Abstract
The TWIST1 bHLH transcription factor controls embryonic development and cancer processes. Although molecular and genetic analyses have provided a wealth of data on the role of bHLH transcription factors, very little is known on the molecular mechanisms underlying their binding affinity to the E-box sequence of the promoter. Here, we used an in silico model of the TWIST1/E12 (TE) heterocomplex and performed molecular dynamics (MD) simulations of its binding to specific (TE-box) and modified E-box sequences. We focused on (i) active E-box and inactive E-box sequences, on (ii) modified active E-box sequences, as well as on (iii) two box sequences with modified adjacent bases the AT- and TA-boxes. Our in silico models were supported by functional in vitro binding assays. This exploration highlighted the predominant role of protein side-chain residues, close to the heart of the complex, at anchoring the dimer to DNA sequences, and unveiled a shift towards adjacent ((-1) and (-1*)) bases and conserved bases of modified E-box sequences. In conclusion, our study provides proof of the predictive value of these MD simulations, which may contribute to the characterization of specific inhibitors by docking approaches, and their use in pharmacological therapies by blocking the tumoral TWIST1/E12 function in cancers.
INTRODUCTION
The epithelial-mesenchymal transition (EMT) is a highly conserved cellular process that allows the transient conversion of epithelial cells into mesenchymal cells and promotes cell migration during embryonic processes. The TWIST1 protein has been associated with this cellular mechanism and was originally described in the developmental process, as an initiator of the formation of the mesoderm. Indeed, this protein is involved in several biological activities via its different functional domains, including the TWIST-box domain (1), which is highly homologous between different eukaryotic species (2), and the basic Helix Loop Helix (bHLH) domains, which enable its dimerization with other bHLH partners (3,4). The TWIST-box domain binds with the runt domain of the RUNX2 protein, and inhibits its transactivation function (2). In parallel, TWIST1 is able to directly repress the expression of Runx2, thus preventing RUNX2 from inducing premature osteoblast differentiation (5,6).
However, the aberrant activation of the EMT in adult cancer cells contributes to the invasion of carcinoma cells and metastasis (7,8), and concomitantly, TWIST1 has also been implicated in tumorigenesis (8,9). In this context, the molecular events triggered by TWIST1 in cancer cells, may be induced through both the TWIST-box domain and the bHLH domain. For example, the TWIST-box domain mediates the formation of a protein complex comprised of TWIST1 and the nuclear factor-kappaB (NF-κB) subunit RELA (p65/NF-κB3), which activates the transcription of the IL8 gene (10). The TWIST-box domain also interacts with p53, which antagonizes p53-dependent activity, especially its pro-apoptotic and transcription functions (11).
Clinically, while the re-expression of TWIST1 in tumors is frequently associated with a poor prognosis in humans, on the one hand (12–14), its repression on the other hand decreases the growth of xenografts (15). Additionally, a decrease in the TWIST1 protein, both in a transgenic mutant Kras lung cancer mouse model and in human lung tumors, restores a senescence program inducing the loss of a neoplastic phenotype (16). Similarly, such a phenomenon is observed using a 8-bromo-5-hydroxy-7-methoxychrysin treatment (15). Finally, the decrease in the level of expression of TWIST1 by small interfering RNA (siRNA) was shown to potentiate the effect of arsenic trioxide chemotherapy (17).
In the present work, we focused on the bHLH domain of a TWIST1 complex, and on its function in the binding of targeted gene promoters. It is well known that TWIST1 is a transcription factor belonging to the bHLH superfamily, which includes TWIST1, TWIST2, E12, E47, HAND1 and HAND2 (3,4). All of them are highly conserved in organisms from yeasts to humans and are composed of two characteristic domains: a HLH domain and a basic domain (4,18). The dimerization carried out via the HLH domains is a prerequisite for the interaction with E-box sequences occurring via basic domains and key residues (3) (this model is depicted in Figure 2C to facilitate its understanding). According to the formed homo- or heterodimer, their transcriptional functions differ. For instance the TWIST1 functions have been largely explored using tethered dimers in several eukaryotic models (19–21). A previous study focusing on the characterization of different prometastatic forms of TWIST1 demonstrated the predominant role of the TWIST1/E2A (E12 or E47, two alternative splicing products of the TCF3 gene) heterodimer in prostate cancer (22). Furthermore, the phosphorylation of TWIST1 at the level of the threonine T121 and the serine S123 residues may enhance the metastatic potential of the TWIST1/E12 (TE) complex (22).
Figure 2.
Description of several canonical recognition cores of E-box sequences (CANNTG) and basic domains of the TE complex. (A-D) The position of important TWIST1/E12 amino acid residues and E-box bases were visualized with the VMD 1.9.1 software. Different views are shown for a better understanding and interpretation of the findings. (A-C) 3D representation of the conserved TWIST1 (T1; grey ribbon)/E12 (green ribbon) complex bound to DNA (light white ribbon) along with the TE-box sequence (CATCTG). The residues are numbered in each strand of the 5′-CATCTG-3′ E-box core. The complementary bases of the opposite strand are highlighted with a star. (A) Representation of the TWIST1 half-site (5′-CAT-3′ highlighted in orange) and the E12 half-site (5′-CAG-3′ highlighted in magenta). (B) (i) Representation of the variable bases and consensus bases highlighted in pink and green, respectively. (ii) Representation of the flanking bases ((≥-2) or (≥-2*)) and proximal flanking bases ((-1) or (-1*)) of E-box shown in yellow and in cyan, respectively. (C) 3D in silico visualization and ClustalW alignment of the basic domains of the TE complex. The underlined residues are able to establish H-bond interactions with DNA and/or other TE residues. The 3D visualization shows the basic TWIST1 and E12 domains in solid grey and green, respectively. The CPK representation highlights the TE residues able to establish H-bonds with DNA. (D) In silico model of ‘specific’ or ‘non-specific’ H-bonds established between TWIST1 and the DNA sequence. (i) The H-bonds are considered as ‘non-specific’ when the lateral chain residues interact with atoms on the phosphate group. In contrast, (ii) the H-bonds are considered as ‘specific’, when the lateral chain residues interact with atoms on the purine or pyrimidine nucleobases. The composition of the atoms varies according to the nucleobase and the number of rings.
A comprehensive classification of these transcription factors has been established according to their tissue distribution, dimerization capabilities (homodimer or heterodimer), and binding specificities to various E-box sequences (23). Since bHLH proteins can form both homo- and heterodimers, they can be further classified into two groups based on their binding preferences with numerous canonical core recognition E-box sequences (CANNTG), namely CACCTG or CAGCTG (group A) and CACGTG or CATGTG (group B) (23). When a bHLH dimer binds to an E-box sequence, the first monomer binds to the forward half-site, while the second monomer binds to the reverse complementary half-site (24). Each monomer, thereby, contributes to making its binding to a particular E-box sequence highly specific, presumably by preferring particular half-sites. For the TE complex, the half-site preferences are the CAT sequence for TWIST1, and the CAT, CAC and CAG sequences for E12 (24). These differences in the composition of the E-box sequences largely modulate the in vivo function of cellular transactivation by closely related transcription factors (24–26) (for brief review see Figure 1A). Recently, Chang demonstrated that a secondary organization might greatly modulate the transactivation function of the bHLH domain (27). The TE complex can dimerize to form a tetramer able to recognize specific double E-box motifs and, in doing so, is able to align two TE complexes on the consecutive DNA grooves, providing a highly stable, bound tetramer (27). Nevertheless, little is known about the molecular characterization of the canonical core recognition of E-box sequences (CANNTG) by the TE complex (23).
Figure 1.
Description of various E-box sequences and the in vitro experiments. (A) Presentation of the different E-box sequences selected in the present study, with the localization of base substitutions and the sequences of the two half-sites. Top, representation of the E-box sequence, with the adjacent bases ((-1) and (-1*)) in blue, conserved bases in green the E-box and the variable bases in pink. The origin of these boxes along with the previous in vitro biochemical findings that prompted the choice of these specific E-box sequences are also reported in the table. The E-box sequences are highlighted in blue in the table. (B) Schematic representation of the different protein constructs used in our study. (i) The cDNA of the tethered TE complex is composed of 896 amino acids and its size is ∼99 kDa. It contains a FLAG-tag at the N-terminal position, TWIST1 cDNA, an amino acid linker and E12 cDNA. (ii) The ∼49 kDa cDNA of the tethered TT complex is composed of 443 amino acids, and includes a FLAG-tag at the N-terminal position, TWIST1 cDNA, an amino acid linker and TWIST1 cDNA. (iii) The cDNA of the E12 protein includes a MYC-tag at the N-terminal position, and the final ∼73 kDa construct is composed of 666 amino acids. (C) Schematic diagram briefly summarizing the Streptavidin/Biotin assay using the TWIST1 antibody. Briefly, in step 1, cells were transfected with a plasmid to produce large amounts of the TWIST1/E12 complex. In step 2, total proteins were extracted. In step 3, proteins were incubated with E-box probes. In step 4, TWIST1/E12 complexes bound to E-box probes were isolated by immunoprecipitation in a Streptavidin/Biotin assay using biotin-labelled E-box probes. In step 5, the protein complexes bound to E-box sequences were visualized by Western blot analysis and the TWIST1/E12 signal was detected using the TWIST1 antibody. A detailed description of these steps is reported in the Materials and Methods section.
The use of molecular dynamics (MD) simulations previously enabled us to study the in silico impact of TWIST1 alterations (insertion or single base mutations) on the binding of the TE complex to DNA (28,29). This approach provided a structural explanation for the loss of function associated with haplo-insufficiency of TWIST1 observed in patients suffering from Saethre-Chotzen syndrome (SCS) (30–34). In the present study, the molecular mechanisms underlying the recognition of the E-box by the TE heterodimer and their stabilization on DNA were investigated using the previously published dimer, initially generated by homology with the murine NEUROD1/E47 crystal structure (28,35). We generated various in silico models of the TE complex bound to putative functional E-box sequences of targeted gene promoters in order to verify the specificity of the binding domains during MD simulations. Consequently we assumed that a positive outcome of our approach, combining in silico models with in vitro assays, may result in the dynamic 3D visualization of a novel predictive model for the identification of specific and functional E-box sequences by the TE complex. This could be used in the future for developing and testing pharmacological TWIST1/E2A blocking molecules by in silico docking prior to in vitro or in vivo studies. Such therapeutic strategies may prove to be invaluable for restoring the pro-senescence and apoptosis responses (9,36–39). Furthermore, identification of a specific interaction between the TWIST1/E2A complex and a distinct E-box sequence, could lead to the determination of a putative pro-oncogenic signature.
