Abstract
The RNase III enzyme Drosha is a key factor in microRNA (miRNA) biogenesis and as such indispensable for cellular homeostasis and developmental processes. Together with its co-factor DGCR8, it converts the primary transcript (pri-miRNA) into the precursor hairpin (pre-miRNA) in the nucleus. While the middle and the C-terminal domain are crucial for pri-miRNA processing and DGCR8 binding, the function of the N-terminus remains cryptic. Different studies have linked this region to the subcellular localization of Drosha, stabilization and response to stress. In this study, we identify alternatively spliced Drosha transcripts that are devoid of a part of the arginine/serine-rich (RS-rich) domain and expressed in a large set of human cells. In contrast to their expected habitation, we find two isoforms also present in the cytoplasm, while the other two isoforms reside exclusively in the nucleus. Their processing activity for pri-miRNAs and the binding to co-factors remains unaltered. In multiple cell lines, the endogenous mRNA expression of the Drosha isoforms correlates with the localization of endogenous Drosha proteins. The pri-miRNA processing efficiency is not significantly different between groups of cells with or without cytoplasmic Drosha expression. In summary, we discovered novel isoforms of Drosha with differential subcellular localization pointing toward additional layers of complexity in the regulation of its activity.
INTRODUCTION
Non-coding RNAs play important roles in the post-transcriptional gene regulation. A prominent subgroup consists of the microRNAs (miRNA), which are single-stranded molecules of about 22 nucleotides (nt).
The canonical maturation of miRNAs involves only two cleavage reactions mediated by the RNase III-type enzymes Drosha and Dicer, but in fact, this is a highly complex and thoroughly regulated mechanism at multiple layers (1,2). First, transcription of human miRNAs is carried out by RNA Polymerase II and controlled by several epigenetic regulators and transcription factors (3,4). Next, the primary miRNA (pri-miRNA) transcript is processed in the nucleus by Drosha. Together with its co-factor DGCR8 (DiGeorge syndrome critical region 8), it forms the microprocessor complex (5–8). This trimeric complex (consisting of two molecules DGCR8 and one Drosha) recognizes long pri-miRNA transcripts and cleaves them close to the base of the stem (9,10). The ∼60–80 nt long product is termed precursor miRNA (pre-miRNA) (11). The pre-miRNA derived in the nucleus is exported into the cytoplasm by Exportin-5 (12–14). There, it is further processed by Dicer to a ∼22 nt long duplex with a two nucleotide overhang at either 3‘-end. Finally, an active (mature) miRNA can be derived from both the 5′- and 3′-strand as well as the loop of a pre-miRNA (15) and is loaded into a protein of the Argonaute (AGO) family. This protein–RNA complex is described as the RNA induced silencing complex (RISC). The RISC suppresses the translation, decreases the mRNA stability or even degrades messenger RNAs (mRNA) by complementary base-pairing of the miRNA to its target mRNA (reviewed in (1,16,17)). Only AGO2 contains a catalytic site to directly cleave the target mRNA (18,19). Other AGO proteins are able to repress translation or induce decay by other proteins or stabilize miRNAs (20–22), reviewed in (23). Thus, miRNAs have a direct influence on the expression of a multitude of proteins involved in many different cellular processes. Deregulation of the miRNA biogenesis pathway can contribute to a variety of diseases, including different forms of cancer.
MiRNA levels can be decreased by either loss or mutation of a gene transcribed into the pri-miRNA or by loss or malfunction of a co-factor involved in the miRNA biogenesis (24–27). The miRNAs itself and regulatory proteins of the miRNA biogenesis, can influence cell survival and cell death either positive or negative, depending on their specific targets (28–30). Particularly, Drosha is linked to cell survival of vascular smooth muscle cells (31) and is highly enriched in the cytoplasm upon stress (32).
The RNase Drosha is part of a second large complex of about 20 different proteins (6), that are involved in the regulation of individual pri-miRNAs, reviewed in (2). Recent publications describe a role of Drosha in response to virus infection through cleavage of circular viral pri-miRNAs in the cytoplasm (33,34). Except for pri-miRNAs, the DGCR8 mRNA is cleaved by Drosha in a feedback mechanism (35,36), which is conserved throughout different species. Beyond miRNA biogenesis, Drosha is involved in further cellular processes: rRNA processing (37–39), alternative splicing of pre-miRNA-like exons (40,41) and regulation of gene expression independently of RNA cleavage (42). Drosha also cleaves mRNAs encoding for inhibitors of myelopoiesis and thus, it regulates the development of dendritic cells completely independent of miRNAs (43). Expression of neurogenin 2 underlies a similar regulatory mechanism (44). Notably, additional target mRNAs are bound by Drosha pointing toward the possibility of additional Drosha functions in the cell (45). Non-canonical functions of Drosha and Dicer are reviewed in (46,47).
While the middle and the C-terminal domains of Drosha are crucial for pri-miRNA recognition and processing as well as DGCR8 binding (7,9), the arginine-/serine-rich (RS-rich) N-terminus has been previously linked to cellular localization (48,49). The domains within Drosha are schematically illustrated in Figure 1A. Generally, RS-rich domains accumulate arginine and serine repeats and are found in Serine-Arginine-rich SR proteins, regulators of alternative splicing as well as proteins involved in other nuclear processes like chromatin remodeling, transcription, cell cycle progression and nuclear export (50–52). Extensive phosphorylation of serines within the RS-rich domain regulates the subcellular localization and activity of such proteins. Exon 6 contains an SPS phosphorylation site, which is involved in the nuclear localization of Drosha (48).
Figure 1.
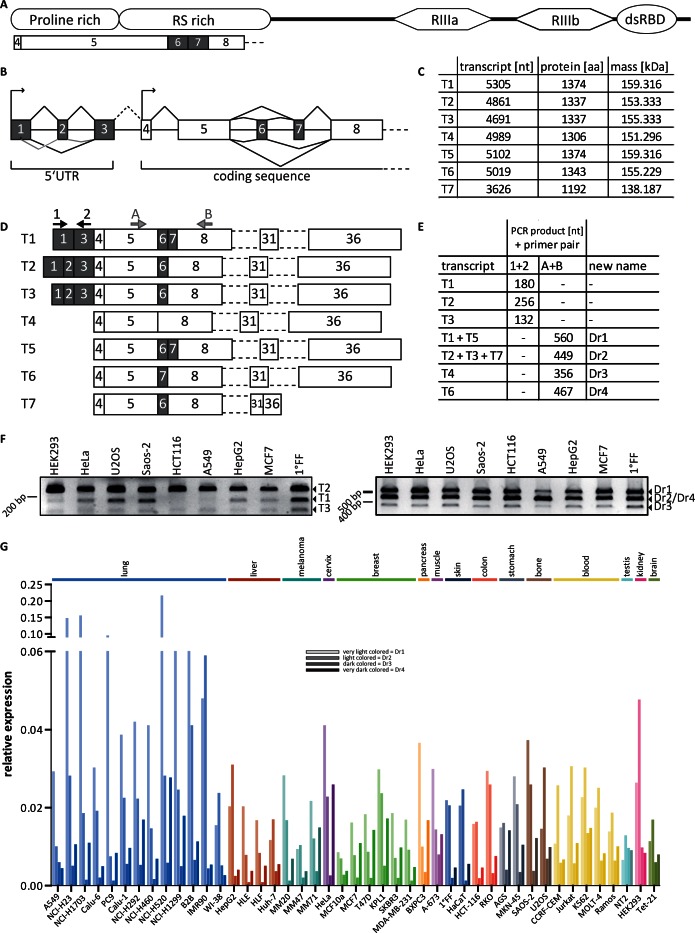
Drosha transcripts and their expression in different cell lines. (A) Schematic representation of the RNase III enzyme Drosha coding region and the position of exons involved in alternative splicing. RIIIa, RNase III domain a; RIIIb, RNase III domain b; dsRBD, double-stranded RNA binding domain. (B) Schematic representation of alternative splincing. (C) Table of transcripts; nt = nucleotides; aa = amino acids; kDa = kilo Dalton. (D) Detail of the alternatively spliced region with combinations of exons both in the 5′UTR and in the N-terminal RS-rich domain in the coding sequence. T7 is additionally spliced between exon 31 and 36. Arrows indicate primer pairs for PCR amplification. (E) Expected PCR products. (F) RT-PCR results from cDNA of the indicated cell lines with primer pairs depicted in D. (G) Relative expression profile of the four identified splice variants in a set of 45 cell lines from different tissue origin, normalized to Cyclophilin A expression (N = 1 with technical duplicates).
