Abstract
Viable constitutive and tamoxifen inducible liver-specific RNase H1 knockout mice that expressed no RNase H1 activity in hepatocytes showed increased R-loop levels and reduced mitochondrial encoded DNA and mRNA levels, suggesting impaired mitochondrial R-loop processing, transcription and mitochondrial DNA replication. These changes resulted in mitochondrial dysfunction with marked changes in mitochondrial fusion, fission, morphology and transcriptional changes reflective of mitochondrial damage and stress. Liver degeneration ensued, as indicated by apoptosis, fibrosis and increased transaminase levels. Antisense oligonucleotides (ASOs) designed to serve as substrates for RNase H1 were inactive in the hepatocytes from the RNase H1 knockout mice and in vivo, demonstrating that RNase H1 is necessary for the activity of DNA-like ASOs. During liver regeneration, a clone of hepatocytes that expressed RNase H1 developed and partially restored mitochondrial and liver function.
INTRODUCTION
RNase H hydrolyzes RNA in RNA–DNA hybrids (1). RNase H enzymes function as endonucleases that requires divalent cations (e.g. Mg2+, Mn2+) to produce cleavage products with 5′-phosphate and 3′-hydroxyl termini (2). RNase H activity is ubiquitous in eukaryotes and bacteria (3–8). Two classes of RNase H enzymes have been identified in mammalian cells (3,8–11). The biological roles for the human enzymes are not fully understood. However, RNase H2 seems to be involved in de novo DNA replication, and RNase H1 has been shown in mice to be important for mitochondrial DNA replication (12,13). Loss of RNase H1 function in humans results in adult-onset mitochondrial encephalomyopathy with chronic progressive external ophthalmoplegia, and multiple muscle weakness conditions (13).
Human RNase H1 has a hybrid binding domain homologous to yeast RNase H1 that is separated from the catalytic domain by a 62-amino acid spacer region (14–16). The catalytic domain is highly conserved from bacteria to humans, and contains the key catalytic residues, as well as lysine residues within the highly basic α-helical substrate binding region found in E. coli RNase H1 (16–20). The spacer region has also been shown to be required for RNase H activity (16). In addition, the catalytic domain has been shown to contain a unique redox switch formed by adjacent cysteine residues (21). Although the RNA binding domain is not required for RNase H activity, this region is responsible for the greater binding affinity of the human enzyme compared to the E. coli enzyme for the heteroduplex substrate, as well as the strong positional preference for cleavage exhibited by the enzyme (16,22).
A previous attempt to generate a constitutive knockout of RNase H1 in mice resulted in embryonic lethality (12). The exons encompassing the catalytic domains of RNase H1 were deleted by gene targeting, and loss of function required the loss of both alleles. Heterozygous RNase H1 knockout mice were viable, fertile and exhibited no obvious abnormalities. Homozygous RNase H1 knockout mice died by mid-gestation (E10.5) due to massive apoptosis (12). Compared to wildtype mice, homozygous RNase H1 knockout embryos exhibited reduced levels of the mitochondrial-encoded COX III DNA. This suggests that RNase H1 is involved in mitochondrial DNA replication. Evidence of mitochondrial dysfunction was also observed in the RNase H1 knockout embryos (12).
Human RNase H1 has been shown to play a dominant role in the activity of DNA-like antisense oligonucleotides (23). Overexpression or reduction of RNase H1 altered the potency of DNA-like ASOs in multiple cell lines, (23) confirming the role of RNase H1 in the activities of DNA-like ASOs. Moreover, overexpression of human RNase H1 in mouse liver increased the potency of a DNA-like ASO targeting Fas after intravenous administration (23).
To better understand the biological roles of RNase H1 and the role of the enzyme in activities of ASOs designed to serve as RNase H1 substrates, we developed liver-specific constitutive and inducible RNase H1 knockout mice expressing an inactive RNase H1 mutant. The RNase H1 knockout mice were viable, allowing us to monitor their liver health and function, and hepatocyte and mitochondria morphology, and function in the absence of functional RNase H1 protein. We found that RNase H1 is essential for proper liver function. RNase H1 is also required for mitochondrial R-loop clearance, transcription, DNA synthesis and mitochondria function. Furthermore, RNase H1 is necessary for the activities of ASOs designed to serve as RNase H1 substrates.
MATERIALS AND METHODS
Generation of RNase H1 liver-specific constitutive and inducible knockout mice
The generation of the RNase H1 floxed mice was performed by GenOway. Briefly, a targeting vector was constructed containing 3′ and 5′ homologous mouse RNase H1 sequences (GenBank accession number AF048992) with an FRT/PGK-neo cassette and flanking LoxP sites positioned at the junctions of exons 3 and 4 and 5 and 6. A detailed description of the targeting vector is available upon request. The correct assembly of the vector was confirmed by DNA sequencing. The targeting vector was linearized and electroporated into endothelial stem cells, followed by neomycin selection of clones positive for homologous recombination. Positive clones confirmed by Southern blot were injected into C57BL/6J blastocysts. C57BL/6J females were bred with ubiquitous Flp recombinase expressing mice (GenOway) to remove the neomycin cassette, which was confirmed by Southern blot. RNase H1 floxed mice were bred to homogeneity and verified by Southern blot. The homozygous RNase H1 floxed mice were bred with the B6.Cg-Tg(Alb-cre)21Mgn/J mouse strain (Jackson Laboratory) to generate homozygous liver-specific RNase H1 constitutive knockout mice. The homozygous RNase H1 inducible knockout mice were generated by breeding the RNase H1 floxed mice with B6.Cg-Tg(UBC-cre/ERT2) mice (IGBMC).
Animal monitoring
Animal experiments were conducted according to American Association for the Accreditation of Laboratory Animal Care guidelines and were approved by the institutions Animal Welfare Committee (Cold Spring Harbor Laboratory's Institutional Animal Care and Use Committee guidelines). Mice were maintained at a constant temperature of 23°C and were allowed standard lab diet and water ad libitum. As indicated, male mice were given a tamoxifen diet (Harlan). Body weights were recorded weekly. Blood was collected weekly or bi-weekly by tail bleed, and plasma was separated by centrifugation at 10 000 x g for 10 min at 4°C. Blood chemistries were determined on an AU400E Clinical Analyzer (Beckman Coulter).
Liver perfusion
Livers were perfused as previously described (24,25). Briefly, mice were anesthetized with an intraperitoneal injection of 0.1 ml per 10 g ketamine/xylazine. The inferior vena cava was catheterized and clamped. The liver was perfused with Hank's Balanced Salt Solution (Life Technologies), and the mesenteric vessel was cut for drainage. The liver was then perfused with collagenase (Roche). Following these perfusions, the liver was removed and gently expressed through sterile nylon mesh. Cells were washed in William's E medium containing 10% fetal calf serum, 10 mM HEPES pH 7.4, 2 mM L-glutamine and 1X antibiotic/antimycotic (all from Thermo Fisher Scientific).
Liver tissue fractionation
A portion of the whole liver cell suspension was centrifuged at 450 × g, washed with PBS containing 0.5% BSA (Sigma) and 2 mM EDTA (Thermo Fisher Scientific) and pelleted. The hepatocyte and non-parenchymal fractions were separated as described previously with the minor modifications (26) and is detailed in Supplemental Experimental Procedures.
RNA purification and PCR analysis of RNase H1 deletion mutant
Pellets were lysed and total RNA was purified using RNeasy Mini Kit (Qiagen). Approximately 1 μg of total RNA was converted to cDNA using M-MLV Reverse Transcriptase (Thermo Fisher Scientific) in accordance to the vender's specifications. A reverse primer, 5′-TCCATGAAGTCCTCCTTGTTG-3′, specific to chromosome 12, exon 7, was used. Approximately 2 μl of cDNA from this reaction was amplified using Taq Polymerase (Thermo Fisher Scientific), forward primer 5′-AGCAGAAGTCACAGGCGAA-3′, which is specific to chromosome 12, exon 3 and the reverse primer to exon 7. PCR products were run on 2% ethidium bromide gels (Lonza).