MATERIALS AND METHODS
In silico study: generation of putative functional models of the TE complex binding to E-box sequences
Selection of X-ray structure and in silico model building
The conserved TE dimer model used throughout this study was generated previously (28). Briefly, sequences of human TWIST1 (Q15672), E12 (P15923-1), NEUROD1 (Q13562) and E47 (P15923-2) were downloaded from the UniProtKB/Swiss-Prot website. Primary sequences, restricted to the bHLH domains, were aligned using the ClustalW 2.0 software (40). In their bHLH domains, human E12 and TWIST1 share 85% and 48% homology with the murine E47 and NEUROD1 sequences, respectively. The X-ray structures of the mouse NEUROD1/E47 complex (PDB code 2ql2) were downloaded from the RCSB protein data bank (35). Two X-ray structures of the mouse NEUROD1/E47 were reported, namely the 2ql2A/2ql2B and the 2ql2C/2ql2D (35), which diverge slightly (root-mean-square deviation evaluated at 0.618 Å) and contain a well-known E-box sequence named the TE-box (5′-TAGGCCATCTGGTCCT-3′). Taking into account the degree of homology between residues in the sequences, in silico models of the conserved TE complex (TE-model) were generated by homology to the 2ql2A/2ql2B structure (28).
The molecular interactions of this TE dimer with several different E-box sequences were then assessed. E-box sequence modifications (or flanking base substitutions) were based on studies evaluating the specificity of the interactions between the canonical 5′-CANNTG-3′ E-box sequences and bHLH transcription factors (20,24,41,42) (Figure 1A). All the DNA base substitutions were carried out in the initial TE-box using the mutation function of the Sybyl-X 1.1 software package. The resulting in silico models studied herein were, therefore, the TE complex bound either to an inactive BMID-box (degenerate sequence with low affinity for TWIST1 complexes), or to various active sequences, namely the initial TE-box, a BMI-box, an E12-box, a TWIST-box and a D-box, or to sequences displaying modifications in their flanking bases, such as the TA-box and the AT-box (Figure 1A).
Molecular dynamics simulations
Prior to conducting molecular dynamics (MD) simulations, which have been described previously (28), minimization of all of the in silico models was carried out using the Sybyl-X 1.1 software package, elaborated by the Tripos Company. The software parameters included a force field using Gasteiger–Marsilli partial charges with a cut off set at 12 Å, and a dielectric constant of 80 to simulate an implicit water phase (the dielectric constant of water being 20.10 at 20°C). No restraints were applied to the in silico models. The established homology model (including DNA sequences) was visualized via the VMD1.9.1 software (43). The resulting model was inserted into a parapipedic TIP3P solvent box using the ‘add solvation box’ module of the VMD 1.9.1 software. A distance of 15 Å was set between the surface of the proteins and the limit of the solvent box (44). For each structure a 10 ns MD simulation was computed using the NaMD software and CHARMM 27 as a force field. The state of equilibrium was reached after approximately 30 ps for all of the models studied.
Analysis of the stability of interactions during MD simulations
Among the interactions occurring between residue-residue and residue-base, we focused our investigation on hydrophobic and hydrogen bond (H-bond) interactions established between donor and acceptor groups. In the first place, hydrophobic interactions are mainly due to the tendency of nonpolar residues to aggregate in aqueous solutions and repulse water molecules. These interactions contribute to the folding of the protein and to the stabilization of the macromolecule. Residues such as alanine, valine, leucine, isoleucine, phenylalanine, tryptophan and methionine can cluster and form hydrophobic interactions. Here, we observed the presence of numerous leucine and phenylalanine residues in the vicinity of the heart of the TE complex. During the MD simulations, the distances in angstrom (Å) between pairs of residues were measured and plotted as a function of time (represented by a frame number: 1 frame per 10 ps). The distances between hydrophobic interactions were represented via GraphPad Prism 5: ‘smooth, differentiate or integrate curve, with eight neighbours’. In the second place, the H-bond interaction is an intermolecular force involving a hydrogen atom and an electronegative atoms such as oxygen or nitrogen. H-bonds can be established between donor atoms (arginine, lysine and tryptophan) and acceptor atoms (aspartic acid, glutamic acid) of side chain residues. We studied the residue-residue or residue-base interactions, which are established between atoms (O−H…:N; O−H…:O; N−H…:N; N−H…:O). Again, the lengths of H-bond interactions were represented via GraphPad Prism 5: ‘smooth, differentiate or integrate curve, with eight neighbours’. These lengths have previously been estimated around 2-2.8 Å depending on the nature of the atoms (45). Using a computer software, we set an arbitrary threshold at 2.1 Å and made the following hypothesis: any interaction occurring under this threshold was deemed ‘true’ and assigned a score of 1, whereas any interaction occurring above this threshold was deemed ‘false’ and given a score of 0 (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 attributed if true and 0 if false; NB.SI function: NB.SI(range;criterion)). The scores were reckoned and the value obtained was designated as the rate of occupancy of H-bond interactions. Thus, for a given H-bond, the higher the rate of occupancy, the greater the number of interactions occurring under 2.10 Å, the higher the level of stability of that H-bond during the MD simulation. All calculations were carried using Excel.
In vitro study: binding of the TE complex to the E-box sequences
Vector constructs
The pBABE-neo expression vector was donated by H. Land & J. Morgenstern & B. Weinberg (Addgene plasmid # 1767). The cDNA of the tethered TE complex composed of 896 amino acids, including a FLAG-tag at the N-terminal position, TWIST1 cDNA, an amino acid linker and E12 cDNA, was cloned into the pBABE-neo expression vector (21). This construct of the ectopic TE protein is approximately 99 kDa in size. The final construct was named the FLAG-TWIST1/E12 pBABE-neo expression vector (TE pBABE; Figure 1Bi).
The cDNA of the tethered TT complex consists of 445 amino acids, including a FLAG-tag at the N-terminal position, TWIST1 cDNA, an amino acid linker and TWIST1 cDNA, was cloned into the pBABE-neo expression vector (21). The final construct (∼49 kDa in size) was named the FLAG-TWIST1/TWIST1 pBABE-neo expression vector (TT pBABE; Figure 1Bii). Finally, the cDNA of the E12 protein (18) was cloned into the pBABE-neo expression vector. The ectopic E12 protein includes a MYC-tag at the N-terminal position, and the final construct is composed of 666 amino acids with a size of 73 kDa. This construct was named the MYC-E12 pBABE-neo expression vector (E12 pBABE; Figure 1Biii). The ectopic E12 homodimer has a size of 147 kDa (E12 pBABE).
Cell culture, transfection and protein extraction
Hela cells were transfected in 10 cm in diameter culture dishes at a density of 80-90% using the FuGENE HD Transfection Reagent for 48 h (Promega #E2311). The transfection reagent contained 19 μg of the empty pBABE-neo expression vector or of the pBABE-neo expression vector (for the FLAG-TWIST1/E12 pBABE vector), as well as 56 μl of FuGENE. Following their transfection, cells were washed with cold PBS (2.7 mM KCl, 1.47 mM KH2PO4, 0.14 M NaCl, 3.4 mM Na2HPO4) and lysed in EBC solution (20 mM Tris pH 8.0, 385 mM NaCl, 2 mM EDTA, 3.5% NP40, PIC 1×, PMSF 1×). After incubation on ice for 20 min, lysates were collected by scrubbing the plates, sonicated (6 cycles of 10 s of ultrasound sonication with a 20 s gap between each cycle) and centrifuged for 20 min at 15 000 rpm at 4°C. The supernatant was collected and the total protein content was quantified using the Bio-Rad Protein Assay (#500-0001).
Streptavidin/Biotin binding assays
The Streptavidin/Biotin binding assay was conducted to isolate proteins (the TE complex) bound to specific DNA sequences (E-box sequences). To do so, total cellular proteins (extracted from cells transfected with the TE pBABE construct or with the empty pBABE control) were incubated with biotin-coupled DNA sequences, and the DNA bound TE complex was then purified by immunoprecipitation (IP) via the recognition of biotin by Streptavidin beads. Hence, this approach decreased the risk of isolating non-specifically bound proteins. Briefly, both strands of DNA probes corresponding to various E-box sequences were labelled in their 3′ extremity with biotin. The probes used were: the inactive BMID-box (5′-CGTAGGCTGGTGTGTCCTCG-3′), the active TE-box (5′-CGTAGGCCATCTGGTCCTCG-3′), the BMI-box (5′-CGTAGGCCAAGTGGTCCTCG-3′), the E12-box (5′-CGTAGGCCACCTGGTCCTCG-3′), the TWIST-box (5′-CGTAGGCCATATGGTCCTCG-3′), the D-box (5′-CGTAGGCCGTCTGGTCCTCG-3′), the TA-box (5′-CGTAGGTCATCTGATCCTCG-3′) and the AT-box (5′-CGTAGGACATCTGTTCCTCG-3′). For probe hybridization, an initial denaturing step at 95°C for 5 min was followed by a hybridization step for 10 min at the specific hybridizing temperature of each probe. Probe hybridizations were then confirmed on 3% agarose gels.
The binding between the DNA hybridized probes (3 μg) and the TE complex (80 μg) was carried out in binding buffer (100 mM KCl, 2% glycerol, 20 mM Hepes, 1 mM dithiothreitol and 0.1 mg/ml BSA) for 1 h at 30°C. Biotinylated DNA probes bound to the TE complex were purified using DynaBeads® M-280 Streptavidin (Invitrogen #11205D). Coupled Streptavidin DynaBeads® bound to the DNA/TE complex were run on a 12% SDS-polyacrylamide gel using a migration buffer (Tris glycine- SDS 1X) at 80 V for 10 min in the stacking gel, followed by the migration phase at 100 V for 120 min. The liquid transfer onto a PVDF membrane was conducted using a transfer buffer (Tris glycine 1× and 20% ethanol) at 200 mA for 60 min. The membrane was then saturated with 5% Tris-buffered saline (TBS) containing 0.05% Tween and 0.5% milk for 45 min, and incubated with the mouse monoclonal TWIST1 antibody (abcam #ab50887) as a primary antibody, and a polyclonal rabbit anti-mouse immunoglobulin conjugated with horseradish peroxidase as the secondary antibody (Dako #P0260). Figures 11B and 11C, the membrane was incubated with the rabbit polyclonal antibody E2A (V-18) as a primary antibody (Santacruz #sc-349), and a polyclonal goat anti-rabbit immunoglobulin conjugated with horseradish peroxidase as the secondary antibody (Dako #P0448). The detection was performed by Western blot analysis using the Luminol Reagent (Santa Cruz #sc-2048). The major steps of this in vitro transfection and Streptavidin/Biotin procedure are summarized in a schematic diagram (Figure 1C).