In this study, we identify novel endogenous alternatively spliced variants of Drosha. Previously, we described the existence of rare processing-deficient Drosha splice variants of the C-terminus (53). Hence, we investigated whether additional, more prevalent isoforms of Drosha may exist and found novel and common isoforms devoid of a part of the N-terminal RS-rich domain affecting cellular localization of Drosha.
MATERIALS AND METHODS
Plasmids
Cloning of wild-type Drosha into pcDNA3.1D-V5/His-TOPO (Thermo Fisher Scientific, Waltham, MA, USA) has been described before (22). The N-terminal coding sequences of the other variants were amplified from cDNA of MCF7 cells using primers, spanning the exons 5–8. Subsequently, the polymerase chain reaction (PCR) products were purified from an agarose gel and cloned into the pCR®II-TOPO vector. From these constructs, subcloning of a fragment containing the alternatively spliced region into a truncated pENTR3C-Dr2-V5/His, via BamHI and SacI restriction sites, was performed. In a next step, the relevant regions were subcloned into pcDNA3.1-Flag-Dr2-V5/His using BamHI and Bsu36I which gave rise to all pcDNA3.1-Flag-Drosha-V5/His splice variant constructs. Finally, Yellow-Fluorescent Protein (YFP) was amplified from pIRESneoFlag/HA-YFP and added N-terminally to all Drosha coding sequences using the restriction enzymes HindIII and BamHI and thereby replacing the Flag-tag. The BamHI site and the Drosha start codon were separated by the nucleotides ACC. Single and multiple nucleotide mutants of every splice variant were generated using primers for site-directed mutagenesis. All primers used for cloning are listed in Supplementary Table S1. The pFlag/HA-DGCR8 plasmid was kindly provided by Thomas Tuschl (Rockefeller University). Generation of HA-DGCR8 and the transdominant negative mutant (TN) with complete processing deficiency was described before (53,54).
Western blot
Protein samples were prepared with RIPA buffer unless otherwise indicated. Therefore, cells were washed with ice cold phosphate buffered saline (PBS), scraped from the culture dish and lysed for 10 min on ice in 250 μl RIPA buffer per well of a 6-well plate. Subsequently, crude lysates were centrifuged for 15 min at 17 000 g. The supernatant was transferred to a fresh tube. Protein concentration was determined with a Bicinchoninic Acid (BCA) assay and samples were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, using 6% self-cast gels (Biorad system; Biorad Laboratories, Hercules CA, USA). After separation, the samples were transferred from the gel to a nitrocellulose (GE Healthcare, Little Chalfont, United Kingdom) or a PVDF membrane (Roche, Basel, Switzerland) at 30 V and 90 mA for 16 h at 4–8°C, using a wet blotting system (Biorad). The following primary antibodies were used according to the manufacturer's recommendations: anti-DGCR8 (1:300, 10 996-1-AP; ProteinTech Group, Chicago IL, USA - http://www.ptglab.com/Products/DGCR8-Antibody-10996-1-AP.htm), anti-Drosha (1:1000 in TBS-T + 5% milk, ab-12286, Abcam, Cambridge, UK), anti-Drosha (D28B1) (1:500, #3364, Cell Signaling, Cambridge, United Kingdom), anti-EWS (1:1000, EWS [G-5], sc-28327; Santa Cruz Biotechnology, Dallas TX, USA), anti -Flag (1:3000, F1804-1MG, Sigma-Aldrich, St. Louis MI, USA), anti-HA.11 (1:500, Mono HA.11 MMS-101P, Covance, Münster, Germany), anti-Lamin A/C (1:2000 in TBS-T + 5% milk, 4C11, Cell Signaling), anti-α/β-Tubulin (1:1000 in TBS-T + 5% milk, #2148, Cell Signaling), anti-V5 (1:1000, R96025, Thermo Fisher Scientific). Detection was performed with HRP-coupled secondary antibodies.
RNA extraction, reverse transcription and qPCR
RNA was isolated from TRI Reagent (Sigma-Aldrich) lysates. Isolation was performed according to the manufacturer's recommendation. Synthesis of cDNA from 1 μg RNA was performed with Random Hexamer Primer and RevertAid Reverse Transcriptase (both Thermo Fisher Scientific). Expression of target genes was detected with qPCR using specific primer pairs and the Power SYBR® Green Master Mix (Thermo Fisher Scientific). Generally, relative expression was determined using the ΔΔCT method. All values were normalized to cylophilin A and compared to a respective control sample. Primers are listed in Supplementary Table S1.
Stem-loop RT-qPCR for mature miRNA detection
RNA was isolated from TRI Reagent (Sigma-Aldrich) lysates. Isolation was performed according to the manufacturer's recommendation. Synthesis of cDNA from 100 ng RNA was performed using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific) according to the provided manual. Expression of mature miRNAs was detected with qPCR using a specific forward primer pair, a universal reverse primer and the Power SYBR® Green Master Mix (Thermo Fisher Scientific). Generally, relative expression was determined using the ΔΔCT method. All values were normalized to the snRNA U6 and compared to a respective control sample. Primers are listed in Supplementary Table S1.
Cell lines
HEK293 and NT2 cells were cultivated in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% Fetal Calf Serum (FCS) (Thermo Fisher Scientific) and 1% L glutamine (Sigma-Aldrich). PC9 and NCI-H1703 cells were cultivated in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10% FCS (Thermo Fisher Scientific). U2OS cells were cultivated in McCoy's 5A medium (Sigma-Aldrich) supplemented with 10% FCS (Thermo Fisher Scientific) and 1% L glutamine (Sigma-Aldrich). All cell lines were cultured at 37°C in a 5% CO2 atmosphere. When a confluence of 80–100% was reached, the cells were passaged as applicable using 0.25% trypsin-ethylenediaminetetraacetic acid (Thermo Fisher Scientific).
Cellular fractionation
Cytoplasmic and total nuclear protein extracts were prepared according to a previously published protocol (55).
Immunofluorescence
Cells were seeded in 12-well plates containing 12 mm cover slips (Glaswarenfabrik Karl Hecht, Sondheim v. d. Rhön, Germany). For transient plasmid transfection, HEK293 cells were seeded in density to reach a confluence of 70–80% 24 h later. Plasmid transfection was performed using TransIT®-LT1 reagent (Mirus Bio LCC, Madison WI, USA) according to the manufacturer's recommendation. Cells were incubated for another 24 h after transfection. For fixation, the medium was removed, the cells were washed with 2 ml PBS and fixed in 1 ml 4% pure formaldehyde (Thermo Fisher Scientific) for 10 min at room temperature. Subsequently, the cells were washed again with 2 ml PBS, permeabilized with 1 ml PBS-Triton-X-100 (0.1%) for 5 min at room temperature, blocked with 2% bovine serum albumin (BSA) in 1 ml PBS-Triton-X-100 (0.1%) for 20 min, and incubated with the primary antibody anti-Drosha (ab-12 286, Abcam; 1:250 in PBS-Triton-X-100 (0.1%) + 2% BSA) for 1 h at room temperature. Cover slips were washed and incubated with the secondary antibody (anti-rabbit Alexa Fluor® 647; Thermo Fisher Scientific; 1:500 in PBS-Triton-X-100 (0.1%) + 2% BSA) and DAPI (5 μg/ml) for 1 h in the dark. Finally, coverslips were mounted in Mowiol (Merck KGaA [EMD], Darmstadt, Germany) on specimen slides (Thermo Fisher Scientific). Images were acquired at 40 or 63× magnification on a Zeiss LSM 700 laser scanning microscope (Carl Zeiss, Oberkochen, Germany) in the DKFZ Light Microscopy Core Facility.