RNase H renaturation gel assay
The RNase H renaturation gel assay was performed as previously described (14). Briefly, cell pellets from whole liver, hepatocytes or non-parenchymal cells were lysed in RIPA buffer containing proteinase inhibitor cocktail. Around 33 μg of total protein was loaded onto a 10% denaturing SDS PAGE gel with approximately 200 000 counts of radio labeled RNA/DNA heteroduplex evenly incorporated throughout the gel. Gels were run for 1.5 h at 150 volts. The gel was washed in Buffer A (25% isopropanol, 50 mM Tris-HCl pH 8.0, 1 mM βME, 0.1 mM EDTA) three times for 20 min. Next, the gel was washed in Buffer B (0.01 M Tris-HCl pH 8.0 and 5 mM βME) twice for 15 min. Then, the gel was washed with Buffer C (50 mM Tris-HCl, pH 8.0, 20 mM βME, 10 mM MgCl2, 10% glycerol and 50 mM NaCl) twice for 20 min. The gel was then soaked in Buffer D (Buffer C with the addition of 2.5% NP-40) for 20 h with 3 buffer changes. The gel was then washed in Buffer E (5% TCA and 1% sodium pyrophosphate) three times for 20 min. Next, the gel was soaked overnight in the Buffer F (5% TCA). The gel was washed with water three times for 20 min and exposed overnight. All washes were performed at room temperature and all reagents were purchased from Sigma.
Analysis of RNase H1 and Cre mRNA expression levels
Hepatocyte cells were collected as described above (N = 4). Due to limited animal availability, there were two exceptions to the number of mice used in each group- week 8 (N = 2) and week 26 (N = 3). Hepatocyte cells were lysed and total RNA was purified using the RNeasy Mini Kit (Qiagen). mRNA was quantified by qRT-PCR as previously described (27). mRNA levels of interest were normalized to total RNA using RiboGreen (Life Technologies). See Supplemental Experimental Procedures for a complete list of primer probe sets.
Transmission electron microscopy
Mouse livers were perfused with primary fixative (3.4% formaldehyde, 1.25% glutaraldehyde and 0.2% picric acid in 0.1 M phosphate buffer, pH 7.2; all from Sigma). Samples were immersed in modified Karnovsky's fixative (2.5% glutaraldehyde and 2% paraformaldehyde in 0.15 M sodium cacodylate buffer, pH 7.4; all from Sigma) for 4 h, post-fixed in 1% osmium tetroxide in 0.15 M cacodylate buffer, pH 7.4 (Sigma) for 1 h and stained en bloc in 2% uranyl acetate (Sigma) for 1 h. Next samples were dehydrated in ethanol, embedded in Durcupan epoxy resin (Sigma), sectioned at 50 to 60 nm on a Leica UCT ultramicrotome and picked up on Formvar and carbon-coated copper grids. Sections were then stained with 2% uranyl acetate (Sigma) for 5 min and Sato's lead stain for 1 min. Grids were viewed using a Tecnai G2 Spirit BioTWIN transmission electron microscope equipped with an Eagle 4k HS digital camera (FEI).
Western blots
Western blots were performed as previously described (28). Blots were incubated with OPA1 antibody (Abcam) diluted 1:1000; DRP1 antibody (Abcam) diluted 1:1000; and GAPDH (Santa Cruz Biotechnology) diluted 1:3000. Signals were detected using horseradish peroxidase-conjugated secondary antibody and Luminata Crescendo Western HRP substrate reagent (Millipore). Blots were imaged using a Chemidoc Imager (Bio-Rad).
Transcriptome analysis
Total RNA was purified from hepatocytes using the miRNeasy mini kit (Qiagen). RNA integrity was evaluated using the 2100 Agilent Bioanalyzer. Samples were depleted of ribosomal RNA and sequenced at Expression Analysis of Q2 Solutions using stranded TruSeq sequencing protocol (http://www.illumina.com/products/truseq_stranded_total_rna_library_prep_kit.html). Sequenced reads were mapped using rnaSTAR to the mouse reference genome GRCm38/mm10 using corresponding refseq gene models (29). Differentially expressed gene expression ratios and significance calls between replicate experiments were calculated with cufflinks using default parameters (30), in which each knockout condition was compared to a Cre-minus control. Pathway analysis was performed using the DAVID Bioinformatics Resource (https://david.ncifcrf.gov) as previously described (31).
Purification and real-time PCR of mitochondrial DNA
Around 10 mg of mouse liver was incubated with Proteinase K (Sigma) overnight at 37°C. The digested lysate was purified using QIAamp DNA Micro spin columns (Qiagen), per manufacturer's instructions. Real-time PCR was performed on the isolated DNA using Express One Step Superscript Kit (Thermo Fisher Scientific) without the Reverse Transcriptase enzyme. See Supplemental Experimental Procedures for a complete list of primer probe sets.
Mitochondrial R-loop detection
Five million hepatocytes were plated per 150-mm dish in William's E medium containing 10% FBS, 2 mM L-glutamine, 1X antibiotic/antimycotic, 10 mM HEPES pH 7.4 (all from Thermo Fisher Scientific) and incubated overnight at 37°C in 5% CO2. Cells were fixed in 1% formaldehyde (Sigma) for 20 min at room temperature, 2 ml of 1 M -glycine (125 mM final) (Sigma) was added and samples were incubated for 10 min. Cells were scraped into tubes and centrifuged at 100 × g for 10 min. Pellets were then washed with PBS, and recovered cells were resuspended and counted. Pellets were frozen and then lysed in 1 ml of IP Buffer (Pierce). Lysates were sonicated using a Branson 450 Sonifier at 10% amplitude for 15 s and allowed to chill for 45 s in between cycles, for a total of 16 cycles. Lysates were cleared by centrifugation at 10 000 × g for 10 min at 4°C.
Proteins in cleared lysates were quantified using the DC Protein Assay (Bio Rad). For samples spiked with RNase H1, 50 μg of total protein from sonicated lysate was incubated with 18 or 90 mg of E. coli RNase H1 (New England BioLabs), 3 mM MgCl2 and 1 × RNase H1 buffer (New England Biolabs) prior to immunoprecipitation. For non-spiked samples, 50 μg of total protein was immunoprecipitated using anti-DNA-RNA hybrid (S9.6) antibody (Kerafast). The remaining steps were performed using the EZ-Magna Chip A/G bead kit (Millipore) beginning at the immunoprecipitation step (step five), per the manufacturer's instructions. See Supplemental Experimental Procedures for a complete list of primer probe sets.
ASO activity
For in vitro analyses of ASO activity, hepatocytes were isolated as described above in ‘liver perfusion.’ Cells were seeded at 12 000 cells/well in wells of a 96-well plate. After incubation at 37°C in 5% CO2 for 2 h, transfection was performed at the indicated doses as described previously (32). To evaluate ASO activity in mice, mice aged 6 weeks, were treated with a single subcutaneous administration of ASO. Inducible mice were treated 1 week post initiation of tamoxifen treatment to induce disruption of RNase H1 expression prior to treatment with ASO. Necropsy was performed as described previously (32). See Supplemental Experimental Procedures for a complete list of primer probe sets and ASOs.
RESULTS
Establishment of liver specific RNase H1 constitutive and inducible knockout mice
Previous studies have shown that site directed mutagenesis of any one of the three catalytic amino acids renders RNase H1 catalytically inactive (16). The codons for the catalytic amino acids are located in exons 3, 4 and 5 of the gene encoding RNase H1. LoxP sites were inserted flanking exons 4 and 5 of the mouse RNase H1 gene, which contain 2 of the 3 key catalytic amino acids (i.e. E186 and D210 Figure 1A). Mice containing the RNase H1 gene with the flanking LoxP sites (RNase H1 floxed) were bred to homogeneity. The RNase H1 floxed mice were then bred with mice hemizygous for the expression of the Cre recombinase gene coupled to an albumin promoter to generate constitutive liver-specific RNase H1 knockout mice. These mice were referred to as cKO mice and the corresponding RNase H1 floxed mice were referred to cControl mice. Due to the albumin promoter, the expression of Cre recombinase coincides with the expression of albumin in the liver, approximately 15 days post fertilization. This resulted in recombination at the LoxP sites of the RNase H1 gene to produce an RNase H1 mutant in which the region containing the catalytic amino acids E186 and D210was deleted after organogenesis (Figure 1A). An inducible liver-specific RNase H1 knockout murine line was also generated by breeding the RNase H1 floxed mice with mice containing hemizygous Cre recombinase gene, coupled to an albumin promoter, controlled by estrogen receptor activation. RNase H1 knockout in these mice was induced by treatment with tamoxifen. These mice were referred to as iKO mice and the corresponding RNase H1 floxed mice were referred to as iControl (Figure 1A). We note that, unless indicated in figure legends, the cKO mice used in this study were 6 weeks old. All iKO mice were treated with tamoxifen at approximately 5 weeks of age to induce RNase H1 deletion. iKO tamoxifen induction lasted for 5 days.