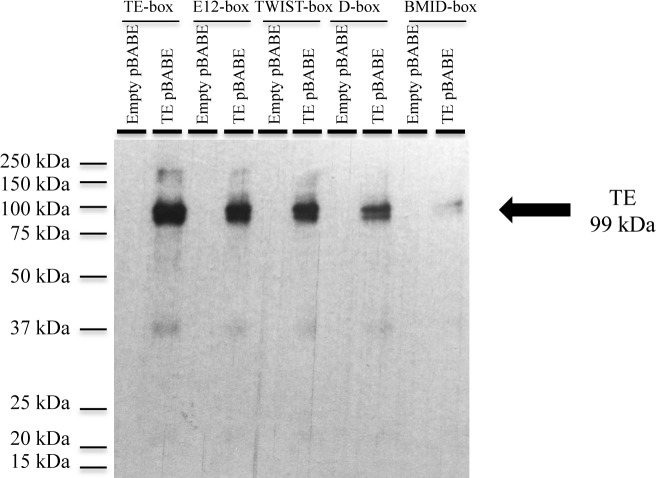
Figure 11.
Streptavidin/Biotin assay of the tethered TWIST1/TWIST1 dimer or of the E12 protein. (A) Streptavidin/Biotin assay using the TWIST1 antibody. Western blot showing the TT and E12 complexes bound to various E-boxes, namely the TE-box, the E12-box, the TWIST-box, the D-box and the BMID-box. The proteins were extracted from cells transfected either with the TT pBABE vector or E12 pBABE vector. The ectopic TT protein is visualized around 49 kDa. The E12 protein was not detected using this antibody. A detailed description of the assay is reported in the Materials and Methods section. (B) Streptavidin/Biotin assay using the E12 antibody. Western blot showing the TT and E12 complexes bound to various E-boxes, namely the TE-box, the E12-box, the TWIST-box, the D-box and the BMID-box. The TT and E12/E12 dimers, as well as the E12 proteins were extracted from cells transfected either with the TT pBABE vector or E12 pBABE vector. The ectopic E12 protein is visualized around 73 kDa and the E12-E12 dimer appears at ∼147 kDa. For the ectopic E12 monomer, we can observe a double band (E12 corresponds to the highest one). The TT dimer was not detected using this antibody. A detailed description of the assay is reported in the Materials and Methods section. (C) Streptavidin/Biotin assay using the E12 antibody. Western blot showing the TE complex bound to various E-box sequences, namely the TE-box, the E12-box, the TWIST-box, the D-box and the BMID-box. The TE dimer was extracted from cells transfected with the TE pBABE vector. The empty pBABE vector served as an assay control. The ectopic TE protein is visualized around 99 kDa as a double-band (TE correspond to the highest one), while a degraded peptide is observed at ∼37 kDa. A detailed description of the assay is reported in the Materials and Methods section.
RESULTS
To characterize the molecular mechanisms underlying the binding and recognition of the E-box sequences (Figure 1A) by the previously generated TWIST1/E12 (TE) complex (28), we submitted all in silico models to 10 ns molecular dynamics (MD) simulations. The resulting binding configuration clearly showed the localization of the TE-box sequence (5′-CATCTG-3′) in the DNA groove, between the basic domains of the TWIST1 and E12 proteins (Figure 2A and 2C). The positions of the consensus and variable bases of the E-box in the DNA groove were also represented (Figure 2Bi). The proximal flanking bases (called adjacent bases) of the E-box are represented in yellow and blue (Figure 2Bii). The basic domains of TWIST1 and E12 proteins are represented in solid grey and green, respectively. To explore the interactions between the TE-complex and the various E-box sequences, we differentiated: (i) the ‘non-specific’ H-bonds (Figure 2Di), established between oxygen elements on the phosphate groups of the DNA bases and an atom of the side chain of the TWIST1 or E12 residues, from (ii) the ‘specific’ H-bonds established between atoms on the purine and pyrimidine nucleobases of the DNA and an atom of the side chain of TWIST1 or E12 residues (Figure 2Dii).
Comparison of the TE complex bound to both active (TE-box, BMI-box) and inactive (BMID-box) E-box sequences
To determine whether our in silico approach was able to predict modifications in the binding of the TE complex to DNA, we studied the binding of the dimer to two active E-box sequences, namely the TE- and BMI-boxes, and to the inactive BMID-box. Our investigations focused on two major interactions occurring in the heart of the TE complex, which may alter its structure, namely the hydrophobic interactions involved in the dimerization of the protein via HLH domains, and the H-bonds established between critical residues. Several leucine (L) and phenylalanine (F) residues close to the heart of the dimer were of particular interest (Figure 3A and 3B). We studied whether they were involved in dimer cohesion by hydrophobic interactions contributing to the dimerization function. In the first instance, the distances between hydrophobic residues localized at the interface of the TE complex (L124-L592, L149-L592, L131-L592, L124-L589, L149-L572, F128-F569) were determined (Figure 3Ci-Cvi). During the MD simulations, L124-L592 and L149-L592 distances were around 2.5 Å, while L131-L592, L149-L572 distances were under 2.3 Å. This strongly indicated that these hydrophobic interactions strongly structured the TE dimer (Figure 3Ci-Civ) irrespective of the E-box sequence included in the model. During the MD simulations the residues (L124-L589) moved closer together in the active BMI and TE models compared to the inactive BMID model, with distances between residues (for example the L124 (T1) and L589 (E12) residues) often under 2.2 Å compared to over 2.6 Å, respectively (Figure 3Cv). The proximity of these residues may largely contribute to the dimerization and to the stability of the TE dimer bound to these active E-boxes. In the second instance, two phenylalanine residues, namely F128 (T1) and F589 (E12) moved closer together during the BMID MD simulation compared to the TE and BMI MD simulations (Figure 3Cvi), inducing a decrease in the level of contact with the E-box bases localized at the heart of the TE complex. The F128 (T1) and F589 (E12) also moved towards adjacent bases of E-box sequences. Among the bHLH proteins, pairs of highly conserved asparagine (N) and lysine (K), namely N125-K145 (T1) and N566-K588 (E12) residues were suspected to contribute directly to the state of conformation of the heart of the TE complex bound to E-box sequences (Figure 3B and 3Di-iii). The H-bonds established between the K145 and N125 residues of TWIST1 and their K588 and N566 counterparts of E12 were largely disrupted in the BMID simulation compared to BMI and TE simulations (Figure 3Di-iii). Taken together, the hydrophobic interactions and H-bond establishments that drive alterations in the structure of the heart of the TE complex, are highly dependent on the E-box sequences used in our in silico models. Furthermore, although modifications arising from hydrophobic interactions seem to impede the binding of the dimer to the E-box sequences, H-bonds appear to play an essential role in the anchoring of the dimer to these boxes. We, therefore, investigated the effect of these bonds in the various models more thoroughly.
Figure 3.
Binding of the TE complex to the active TE-box and BMI-box sequences, as well as to the inactive BMID-box sequences. (A and B) 3D visualization of the localization of the putative hydrophobic bonds in the heart of the TE dimer. TWIST1 and E12 are represented in grey and green ribbons, respectively. The position of important TWIST1/E12 amino acid residues and E-box bases were visualized with the VMD 1.9.1 software. Different views are shown for a better understanding and interpretation of the findings. (A) Two phenylalanine residues (F128 of TWIST1 and F569 of E12), represented in solid green and six leucine residues may strongly contribute to the structure of the heart of the TE complex via hydrophobic interactions. Pairs of leucine residues named L124-L589, L131-592 and L149-L572 are shown in solid yellow, blue and red colours, respectively. (B) Pairs of leucine L124-L589 and phenylalanine residues F128-F569 are represented in solid yellow and green, respectively. The H-bond interactions between asparagine and lysine residues (N125 and K145 of TWIST1 and N566 and K588 of E12) are shown as grey and green CPK representations, respectively. Since the side chain of the lysine has three donors atom (NZ1, NZ2, NZ3), it can make alternatively three H-bonds with the same hydrogen acceptor atom (OD1) of asparagine. This case occurred when we studied the H-bond stability established between N125-K145 and N566-K588. (C) Plots representing the distances (Å) between pairs of residues close to the heart of the dimer as a function of time (total time = 10 ns, 1 frame per 10 ps), during the 10 ns MD simulations of the three TE (blue line), BMI (orange line) and BMID (grey line) in silico models. These distances were estimated during the MD simulations using the VMD 1.9.1 software (Graph of Labels Bonds). The lengths of H-bond interactions were represented via GraphPad Prism 5: ‘smooth, differentiate or integrate curve, with eight neighbours’. The pairs of leucine residues studied were (i) L124-L592, (ii) L149-L592, (iii) L131-L592, (iv) L149-L572 and (v) L124-L589, while the pair of phenylalanine residues was (vi) F128-F569. (D) Plots representing the shortest distances (Å) between the OD1 atom of asparagine N125 and the NZ1, NZ2 or NZ3 atoms of the lysine K145 residues (i), and between N566 and K588 residues (ii) as a function of time (total time = 10 ns, 1 frame per 10 ps), during the TE (blue line), BMI (orange line) and BMID (grey line) simulations. The distances were estimated as described in Figure 3C. (iii) The horizontal bar chart shows the cumulated occupancy values for the H-bond interactions between the N125-K145 residues of TWIST1 (grey) and the N566-K588 residues of E12 (green) during the TE, BMI and BMID MD simulations. The cumulated occupancy values were calculated as described in the Materials and Methods section. Briefly, H-bond interactions are assigned a value according to the distance between their atomic donors/acceptors (interactions score 1 if their distance is under 2.10 Å, and 0 if above). Higher occupancy values being obtained for shorter and, therefore, more stable interactions. (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 if true and 0 if false; NB.SI function: NB.SI(range;criterion)). Calculations were carried using Excel.