Immunoprecipitation
This experiment was performed as described previously (53,56). Additionally, ANTI-FLAG M2® Affinity Gel (Thermo Fisher Scientific) was used for the pull down of Flag-tagged protein constructs.
In vitro pri-miRNA processing assay
HEK293 cells were reverse transfected with a Drosha-siRNA: 5′ GCAUGCAAGCGCGCAGUAU(dTdT) or the non-targeting siRNA control siAllstars using Lipofectamine RNAiMAX in 6-well plates, according to the manufacturer's recommendation. After 24 h, cells were transfected each with 1.5 μg of plasmid encoding for YFP-Drosha-V5/His as wild-type or phosphorylation triple mutant (tripleA and tripleD), pFlag/HA-DGCR8, a processing-deficient TN mutant Drosha construct (TN; both catalytic residues mutated: E1045Q, E1222Q), or an empty vector (EV) using 6-μl Polyethylenimine (PEI) in 100 μl Tris-Buffered Saline (TBS) buffer per well. Forty-eight hours later, whole cell extracts were prepared in a buffer containing 20 mM Tris–HCl (pH 8.0), 100 mM KCl by sonication (3 × 30 s at low intensity; 30 s break in-between each sonication step), followed by centrifugation at 17 000 g for 15 min. Supernatants were transferred to a fresh tube. The T7 template for the primary transcript was amplified by PCR, gel purified from a 6% TBE-Urea gel, precipitated and allowed to fold for 10 min at 60°C before further usage (as described in (57)). The processing reaction (total volume 35 μl) contained 40 μg of total protein, 0.5 mM ATP, 20 mM creatine phosphate, 4 mM MgCl2, 20 U RiboLock RNase Inhibitor (Thermo Fisher Scientific) and 104 to 105 counts per minute of the [32P]-labeled transcript containing pri-miR-15b∼16-2. The reaction was incubated for 40 min at 37°C, extracted with phenol–chloroform, precipitated with 100% ethanol and the RNA pellet was resuspended in RNA sample buffer. The samples were loaded onto a precast 10% TBE-urea gel (Biorad) and separated by electrophoresis. Finally, the gel was used for direct exposure of x-ray films (Fujifilm, Tokio, Japan) for up to 96 h.
In vivo pri-miRNA processing assay
HEK293 cells were reverse transfected with a Drosha-siRNA: 5′-GCAUGCAAGCGCGCAGUAU(dTdT)-3′ or the scrambled siRNA control siAllstars (Qiagen, Venlo, Netherlands) using Lipofectamine RNAiMAX in 6-well plates. After 24 h, cells were transfected each with 1.5 μg of plasmid encoding for YFP-Drosha-V5/His as wild-type or phosphorylation triple mutant (tripleA), pFlag/HA-DGCR8, a processing deficient TN mutant Drosha construct (TN), YFP or an EV using 6-μl PEI in 100-μl TBS buffer per well. Forty-eight hours later, RNA was isolated using TRI Reagent (Sigma-Aldrich), digested with DNaseI (Roche) and reverse transcribed. The samples were analyzed for pri-miRNA levels and endogenous Drosha mRNA levels by RT-qPCR and protein levels by western blot.
RESULTS
Drosha splice variants are expressed in a variety of human cell lines
In a first approach, we searched for potentially alternatively spliced transcripts of key human miRNA processing factors in databases of expressed sequence tags or in transcriptome datasets. We found seven different variants of Drosha that were cataloged at that time (Figure 1B and C), none of which had been validated, annotated or characterized. Three of them were listed in GenBank (NM_013235.4 [T1], NM_001100412.1 [T2] and XM_005248294.2 [T3]), three in Ensembl (ENST00000265075 [T4], ENST00000511367 [T5] and ENST00000382188 [T7]), and one was identified in a transcriptome-wide sequencing study [T6] (58). Today, Ensembl also lists T1 and T2, whereas GenBank included T7. The transcript T3 and T4 were excluded and still, T6 is neither listed in GenBank nor Ensemble. Splice variants for other key miRNA processing factors were also found but in lower abundance than for Drosha.
According to the identified isoforms, Drosha transcripts can have different start sites. Isoforms T4–T7 lack exons 1–3 and have a shorter 5′UTR, contained in exon 4. T1–T3 share the same upstream start site but differ in their sequence of the 5′UTR by alternative splicing of exon 2 or exon 2 and part of exon 1. Furthermore, exon 6 and 7 in the N-terminal region of the transcript are affected by alternative splicing, which lie in the RS-rich domain of the protein that bares no known catalytic site but has previously been shown to play a role in subcellular localization of Drosha (32,48,59). Hence, we hypothesized that an alteration of the sequence in that area might have an influence on the localization or the functionality of Drosha itself or on the regulation of the miRNA biogenesis pathway.
To confirm the existence of the identified alternative splice variants, we performed an RT-PCR from cDNA of different cancer cell lines. Amplification of the 5′UTR with a primer pair spanning the area between exon 1 and 3 (Figure 1D and E) resulted in three bands of the expected size in all analyzed cell lines (Figure 1F, left panel). Gel purification, cloning into pCR®II and sequencing verified the existence of all splice variants of the 5′UTR. However, the expression level of the variants differed and was strongest for the combination of exon 1, 2 and 3 (as in T2). Since we found one clearly predominant isoform for this region, we did not further pursue the analysis of these isoforms.
For verification of the N-terminal splice variants differing in the presence of exon 6 and 7 (from here on called Dr1–Dr4), we amplified the region between exon 5 and 8 (Figure 1D, E and F, right panel). The PCR resulted in the three expected bands as Dr2 and Dr4 (containing either exon 6 or exon 7) are very similar in length (Figure 1F; right panel). All isoforms were clearly detectable in all cell lines with some differences in their relative intensities among the human cell lines analyzed. Again, the PCR products were gel purified, cloned into pCR®II and sequenced. The sequencing results verified the existence of all four possible exon combinations (Supplementary Figure S1). Furthermore, the expression was quantitatively confirmed. We determined the expression pattern of the Drosha splice variants in cells from different origin through qPCR in cDNA from 45 cell lines including both cancerous and non-malignant cells. To distinguish the splice variants, we used specific primer pairs that recognized exon–exon junctions which were unique for the respective transcript. In accordance with the intensities of the bands obtained with the PCR from cDNA, strongest expression was observed for variants Dr1 and Dr2, whereas levels for Dr3 were generally lowest in almost every analyzed cell line (Figure 1G). Variant Dr4 showed a more heterogeneous expression pattern, but was lower than Dr1 as well as Dr2 and higher than Dr3 in most cell lines.
All Drosha splice variants form the microprocessor complex with DGCR8 and bind additional interaction partners
To evaluate the binding of Drosha to its co-factors (Figure 2A), we performed co-immunoprecipitation experiments and subsequent western blot analysis.
Figure 2.