Figure 1.
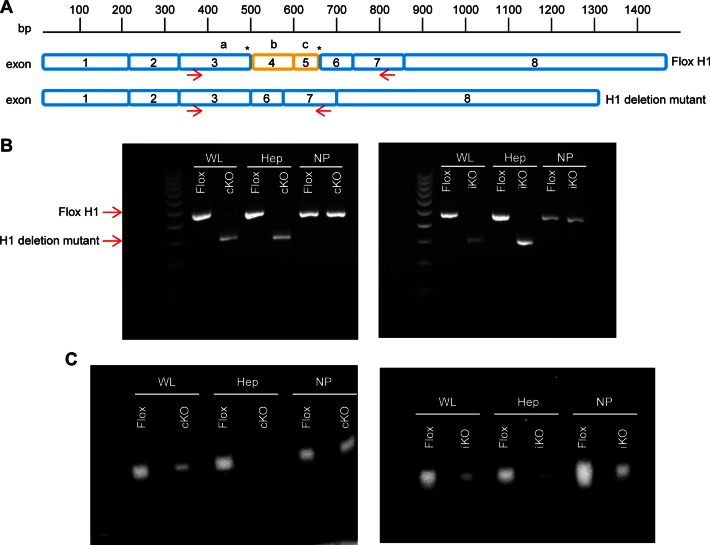
RNase H1 deletion mutant is expressed in hepatocytes from knockout mice and is catalytically inactive. (A) Schematic of RNase H1 mRNA with LoxP sites indicated by asterisks, and schematic of deletion mutant mRNA missing exons 4 and 5. Arrows indicate the binding sites of PCR primers. The relative positions of the three catalytic amino acids D145, E186 and D210 are indicated by a, b and c, respectively. (B) PCR products and (C) RNase H1 activity in whole liver (WL), hepatocytes (Hep) and non-parenchymal cells (NP) from RNase H1 floxed mice (Flox), constitutive knockout (cKO) mice and inducible knockout (iKO) mice. Mice were sacrificed at 6 weeks of age. iKO mice were treated with tamoxifen for 1 week prior to sacrifice. WL, Hep and NP fractions were separated from one mouse liver. The experiments were performed three times and representative results are shown.
We determined the expression levels of the RNase H1 for all strains in total liver tissue, hepatocytes and non-parenchymal cells by PCR using forward and reverse primers targeting, respectively, exons 3 and 7 (Figure 1A). We note that, unless indicated in figure legends, the mice used in this study were 6-week old.
PCR products consistent with full-length floxed RNase H1 mRNA were observed in liver tissue from the cControl and iControl mice. However, the PCR products observed with samples from the 6 week cKO mice were consistent with the deletion of exons 4 and 5 (Figure 1B). Expression of the RNase H1 deletion mutant appeared to occur in the hepatocytes and not the non-parenchymal cells from the cKO mice (Figure 1B). A similar expression pattern was observed 1 week following tamoxifen treatment of the iKO mice (Figure 1B). Importantly, the efficacy of the inducible knockout model was demonstrated by the exclusive hepatocyte expression of the mRNA corresponding to the RNase H1 deletion mutant instead of full-length floxed RNase H1 mRNA in tamoxifen-treated iKO mice (Figure 1B).
RNase H1 activity was determined in total liver tissue, hepatocytes and non-parenchymal cells from the control and knockout mice (Figure 1C) using a previously described renaturation assay (33). The denaturant was removed allowing RNase H1 to renature in the gel, and the presence of active enzyme was detected by the in situ degradation of the radio labeled RNA/DNA heteroduplex. RNase H1 activity was observed in samples from the total liver tissue, hepatocytes and non-parenchymal cells from the control mice (Figure 1C). RNase H1 activity was also observed in the non-parenchymal cells from the cKO mice, consistent with the expression of full-length floxed RNase H1 mRNA in the non-parenchymal cells (Figure 1B and C). In contrast, no RNase H1 activity was observed in the hepatocytes from the cKO mice demonstrating that the RNase H1 deletion mutant was catalytically inactive (Figure 1C). Thus, the modest RNase H1 activity observed in the total liver tissue from the cKO mice was likely due to the expression of full-length floxed RNase H1 mRNA in the non-parenchymal cells, which make up approximately 20% of the total liver cell mass. This is also consistent with the observations in Figure 1B that in the whole live sample of KO mice, trace amount of WT mRNA could be detected.
A similar RNase H1 activity profile was observed in the tamoxifen-treated iKO mice. No RNase H1 activity was observed in the hepatocytes (Figure 1C). Reduced RNase H1 activity was observed as early as 2 days post tamoxifen treatment and complete loss of RNase H1 activity was observed at day 5 (data not shown).
RNase H1 knockout results in mitochondrial dysfunction and liver degeneration
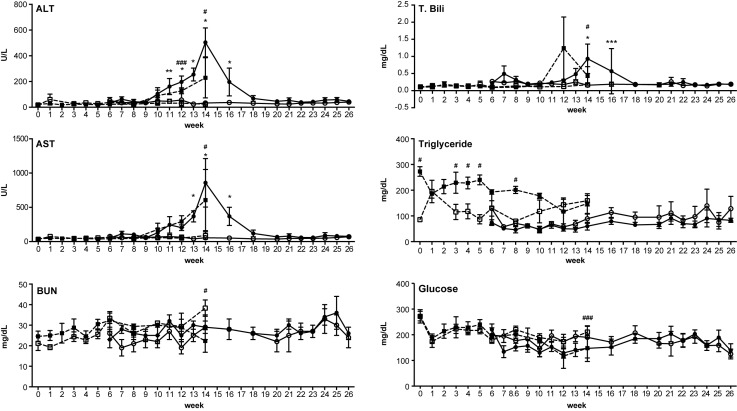
Liver function and health were monitored in the cKO mice ranging in age from 6 to 26 weeks, and in 6 week iKO mice ranging from 1 to 14 weeks post tamoxifen treatment (Figure 2, Supplementary Figure S1A–F). Normal transaminase levels were observed in both cKO and iKO mice up to week 10 (Figure 2). Elevated transaminase levels were observed in the tamoxifen-treated iKO mice from weeks 11 to 14, and in the cKO mice from weeks 11 to 16, suggesting impaired liver function (Figure 2). After week 14, the transaminase levels observed in the cKO mice appeared to return to normal, suggesting that the impaired liver function was transient (Figure 2, Supplementary Figure S1A–F). No significant changes in glucose, or blood urea nitrogen were observed in the cKO mice (Figure 2 and data not shown). Relatively transient, but statistically significant changes in total bilirubin were observed at week 14 and 16 in cKO mice, and a slight increase in triglyceride serum levels was observed in the tamoxifen-treated iKO mice (Figure 2).
Figure 2.
RNase H1 knockout impairs liver function. Plasma levels of alanine transaminase (ALT), aspartate transaminase (AST), blood urea nitrogen (BUN), total bilirubin (T. Bili), triglycerides and glucose were determined for cKO mice (solid circles, solid lines, N = 4), iKO mice treated with tamoxifen beginning at week 6 (solid boxes, dashed lines, N = 4), untreated floxed mice (open circles, solid lines, N = 4) and tamoxifen-treated floxed mice (open boxes, dashed lines, N = 4). The cKO mice were monitored from ages 6 to 26 weeks of age, and the iKO mice were continuously monitored 1 week prior to tamoxifen treatment, beginning at 6 weeks of age, until death or extreme illness, which occurred at 14 weeks of age. Data are the means of four animals and ± SEM. Also see Supplementary Figure S1. GraphPad two-way ANOVA calculations were used to determine significance. * (cKO) or # (iKO) P < 0.0001, ** (cKO) or ##(iKO) P < 0.001, *** (cKO) or ###(iKO) P < 0.01.