The cumulated rates of occupancy were calculated (as described in the Materials and Methods section), focusing on the ‘non-specific’ and ‘specific’ H-bond interactions (Figure 2D), in order to evaluate the overall stability of H-bond interactions between the TE complex and DNA during the in silico MD simulations. H-bond interactions were predominantly established between the dimer and E-box bases compared to adjacent bases, and these essentially occurred with conserved bases rather than with variable bases of the half-sites of the E-boxes (Figure 4A). However, while the cumulated occupancy values of H-bonds established with the different E-box bases were similar, this value was markedly higher between residues and adjacent bases in the BMI (+114%) and BMID (+83%) simulations compared to the TE MD simulations (Figure 4A). These findings indicate that there is an increase in H-bond interactions between the dimer and adjacent bases in the modified E-box sequences. Next, since the specificity of E-box recognition is likely due to these specific H-bond interactions, we focused on the specific H-bond binding affinities of the lateral chain residues either of TWIST1 or of E12. Of the H-bonds established between the dimer and E-box bases in the TE-model (Figure 4A), the specific H-bonds were principally attributed to TWIST1 residues (Figure 4Bi). Interestingly, few specific H-bond interactions were established with the adjacent bases in the TE simulation, indicating that they had a lower impact on the E-box recognition specificity. In contrast, of the elevated specific H-bond interactions observed between the dimer and adjacent bases in the BMI and BMID simulations (Figure 4A), the specific H-bond interactions were predominantly related to the E12 residues (Figure 4Bi). Furthermore, these specific H-bond interactions mainly occurred with the flanking ((-1) and (-1*)) bases (Figure 4Bi and ii), representing 50% of the total specific H-bond interactions. Specific H-bond interactions were, overall, much lower in the non-functional BMID model than in the BMI MD simulation (Figure 4Bi). This suggests that adjacent E-box bases, play an important role in the binding affinity of the TE dimer to less functional E-box sequences, such as the BMI-box. Taken together, our data on the stability of specific H-bond interactions, suggest that the E-box composition largely participates in the selectivity of the TE binding, this may influence the panel of regulatory targeted genes for the various TWIST1 complexes.
Figure 4.
H-bond interactions between the TWIST1/E12 complex and active or inactive E-box sequences. (A–C) All cumulated occupancy values of the H-bonds were calculated as described in the Materials and Methods section. Briefly, H-bond interactions are assigned a value according to the distance between their atomic donors/acceptors during the time (the 10 ns of the MD) (interactions score 1 if their distance is under 2.10 Å, and 0 if above). Higher occupancy values being obtained for shorter and, therefore, more stable interactions. (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 if true and 0 if false; NB.SI function: NB.SI(range;criterion)). The calculations were carried out using Excel. (A) Top, representation of the E-box sequence, with the forward hal-site and the reverse complementary half-site in orange and pink, respectively. Table showing the rate of cumulated occupancy of H-bonds established between residues and DNA, during MD simulations of the TE complex binding to the active TE and BMI or inactive BMID E-box sequences. We focused on H-bonds established between the TE complex and either adjacent bases or E-box sequences (consensus and variable bases). The percentage of variation of the BMI- and BMID-boxes compared to the TE-box is reported in brackets. (B) (i) Bar chart representing the cumulated rate of occupancy of ‘specific’ H-bonds (as defined in Figure 2D) established during MD simulations either between E12 (green stippling) and TWIST1 (grey stippling) and adjacent bases of E-box sequences, or between E12 (solid green) and TWIST1 (solid grey) and E-box bases, in the case of the TE-, BMI- and BMID-box models. (ii) 3D in silico model showing the binding of the TE complex to DNA, carried out via the VMD 1.9.1 software. The proximal flanking ((-1) and (-1*)) bases and the first base of each half-site of the E-box are represented in grey and green VDW, respectively. (C) Distribution of ‘specific’ H-bonds established between residues and E-box bases in the (i) TE, (ii) BMI and (iii) BMID molecular dynamics simulations. The pie charts show percentages of the total rate of occupancy of specific H-bonds established during the MD simulation. Consensus bases and variable bases of the E-box sequences are represented in grey and pink, respectively. The bar chart highlights the specific residues binding to Cytosine ((+1) and (+1*)) bases, and shows their implication in the DNA binding affinity. (D) 3D representation of the TE complex within the DNA groove of the TE-, BMI- and BMID-boxes sequences. CPK representation of the glutamic acid E558 and asparagine N554 residues of E12 (shown in green), and the E117 and arginine R118 residues of TWIST1 (shown in grey). The red arrow points to the interactions of the TE complex with E-box sequences according to the DNA substitutions. (i) CPK representation of the Cytosine ((+1), (+1*) and (+4*)) and Thymine (+3) bases of the TE-box, (ii) Cytosine ((+1) and (+1*)) and Guanine (+6*) of the BMI-box, and (iii) Adenine (+1*) and Cytosine (+5*) of the BMID-box.
Having addressed the importance of the E-box composition in the binding of the dimer, we focused our investigations on the exact mechanisms underlying this binding process. According to the literature (42), the first conserved Cytosine bases ((+1) and (+1*)) on each half-site of the E-box are important for the recognition of the E-box by a bHLH complex. Consequently, we studied the distribution of the specific H-bonds established between particular residues of the TE complex (two glutamic acids: E558 and E117, an arginine: R118, and an asparagine: N554) and bases of the E-box sequences (Figure 4Ci and Cii for better visualization). Specific H-bond occupancies with the first Cytosine bases ((+1) and (+1*)) of the half-sites of the E-box sequences represented 65%, 75% and 87% of the total occupancies in the TE, BMI, and BMID MD simulations, respectively (Figure 4Ci, Cii, Ciii). Interestingly, in both of the functional TE and BMI MD simulations, specific H-bonds were established between Cytosine (+1) and E558, while this was completely disrupted in the BMID simulation, where the Cytosine (+1) base was replaced by a Thymine. However, the specific H-bond established between base (+1*) and E117 was conserved in all of the simulations (Figure 4Ci-Ciii). Of note, in the TE simulation, numerous additional specific H-bond interactions were established between TE residues and the ((+3) and (+4*)) variable bases of the E-box sequence (represented in pink, Figure 4Ci and Di). In contrast, the H-bond interactions established with central variable bases were completely disrupted in BMI and BMID MD simulations (Figure 4Cii-iii and 4Dii-iii). Nevertheless, there was a clear shift towards the conserved base (+6*) in the BMI simulation (Figure 4Cii), and a slight shift towards base (+5*) during the BMID simulation (Figure 4Ciii and 4Diii). The 3D representations of the position of these H-bond interactions showed the binding of the TE-complex to different areas of the DNA groove (Figure 4Di-Diii). During the TE model simulation, these H-bond interactions occurred in four areas and acted as anchors allowing the specific recognition of the E-box sequence (Figure 4Di). Specific H-bond interactions with conserved central bases were partially lost, during the BMI and BMID model simulations, with only three (Figure 3Dii) and two (Figure 3Diii) areas of interactions remaining, respectively, resulting in a less stable anchoring of the dimer. To corroborate these in silico findings, we clearly observed in vitro that the TE complex was able to bind to the active TE-box and BMI-box sequences (Figure 5). Having demonstrated an impaired in silico binding during the BMID MD simulations, we expected a similar non-functional binding in vitro. As expected, a residual signal was observed with the inactive BMID-box sequence (Figure 5), which can likely be explained by the fact that bHLH complexes scan gene promoters to look for E-box sequences in the absence of stabilization of amphipathic helix conformations (46). No binding was observed in cells transfected with the empty pBABE-neo vector.
Figure 5.
Streptavidin/Biotin assay of DNA bound to the TWIST1/E12 complex, in the case of active TE and BMI or inactive BMID E-box sequences. Streptavidin/Biotin assay using the TWIST1 antibody. Western blot showing the TE complex bound to various E-boxes, namely the TE-box, the BMI-box, and the BMID-box. The TE dimer was extracted from cells transfected with the TE pBABE vector. The empty pBABE vector was used as an assay control. The ectopic TE protein is visualized around 99 kDa, while a degraded peptide is observed at ∼37 kDa. A detailed description of the assay is reported in the Materials and Methods section.
Deciphering of the key elements involved in the binding specificity of the TE complex to various functional E-boxes
Next, we determined whether the MD simulations accurately predicted the binding specificity of the TE dimer to various functional E-box sequences. We, thus, explored how the previously published TE complex (28) (Figure 1A) interacts with modified E-box sequences during the MD simulations. We substituted several bases in the DNA sequence of the TE-box (CATCTG), which was previously described by Firulli to bind with high affinity to heterodimers including to the TE complex (20), and compared the stabilities of the TE/modified E-box interactions (Figure 1A). The modified E-box sequences were as follows: the CACCTG sequence (named E12-box), described to bind to the E12 homodimer (24,35), the CATATG sequence (named TWIST-box) reported to bind to the TWIST1 homodimer (20,24,47) and finally the CGTCTG sequence (named D-box) known to bind to a lesser extent to the TE complex (20,48). We considered interactions with adjacent, conserved and variable bases of the E-box sequences separately.
Overall, we did not observe any major modifications in the positions of the leucine and phenylalanine residues during these MD simulations, suggesting that the establishment of hydrophobic interactions was not modified in the presence of modified E-box sequences (data not shown). In contrast, the stability of the H-bonds created between the asparagine and lysine N125-K145 residues (Figure 6Ai-Aiv; Bi-Bii) was completely disrupted in TE simulations bound to the E12- (Figure 6Aii and Bii), the TWIST- (Figure 6Aiii and Bii) and the D-box sequences (Figure 6Aiv and Bii). The H-bond interactions established between the N566-K588 of the E12 protein were conserved, irrespective of the E-box sequence chosen (Figure 6Ai-iv and Bii). These findings indicate that the heart of the dimer had a slightly altered stability according to the modified E-box sequence. To further explore the molecular mechanisms underlying this binding specificity, the overall stability of the H-bond interactions was then investigated during in silico MD simulations, focusing particularly on interactions occurring with all DNA bases. Total occupancy rates were calculated and revealed that the dimer preferentially bound to the conserved bases of the E-box sequences (Figure 6C). However, in the case of the modified E-box sequences, a shift was observed towards adjacent bases, in particular in the D-box with an increase in 90% of H-bond interactions with adjacent bases, confirming a decrease in the DNA binding specificity and the importance of E-box bases in the sequence recognition (Figure 6C) (20). We then identified the residues that created H-bonds with the E-box or with the adjacent ((-1), (+1), (-1*) and (+1*)) bases (Figure 6Dii-v). We observed that these four bases were close to the heart of the TE complex (Figure 6Di), and that H-bond interactions with these bases were evenly distributed between the TWIST1 and E12 proteins during the MD simulations, irrespective of the E-box model studied (Figure 6Dii-v). While the H-bond interactions between the adjacent (-1) base and the dimer represented only 2% of the total rate of occupancy during the MD simulations of the TE-box (Figure 6Dii), this percentage increased to 16%, 19% and 30% in the case of the E12- (Figure 6Diii), TWIST- (Figure 6Div) and D-box sequences (Figure 6Dv), respectively. The TWIST1 protein being the only one to interact with the adjacent (-1*) base of the TWIST-box, albeit at a very low rate (1%) (Figure 6Div). The shift of these H-bond establishments towards adjacent bases was concomitant with the experimental binding affinities of these E-box sequences (TE-box <<< E12-box ≈ TWIST-box << D-box) (Figure 6Di-v) (20,24,47,48). The pivotal residues were the serine S144 on TWIST1 and the arginine R561 on E12 for the DNA binding function, since they are located at the vicinity of the proximal flanking ((-1) and (-1*)) bases (Figure 6Di). Indeed, S144 is localized at the bottom of the TWIST1 loops, while R561 is present in the C-alpha HLH domain of E12 (Figure 6Di). Overall, the stability of the TE dimer binding to modified E-box sequences is dependant of H-bond interactions with ((-1) and (-1*)) bases.
