. As stated above, the functional attributes of BRC5–8-DBD may include the protection of stalled replication forks as proposed by Schlacher et al. (7).
In the absence of BRCA2 protein (see Empty Vector in Supplementary Figure S8), IR-induced RAD51 foci are not detectable under our conditions. All of the BRCn-DBD proteins in our study were capable of restoring RAD51 foci after IR damage to some extent (Figure 6C). In concordance with re-engagement of HDR repair, residual DNA damage measured by nuclear γ-H2AX foci was significantly reduced in BRCn-DBD and full-length BRCA2 complemented cells (see 48 h timepoints in Figure 6D). Complementation with full-length BRCA2 resulted in the majority of nuclei examined to exhibit distinct RAD51 foci with minimal diffuse (pan-nuclear) RAD51 staining (see bottom panel in Supplementary Figure S8). The majority of nuclei from BRCn-DBD expressing cells exhibited distinct foci upon a background of diffuse RAD51 signal (Supplementary Figure S8). This diffuse RAD51 staining pattern could represent a free pool of RAD51 that has not been mobilized to sites of DNA DSBs within the nucleus. Recent studies in live cells using fluorescence correlation spectroscopy (FCS) and single particle tracking (SPT) suggest full-length BRCA2 protein engages the majority of RAD51 protein within the nucleus (59). The residual diffuse RAD51 staining we observe in BRCn-DBD expressing cells may indicate disruption of this functional association inherent only to the full-length BRCA2 protein.
The repair kinetics of IR-induced DNA damage foci, complementation in response to MMC damage, and increased HDR of a nuclease-induced DSB collectively demonstrate the important cellular role the BRC5–8 module plays in responding appropriately to DNA damage. Our data utilizing purified proteins in biochemical assays provide mechanistic evidence that RAD51 filament stabilization and stimulation of RAD51-mediated DNA strand pairing are the basis for the ability of BRC5–8-DBD to engage and repair DNA DSBs. In addition to BRC repeats 1–4 and the DBD influencing RAD51 behavior, we have found that BRC5–8 supplies an additional level of stability and maintenance over the RAD51 filament. By placing several layers of regulatory control over RAD51, we envision that BRCA2 prevents RAD51 from engaging in non-productive DNA interactions, stabilizes the pre-synaptic filament, and promotes strand invasion and the subsequent search for homologous sequences. These concerted actions prevent inappropriate recombination events leading to chromosomal rearrangements, loss-of-heterozygosity, or other deleterious genomic structural variations. The mobilization and loading of RAD51 onto resected DNA DSBs is carefully orchestrated by BRCA2 to ensure faithful HDR and to protect the genome against the harbingers of neoplastic transformation.
Supplementary Material
Acknowledgments
We thank Sharon Cantor, Joann Sweasy, Carolyn Marsden, Mark Baker, Faye Rogers and all members of the Jensen lab for valuable comments and insight on the manuscript.