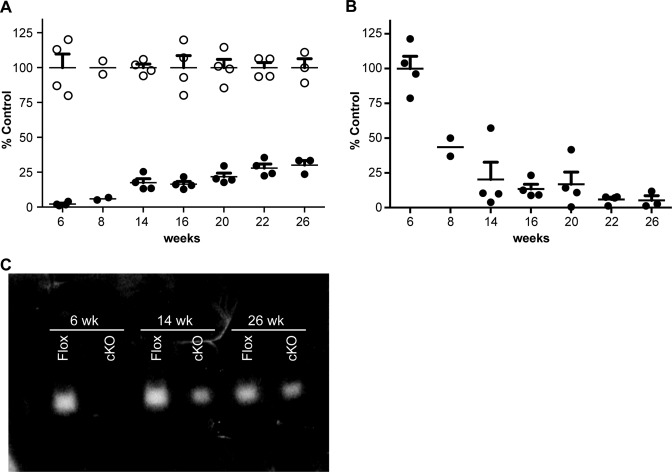
Given the transient elevation in transaminase levels observed in the cKO mice, the expression levels of floxed RNase H1 mRNA in hepatocytes were monitored from week 6 to 26 (Figure 3A). Little to no floxed RNase H1 mRNA expression was observed at weeks 6 and 8, but an increase in expression was observed from weeks 14 to 26, with the maximum floxed RNase H1 mRNA levels reaching 25% of the levels in cControl mice (Figure 3A). The increase in floxed RNase H1 mRNA expression in the hepatocytes from the cKO mice coincided with a reduction in Cre mRNA expression (Figure 3B). Consistent with the increase in floxed RNase H1 mRNA expression, RNase H1 activity was observed in the hepatocytes from the cKO mice at weeks 14 and 26, although at a fraction of the activity observed for the cControl (Figure 3C). Taken together, these data suggest that elevated transaminase levels indicative of impaired liver function correlated with loss of RNase H1 activity, and that improved liver function coincided with the expression of active floxed RNase H1 (Figures 2 and 3). These data further suggest that impaired liver function leads to proliferation of hepatocytes [regeneration]. During this process, hepatocyte clones that do not express Cre, thus do express RNase H1, have survival advantages over hepatocytes with no RNase H1. These clones probably repopulate the liver.
Figure 3.
RNase H1 expression and activity in constitutive RNase H1 knockout mice as a function of age. (A) RNase H1 mRNA expression levels in hepatocytes from RNase H1 floxed (open circles) and cKO mice (solid circles) were determined by qRT-PCR. (B) Cre mRNA expression levels in hepatocytes from cKO mice (solid circles) were determined by qRT-PCR. Data are the means of four animals per group, with the exception of week 8 (n = 2) and week 26 (n = 3). The error bars represent ± SEM. (C) Gel renaturation assay showing RNase H1 activity in hepatocytes from 6, 14 and 26 week old RNase H1 floxed (Flox) and cKO mice. Each lane is representative of hepatocytes RNase H activity from one mouse. The experiments were performed three times and representative results are shown.
Because the iKO mice died or were in extremis and were sacrificed, we cannot comment on whether the liver function in these mice might have recovered. However, just as in the cKO mice, the tamoxifen induced 14 week iKO mice had reduced Cre and elevated floxed RNase H1 mRNA expression levels (data not shown). We speculate, therefore, that liver regeneration in iKO mice would have developed had they not been sacrificed.
To better understand the roles of RNase H1 in liver function, histological analyses were performed on the liver tissue from cKO mice at weeks 6, 14 and 26, and 1 week after tamoxifen treatment for the iKO mice. The livers from the cControl mice at weeks 6, 14 and 26 exhibited normal morphology with no signs of liver disease (Supplementary Figure S1A3–5). The livers from the iControl and iKO mice treated with tamoxifen for 1 week appeared normal, aside from signs of minor hepatocyte swelling, consistent with the effects of tamoxifen treatment (Supplementary Figure S1A1–2). In contrast, morphology of the liver in cKO mice at 6 and 14 weeks exhibited variable levels of degenerative hepatocytes, manifested by hepatocellular polyploidy, hepatocellular hypertrophy, single cell apoptosis and megalocytosis with marked nuclear pleomorphism, often with cytoplasmic hypereosinophilia (Supplementary Figure S1A6–7). The week 14 cKO mice exhibited severe liver degeneration, including hypereosinophilic coagulative cytoplasm with marked nuclear pleomorphism and islet necrosis (Supplementary Figure S1A7). Other than mild hepatocellular swelling and increased signs of mitosis, the liver appearance in cKO mice at 26 weeks was relatively normal (Supplementary Figure S1A8). These observations are consistent with the peak transaminase levels observed in these animals (Figure 2), demonstrating both histological and functional evidence of severe liver injury at week 14.
A modest decrease in glycogen levels was observed in the livers from the cKO mice relative to cControl at week 6 and 14 (Supplementary Figure S1B6–7), whereas a marginal decrease in glycogen levels was observed at week 26 (Supplementary Figure S1B8). Although no evidence of hepatocyte proliferation was observed in the cKO mice at week 26 (Supplementary Figure S1C8), at weeks 6 and 14 a modest increase in hepatocyte proliferation consistent with liver regeneration was observed (Supplementary Figure S1C6–7). cKO mice had reduced COX IV levels at weeks 6 and 14, suggesting mitochondrial dysfunction, but by week 26, COX IV levels returned to normal (Supplementary Figure S1D6–8). Signs of modest apoptosis were also observed in the livers from the cKO mice at weeks 6, 14 and 26 (Supplementary Figure S1E6–8). Finally, staining consistent with mild fibrosis was observed in livers from the cKO mice at week 26 (Supplementary Figure S1F8). This suggests that liver damage was substantial enough to lead to cirrhosis, which would have progressed had repopulation of the liver with hepatocytes expressing RNase H1 not occurred.
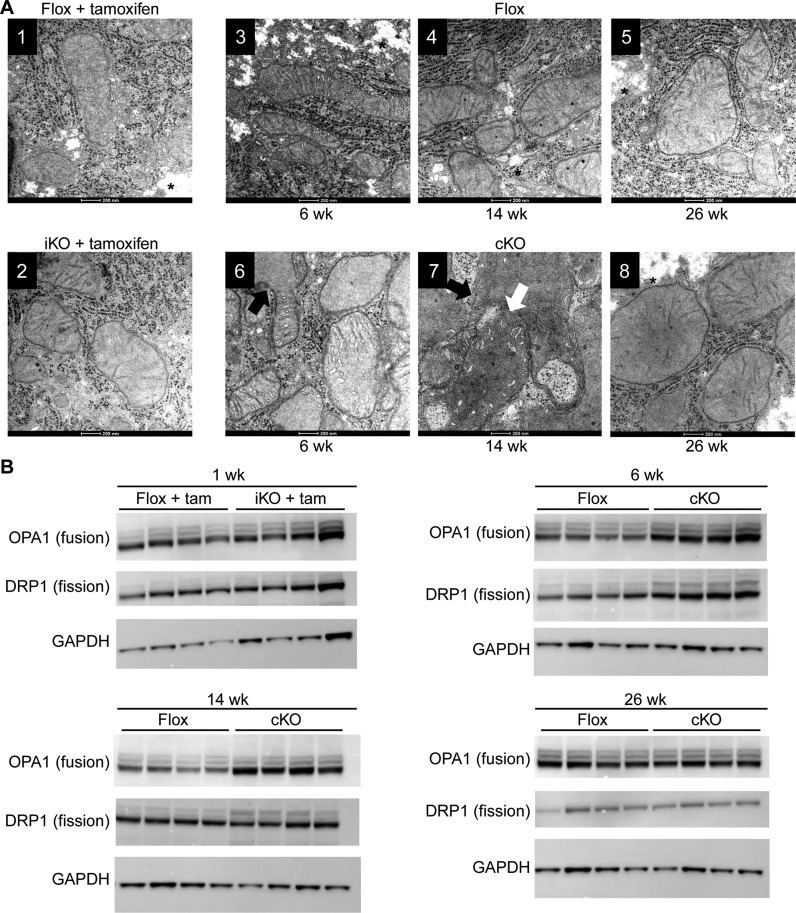
Next, we performed electron microscopy to evaluate the morphology of the mitochondria in livers of the iKO mice 1 week post tamoxifen treatment, and the cKO mice at weeks 6, 14 and 26 (Figure 4A). Normal mitochondria were observed in the hepatocytes of the control mice (Figure 4A1, 3–5, Supplementary Figure S1D). After one week of tamoxifen treatment, the hepatocytes of the tamoxifen-treated iKO mice showed mild to moderate mitochondrial swelling (Figure 4A2). A significant increase in mitochondrial size and disorganization, including branching, fusion and loss of cristae, was observed in both 6 and 14 week cKO time points (Figure 4A6, 7, Supplementary Figure S1D). Consistent with light microscopy findings, reduction of glycogen was also observed in the hepatocytes of the cKO mice at week 6 and 14 (Figure 4A6, 7, Supplementary Figure S1B). A modest increase in mitochondria number was also observed for the hepatocytes from cKO mice at week 6 and14 (Figure 4A6, 7), suggesting an ongoing fission process. The mitochondria from the livers of the 26 week cKO mice exhibited slight swelling but otherwise the structures and number appeared normal compared to the cControl mice (Figure 4A8).