Figure 6.
Binding of the TE complex to various active and modified E-box sequences, namely the E12-, TWIST- and D-boxes. (A) Plots represent the distances (Å) between the asparagine and lysine N125-K145 residues of TWIST1 and the N566-K588 residues of E12 as a function of time (total time = 10 ns, 1 frame per 10 ps) for the four (i) TE, (ii) E12, (iii) TWIST and (iv) D models during the MD simulations. These distances were estimated during the MD simulations using the VMD 1.9.1 software (Graph of Labels Bonds). The lengths of H-bond interactions were represented via GraphPad Prism 5: ‘smooth, differentiate or integrate curve, with 8 neighbours’. (B and C) All cumulated occupancy values of the H-bonds were calculated as described in the Materials and Methods section. Briefly, H-bond interactions are assigned a value according to the distance between their atomic donors/acceptors during the time (the 10 ns of the MD)(interactions score 1 if their distance is under 2.10 Å, and 0 if above). Higher occupancy values being obtained for shorter and, therefore, more stable interactions. (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 if true and 0 if false; NB.SI function: NB.SI(range;criterion)). The calculations were carried out using Excel. (B) (i) 3D representation of the TE dimer highlighting the position of the interaction between the asparagine and lysine residues N125-K145. (ii) The horizontal bar chart represents the rate of cumulated occupancy of the N125-K145 of TWIST1 and the N566-K588 of E12, in grey and green, respectively, for the four E-box sequences. (C) Top, representation of the E-box sequence, with the forward half-site and the reverse complementary half-site in orange and pink, respectively. Table showing the rate of cumulated occupancy of H-bonds established between the TE complex and either adjacent bases, or consensus and variable bases of TE-, E12-, TWIST-, and D-box sequences. The percentage of variation compared to the TE-box is reported in brackets. (D) (i) 3D in silico representation of the TE complex bound to DNA. The proximal flanking ((-1) and (-1*)) and first ((+1*) and (+1*)) bases of the E-box are represented in green and grey VDW, respectively. The serine S144 and arginine R561 residues are highlighted in magenta and orange, respectively. (ii-v) The pie charts show the percentages of cumulated occupancy of H-bonds established between residues of TWIST1 (grey) and E12 (green) and DNA bases during the (ii) TE-, (iii) E12-, (iv) TWIST- and (v) D-box MD simulations. Occupancies of H-bonds established with the proximal flanking ((-1) and (-1*)) bases are represented in yellow, while the bar charts highlight the exact residues binding to those bases. The H-bonds established with the first consensus ((+1) and (+1*)) bases of the E-box are represented in grey and green for the TWIST1 and the E12 residues, respectively.
The analysis of the distribution of the specific H-bond interactions between all of the conserved or adjacent bases with the dimer, confirmed the predominant binding to the active E-box sequences compared to adjacent bases in all four models (Figure 7A). Surprisingly, the cumulated specific H-bond interactions in the D-box model were higher than the others (Figure 7A). In order to unravel this phenomenon, we studied the distribution of specific H-bonds with all of the bases. This binding was principally observed between the dimer and the conserved E-box bases with rates of cumulated occupancies of 65%, 77%, 69% and 100% in the case of the TE- (Figure 7Bi), E12- (Figure 7Bii), D- (Figure 7Biii) and TWIST-boxes (Figure 7Biv), respectively. The TE complex created close contacts and specific interactions with the four ((+1), (+1*), (+3) and (+4*)) bases in the TE-box (Figure 7Bi, Ci and Di). These interactions mainly relied on the binding of the glutamic acid residue E117 of TWIST1 and the glutamic acid residue E558 of E12 to the conserved and functional Cytosine ((+1*) and (+1)), driving the E-box recognition (Figure 7Bi-Biv), as well as the binding of the arginine residue R118 of TWIST1 and the glutamic acid residue E558 of E12 to the conserved ((+3) and (+4*)) bases (Figure 7Bi). These residues, therefore, contributed to the binding specificity of the dimer to the E-box sequence. In the modified models, the H-bond interactions established between the glutamic acid residue E117 of TWIST1 and the (+1*) base were partially preserved, while those between the (+4*) base and the glutamic acid residue E558 of E12 were completely lost (Figure 7B-D). Furthermore, in these models a clear shift towards the conserved (+1) base was observed, particularly in the TWIST-box simulation (Figure 7Biv, Civ, Div), which may explain the low specificity of the TE heterodimer for this box.
Figure 7.
Predictive functional effect of base substitutions of E-box sequences on the H-bond establishment between the TE complex and DNA. (A and B) All cumulated occupancy values of the H-bonds were calculated as described in the Materials and Methods section. Briefly, H-bond interactions are assigned a value according to the distance between their atomic donors/acceptors during the time (the 10 ns of the MD)(interactions score 1 if their distance is under 2.10 Å, and 0 if above). Higher occupancy values being obtained for shorter and, therefore, more stable interactions. (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 if true and 0 if false; NB.SI function: NB.SI(range;criterion)). The calculations were carried out using Excel. (A) Bar chart showing the rate of cumulated occupancy of the ‘specific’ H-bonds (see Figure 2D) established between E12 (green) and TWIST1 (grey) and adjacent bases (stippling) or E-box sequences (solid colouring). The values are given for the four E-box models (TE-, E12-, TWIST-, and D-box sequences). (B-D) Pie-charts depicting the percentages of cumulated occupancy of ‘specific’ H-bonds established between specific residues of the TE complex and variable (pink) or conserved (grey) bases of the E-box sequences, during MD simulations of the (Bi) TE-, (Bii) E12-, (Biii) D- and (Biv) TWIST-models. The bar charts highlight the specific residue interactions with conserved Cytosine ((+1) and (+1*)) bases. The 3D representations show the positions of the TE complex in the DNA groove of the various (Ci) TE-, (Cii) E12-, (Ciii) D- and (Civ) TWIST-box sequences. The glutamic acid residue E558 of the E12 protein and the arginine R118 and E117 of the TWIST1 protein are represented in green and grey CPK shapes, respectively. The red arrows point to the major H-bond interactions of the TE complex with bases of the DNA groove, according to the DNA base substitutions listed opposite. Active bases in the (Di) TE-box: Cytosine ((+1) and (+1*)), Thymine (+3) and Adenine (+4*) bases (purple), (Dii) E12-box: Cytosine ((+1), (+4) and (+1*)) bases (pink), (Diii) D-box Cytosine ((+1), (+4) and (+1*)) and Adenine (+2*) bases (orange), and (Div) TWIST-box: Cytosine ((+1) and (+1*)) bases (green).
Interestingly, in the functional TE simulations, specific H-bonds established between Cytosine (+1) and E558, and between Cytosine (+1*) and E117, were conserved in all of the simulations (Figure 7Bi). Of note, in the TE-box and E12-box simulations, numerous additional specific H-bond interactions were established between TE residues and the (+3) and (+4*) variable bases of E-box sequences (represented in pink, Figure 7Bi and 7Biii). In contrast, the H-bond interactions established with central variable bases were partially disrupted in TWIST-box simulations (Figure 7Biv and Civ). There was, however, a clear shift towards the conserved base in the D-box and TWIST-box simulations (Figure 7Ciii). The 3D representations of the position of these H-bond interactions showed the binding of the TE-complex to different areas of the DNA groove (Figure 7Di-Div). During the TE MD simulations, these H-bond interactions occurred in four areas and acted as anchors allowing the specific recognition of the E-box sequence (Figure 7Di). Specific H-bond interactions with conserved central bases were partially lost, during the modified E12- and D-box simulations, with only three (Figure 7Diii) and two (Figure 7Div) areas of interactions remaining, respectively, resulting in a less stable anchoring of the dimer. The in vitro experiments conducted in parallel using the Streptavidin/Biotin binding assay, corroborated these in silico findings (Figure 8). Indeed, the binding of the TE complex to the TE-box probes was stronger than that obtained with E12-box, TWIST-box and D-box probes. Moreover, the residual signal observed when using the degenerate BMID-box probe confirmed that the modified boxes were truly active (Figure 8). This result was further confirmed by running a blot using the E12 antibody, instead of the TWIST1 antibody (Figure 11C). These findings highlight the functional importance of the variable bases of the E-boxes. The correlation obtained between the in silico prediction and the in vitro findings could lead to the screening of gene promoters in order to cluster targeted genes according to the E-box sequences they contain.
Figure 8.
Streptavidin/Biotin assay of DNA bound to TE complex, in the case of modified E-box sequences. Streptavidin/Biotin assay using the TWIST1 antibody. Western blot showing the TE complex bound to various E-boxes, namely the TE-box, the E12-box, the D-box, the TWIST-box, as well as to the degenerate BMID-box. The TE dimer was extracted from cells transfected with the TE pBABE vector. The empty pBABE vector was used as an assay control. The ectopic TE protein is visualized around 99 kDa, while a degraded peptide is observed at ∼37 kDa. A detailed description of the assay is reported in the Materials and Methods section.
Prediction of the contribution of the flanking proximal bases to the recognition of the E-box sequence
Having studied both the impact of using active versus inactive box-sequences and non-modified versus modified active box sequences on the binding of the TE complex, we next focused on the contribution of adjacent bases to its binding affinity. Indeed, following a study on the influence of proximal flanking sequences on E-box recognition by bHLH dimers (49), and in light of our own findings (see sections above), we investigated the contribution of the ((-1) and (-1*)) adjacent bases to the binding affinity of the TE-complex during MD simulations. We then exchanged the proximal flanking Cytosine (-1), Guanine (+7), Cytosine (-1*) and Guanine (+7*) bases by Adenine (-1), Thymine (+7), Adenine (-1*) and Thymine (+7*) in the AT-box, respectively, and proceeded to the opposite substitutions for the TA-box (Figure 1A, Figure 2Bii and Figure 9Aii).