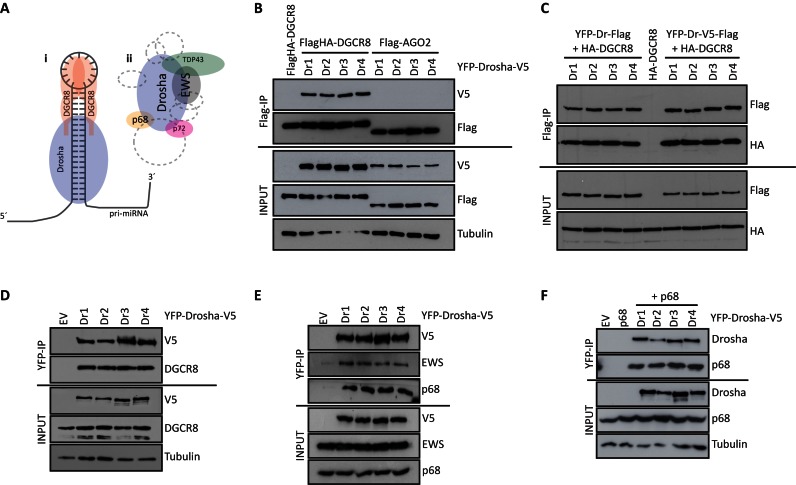
All Drosha splice variants form the microprocessor complex with DGCR8 and bind additional interaction partners. (A) Schematic representations of the microprocessor complex (i) and an additional Drosha-containing complex (ii). A selection of interacting partners is represented. Additional proteins are indicated with dashed lines. (B) Immunoprecipitation of Flag-tagged DGCR8 pulled down its interaction partner V5-tagged Drosha. (C) Immunoprecipitation of Flag-tagged Drosha pulled down its HA-tagged microprocessor co-factor DGCR8. (D) Immunoprecipitation of YFP-tagged Drosha pulled down endogenous DGCR8, (E) endogenous EWS and p68 and (F) exogenous p68. All co-immunoprecipitation experiments were performed from cell lysates of HEK293 cells transiently transfected in at least two biological replicates with the respective Drosha splice variant (YFP-Drosha-V5) construct as well as Flag/HA-DGCR8 or Flag-AGO2 (B), HA-DGCR8 (C) or p68 (F), respectively.
First, we tested whether all four isoforms were able to interact with the microprocessor co-factor DGCR8 (Figure 2Ai). First, HEK293 cells were co-transfected with Flag-tagged DGCR8 and one of the four V5-tagged Drosha splice variants. Flag-tagged AGO2 was used as negative control. All Drosha variants were equally expressed and were pulled down with DGCR8 but not with AGO2 (Figure 2B). The same result was obtained when Flag-tagged Drosha was used to pull down DGCR8 (Figure 2C). Lastly, all four Drosha isoforms were also binding to endogenous DGCR8 (Figure 2D). In the input, both DGCR8 in unmodified (between 82 and 86 kDa) and post-modified state (extensively phophorylated, around 120 kDa) was visible. In the pull down sample, only post-modified DGCR8 was detected (60).
Next, we determined the interaction of the Drosha isoform with selected members of a second interaction complex (6) (Figure 2Aii). Endogenously expressed interaction partners p68 and EWS were efficiently co-immunoprecipitated with all exogenous Drosha variants (Figure 2E). Also exogenous p68 was efficiently pulled down by Drosha (Figure 2F).
All Drosha splice variants efficiently process pri-miRNAs and the DGCR8 mRNA
All four splice variants bind to DGCR8 and possess the RNase III and the dsRNA binding domains. Hence, we tested whether they would also all be capable of processing pri-miRNAs and the DGCR8 mRNA.
To test whether the Drosha isoforms processed pri-miRNAs, we applied a knockdown and rescue approach. HEK293 cells were transfected with an siRNA, targeting the 3′UTR of Drosha to knockdown endogenous Drosha. Then, one of the Drosha splice variants was co-transfected together with DGCR8. We tested four different siRNAs in the limited sequence space of the 3′UTR and si4491 showed the best knockdown efficiency at mRNA level for endogenous Drosha compared to the non-targeting control siRNA (Figure 3A). At the protein level, the knockdown was much more prominent and Drosha expression was replenished to a physiological level by the co-transfection with the Drosha variants (Figure 3B). As additional control, a TN mutant of Drosha was used, which should abolish pri-miRNA processing (54).
Figure 3.
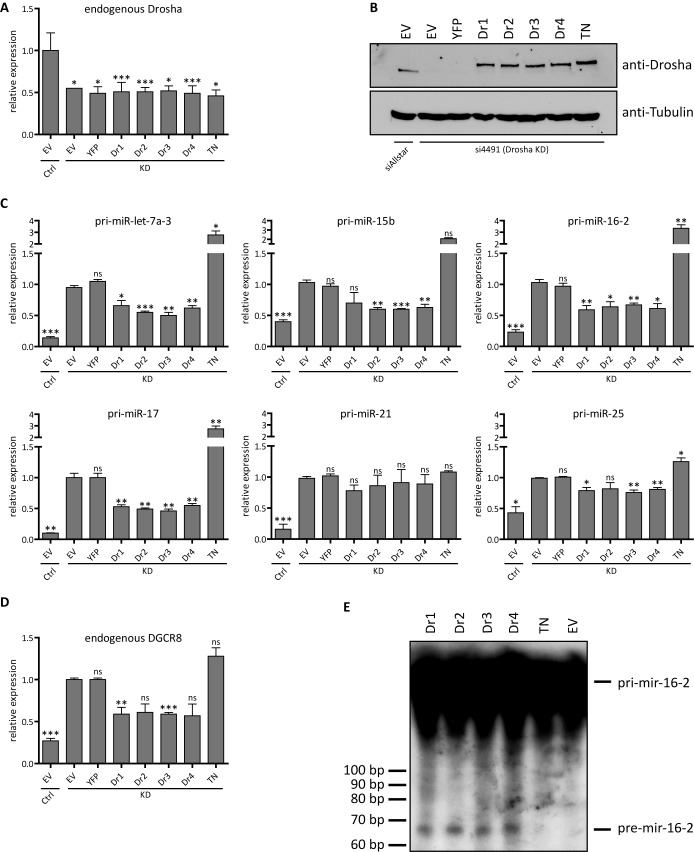
All Drosha splice variants efficiently process pri-miRNAs and the DGCR8 mRNA in vitro and in vivo. HEK293 cells were transfected with an siRNA targeting the 3′-UTR of endogenous Drosha for 72 h and after 24 h additionally with a plasmid for a Drosha splice variant or an empty vector (EV) or a transdominant negative mutant (TN) for 48 h. The cells were co-transfected with pFlag/HA-DGCR8. (A) RT-qPCR results for endogenous Drosha levels normalized to cyclophilin A and compared to the EV control. Statistical significance calculated with student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001; N = 3. (B) Western blot verification of efficient knock-down and co-overexpression of Drosha protein. (C) RT-qPCR results for the in vivo processing efficiency as measured by the abundance of selected pri-miRNAs normalized to cyclophilin A and compared to the EV KD control. Statistical significance calculated with student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001; ns = not significant. Values are presented in Supplemental Table S2; N = 3. (D) RT-qPCR results of DGCR8 mRNA levels indicate Drosha-mediated processing by the different isoforms. (E) In vitro processing assay results of pri-mir-16-2. Detection of pri- and pre-miR-16-2 by autoradiography after incubation with Drosha-transfected cell lysates. Error bars indicate SEM. KD = knockdown; Ctrl = control; bp = base pair.