Figure 4.
Morphology and biochemical characteristics of hepatocyte mitochondria from RNase H1 knockout mice. Liver tissue was harvested from control RNase H1 floxed mice (flox + tam) and inducible RNase H1 knockout mice (iKO + tam) 1 week post tamoxifen treatment and control RNase H1 floxed mice (flox) and constitutive RNase H1 knockout mice (cKO) at weeks 6, 14 and 26. (A) Transmission electron micrograph. The white arrow indicates mitochondrial fusion and the black arrow indicates mitochondrial branching. * indicates glycogen appearance. (B) Western blots of proteins involved in mitochondrial fusion (OPA1) and fission (DRP1). The house-keeping gene GAPDH was used as a loading control for the western analysis. Each lane shows the expression levels of these proteins in individual mice (N = 4). Also see Supplementary Figure S1D.
The morphology of the mitochondria in the livers from the cKO mice indicated that the mitochondria were undergoing both fusion and fission processes. To confirm this, we determined the expression levels of the key proteins involved in mitochondrial fusion and fission (Figure 4B). Consistent with the observed morphology of the mitochondria, a modest increase in the levels of the mitochondrial fusion protein OPA1 and fission protein DRP1 were observed for the livers in samples from the iKO mice at 1 week post tamoxifen treatment, compared to the livers from the iControl mice (Figure 4B) (34). A greater increase in the level of OPA1 was observed in the livers from the cKO mice at weeks 6 and 14, with the greatest increase in DRP1 levels observed at week 6 (Figure 4B). Again, consistent with the electron microscopic observations, by week 26 the levels of the fusion and fission proteins in livers from the cKO mice were comparable to the cControl mice (Figure 4B).
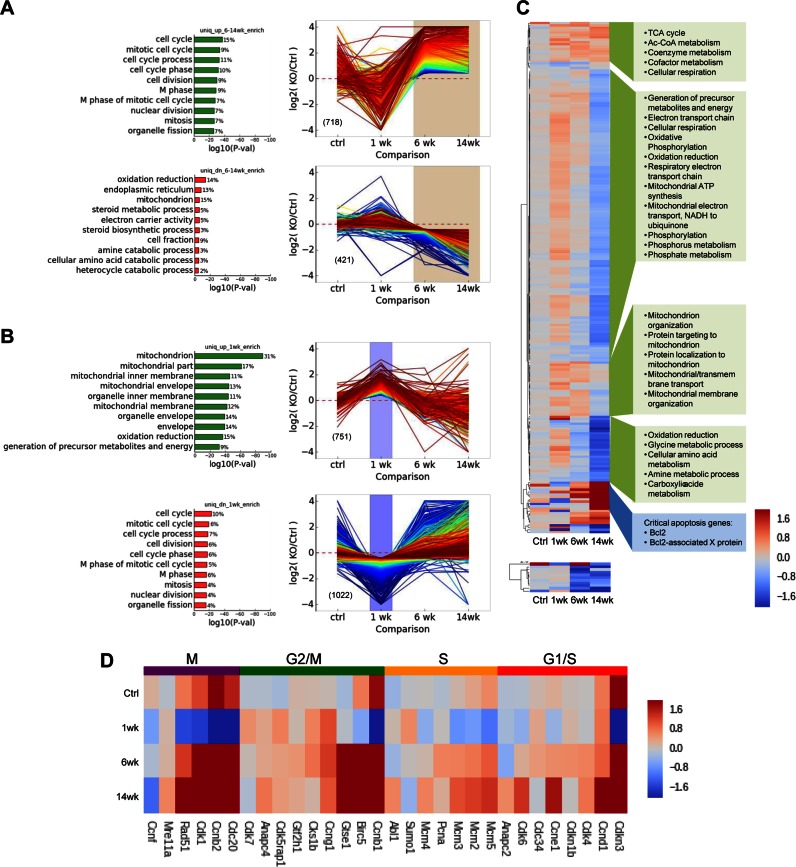
Finally, transcriptome-wide analyses were performed on the purified hepatocytes from the tamoxifen-treated iControl mice, the tamoxifen-treated iKO mice, the cControl mice and the cKO mice (Figure 5). Within 1 week following tamoxifen treatment, the hepatocytes from the iKO mice exhibited lowered expression of the mRNAs associated with cell replication, compared to the tamoxifen-treated iControl mice (Figure 5A top left and D). In addition, increased expression of nucleus encoded mitochondrial mRNAs was observed in the hepatocytes from these animals compared to the iControl mice (Figure 5C). However, expression of the mitochondrially encoded mRNAs of the hepatocytes from the tamoxifen-treated iKO mice was comparable to the iControl mice (Figure 5C). Increased expression of mRNAs associated with cell cycle M-phase, and reduced expression of nuclear and mitochondria-encoded mitochondrial mRNAs were observed for the hepatocytes from the 6 and 14 week cKO mice, compared to the cControl mice, indicating compromised mitochondrial function (Figure 5B and D). Note also that the expression of transcripts for electron transport proteins declined steadily in the iKO mice (Figure 5A lower panel) and in the cKO (Figure 5B).
Figure 5.
Transcriptome analysis of RNaseH1 knockout in hepatocytes. Differential gene expression patterns and geneset enrichment analysis (A) Expression of transcripts uniquely enriched or depleted after 1 week tamoxifen induction in iKO hepatocytes relative to inducible control. (B) Transcripts uniquely enriched or depleted in 6 and 14 week cKO hepatocytes relative to constitutive 6 and 14 week controls respectively. The top 10 GO genesets (Biological Process) with minimum P value of 0.001 are shown (P values are adjusted for multiple hypothesis testing); percentage of differentially expressed genes within the specified class is given to the right of each bar. Gene expression profiles (Log2 fold change relative to appropriate control conditions) are colored by rank order. (C) Expression heatmap of genes encoded in the nucleus (upper) and the mitochondria (lower). Hierarchical clustering identified five major gene expression clusters among nuclear-encoded genes. Expression is shown as Log2 fold change relative to appropriate control condition. Enriched for GO Biological Processes are shown in green boxes (P < 0.005) or individual critical pathways (blue box). (D) Representative genes were selected for each of the indicated phases of the cell cycle and those for which we observed at least one knockout condition exhibiting significant differential expression are included. Expression is shown as Log2 fold change relative to appropriate control condition.