Figure 9.
Predictive functional effect of the substitution of the proximal adjacent bases on the binding of the TE complex to DNA. (A) (i) Chemical structure of the purine and pyrimidine bases, showing the aromatic rings. (ii) The table depicts the base substitution generated in the TE-box, resulting in the TA- and AT-boxes. The latter substitutions led to the presence of two aromatic rings in the crucial ((-1) and (-1*)) base locations. (iii) The 3D representation of the TE complex (grey ribbon) in the DNA groove highlighting the location of the ((-1) and (-1*)) and ((+1) and (+1*)) base pairs in solid grey and green, respectively. (B) (i) in silico 3D model of the TWIST1 (grey ribbon)/E12 (green ribbon) dimer bound to DNA (white ribbon). Serine S144 and arginine R561 residues are shown in solid grey and green colouring, respectively, while CPK representation is used to highlight the first adjacent base of the E-box. (ii-iii) Plots representing the distances (Å) between residues and the (-1) base of the E-box sequences according to time (total time = 10 ns, 1 frame per 10 ps), focusing on (ii) S144 and (-1) base interactions, and (iii) R561 and (-1) base interactions, for all of the TE-box (in blue), TA-box (in green) and AT-box models (in purple). These distances were estimated during the MD simulations using the VMD 1.9.1 software (Graph of Labels Bonds). The lengths of H-bond interactions were represented via GraphPad Prism 5: ‘smooth, differentiate or integrate curve, with 8 neighbours’. (C and D) All cumulated occupancy values of the H-bonds were calculated as described in the Materials and Methods section. Briefly, H-bond interactions are assigned a value according to the distance between their atomic donors/acceptors during the time (the 10 ns of the MD) (interactions score 1 if their distance is under 2.10 Å, and 0 if above). Higher occupancy values being obtained for shorter and, therefore, more stable interactions. (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 if true and 0 if false; NB.SI function: NB.SI(range;criterion)). The calculations were carried out using Excel. (C) Pie-charts representing the percentages of cumulated occupancy of H-bonds established between residues of the TWIST1 (grey) and E12 (green) proteins and ((-1), (-1*), (+1) and (+1*)) bases. Percentage occupancies of H-bonds established with ((-1) and (-1*)) bases are represented in yellow, the bar charts highlight the exact residues interacting with these bases, in the (i) TE-, (ii) TA-, and (iii) AT-models. (D) Plots representing the distances (Å) between (i) N125-K145 residues of TWIST1, and (ii) N566-K588 residues of E12 as a function of time (total time = 10 ns, 1 frame per 10 ps), during TE- (in blue), TA- (in green) and AT-box (in purple) simulations. These distances were estimated during the MD simulations using the VMD 1.9.1 software (Graph of Labels Bonds). The lengths of H-bond interactions were represented via GraphPad Prism 5: ‘smooth, differentiate or integrate curve, with eight neighbours’. (iii) The horizontal bar chart shows the rate of cumulated occupancy of the interactions between the N125-K145 residue pairs of TWIST1 (in grey) and the N566-K588 residue pairs of E12 (in green) for each of the TE-, TA- and AT-box models.
The critical S144 (TWIST1) and R561 (E12) residues (described in the previous section) were localized close to the ((-1) and (-1*)) substituted bases (Figure 6Di and 9Aiii). In the modified TE-box sequences, and particularly in the case of the AT-box, base substitutions led to the presence of two aromatic rings, namely an Adenine (-1) and a Guanine (-1*), in these critical base locations (Figure 9Ai and Aii). These aromatic double rings have a higher molecular volume compared to the Thymine and Cytosine bases, which likely modifies the H-bond interactions at the interface of the DNA and the TE complex. Indeed, the aromatic rings on the Adenine ((-1) and (-1*)) bases strongly contributed to the stability of H-bond interactions with neighbouring S144 and R561 (Figure 9Bi and Biii) residues during the AT-box simulation. Furthermore, in the case of this model, of the 28% H-bond interactions occurring with adjacent bases, 3% were associated with the Adenine (-1*) base (Figure 9Ciii), whereas these interactions were non-existent in the 10% and in the 2% of adjacent H-bond interactions established in the TA-box (Figure 9Cii) and TE-box simulations (Figure 9Ci), respectively. These interactions with the flanking Adenine ((-1) and (-1*)) bases likely contributed to stabilizing the H-bond interactions with adjacent sequences. They had a detrimental effect on the H-bond interaction between the N125-K145 residue pair of the TWIST1 protein that are at heart of the dimer, which may have resulted in a decrease in the stability and affinity of the TE complex for the TE-box (Figure 9Di and Diii). The H-bonds created between the asparagine and lysine N125-K145 residues were partially and completely disrupted in TA- and AT-box simulations, respectively, while the H-bond interactions occurring in the N566-K588 residue pair remained stable in all three models (Figure 9Di-iii). These findings indicate that the heart of the dimer had an altered stability according to the proximal flanking base included in the models. Overall our findings demonstrate that the stability of the H-bonds established between residues of the TE complex and adjacent ((-1) and (-1*)) bases is higher during the molecular dynamics of the TA- and AT-box simulations compared to the TE simulation, and that it strongly impacts the establishment of a stable H-bond interaction between the N125-K145 residue pair of TWIST1. In conclusion, to screen gene-targets of the TE complex, the flanking proximal bases should be taken into account, since they modulate the function of key residues, such as K145. The latter is reported as a mutation of TWIST1, linked to the SCS syndrome.
When examining the rate of cumulated occupancy of H-bond interactions between the dimer and E-box sequences, taking into account all of the adjacent bases and not only the ((-1) and (-1*)) bases, the AT- and TA-box simulations displayed higher rates of binding with adjacent bases, with 82% and 32%, respectively, compared to the TE-box (Figure 10A). Moreover, a decrease (by 21%) in the rate of cumulated occupancies of H-bond interactions with central bases of the TA-box was observed (Figure 10A). To identify key structural modifications in altered MD simulations, we then studied the specific H-bond interactions (as defined in Figure 2D) focusing on the variable and conserved bases, in particular ((+1) and (+1*)) bases (Figure 10Bi). While specific H-bond interactions with variable ((+3), (+4), (+3*) and (+4*)) bases represented 32% of all of the interactions with the TE-box sequence (Figure 10Bii), these were largely redistributed to the conserved ((+1) and (+1*)) bases in the AT- (Figure 9Biii) and TA-box simulations (Figure 10Biv). These data denote a decrease in the interactions with central bases in the TA-box simulation, which are highly involved in the specific recognition of the E-box by the TE complex. These findings were confirmed using the Streptavidin/Biotin assay, since the binding of TE complex to the TE-box probe was stronger than the binding obtained with AT-box and TA-box probes (Figure 10C). The altered TE binding to these modified E-boxes is in agreement with Chip experiments, in which authors observed a clear decease in the binding of the yeast Tye7 and Cbf1 bHLH to a functional E-box, by modifying the flanking proximal sequence (49). In conclusion, these findings confirmed the critical function of the flanking proximal bases in the binding of the TE complex to DNA. Furthermore, the MD simulations were able to predict and identify the molecular mechanism of TE complex binding on these modified DNA sequences.
Figure 10.
Prediction of the impact of proximal adjacent base substitutions on the specific interactions of the TE complex with E-box sequences. (A and B) All cumulated occupancy values of the H-bonds were calculated as described in the Materials and Methods section. Briefly, H-bond interactions are assigned a value according to the distance between their atomic donors/acceptors during the time (the 10 ns of the MD)(interactions score 1 if their distance is under 2.10 Å, and 0 if above). Higher occupancy values being obtained for shorter and, therefore, more stable interactions. (SI function: SI(test_logic; value_if_true;value_if_false) with logic test:‘<2.10’, value of 1 if true and 0 if false; NB.SI function: NB.SI(range;criterion)). The calculations were carried out using Excel. (A) Table showing the rate of cumulated occupancy of H-bonds established between the TE complex and either adjacent bases, or consensus and variable bases of TE-, AT-, and TA-box sequences. The percentage of variation compared to TE-box is reported in brackets. (B) (i) 3D representation showing the position of the TE complex in the DNA groove. The TWIST1 and E12 dimers are represented in grey and green ribbons, respectively, while the variable base interactions, close to the ((+1) and (+1*)) bases, are shown in magenta CPK representations. (ii-iv) Pie-charts representing the percentage of cumulated occupancies of specific H-bonds established between the TE complex and either conserved (grey) or variable (magenta) bases. The bar charts highlight the exact residues interacting with the conserved ((+1) and (+1*)) bases, in the (ii) TE-, (iii) TA-, and (iv) AT-models. (C) Streptavidin/Biotin assay using the TWIST1 antibody. Western blot showing the TE complex bound to various E-boxes, namely the TE-box, the AT-box, and the TA-box. The TE dimer was extracted from cells transfected with the TE pBABE vector. The empty pBABE vector was used as an assay control. The weaker intensity of the signal was due to an unintentional shorter exposure-time of the radiographic film. The ectopic TE protein is visualized around 99 kDa, while a degraded peptide is observed at ∼37 kDa. A detailed description of the assay is reported in the Materials and Methods section.
DISCUSSION
Our study provides an original proof that the in silico approach we used to determine the impact of base mutations on the binding of the oncogenic TE complex to E-box sequences, during MD simulations, could accurately predict the functional binding affinity of the dimer. We first addressed the molecular role of the arginine R118, serine S144 and lysine K145 residues of TWIST1. Several variants of the TWIST1 protein have been found in patients suffering from TWIST1 haplo-insufficiency. This is the case for example in patients with SCS syndrome, where TWIST1 mutations occur in the R118H/Q/C (arginine118histidine/glutamine/cysteine), R120P (arginine120proline), S144R, and K145E/Q residues (30,32–34,50–52). These variations may modify the binding affinity of TWIST1 with the E-box sequences during the developmental process leading to SCS syndrome (32,53,54). A previous study using similar MD simulations focusing on the effect of structural changes between wild-type versus three alterations in the R118C, S144R and K145 residues, showed an increase in the instability of the mutated dimers (29). In the present work, we completed this study by showing that the H-bond interactions between the N125-K145 residue pair is an important element involved in stabilizing the structure of the heart of the TE dimer, enabling the establishment of specific H-bonds of R118 of TWIST1 and E558 of E12 with central variable bases of the E-box sequence. We also highlighted the crucial function of R118 and E558 in the recognition and binding specificity, since we observed that the higher the affinity between the TE complex and the E-box, the greater the specific H-bond interactions between R118 and E558 residues and central variable bases of the E-box (Figure 7). The third S144 residue, close to the hydroxyl group, is also involved in the anchoring of the TE dimer (i) to E-box sequences with low binding affinity or (ii) when the flanking proximal bases are modified. By doing so, it appears to compensate for the low binding specificity to such sequences. Overall, we showed that these residues are differentially involved in H-bond interactions and likely participate in the diverse functions of the TWIST1 complexes.