For the analysis of pri-miRNA processing after knockdown of the endogenous Drosha and rescue with the different Drosha isoforms, we selected a set of well-studied pri-miRNAs. Pri-miR-let-7a-3, pri-miR-15b, pri-miR-16-2, pri-miR-17, pri-miR-21 and pri-miR-25 were processed to a similar extent by all four splice variants as indicated by a decrease of pri-miRNA abundance after Drosha expression compared to the EV knockdown control. However, the respective turnover rates of pri-miRNAs were lower than in the non-targeting siRNA control sample. Knockdown of endogenous Drosha and overexpression of the TN mutant led to a decrease in processing efficiency compared to the EV and knockdown alone, which was evident in highly increased levels of pri-miRNAs (Figure 3C). For pri-miR-21, the reduction upon Drosha expression was less pronounced, and not significant, but again similar for all isoforms—here, the TN mutant had no additional effect (Figure 3C and Supplementary Table S2). Furthermore, levels of the endogenous DGCR8 mRNA—which is regulated by Drosha (36)—increased 4-fold upon Drosha knockdown and decreased upon overexpression of all isoforms to a similar extent (Figure 3D).
To further analyze processing activity also in vitro, cell extracts after knockdown of endogenous Drosha and co-transfection of one Drosha variant were incubated with [32P]-labeled pri-miR-16-2 transcripts. Autoradiography demonstrated similar processing activity for all isoforms from the primary transcript into the pre-miRNA (Figure 3E). The level of pre-miRNA-16-2 was not determined by RT-qPCR since it would have been impossible to distinguish this intermediate from the primary transcript. Instead, we determined the expression of mature miRNAs derived in vivo upon endogenous Drosha knockdown and subsequent rescue. The overexpression of a respective Drosha variant led to an increase of mature miRNA levels compared to the EV knockdown control. However, the expression of mature miRNAs was not restored entirely. Clear differences were not found between the four variants (Supplementary Figure S2). These results supported the effect observed on the pri-miRNA level as well as the lack of isoform-specific differences (Figure 3C).
In summary, all Drosha splice variants efficiently processed pri-miRNAs and the DGCR8 mRNA.
Alternative splicing affects the localization of Drosha
Since Drosha contains several phosphorylation sites (32,48,49) and predicted nuclear localization signals (NLS) in the RS-rich domain (Figure 4A), we hypothesized that alternative splicing in this domain might have an influence on the subcellular localization.
Figure 4.
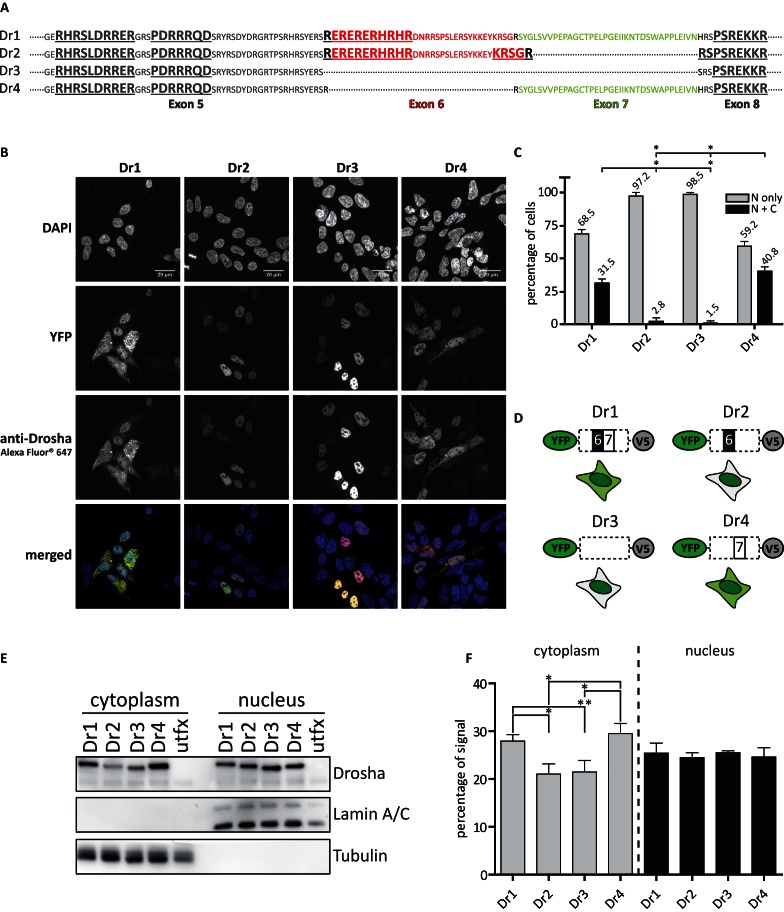
Alternative splicing affects the localization of Drosha. (A) Sequence of the alternatively spliced region of Drosha. Bold and underlined sequences indicate predicted nuclear localization sequences (NLS). (B) Microscopic results of HEK293 cells transiently transfected with the respective YFP-tagged Drosha splice variants. Nuclei were stained with DAPI, Drosha was co-stained with a secondary antibody labeled with Alexa Fluor® 647. Images were acquired in 63× magnification. Scale bars represent 20 μM. (C) Quantitative analysis of immunofluorescence experiments performed on HEK293 cells transiently transfected with the respective Drosha splice variants on images acquired with 40× magnification. A number of at least 40 cells were counted per image. N: nucleus; C: cytoplasm. Statistical significance calculated with student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001. N = 3. Error bars indicate SEM. (D) Schematic representation of YFP-tagged constructs of the Drosha splice variants and their detected cellular distribution in the cell. (E) Western blot results of the fractionation of cellular compartments from HEK293 cells transiently transfected with the respective YFP-tagged Drosha splice variants. (F) Quantitation of chemoluminescent signals derived from E. Percentages of signals were calculated for each splice variant based on the sum of signal volume from the respective cellular compartment. Statistical significance was calculated with student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001. Error bars indicate SEM. N = 3.
To elucidate whether the four different coding sequence splice variants differed in their localization pattern we transfected HEK293 cells with YFP-Drosha-V5 constructs. Interestingly, confocal microscopy revealed indeed a difference in subcellular localization (Figure 4B): while the alternative splice variants lacking exon 7 (Dr2 and Dr3) were localized almost exclusively nuclear, variants containing exon 7 (Dr1 and Dr4) were also detected in the cytoplasm at a much higher rate (Figure 4C). Compared to Dr2 and Dr3, splice variants Dr1 and Dr4 were significantly enriched in the cytoplasm and decreased in the nucleus, respectively (Figure 4C). The subcellular localization of the isoforms is schematically illustrated (Figure 4D). These results were confirmed through the biochemical fractionation of cellular compartments. Although all Drosha variants were found at a detectable level in the cytoplasm of transfected HEK293 cells, the results resembled the observations from microscopy. While nuclear expression levels were equal, the signals for cytoplasmic Dr1 and Dr4 were significantly stronger compared to Dr2 and Dr3 in the same cellular compartment (Figure 4E and F).
Since splice variants containing exon 7 showed a pool of cells with nuclear and cytoplasmic localization, we searched for motifs potentially responsible for the cytoplasmic localization. One possibility could be a weak nuclear export signal (NES) in exon 7 (as predicted with the NetNES 1.1 server (61)). Variant Dr4, which was devoid of exon 6, additionally lacked two NLS sequences which might have supported the observed distribution pattern. However, transfection of HEK293 cells with deletion constructs devoid of the NES or N-/C-terminal parts of exon 7, respectively, and subsequent confocal laser scanning microscopy revealed no significant difference between Dr1 as well as Dr4 wild-type and the respective deletion mutants (data not shown).
Several studies implicated phosphorylation sites in Drosha (32,48,49), including three Serine residues that are also predicted and listed in the PHOSIDA database (62). All three sites were located at the borders of the alternatively spliced exons 6 and 7 at the positions 284, 355 and 357 (Supplementary Figure S3A). The latter two represent an SPS phosphorylation site, just like serine 300 and 302 positioned in exon 6 (48). Phosphorylation of SPS sequences in SMAD3 or MEK1 induced nuclear accumulation and hence SPS is considered as a general nuclear transport signal (63). We mutated these three phosphorylation sites to alanine, which prevented phosphorylation, or to aspartate, which acted as a phosphorylation mimic due to its negative charge.