FUNDING
American Cancer Society (#IRG 58-012-55 to R.B.J.); BreastCancer Alliance (to R.B.J.); Pilot Project Program grant from Women's HealthResearch at Yale and the Yale Comprehensive Cancer Center (to R.B.J.). Fundingfor open access charge: institutional funds.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yu D.S., Sonoda E., Takeda S., Huang C.L., Pellegrini L., Blundell T.L., Venkitaraman A.R. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol. Cell. 2003;12:1029–1041. doi: 10.1016/s1097-2765(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 2.Davies A.A., Masson J.Y., McIlwraith M.J., Stasiak A.Z., Stasiak A., Venkitaraman A.R., West S.C. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 3.Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J., Doty T., Gibson B., Heyer W.D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang H., Li Q., Fan J., Holloman W.K., Pavletich N.P. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 7.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto Y., Ray Chaudhuri A., Lopes M., Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomonosov M., Anand S., Sangrithi M., Davies R., Venkitaraman A.R. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esashi F., Christ N., Gannon J., Liu Y., Hunt T., Jasin M., West S.C. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 13.Esashi F., Galkin V.E., Yu X., Egelman E.H., West S.C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 14.Richardson C., Stark J.M., Ommundsen M., Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- 15.Klein H.L. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair. 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies O.R., Pellegrini L. Interaction with the BRCA2 C terminus protects RAD51-DNA filaments from disassembly by BRC repeats. Nat. Struct. Mol. Biol. 2007;14:475–483. doi: 10.1038/nsmb1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bignell G., Micklem G., Stratton M.R., Ashworth A., Wooster R. The BRC repeats are conserved in mammalian BRCA2 proteins. Hum. Mol. Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- 18.Bork P., Blomberg N., Nilges M. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 19.Chen P.L., Chen C.F., Chen Y., Xiao J., Sharp Z.D., Lee W.H. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5287–5292. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong A.K.C., Pero R., Ormonde P.A., Tavtigian S.V., Bartel P.L. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 21.Sharan S.K., Morimatsu M., Albrecht U., Lim D.S., Regel E., Dinh C., Sands A., Eichele G., Hasty P., Bradley A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 22.Mizuta R., LaSalle J.M., Cheng H.L., Shinohara A., Ogawa H., Copeland N., Jenkins N.A., Lalande M., Alt F.W. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 24.Ochiai K., Yoshikawa Y., Yoshimatsu K., Oonuma T., Tomioka Y., Takeda E., Arikawa J., Mominoki K., Omi T., Hashizume K., et al. Valine 1532 of human BRC repeat 4 plays an important role in the interaction between BRCA2 and RAD51. FEBS Lett. 2011;585:1771–1777. doi: 10.1016/j.febslet.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 25.Lo T., Pellegrini L., Venkitaraman A.R., Blundell T.L. Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair. 2003;2:1015–1028. doi: 10.1016/s1568-7864(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 26.Barber L.J., Rosa Rosa J.M., Kozarewa I., Fenwick K., Assiotis I., Mitsopoulos C., Sims D., Hakas J., Zvelebil M., Lord C.J., et al. Comprehensive genomic analysis of a BRCA2 deficient human pancreatic cancer. PLoS One. 2011;6:e21639. doi: 10.1371/journal.pone.0021639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oddoux C., Struewing J.P., Clayton C.M., Neuhausen S., Brody L.C., Kaback M., Haas B., Norton L., Borgen P., Jhanwar S., et al. The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1% Nat. Genet. 1996;14:188–190. doi: 10.1038/ng1096-188. [DOI] [PubMed] [Google Scholar]
- 28.Szabo C., Masiello A., Ryan J.F., Brody L.C. The breast cancer information core: database design, structure, and scope. Hum. Mutat. 2000;16:123–131. doi: 10.1002/1098-1004(200008)16:2<123::AID-HUMU4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 29.Carreira A., Kowalczykowski S.C. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carreira A., Hilario J., Amitani I., Baskin R.J., Shivji M.K., Venkitaraman A.R., Kowalczykowski S.C. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siaud N., Barbera M.A., Egashira A., Lam I., Christ N., Schlacher K., Xia B., Jasin M. Plasticity of BRCA2 function in homologous recombination: genetic interactions of the PALB2 and DNA binding domains. PLoS Genet. 2011;7:e1002409. doi: 10.1371/journal.pgen.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San Filippo J., Chi P., Sehorn M.G., Etchin J., Krejci L., Sung P. Recombination mediator and Rad51 targeting activities of a human BRCA2 polypeptide. J. Biol. Chem. 2006;281:11649–11657. doi: 10.1074/jbc.M601249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao W., Vaithiyalingam S., San Filippo J., Maranon D.G., Jimenez-Sainz J., Fontenay G.V., Kwon Y., Leung S.G., Lu L., Jensen R.B., et al. Promotion of BRCA2-dependent homologous recombination by DSS1 via RPA targeting and DNA mimicry. Mol. Cell. 2015;59:176–187. doi: 10.1016/j.molcel.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jensen R. Purification of recombinant 2XMBP tagged human proteins from human cells. Methods Mol. Biol. 2014;1176:209–217. doi: 10.1007/978-1-4939-0992-6_17. [DOI] [PubMed] [Google Scholar]