Our work clearly demonstrated that the MD simulations allowed us to differentiate active and inactive E-box sequences, and revealed the critical role of the consensus ((+1) and (+1*)) bases in the E-box recognition process. The affinity of TE complex for an E-box is partially defined by the specific H-bond interactions between residues and DNA bases. We determined their localization and uncovered their role in stabilizing the binding of TE complex to various active and modified E-box sequences during MD simulations. We found that specific H-bond interactions are principally established with the ((+1) or (+1*)) bases of the half-sites of the active E-box sequences. This was clearly reduced in the MD simulation of the inactive BMID model, confirming the pivotal function of these consensus Cytosine ((+1) and (+1*)) in E-box recognition by bHLH complexes. Indeed, the substitution of Cytosine to Thymine in BMID sequence was sufficient to impair the interaction between the E-box sequence and the bHLH complex (42,55) as shown by the loss of E558-(+1) specific H-bonds during the dynamic simulation of BMID simulation. These in silico findings were supported by in vitro data, in which the binding of the TE complex was higher in the TE- and BMI-boxes, while only a residual signal was observed in the negative control and in the BMID-box (Figure 5). Our predictions are in agreement with previously published biochemical findings (42), which demonstrated the ability of different TWIST1 complexes to bind various E-box sequences using EMSA assays, and measured the transactivation function of these complexes using reporter gene assays. These studies found that the higher the affinity between the TE-box and the TE complex, the stronger the binding (20,24,42,47,48,55). In the present study, this observation was verified for different complexes, since we also obtained a stronger binding between the TWIST1/TWIST1 homodimer and the TWIST-box using the TWIST1 antibody (Figure 11A), and between the E12/E12 homodimer and the E12-box using the E12 antibody (Figure 11B).
We also evaluated whether our MD simulations were able to predict the degree of affinity of E-box sequences, by studying the distribution of specific H-bonds established inside various E-box sequences with modified central or variable bases. We observed a shift in the specific interactions to conserved bases of the E-box and a decrease in the binding affinity of the TE-complex in Streptavidin/Biotin assays (TE-box <<< E12-box ≈ TWIST-box << D-box). The pivotal function of the variable third base of the half-site of the E-box was highlighted, since the substitution of the complementary half-site CAG by the CAT sequence of TWIST-box abolished the specific H-bond interactions between the TE complex and the TWIST-box sequence. The half-site CGT of the D-box and CAC of the E12-box were, however, still recognized by TWIST1 (20,24). This is supported by the fact that the CAC half-site is specific to the E12 homodimers and its specificity is decreased in the TE heterodimers, while CAT and CAG half-sites are normally recognized with high affinity by the TE complex (24). These findings are in agreement with the in vitro Streptavidin/Biotin assays conducted with the TE heterodimer, and the TWIST1 and E12 homodimers (Figure 11). In the present study, we clearly highlighted a redistribution of the specific H-bonds during the simulations according to the biological binding affinity of the TE complex to a given E-box sequence, with central variable bases of the E-box largely contributing to the TE complex binding specificity. Overall, this in silico approach may provide a valuable predictive tool to select E-box sequences with flanking proximal sequences with a higher probability of binding affinity to the TE complex. Lastly, it will enable the screening of gene promoters for highly specific E-box sequences in order to establish a clustering of targeted gene sets. Such an approach may also be applicable to other proteins (or complexes) bound to their ligands. Furthermore, the identification of a specific interaction between the oncogenic TE complex and a distinct E-box sequence could lead to the determination of a putative pro-oncogenic signature.
To summarize our findings and based on Sauve's model (46), we described three different states of binding of the bHLH complex to functional and degenerate E-box sequences based on molecular dynamics simulations. The first conformation is represented by the TE-box dynamics simulation, in which specific H-bond interactions are established with the variable bases of the E-box (Figure 12). This ‘ideal’ conformational state favours stability and complete transactivation of the TE complex. The second or ‘intermediate’ conformation includes all of the active, modified E-box sequences, which are not highly specific for the TE complex. Although, these sequences are recognized (Figure 7B) by the TE complex, the binding specificity and efficiency to these BMI-, E12-, D- and TWIST-boxes is not ideal (Figure 12). The third state is a non-specific conformation that enables the TE complex to scan DNA to look for E-box sequences. This state is, represented by the BMID simulation. In this context, only the specific E117-(+1*) interaction is preserved and a shift of the interactions towards adjacent bases of the E-box sequence occurs. The in vitro binding also decreases, since the binding affinity is partially impaired. This strongly suggests that there is an increase in the binding flexibility of the TE complex to DNA, allowing the proper promoter scanning function (Figure 12).
Figure 12.
Synopsis of the three conformational states of the TE complex bound to the E-box sequences based on Sauve's model. Three protein structures are depicted on this diagram. The first state, represented by TE-box simulation, demonstrates the complete stabilization of the alpha-helical conformation of the basic region. The second state, represented by BMI-box, E12-box, TWIST-box and D-box simulations, shows the partial stabilization of the alpha-helical conformation of the basic region. The third state, represented by BMID-box simulation, reveals that there is no stabilization of the alpha-helical conformation of the basic region.
In conclusion, our study is a proof of concept of the predictive value of these molecular modeling approaches on deciphering molecular mechanisms underlying the binding of the putative oncogenic TE form to E-box sequences. In the future, it may be possible to identify specific transcriptional signatures according to the TWIST1 heterodimers generated, for example TWIST1/E2A versus TWIST1/TWIST1 or TWIST1/HANDs, and to their E-box binding specificities. Furthermore, these validated in silico models of the TE complex may contribute to the characterization of specific inhibitors by docking approaches of the putative oncogenic form of TE complex targeting the heart of the dimer. This could be used in the future for developing and testing pharmacological TWIST1/E12 blocking molecules by in silico docking prior to in vitro or in vivo studies.
Acknowledgments
The pBABE-neo expression vector was donated by H. Land & J. Morgenstern & B. Weinberg (Addgene plasmid # 1767). Our team is accredited by the Ligue Nationale contre le Cancer.
Footnotes
Present address: Pr. Lea Payen, Inserm UMR-S1052, Centre de Recherche en Cancérologie de Lyon, Lyon 69008, France.
FUNDING
LabEX DEVweCAN [ANR-10-LABX-61]; LyRIC (Lyon Recherche Intégrée en Cancérologie, Institut National contre le Cancer [INCa_4664]; Ligue Nationale contre le Cancer (to C.B.). Funding for open access charge: LabEX DEVweCAN [ANR-10-LABX-61]; LyRIC (Lyon Recherche Intégrée en Cancérologie, Institut National contre le Cancer [INCa_4664]; Ligue Nationale contre le Cancer (to C.B.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bialek P., Kern B., Yang X., Schrock M., Sosic D., Hong N., Wu H., Yu K., Ornitz D.M., Olson E.N., et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 2.Seto M.L., Hing A.V., Chang J., Hu M., Kapp-Simon K.A., Patel P.K., Burton B.K., Kane A.A., Smyth M.D., Hopper R., et al. Isolated sagittal and coronal craniosynostosis associated with TWIST box mutations. Am. J. Med. Genet. A. 2007;143:678–686. doi: 10.1002/ajmg.a.31630. [DOI] [PubMed] [Google Scholar]
- 3.Massari M.E., Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teng Y., Li X. The roles of HLH transcription factors in epithelial mesenchymal transition and multiple molecular mechanisms. Clin. Exp. Metast. 2014;31:367–377. doi: 10.1007/s10585-013-9621-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu T.M., Lee E.H. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng. B, Rev. 2013;19:254–263. doi: 10.1089/ten.teb.2012.0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosty J., Vogel T.W. Insights into the development of molecular therapies for craniosynostosis. Neurosurg. Focus. 2015;38:E2. doi: 10.3171/2015.2.FOCUS155. [DOI] [PubMed] [Google Scholar]
- 7.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Ansieau S., Bastid J., Doreau A., Morel A.P., Bouchet B.P., Thomas C., Fauvet F., Puisieux I., Doglioni C., Piccinin S., et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Li S., Kendall S.E., Raices R., Finlay J., Covarrubias M., Liu Z., Lowe G., Lin Y.H., Teh Y.H., Leigh V., et al. TWIST1 associates with NF-kappaB subunit RELA via carboxyl-terminal WR domain to promote cell autonomous invasion through IL8 production. BMC Biol. 2012;10:73. doi: 10.1186/1741-7007-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Piccinin S., Tonin E., Sessa S., Demontis S., Rossi S., Pecciarini L., Zanatta L., Pivetta F., Grizzo A., Sonego M., et al. A "twist box" code of p53 inactivation: twist box: p53 interaction promotes p53 degradation. Cancer Cell. 2012;22:404–415. doi: 10.1016/j.ccr.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Kwok W.K., Ling M.T., Lee T.W., Lau T.C., Zhou C., Zhang X., Chua C.W., Chan K.W., Chan F.L., Glackin C., et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 13.Wushou A., Hou J., Zhao Y.J., Shao Z.M. Twist-1 up-regulation in carcinoma correlates to poor survival. Int. J. Mol. Sci. 2014;15:21621–21630. doi: 10.3390/ijms151221621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raatikainen S., Aaltomaa S., Palvimo J.J., Karja V., Soini Y. TWIST overexpression predicts biochemical recurrence-free survival in prostate cancer patients treated with radical prostatectomy. Scand. J. Urol. 2015;49:51–57. doi: 10.3109/21681805.2014.909529. [DOI] [PubMed] [Google Scholar]