RNase H1 knockout mice exhibit impaired mitochondrial DNA replication and R-loop processing
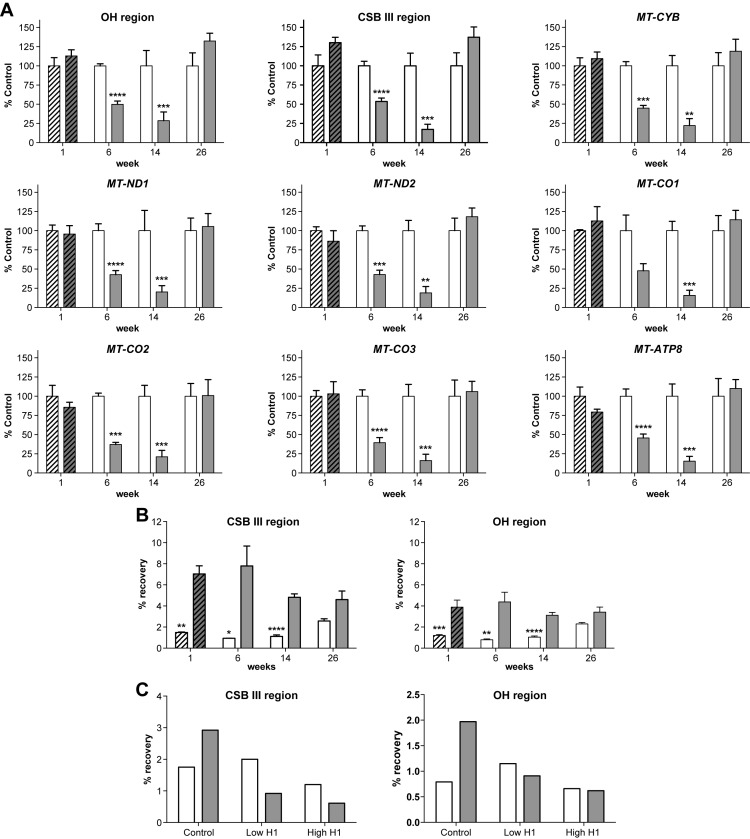
To better understand the nature of the observed changes in mitochondrial morphology and function, the levels of the seven mitochondrial-encoded genes were determined in the livers of the control and knockout mice by PCR (Figure 6A). PCR was also performed on the conserved sequence block III (CSB III) and OH regions of the mitochondrial genome corresponding to, respectively, the S7 R-loop and S7 D-loop regions (35–37). With the exception of CSB III, the DNA levels observed in the livers from the tamoxifen-treated iKO mice at one week were comparable to, or slightly lower than iControl mice (Figure 6A). This is consistent with the modest effects on mitochondrial morphology observed after one week of tamoxifen treatment. Consistent with the transcriptome analysis, an approximate 50% reduction in the levels of the transcripts from all the mitochondrial genes tested in samples from the livers of the 6 week cKO mice was observed (Figures 5C and 6A). A greater reduction in levels of these genes was observed in the livers from the cKO mice at week 14 (Figure 6A). By week 26, the mitochondrial DNA levels for the livers from the cKO mice were comparable to the cControl livers (Figure 6A). Finally, the reduction in mitochondrial DNA levels observed for the cKO mice was consistent with the reduction in the mitochondrially-encoded mRNA levels observed with the transcriptome analysis, suggesting that RNase H1 plays a role in mitochondrial DNA replication (Figure 6).
Figure 6.
Levels of mitochondrial DNA reduced and R-loop processing impaired in knockout livers. (A) Levels of mitochondrial DNA were determined by PCR. The liver tissue from the tamoxifen-treated RNase H1 floxed (hashed open bars) and iKO (hashed solid bars) mice were harvested 1 week after initiation of tamoxifen treatment. The liver tissue from RNase H1 floxed (open bars) and cKO (solid bars) mice were harvested at 6, 14 and 26 weeks of age. Data are the means of four animals per group and error bars represent ±SEM. (B) S7 R-Loop levels determined in hepatocytes from the tamoxifen-treated RNase H1 floxed (hashed open bars) and iKO (hashed solid bars) mice were harvested 1 week after initiation of tamoxifen treatment. Hepatocytes from RNase H1 floxed (open bars) and cKO (solid bars) mice were harvested at 6, 14 and 26 weeks of age. Data are the means of four animals per group and error bars represent ±SEM. (C) S7 R-Loop levels determined after treatment of the hepatocyte samples from 6-week-old RNase H1 floxed (open bars) and cKO (solid bars) mice with 90 mg (high) or 18 mg (low) of E. coli RNase H1 prior to immunoprecipitation with the heteroduplex antibody. The figure shows results from a single experiment. GraphPad two-way ANOVA calculations were used to determine significance.* P < 0.0001 ** P < 0.001, *** P < 0.01 and **** P < 0.05.
To determine whether RNase H1 also plays a role in mitochondrial R-loop processing, RNA/DNA hybrids were immunoprecipitated from the hepatocytes of the RNase H1 knockout mice. S7 R-loop levels were determined using PCR primers targeting the CSB III or OH regions of the mitochondrial genome, which are complementary to or immediately downstream of the S7 RNA, respectively. Elevated levels of the S7 R-loop were observed in the hepatocytes from the tamoxifen-treated iKO mice 1 week post induction and the cKO mice at weeks 6, 14 and 26 compared to the levels in hepatocytes from iControl and cControl mice (Figure 6B). In addition, the signals for S7 R-loop using the CSB III PCR primer set targeting the region adjacent to the S7 RNA were higher compared to the downstream OH PCR primer set (Figure 6B). To ensure that the PCR products were derived from immunoprecipitated RNA/DNA heteroduplex, samples were treated with E. coli RNase H1 prior to immunoprecipitation (Figure 6C). The control cKO mice samples not treated with E. coli RNase H1 again showed elevated levels of S7 R-loop compared to the cControl mice (Figure 6C). In contrast, the immunoprecipitated samples from the cKO mice digested with the E. coli enzyme showed S7 R-loop levels comparable to the cControl samples (Figure 6C). These data demonstrate that the PCR products correspond to immunoprecipitated heteroduplex. In addition, the modest S7 R-loop levels observed for cControl mice were similar to the E. coli RNase H1 treated samples, suggesting that the control signal was not from immunoprecipitated heteroduplex. The accumulation of S7 R-loop observed for the iKO and cKO mice suggests that RNase H1 is involved in degrading S7 RNA.
RNase H1 is required for the activity of ASOs designed to serve as RNase H1 substrates
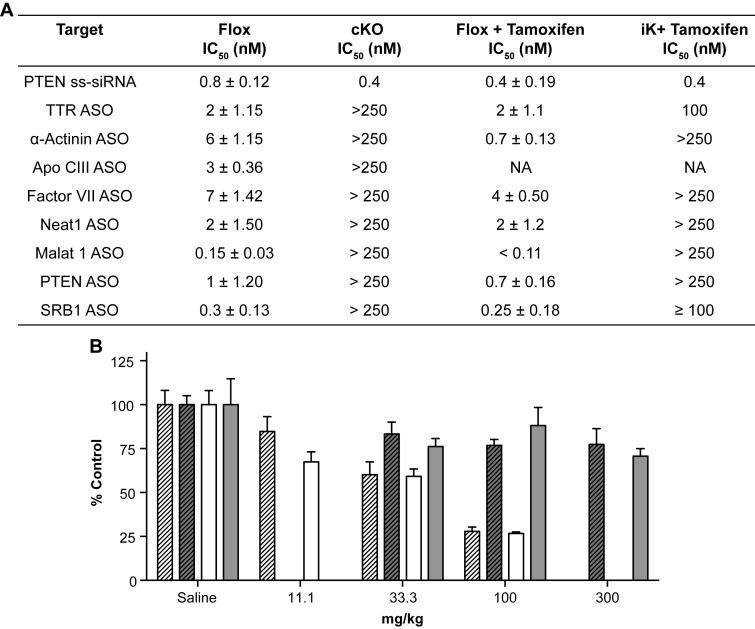
To better understand the contribution of the RNase H1 to the activities of ASOs designed to serve as RNase H1 substrates, hepatocytes from the control and knockout mice were transfected with ASOs targeting a diverse set of mRNAs (Figure 7A). Potencies ranging from the sub-nM to low-nM were observed in both iControl and cControl hepatocytes transfected with the various ASOs (Figure 7A). Conversely, no ASO activity was observed in the hepatocytes from either the cKO mice or tamoxifen-treated iKO mice (Figure 7A).