First, we tested whether the loss of phosphorylation would affect the pri-miRNA processing efficiency of the Drosha isoforms. All phosphorylation site mutants showed an effect similar to their corresponding wild-type isoforms, although with a slightly lower significance (Supplementary Table S2). Generally, we did not find a distinct tendency to a decreased or increased processing for any of the mutant variants (Supplementary Figure S3B). Next, we investigated the localization of the serine mutants by confocal lasers scanning microscopy of HEK293 cells. The splice variants that showed an exclusively nuclear signal for wild-type Drosha (Dr2, Dr3) exhibited almost no change when mutated to tripleA or tripleD (Supplementary Figure S3C and D). However, for tripleA mutants of Dr1 and Dr4, the number of cells with a nuclear and cytoplasmic signal further increased to over 50% with a significant result for Dr1 (Supplementary Figure S3D). The tripleD mutants of Dr1 and Dr4 gave rise to similar results as their corresponding wild-type.
Cells with differential endogenous expression of Drosha isoforms show a distinctive cellular distribution pattern
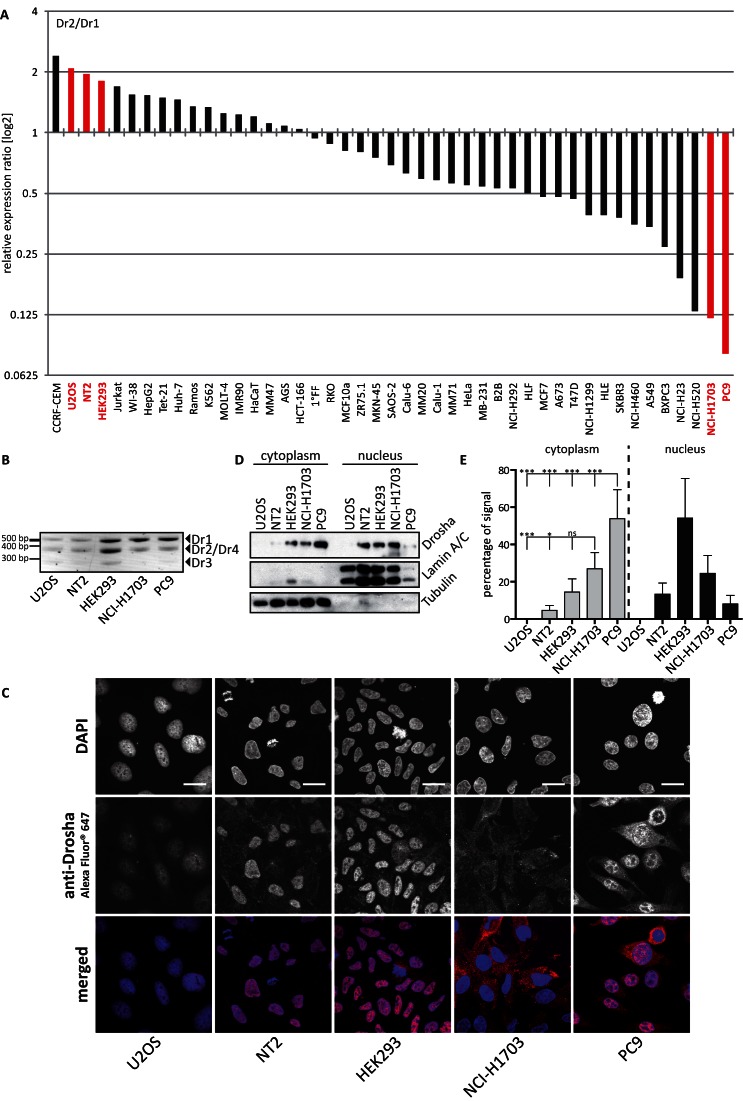
After we documented differential localization of the Drosha isoforms in an exogenous setting, we wanted to determine whether this could also be observed for the endogenous proteins. Since the overlap between the isoforms did not allow the generation of specific antibodies or detection in mass spectrometry, we used the differential expression patterns observed in various cell lines (Figure 1G). We first calculated the ratio of relative expression of the two most abundant isoforms at mRNA level, Dr2 (only nuclear) over Dr1 (nuclear and cytoplasmic) (Figure 5A and Supplementary Table S3). We selected the cell lines U2OS, NT2 and HEK293 that showed the highest ratio of Dr2 over Dr1 among the adherent cell lines. In contrast, NCI-H1703 and PC9 had the lowest ratio (Figure 5A and Supplementary Table S3). RT-PCR results further validated the observations from the RT-qPCR with an increased Dr1 expression compared to the other isoforms in NCI-H1703 and PC9 lung cancer cells (Figure 5B).
Figure 5.
Cells with differential endogenous expression of Drosha isoforms show a distinctive distribution pattern. (A) Expression ratio of the indicated cell lines calculated from the normalized relative expression values (Figure 1G). Highlighted are adherent cell lines selected for further experiments. (B) RT-PCR results of cDNA from the indicated cell lines and their respective expression of Drosha splice variants. N = 3. (C) Microscopic results of cell lines differing in Dr1/Dr2 expression ratios. Nuclei were stained with DAPI, endogenous Drosha was co-stained with Alexa Fluor® 647. Images were acquired with 63× magnification. Scale bars represent 20 μm. N = 3. (D) Western blot results of the fractionation of cellular compartments from the indicated cells for endogenous Drosha. LaminA/C served as a marker for the nuclear compartment, while Tubulin served as a marker for the cytoplasm. N = 3. (E) Quantitation of chemoluminescent signals derived from D. Percentages of signal were calculated for each splice variant based on the sum of signal volume from the corresponding cellular compartment and normalized to the compartment marker α/β-tubulin or lamin A/C. Statistical significance was calculated with student's t-test: *P < 0.05; **P < 0.01; ***P < 0.001; ns = not significant. Error bars indicate SEM. N = 3.
To investigate whether the subcellular distribution of the Drosha protein varied between the cell lines, the cells were fixed and stained with an anti-Drosha antibody. Confocal laser scanning microscopy indeed revealed a differential distribution pattern of the splice variants: while U2OS, NT2 and HEK293 showed only a nuclear staining for Drosha (higher Dr2 expression), NCI-H1703 and PC9 were stained both in the nucleus as well as in the cytoplasm (higher Dr1 expression) (Figure 5C). Knockdown experiments of Drosha proved the specificity of the used antibody (Supplementary Figure S4A). Fractionation of cellular compartments from the five cell lines and subsequent western blot confirmed the distribution pattern observed in microscopy with an independent approach (Figure 5D). As evident especially in the three replicates of this experiment (Supplementary Figure S4B), the cytoplasmic expression of Drosha was significantly enriched in NCI-H1703 and PC9 cells (Figure 5E). Unfortunately, the overall levels of Drosha in U2OS were very low and not detectable by western blot. This is in line with the comparably low nuclear Drosha signal and the hardly detectable cytoplasmic staining in the microscopy images.
Taken together, these results showed that endogenous Drosha was found also in the cytoplasm in cell lines with a high relative expression of the Dr1 isoform as verified by microscopy as well as biochemical fractionation.