- 15.Ren K.Q., Cao X.Z., Liu Z.H., Guo H., Quan M.F., Liu F., Jiang L., Xiang H.L., Deng X.Y., Cao J.G. 8-bromo-5-hydroxy-7-methoxychrysin targeting for inhibition of the properties of liver cancer stem cells by modulation of Twist signaling. Int. J. Oncol. 2013;43:1719–1729. doi: 10.3892/ijo.2013.2071. [DOI] [PubMed] [Google Scholar]
- 16.Tran P.T., Shroff E.H., Burns T.F., Thiyagarajan S., Das S.T., Zabuawala T., Chen J., Cho Y.J., Luong R., Tamayo P., et al. Twist1 suppresses senescence programs and thereby accelerates and maintains mutant Kras-induced lung tumorigenesis. PLoS Genet. 2012;8:e1002650. doi: 10.1371/journal.pgen.1002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo S.K., Kim J.H., Choi H.N., Choe T.B., Hong S.I., Yi J.Y., Hwang S.G., Lee H.G., Lee Y.H., Park I.C. Knockdown of TWIST1 enhances arsenic trioxide- and ionizing radiation-induced cell death in lung cancer cells by promoting mitochondrial dysfunction. Biochem. Biophys. Res. Commun. 2014;449:490–495. doi: 10.1016/j.bbrc.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 18.Murre C., Bain G., van Dijk M.A., Engel I., Furnari B.A., Massari M.E., Matthews J.R., Quong M.W., Rivera R.R., Stuiver M.H. Structure and function of helix-loop-helix proteins. Biochim. Biophys. Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 19.Castanon I., Von Stetina S., Kass J., Baylies M.K. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128:3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- 20.Firulli B.A., Redick B.A., Conway S.J., Firulli A.B. Mutations within helix I of Twist1 result in distinct limb defects and variation of DNA binding affinities. J. Biol. Chem. 2007;282:27536–27546. doi: 10.1074/jbc.M702613200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connerney J., Andreeva V., Leshem Y., Muentener C., Mercado M.A., Spicer D.B. Twist1 dimer selection regulates cranial suture patterning and fusion. Dev. Dyn. 2006;235:1345–1357. doi: 10.1002/dvdy.20717. [DOI] [PubMed] [Google Scholar]
- 22.Gajula R.P., Chettiar S.T., Williams R.D., Nugent K., Kato Y., Wang H., Malek R., Taparra K., Cades J., Annadanam A., et al. Structure-function studies of the bHLH phosphorylation domain of TWIST1 in prostate cancer cells. Neoplasia. 2015;17:16–31. doi: 10.1016/j.neo.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones S. An overview of the basic helix-loop-helix proteins. Genome Biol. 2004;5:226. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Masi F., Grove C.A., Vedenko A., Alibes A., Gisselbrecht S.S., Serrano L., Bulyk M.L., Walhout A.J. Using a structural and logics systems approach to infer bHLH-DNA binding specificity determinants. Nucleic Acids Res. 2011;39:4553–4563. doi: 10.1093/nar/gkr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong A.P., Yao Z., Zhong J.W., Cao Y., Ruzzo W.L., Gentleman R.C., Tapscott S.J. Genetic and epigenetic determinants of neurogenesis and myogenesis. Dev. Cell. 2012;22:721–735. doi: 10.1016/j.devcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grove C.A., De Masi F., Barrasa M.I., Newburger D.E., Alkema M.J., Bulyk M.L., Walhout A.J. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. 2009;138:314–327. doi: 10.1016/j.cell.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang A.T., Liu Y., Ayyanathan K., Benner C., Jiang Y., Prokop J.W., Paz H., Wang D., Li H.R., Fu X.D., et al. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev. 2015;29:603–616. doi: 10.1101/gad.242842.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouard C., Terreux R., Hope J., Chemelle J.A., Puisieux A., Ansieau S., Payen L. Interhelical loops within the bHLH domain are determinant in maintaining TWIST1-DNA complexes. J. Biomol. Struct. Dyn. 2014;32:226–241. doi: 10.1080/07391102.2012.762722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maia A.M., da Silva J.H., Mencalha A.L., Caffarena E.R., Abdelhay E. Computational modeling of the bHLH domain of the transcription factor TWIST1 and R118C, S144R and K145E mutants. BMC Bioinformatics. 2012;13:184. doi: 10.1186/1471-2105-13-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gripp K.W., Kasparcova V., McDonald-McGinn D.M., Bhatt S., Bartlett S.P., Storm A.L., Drumheller T.C., Emanuel B.S., Zackai E.H., Stolle C.A. A diagnostic approach to identifying submicroscopic 7p21 deletions in Saethre-Chotzen syndrome: fluorescence in situ hybridization and dosage-sensitive Southern blot analysis. Genet. Med. 2001;3:102–108. doi: 10.1097/00125817-200103000-00003. [DOI] [PubMed] [Google Scholar]
- 31.Corsi A.K., Brodigan T.M., Jorgensen E.M., Krause M. Characterization of a dominant negative C. elegans Twist mutant protein with implications for human Saethre-Chotzen syndrome. Development. 2002;129:2761–2772. doi: 10.1242/dev.129.11.2761. [DOI] [PubMed] [Google Scholar]
- 32.el Ghouzzi V., Le Merrer M., Perrin-Schmitt F., Lajeunie E., Benit P., Renier D., Bourgeois P., Bolcato-Bellemin A.L., Munnich A., Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat. Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 33.El Ghouzzi V., Lajeunie E., Le Merrer M., Cormier-Daire V., Renier D., Munnich A., Bonaventure J. Mutations within or upstream of the basic helix-loop-helix domain of the TWIST gene are specific to Saethre-Chotzen syndrome. Eur. J. Hum. Genet.: EJHG. 1999;7:27–33. doi: 10.1038/sj.ejhg.5200240. [DOI] [PubMed] [Google Scholar]
- 34.El Ghouzzi V., Legeai-Mallet L., Benoist-Lasselin C., Lajeunie E., Renier D., Munnich A., Bonaventure J. Mutations in the basic domain and the loop-helix II junction of TWIST abolish DNA binding in Saethre-Chotzen syndrome. FEBS Lett. 2001;492:112–118. doi: 10.1016/s0014-5793(01)02238-4. [DOI] [PubMed] [Google Scholar]
- 35.Longo A., Guanga G.P., Rose R.B. Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry. 2008;47:218–229. doi: 10.1021/bi701527r. [DOI] [PubMed] [Google Scholar]
- 36.Nardella C., Clohessy J.G., Alimonti A., Pandolfi P.P. Pro-senescence therapy for cancer treatment. Nat. Rev. Cancer. 2011;11:503–511. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- 37.Acosta J.C., Gil J. Senescence: a new weapon for cancer therapy. Trends Cell Biol. 2012;22:211–219. doi: 10.1016/j.tcb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Valsesia-Wittmann S., Magdeleine M., Dupasquier S., Garin E., Jallas A.C., Combaret V., Krause A., Leissner P., Puisieux A. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 39.Morel A.P., Hinkal G.W., Thomas C., Fauvet F., Courtois-Cox S., Wierinckx A., Devouassoux-Shisheboran M., Treilleux I., Tissier A., Gras B., et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PLoS Genet. 2012;8:e1002723. doi: 10.1371/journal.pgen.1002723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., Hassan M.Q., Li Z.Y., Stein J.L., Lian J.B., van Wijnen A.J., Stein G.S. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J. Cell. Biochem. 2008;105:487–496. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M.H., Hsu D.S., Wang H.W., Wang H.J., Lan H.Y., Yang W.H., Huang C.H., Kao S.Y., Tzeng C.H., Tai S.K., et al. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat. Cell Biol. 2010;12:982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- 43.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Slovak J., Tanaka H. Computer simulation study of metastable ice VII and amorphous phases obtained by its melting. J. Chem. Phys. 2005;122:204512. doi: 10.1063/1.1906217. [DOI] [PubMed] [Google Scholar]
- 45.Song X.-J., Rienstra C.M., McDermott A.E. N—H bond stretching in histidine complexes: a solid-state NMR study. Magn. Reson. Chem. 2001;39:S30–S36. [Google Scholar]
- 46.Sauve S., Naud J.F., Lavigne P. The mechanism of discrimination between cognate and non-specific DNA by dimeric b/HLH/LZ transcription factors. J. Mol. Biol. 2007;365:1163–1175. doi: 10.1016/j.jmb.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 47.Ozdemir A., Fisher-Aylor K.I., Pepke S., Samanta M., Dunipace L., McCue K., Zeng L., Ogawa N., Wold B.J., Stathopoulos A. High resolution mapping of Twist to DNA in Drosophila embryos: Efficient functional analysis and evolutionary conservation. Genome Res. 2011;21:566–577. doi: 10.1101/gr.104018.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knofler M., Meinhardt G., Bauer S., Loregger T., Vasicek R., Bloor D.J., Kimber S.J., Husslein P. Human Hand1 basic helix-loop-helix (bHLH) protein: extra-embryonic expression pattern, interaction partners and identification of its transcriptional repressor domains. Biochem. J. 2002;361:641–651. doi: 10.1042/0264-6021:3610641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordan R., Shen N., Dror I., Zhou T., Horton J., Rohs R., Bulyk M.L. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 2013;3:1093–1104. doi: 10.1016/j.celrep.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson D., Horsley S.W., Moloney D.M., Oldridge M., Twigg S.R., Walsh S., Barrow M., Njolstad P.R., Kunz J., Ashworth G.J., et al. A comprehensive screen for TWIST mutations in patients with craniosynostosis identifies a new microdeletion syndrome of chromosome band 7p21.1. Am. J. Hum. Genet. 1998;63:1282–1293. doi: 10.1086/302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose C.S., Patel P., Reardon W., Malcolm S., Winter R.M. The TWIST gene, although not disrupted in Saethre-Chotzen patients with apparently balanced translocations of 7p21, is mutated in familial and sporadic cases. Hum. Mol. Genet. 1997;6:1369–1373. doi: 10.1093/hmg/6.8.1369. [DOI] [PubMed] [Google Scholar]
- 52.Paznekas W.A., Cunningham M.L., Howard T.D., Korf B.R., Lipson M.H., Grix A.W., Feingold M., Goldberg R., Borochowitz Z., Aleck K., et al. Genetic heterogeneity of Saethre-Chotzen syndrome, due to TWIST and FGFR mutations. Am. J. Hum. Genet. 1998;62:1370–1380. doi: 10.1086/301855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howard T.D., Paznekas W.A., Green E.D., Chiang L.C., Ma N., Ortiz de Luna R.I., Garcia Delgado C., Gonzalez-Ramos M., Kline A.D., Jabs E.W. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat. Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 54.Firulli B.A., Krawchuk D., Centonze V.E., Vargesson N., Virshup D.M., Conway S.J., Cserjesi P., Laufer E., Firulli A.B. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat. Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohno K., Sadeh M., Blatt I., Brengman J.M., Engel A.G. E-box mutations in the RAPSN promoter region in eight cases with congenital myasthenic syndrome. Hum. Mol. Genet. 2003;12:739–748. doi: 10.1093/hmg/ddg089. [DOI] [PubMed] [Google Scholar]