Figure 7.
ASO activity impaired in RNase H1 knockout mice. (A) ASO activities in hepatocytes from RNase H1 floxed or knockout mice transfected with ASO at doses ranging from 40 pM to 250 nM. Hepatocytes transfected with ASO, and mRNA levels were evaluated 48-h post transfection. IC50 correspond to the ASO concentrations resulting in 50% reduction of the target mRNA levels compared to the lipid-only treated cells. Data are the mean of triplicate determination and ± SEM. All changes between flox and knockout animals were significant, P < 0.001 and calculated using standard t-test in GraphPad software. (B) ASO activity was determined following a single subcutaneous administration at the indicated doses in mg/kg. F7 mRNA levels from the tamoxifen-treated RNase H1 floxed (hashed open bars), tamoxifen-treated iKO (hashed solid bars), RNase H1 floxed (open bars) and cKO (solid bars) at 6 weeks of age are reported as a percentage of levels in the saline-treated control group. Data are the means of four animals and ±SEM. Changes between flox and knockout animals were significant, P < 0.001 and calculated using standard t-test in GraphPad software.
Next, we determined the in vivo activity for an ASO designed to serve as an RNase H1 substrate targeting the hepatocyte specific Factor VII mRNA (Figure 7B). The cControl and cKO mice were treated with a single subcutaneous dose of the ASO and sacrificed 48 h post treatment (Figure 7B). The iControl and iKO mice were administered the ASO 1 week following the tamoxifen treatment and sacrificed 48h later. A dose dependent reduction in the target mRNA levels was observed in both iControl and cControl mice treated with the ASO, resulting in an approximate 75% reduction of the target mRNAs at the 100 mg/kg dose (Figure 7B). However, no ASO activity was observed for either the cKO or iKO mice at the 300 mg/kg dose (Figure 7B). These data unequivocally demonstrate that RNase H1 is necessary to degrade the RNA targeted by such ASOs.
DISCUSSION
In this study, we developed and characterized viable liver-specific RNase H1 constitutive and inducible knockout mice. The inducible knockout system was remarkably efficient, with reduced expression and activity of RNase H1 in the liver as early as 2 days after initiation of tamoxifen treatment and complete loss of RNase H1 floxed expression and activity by day 5 through week 14 (Figure 1 and data not shown). The cKO mice also displayed liver-specific loss of RNase H1 expression approximately 15 days post fertilization to week 14, at which point a rebound in RNase H1 floxed mRNA expression was observed, reaching approximately 25% of the cControl mice by week 26 (Figures 1 and 3). The rebound in floxed RNase H1 expression in the livers from the cKO mice coincided with a decline in Cre mRNA expression and with peak transaminase levels. This suggests liver degeneration and regeneration likely contributed to the observed drop in the Cre expression (Figures 2, 3 and Supplementary Figure S1A6, 7). During the period in which the peak of degenerative changes was detected in the cKO mice, signs of liver regeneration were also observed (Supplementary Figure S1C6, 7). We speculate that the proliferation of a clone or clones of hepatocytes that do not express Cre, and therefore do express RNase H1, may have contributed to the return in RNase H1 floxed expression (Supplementary Figure S1C and D). Interestingly, the rebound in RNase H1 floxed expression was sufficient to restore liver function, but not fully reverse liver degeneration (Figures 2, 3 and Supplementary Figure S1A–F).
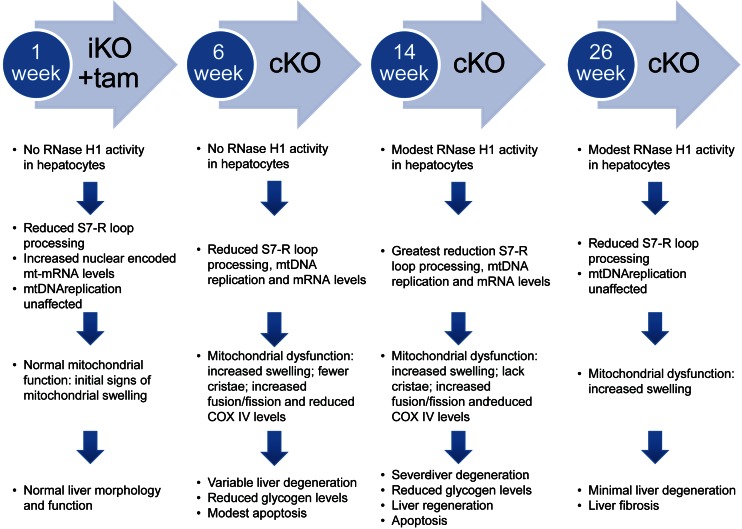
Taken together, our analysis of the liver-specific viable RNase H1 knockout mice supports a more detailed understanding of the roles of mammalian RNase H1 in hepatocytes and the molecular events that result in liver toxicity and degeneration (Figure 8). The primary injury appeared to be in mitochondria and most likely resulted from the inability to clear S7 R-loops. Initially the liver attempted to compensate for mitochondrial dysfunction by increasing mitochondrial mRNA to maintain homeostasis. Despite these efforts, there were moderate increases in mitochondrial fission and fusion. As S7 R-loops increased, mitochondrial dysfunction, reductions in mitochondrial DNA synthesis and transcription led to changes in mitochondrial number and morphology. Reduced COX IV levels led to modest apoptosis and early liver degeneration ensued (38). As this process continued, hepatocyte morphology and performance worsened, resulting in death of hepatocytes, reduced glycogen levels, severe liver degeneration and increased serum transaminases. By week 26, evidence of fibrosis was present despite liver regeneration.
Figure 8.
Progression of mitochondrial dysfunction and liver disease absent functional RNase H1. Summary of the observations of RNase H1 iKO and cKO mice.
Post week 14, Cre expression declined and RNase H1 floxed expression in hepatocytes increased. We speculate that during liver regeneration a clone(s) of hepatocytes that regained RNase H1 floxed expression developed as demonstrated by the increased levels of RNase H1 floxed mRNA transcript and RNase H1 activity. This fortuitous event supported evaluation of the molecular events underlying recovery of the hepatocytes from injury. In this case, recovery appeared to be simply the reverse of processes that led to liver injury.
For the first time, an evaluation of viable RNase H1 knockout mice clearly demonstrated in vivo that mammalian RNase H1 is required for the effective removal of mitochondrial S7 R-loops. Additionally, the failure to transcribe essential mitochondrial DNA was observed. This in turn led to mitochondrial dysfunction, loss of mitochondria and apoptosis.
RNase H1 has been implicated in the activities of ASOs designed to serve as the RNase H1 substrates (23). Specifically, overexpression of RNase H1 enhanced the potency of ASOs in human cells. Conversely, when RNase H1 was reduced in cells by approximately 90%, a 2- to 3-fold reduction in ASO potency was observed (23). Although these data demonstrated that RNase H1 contributed to the activities of ASOs, the possibility that other enzymes may also contribute to the activities of ASOs could not be excluded. For example, RNase H2 and Flap endonuclease 1 (FEN1) have been shown to degrade RNA/ DNA hybrids (39,40). The RNase H1 knockout mice clearly demonstrate that RNase H1 is necessary for the activity of such ASOs.
Supplementary Material
Acknowledgments
We thank Tracy Reigle for her graphic expertise, and Donna Parrett and Tori Kniss for their administrative support who are employees and shareholders of Ionis Pharmaceuticals. This study was supported by an internal funding from Ionis Pharmaceuticals.
FUNDING
This was funded by Ionis Pharmaceuticals internal funding.
Conflict of interest statement. All authors are employees and shareholders of Ionis Pharmaceuticals.