The substrate processing is unaffected by the distinctive cellular distribution pattern of endogenous Drosha splice variants
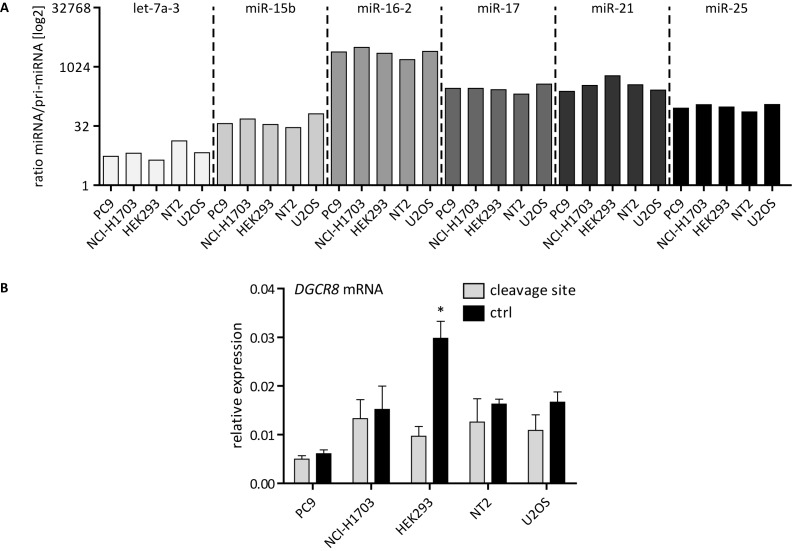
Finally, we wanted to determine the processing efficiency of pri-miRNAs in the five cell lines PC9, NCI-H1703, HEK293, NT2 and U2OS. To allow the direct comparison to the results obtained before (Figure 3C), the same primary and mature miRNAs were used in this experiment. As the different cell lines could exhibit varying expression levels of pri-miRNAs and mature miRNAs, the ratios of both transcripts were calculated, which ultimately provided information about the substrate processing efficiency. Potential effects onto substrate processing would be reflected by an increase or decrease of this ratio. For the six analyzed miRNAs, we could not find differences in the ratio between the respective mature and pri-miRNA indicating a full processing efficiency in the five cell lines used (Figure 6A). The processing of the DGCR8 mRNA was determined through RT-qPCR using specific primer pairs covering the known cleavage site or a fragment located upstream of that region (control fragment = ctrl). While a significantly higher expression of the control fragment than the fragment covering the cleavage site was visible in HEK293 cells, there was no general difference between the two groups of cell lines with and without cytoplasmic Drosha expression, respectively (Figure 6B). Hence, the differential expression of Drosha splice variants did not seem to affect the metabolism of pri-miRNAs or DGCR8.
Figure 6.
The substrate processing is generally unaffected by the distinctive cellular distribution pattern of endogenous Drosha splice variants. (A) Relative expression ratio of mature miRNAs over pri-miRNAs in log2 scale normalized to snRNA U6 or cyclophilin A, respectively. (B) Relative expression of a fragment located upstream (ctrl) to the region including the cleavage site normalized to cyclophilin A. Ctrl = control. Statistical significance calculated with student's t-test, performed for normalized values: *P < 0.05; **P < 0.01; ***P < 0.001. N = 3. Error bars indicate SEM.
DISCUSSION
Drosha is a highly versatile enzyme, which acts in different processes with the most prominent example being the maturation of miRNAs. Many targets of miRNAs are involved in essential cellular processes (64). A deregulation of such processes can lead to different diseases including cancer.
For Drosha, we previously proved the existence of rare processing-deficient splice variants in melanoma cells (53). In the current study, we identified novel and abundant isoforms in the 5′UTR and the N-terminal coding region of Drosha and further characterized the latter. In contrast to the previously described rare C-terminal isoforms, the now discovered N-terminal Drosha isoforms were abundantly expressed, efficiently bound all analyzed co-factors and were catalytically active. The localization experiments proved the presence of Drosha protein in both the nucleus and cytoplasm of Drosha variants harboring exon 7. The interpretation of these results is impeded by the extensive and complex phosphorylation of Drosha which is beyond the scope of this splicing-centered study. Several serine and threonine residues are linked to the regulation of Drosha's subcellular localization in our own and previous studies with sometimes non-overlapping results (32,48,49). This illustrates the elusive picture of the mechanism of Drosha phosphorylation in the context of localization and other cellular processes. Additionally, deletion mutants devoid of the NES or parts of exon 7, respectively, did not exhibit the expected difference between Dr1 as well as Dr4 wild-type and mutant form.
The exact role of the herein discovered splice variants remains to be elucidated. It is tempting to speculate that the alternative splicing and changes in localization could be linked to differential functions due to different Drosha interaction partners or substrates in the nucleus and the cytoplasm. According to previous findings, it could also be involved in response to different stressors like heat or reactive oxygen species. For instance, heat and peroxidative stress leads to a p38 MAPK dependent phosphorylation and subsequent cytoplasmic re-localization of Drosha where it is rapidly degraded by the protease calpain (32). p38 MAPK seems to be capable to directly or indirectly modulate Drosha function to either compromise or increase miRNA biogenesis. The functions and changes of miRNAs and co-factors upon stress are reviewed in (65). While it appears plausible that cytoplasmic Drosha may have different substrates than nuclear Drosha, it could also be possible that selected pri-miRNAs may be exported from the nucleus and processed in the cytoplasm. This in turn would also require a significant expression of DGCR8 protein in the cytoplasm which has not been documented thus far.
In summary, we discovered the first Drosha splice isoforms with abundant expression altering the N-terminal RS-rich domain. Alternative splicing did not affect the binding to co-factors and pri-miRNA or DGCR8 mRNA processing efficiency was equal for all splice variants. However, two of these variants were strongly present in the cytoplasm, whereas two others remained exclusively nuclear. Future investigations are needed to elucidate the connection between alternative splicing of Drosha, its subcellular localization and functional consequences potentially depending on cytoplasmic interaction partners, substrate RNAs or regulatory mechanisms. Hence, this study lays the foundation for a deeper exploration of the N-terminal Drosha splice variants and their role inside and outside of the miRNA biogenesis pathway. Importantly, our study emphasizes the urgent need to systematically identify splicing isoforms also for well-studied proteins and critically analyze their functional and regulatory differences.
Supplementary Material
Acknowledgments
We thank Sarina Keller, Robert Krautz and Hannah Uckelmann for their support in performing experiments as well as all members of the Diederichs lab for helpful discussions. The authors also thank Thomas Tuschl, Rockefeller University, for providing expression plasmids as well as Manuela Brom and Dr Damir Krunic from the DKFZ Light Microscopy Facility for help in image acquisition.
Footnotes
Present address: Stefanie E. Grund, CureVac AG, Paul-Ehrlich-Strasse 15, 72076 Tübingen, Germany.
FUNDING
German Research Foundation [DFG; Di 1421/7-1, SFB 850]; Cell Networks ExcellenceCluster [DFG EXC81 EcTop 5]; RNA@DKFZ Cross Program Topic.
Conflict of interest statement. None declared.