- 35.Hucl T., Rago C., Gallmeier E., Brody J.R., Gorospe M., Kern S.E. A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res. 2008;68:5023–5030. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H., Jeffrey P.D., Miller J., Kinnucan E., Sun Y., Thoma N.H., Zheng N., Chen P.L., Lee W.H., Pavletich N.P. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Q., Kojic M., Holloman W.K. DNA-binding domain within the Brh2 N terminus is the primary interaction site for association with DNA. J. Biol. Chem. 2009;284:8265–8273. doi: 10.1074/jbc.M809226200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kraakman-van der Zwet M., Overkamp W.J., van Lange Essers R.E. J., van Duijn-Goedhart A., Wiggers I., Swaminathan S., van Buul P.P., Errami A., Tan R.T., et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Mol. Cell. Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spain B.H., Larson C.J., Shihabuddin L.S., Gage F.H., Verma I.M. Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13920–13925. doi: 10.1073/pnas.96.24.13920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouet P., Smih F., Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., Munro M., Pinder J., Salsman J., Dellaire G., et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature. 2015;528:422–426. doi: 10.1038/nature16142. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Chun J., Buechelmaier E.S., Powell S.N. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 2013;33:387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol: CB. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 44.Rogakou E.P., Boon C., Redon C., Bonner W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 46.Haaf T., Golub E.I., Reddy G., Radding C.M., Ward D.C. Nuclear foci of mammalian Rad51 recombination protein in somatic cells after DNA damage and its localization in synaptonemal complexes. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rothkamm K., Barnard S., Moquet J., Ellender M., Rana Z., Burdak-Rothkamm S. DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 2015;56:491–504. doi: 10.1002/em.21944. [DOI] [PubMed] [Google Scholar]
- 48.Saeki H., Siaud N., Christ N., Wiegant W.W., van Buul P.P., Han M., Zdzienicka M.Z., Stark J.M., Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. U.S.A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkitaraman A.R. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu. Rev. Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 50.Rajendra E., Venkitaraman A.R. Two modules in the BRC repeats of BRCA2 mediate structural and functional interactions with the RAD51 recombinase. Nucleic Acids Res. 2010;38:82–96. doi: 10.1093/nar/gkp873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 52.Hughes-Davies L., Huntsman D., Ruas M., Fuks F., Bye J., Chin S.F., Milner J., Brown L.A., Hsu F., Gilks B., et al. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–535. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- 53.Sakai W., Swisher E.M., Karlan B.Y., Agarwal M.K., Higgins J., Friedman C., Villegas E., Jacquemont C., Farrugia D.J., Couch F.J., et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Andrea A.D., Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 55.Dray E., Etchin J., Wiese C., Saro D., Williams G.J., Hammel M., Yu X., Galkin V.E., Liu D., Tsai M.S., et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buisson R., Dion-Cote A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., Masson J.Y. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiese C., Collins D.W., Albala J.S., Thompson L.H., Kronenberg A., Schild D. Interactions involving the Rad51 paralogs Rad51C and XRCC3 in human cells. Nucleic Acids Res. 2002;30:1001–1008. doi: 10.1093/nar/30.4.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiese C., Dray E., Groesser T., San Filippo J., Shi I., Collins D.W., Tsai M.S., Williams G.J., Rydberg B., Sung P., et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell. 2007;28:482–490. doi: 10.1016/j.molcel.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reuter M., Zelensky A., Smal I., Meijering E., van Cappellen W.A., de Gruiter H.M., van Belle G.J., van Royen M.E., Houtsmuller A.B., Essers J., et al. BRCA2 diffuses as oligomeric clusters with RAD51 and changes mobility after DNA damage in live cells. J. Cell Biol. 2015;208:857. doi: 10.1083/jcb.20140501402182015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.