REFERENCES
- 1.Stein H., Hausen P. Enzyme from calf thymus degrading the RNA moiety of DNA-RNA Hybrids: effect on DNA-dependent RNA polymerase. Science (Wash DC) 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 2.Crouch R.J., Dirksen M.L. Ribonuclease H. In: Linn SM, Roberts RJ, editors. Nucleases. NY: Cold Spring Harbor Laboratory Press; 1982. pp. 211–241. [Google Scholar]
- 3.Busen W. Purification, subunit structure, and serological analysis of calf thymus ribonuclease H I. J. Biol. Chem. 1980;255:9434–9443. [PubMed] [Google Scholar]
- 4.Rong Y.W., Carl P.L. On the molecular weight and subunit composition of calf thymus ribonuclease H1. Biochemistry. 1990;29:383–389. doi: 10.1021/bi00454a012. [DOI] [PubMed] [Google Scholar]
- 5.Itaya M., Kondo K. Molecular cloning of a ribonuclease H (RNase HI) gene from an extreme thermophile Thermus thermophilus HB8: a thermostable RNase H can functionally replace the Escherichia coli enzyme in vivo. Nucleic Acids Res. 1991;19:4443–4449. doi: 10.1093/nar/19.16.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itaya M., McKelvin D., Chatterjie S.K., Crouch R.J. Selective cloning of genes encoding RNase H from Salmonella typhimurium, Saccharomyces cerevisiae and Escherichia coli rnh mutant. Mol. Gen. Genet. 1991;227:438–445. doi: 10.1007/BF00273935. [DOI] [PubMed] [Google Scholar]
- 7.Kanaya S., Itaya M. Expression, purification, and characterization of a recombinant ribonuclease H from Thermus thermophilus HB8. J. Biol. Chem. 1992;267:10184–10192. [PubMed] [Google Scholar]
- 8.Eder P.S., Walder R.T., Walder J.A. Substrate specificity of human RNase H1 and its role in excision repair of ribose residues misincorporated in DNA. Biochimie. 1993;75:123–126. doi: 10.1016/0300-9084(93)90033-o. [DOI] [PubMed] [Google Scholar]
- 9.Masutani C., Enomoto T., Suzuki M., Hanaoka F., Ui M. DNA primase stimulatory factor from mouse FM3A cells has an RNase H activity. Purification of the factor and analysis of the stimulation. J. Biol. Chem. 1990;265:10210–10216. [PubMed] [Google Scholar]
- 10.Frank P., Albert S., Cazenave C., Toulme J.J. Purification and characterization of human ribonuclease HII. Nucleic Acids Res. 1994;22:5247–5254. doi: 10.1093/nar/22.24.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H., Lima W.F., Crooke S.T. Molecular Cloning and Expression of cDNA for Human RNase H. Antisense Nucleic Acid Drug Dev. 1998;8:53–61. doi: 10.1089/oli.1.1998.8.53. [DOI] [PubMed] [Google Scholar]
- 12.Ceritelli S.M., Frolova E.G., Feng C., Grinberg A., Love P.E., Crouch R.J. Failure to produce mitochondrial DNA results in embryonic lethality in Rnaseh1 null mice. Mol. Cell. 2003;11:807–815. doi: 10.1016/s1097-2765(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 13.Reyes A., Melchionda L., Nasca A., Carrara F., Lamantea E., Zanolini A., Lamperti C., Fang M., Zhang J., Ronchi D., et al. RNase H1 mutations impair mtDNA replication and cause adult-onset mitochondrial encephalomyopathy. Am. J. Hum. Genet. 2015;97:186–193. doi: 10.1016/j.ajhg.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerritelli S.M., Crouch R.J. The non-RNase H domain of Saccharomyces cerevisiae RNase H1 binds double-stranded RNA: magnesium modulates the switch between double-stranded RNA binding and RNase H activity. RNA. 1995;1:246–259. [PMC free article] [PubMed] [Google Scholar]
- 15.Evans S.P., Bycroft M. NMR structure of the N-terminal domain of Saccharomyces cerevisiae RNase HI reveals a fold with a strong resemblance to the N-terminal domain of ribosomal protein L9. J. Mol. Biol. 1999;291:661–669. doi: 10.1006/jmbi.1999.2971. [DOI] [PubMed] [Google Scholar]
- 16.Wu H., Lima W.F., Crooke S.T. Investigating the structure of human RNase H1 by site-directed mutagenesis. J. Biol. Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 17.Katayanagi K., Miyagawa M., Matsushima M., Ishkiawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature (Lond) 1990;347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 18.Yang W., Hendrickson W.A., Crouch R.J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 19.Kanaya S., Katsuda-Kakai C., Ikehara M. Importance of the positive charge cluster in Escherichia coli ribonuclease HI for the effective binding of the substrate. J. Biol. Chem. 1991;266:11621–11627. [PubMed] [Google Scholar]
- 20.Nakamura H., ODA Y., Iwai S., Inoue H., Ohtsuka E., Kanaya S., Kimura S., Katsuda C., Katayanagi K., Morikawa K., et al. How does RNase H recognize a DNA.RNA hybrid? Proc. Natl. Acad. Sci. U.S.A. 1991;88:11535–11539. doi: 10.1073/pnas.88.24.11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima W.F., Wu H., Nichols J.G., Manalili S.M., Drader J.J., Hofstadler S.A., Crooke S.T. Human RNase H1 activity is regulated by a unique redox switch formed between adjacent cysteines. J. Biol. Chem. 2003;278:14906–14912. doi: 10.1074/jbc.M211279200. [DOI] [PubMed] [Google Scholar]
- 22.Lima W.F., Wu H., Nichols J.G., Prakash T.P., Ravikumar V., Crooke S.T. Human RNase H1 uses one tryptophan and two lysines to position the enzyme at the 3′-DNA/5′-RNA terminus of the heteroduplex substrate. J. Biol. Chem. 2003;278:49860–49867. doi: 10.1074/jbc.M306543200. [DOI] [PubMed] [Google Scholar]
- 23.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 24.Graham M.J., Crooke S.T., Monteith D.K., Cooper S.R., Lemonidis K.M., Stecker K.K., Martin M.J., Crooke R.M. In vivo distribution and metabolism of a phosphorothioate oligonucleotide within rat liver after intravenous administration. J. Pharmacol. Exp. Ther. 1998;286:447–458. [PubMed] [Google Scholar]
- 25.Graham M.J., Crooke S.T., Lemonidis K.M., Gaus H.J., Templin M.V., Crooke RM. Hepatic distribution of a phosphorothioate oligodeoxynucleotide within rodents following intravenous administration. Biochem. Pharmacol. 2001;62:297–306. doi: 10.1016/s0006-2952(01)00669-4. [DOI] [PubMed] [Google Scholar]
- 26.Liu W., Hou Y., Chen H., Wei H., Lin W., Li J., Zhang M., He F., Jiang Y. Sample preparation method for isolation of single-cell types from mouse liver for proteomic studies. Proteomics. 2011;11:3556–3564. doi: 10.1002/pmic.201100157. [DOI] [PubMed] [Google Scholar]
- 27.Swayze E.E., Siwkowski A.M. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acid Res. 2007;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen W., Liang X.H., Crooke S.T. Phosphorothioateoligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42:8648–8662. doi: 10.1093/nar/gku579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Hendrickson D., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2012;1:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009;1:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 32.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P., et al. Single-stranded siRNAs activate RNAi in animals. Cell. 2012;150:883–894. doi: 10.1016/j.cell.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 33.Han L.Y., Ma W.P., Crouch R.J. Ribonuclease H renaturation gel assay using a fluorescent-labeled substrate. Biotechniques. 1997;23:920–926. doi: 10.2144/97235rr01. [DOI] [PubMed] [Google Scholar]
- 34.Yule R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taanman J-W. The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta. 1999;1410:103–123. doi: 10.1016/s0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 36.Nicholls J., Minczuk M. In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol. 2014;56:175–181. doi: 10.1016/j.exger.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 38.Crary G.S., Albrecht J.H. Expression of cyclin-dependent kinase inhibitor p21 in human liver. Hepatology. 1998;28:738–743. doi: 10.1002/hep.510280320. [DOI] [PubMed] [Google Scholar]
- 39.Lima W.F., Wu H., Crooke S.T. The RNase H Mechanism. In: Crooke ST, editor. In Antisense Drug Technology: Principles, Strategies, and Applications. 2nd edn. Boca Raton: CRC Press; 2008. pp. 47–74. [Google Scholar]
- 40.Liu Y., Kao H-I., Bambara R.A. Flap endonuclease 1: a central component of DNA metabolism. Ann. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.