REFERENCES
- 1.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 2.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 7.Landthaler M., Yalcin A., Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Yeom K.H., Lee Y., Han J., Suh M.R., Kim V.N. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen T.A., Jo M.H., Choi Y.G., Park J., Kwon S.C., Hohng S., Kim V.N., Woo J.S. Functional anatomy of the human microprocessor. Cell. 2015;161:1374–1387. doi: 10.1016/j.cell.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Kwon S.C., Nguyen T.A., Choi Y.G., Jo M.H., Hohng S., Kim V.N., Woo J.S. Structure of human DROSHA. Cell. 2016;164:81–90. doi: 10.1016/j.cell.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 12.Yi R., Qin Y., Macara I.G., Cullen B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 15.Winter J., Link S., Witzigmann D., Hildenbrand C., Previti C., Diederichs S. Loop-miRs: active microRNAs generated from single-stranded loop regions. Nucleic Acids Res. 2013;41:5503–5512. doi: 10.1093/nar/gkt251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 17.Ghildiyal M., Zamore P.D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 20.Winter J., Diederichs S. Argonaute proteins regulate microRNA stability: Increased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol. 2011;8:1149–1157. doi: 10.4161/rna.8.6.17665. [DOI] [PubMed] [Google Scholar]
- 21.Diederichs S., Jung S., Rothenberg S.M., Smolen G.A., Mlody B.G., Haber D.A. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diederichs S., Haber D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 23.Hutvagner G., Simard M.J. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 24.Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 25.Ozen M., Creighton C.J., Ozdemir M., Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 26.Karube Y., Tanaka H., Osada H., Tomida S., Tatematsu Y., Yanagisawa K., Yatabe Y., Takamizawa J., Miyoshi S., Mitsudomi T., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 28.Afanasyeva E.A., Mestdagh P., Kumps C., Vandesompele J., Ehemann V., Theissen J., Fischer M., Zapatka M., Brors B., Savelyeva L., et al. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18:974–984. doi: 10.1038/cdd.2010.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Formosa A., Markert E.K., Lena A.M., Italiano D., Finazzi-Agro E., Levine A.J., Bernardini S., Garabadgiu A.V., Melino G., Candi E. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33:5173–5182. doi: 10.1038/onc.2013.451. [DOI] [PubMed] [Google Scholar]
- 30.Jovanovic M., Hengartner M.O. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- 31.Fan P., Chen Z., Tian P., Liu W., Jiao Y., Xue Y., Bhattacharya A., Wu J., Lu M., Guo Y., et al. miRNA biogenesis enzyme Drosha is required for vascular smooth muscle cell survival. PLoS One. 2013;8:e60888. doi: 10.1371/journal.pone.0060888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Q., Li W., She H., Dou J., Duong D.M., Du Y., Yang S.H., Seyfried N.T., Fu H., Gao G., et al. Stress induces p38 MAPK-mediated phosphorylation and inhibition of Drosha-dependent cell survival. Mol. Cell. 2015;57:721–734. doi: 10.1016/j.molcel.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro J.S., Langlois R.A., Pham A.M., Tenoever B.R. Evidence for a cytoplasmic microprocessor of pri-miRNAs. RNA. 2012;18:1338–1346. doi: 10.1261/rna.032268.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shapiro J.S., Schmid S., Aguado L.C., Sabin L.R., Yasunaga A., Shim J.V., Sachs D., Cherry S., tenOever B.R. Drosha as an interferon-independent antiviral factor. Proc. Natl. Acad. Sci. U.S.A. 2014;111:7108–7113. doi: 10.1073/pnas.1319635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triboulet R., Chang H.M., Lapierre R.J., Gregory R.I. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA. 2009;15:1005–1011. doi: 10.1261/rna.1591709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukuda T., Yamagata K., Fujiyama S., Matsumoto T., Koshida I., Yoshimura K., Mihara M., Naitou M., Endoh H., Nakamura T., et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 38.Liang X.H., Crooke S.T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Res. 2011;39:4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu H., Xu H., Miraglia L.J., Crooke S.T. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- 40.Havens M.A., Reich A.A., Hastings M.L. Drosha promotes splicing of a pre-microRNA-like alternative exon. PLoS Genet. 2014;10:e1004312. doi: 10.1371/journal.pgen.1004312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karginov F.V., Cheloufi S., Chong M.M., Stark A., Smith A.D., Hannon G.J. Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gromak N., Dienstbier M., Macias S., Plass M., Eyras E., Caceres J.F., Proudfoot N.J. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep. 2013;5:1499–1510. doi: 10.1016/j.celrep.2013.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johanson T.M., Keown A.A., Cmero M., Yeo J.H., Kumar A., Lew A.M., Zhan Y., Chong M.M. Drosha controls dendritic cell development by cleaving messenger RNAs encoding inhibitors of myelopoiesis. Nat. Immunol. 2015;16:1134–1141. doi: 10.1038/ni.3293. [DOI] [PubMed] [Google Scholar]
- 44.Knuckles P., Vogt M.A., Lugert S., Milo M., Chong M.M., Hautbergue G.M., Wilson S.A., Littman D.R., Taylor V. Drosha regulates neurogenesis by controlling neurogenin 2 expression independent of microRNAs. Nat. Neurosci. 2012;15:962–969. doi: 10.1038/nn.3139. [DOI] [PubMed] [Google Scholar]
- 45.Kadener S., Rodriguez J., Abruzzi K.C., Khodor Y.L., Sugino K., Marr M.T. 2nd, Nelson S., Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burger K., Gullerova M. Swiss army knives: non-canonical functions of nuclear Drosha and Dicer. Nat. Rev. Mol. Cell Biol. 2015;16:417–430. doi: 10.1038/nrm3994. [DOI] [PubMed] [Google Scholar]
- 47.Macias S., Cordiner R.A., Caceres J.F. Cellular functions of the microprocessor. Biochem. Soc. Trans. 2013;41:838–843. doi: 10.1042/BST20130011. [DOI] [PubMed] [Google Scholar]
- 48.Tang X., Zhang Y., Tucker L., Ramratnam B. Phosphorylation of the RNase III enzyme Drosha at Serine300 or Serine302 is required for its nuclear localization. Nucleic Acids Res. 2010;38:6610–6619. doi: 10.1093/nar/gkq547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang X., Li M., Tucker L., Ramratnam B. Glycogen synthase kinase 3 beta (GSK3beta) phosphorylates the RNAase III enzyme Drosha at S300 and S302. PLoS One. 2011;6:e20391. doi: 10.1371/journal.pone.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twyffels L., Gueydan C., Kruys V. Shuttling SR proteins: more than splicing factors. FEBS J. 2011;278:3246–3255. doi: 10.1111/j.1742-4658.2011.08274.x. [DOI] [PubMed] [Google Scholar]
- 51.Long J.C., Caceres J.F. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 52.Shepard P.J., Hertel K.J. The SR protein family. Genome Biol. 2009;10:242. doi: 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grund S.E., Polycarpou-Schwarz M., Luo C., Eichmuller S.B., Diederichs S. Rare Drosha splice variants are deficient in microRNA processing but do not affect general microRNA expression in cancer cells. Neoplasia. 2012;14:238–248. doi: 10.1593/neo.111586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 55.Gagnon K.T., Li L., Janowski B.A., Corey D.R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014;9:2045–2060. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rothbauer U., Zolghadr K., Muyldermans S., Schepers A., Cardoso M.C., Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Allegra D., Mertens D. In-vivo quantification of primary microRNA processing by Drosha with a luciferase based system. Biochem. Biophys. Res. Commun. 2011;406:501–505. doi: 10.1016/j.bbrc.2011.02.055. [DOI] [PubMed] [Google Scholar]
- 58.Sultan M., Schulz M.H., Richard H., Magen A., Klingenhoff A., Scherf M., Seifert M., Borodina T., Soldatov A., Parkhomchuk D., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 59.Zhou H., Di Palma S., Preisinger C., Peng M., Polat A.N., Heck A.J., Mohammed S. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J. Proteome Res. 2013;12:260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 60.Herbert K.M., Pimienta G., DeGregorio S.J., Alexandrov A., Steitz J.A. Phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep. 2013;5:1070–1081. doi: 10.1016/j.celrep.2013.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.la Cour T., Kiemer L., Molgaard A., Gupta R., Skriver K., Brunak S. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 62.Gnad F., Ren S., Cox J., Olsen J.V., Macek B., Oroshi M., Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chuderland D., Konson A., Seger R. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol. Cell. 2008;31:850–861. doi: 10.1016/j.molcel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 64.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 65.Leung A.K., Sharp P.A. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.