Abstract
Artemis is a vertebrate nuclease with both endo- and exonuclease activities that acts on a wide range of nucleic acid substrates. It is the main nuclease in the non-homologous DNA end-joining pathway (NHEJ). Not only is Artemis important for the repair of DNA double-strand breaks (DSBs) in NHEJ, it is essential in opening the DNA hairpin intermediates that are formed during V(D)J recombination. Thus, humans with Artemis deficiencies do not have T- or B-lymphocytes and are diagnosed with severe combined immunodeficiency (SCID). While Artemis is the only vertebrate nuclease capable of opening DNA hairpins, it has also been found to act on other DNA substrates that share common structural features. Here, we discuss the key structural features that all Artemis DNA substrates have in common, thus providing a basis for understanding how this structure-specific nuclease recognizes its DNA targets.
INTRODUCTION
Pathological DNA DSBs can be the most deleterious forms of DNA damage. These breaks can result in cell death from the deletion of a chromosomal arm or by promoting the p53-mediated apoptosis pathway (1). In mammalian cells, DSBs are repaired predominantly by the non-homologous DNA end-joining pathway (NHEJ) pathway. Non-pathological DSBs are created during V(D)J recombination and immunoglobulin heavy chain class switch recombination (CSR), which both contribute to the adaptive immune response. These DSBs also require the NHEJ pathway to be resolved. Any defects in the NHEJ pathway can result in marked sensitivity to ionizing radiation and lead to the ablation of all lymphocytes. However, NHEJ is often imprecise, a characteristic that is useful for immune diversification in lymphocytes, but which might also contribute to deleterious genetic alterations that lead to cancer and perhaps aging. The Artemis nuclease plays a critical role in NHEJ in processing various DNA end configurations at DSBs and is required to open the DNA hairpin intermediates in V(D)J recombination. Artemis is activated by physical contact with DNA-PKcs (DNA–protein kinase catalytic subunit) (2), which must be autophosphorylated in order to stimulate Artemis activity (2–4).
SCID is a genetic disorder characterized by an impairment in the adaptive immune system (5). Humans with mutations in either Artemis or DNA-PKcs are both diagnosed with T−B−NK+ SCID (6,7). Although T−B−NK+ SCID is rare in the general population (1 in 50 000 to 500 000 live births), there is a high incidence (1 in 2000 live births) in the Navajo and Apache Native Americans (8,9). The subset of SCID found in in these Athabascan-speaking Native Americans is due to an autosomal recessive nonsense Artemis mutation in exon 8, which causes a truncation of the 692 aa protein at the 192nd aa (6). As expected, these patients suffer from the early onset of serious infections due to their lack of an adaptive immune system (10). Thus, the biomedical importance of Artemis is substantial.
UNIFYING ELEMENTS OF THE ARTEMIS SUBSTRATES
Hairpins and overhangs were initially identified as Artemis:DNA-PKcs substrates (2). Subsequently, a wider range of DNA structures with single- and double-strand transitions were also identified as substrates (11). For example, blunt-ended DNA molecules were shown to be weaker substrates (12). However, it was unclear what similarities and differences distinguish these substrates.
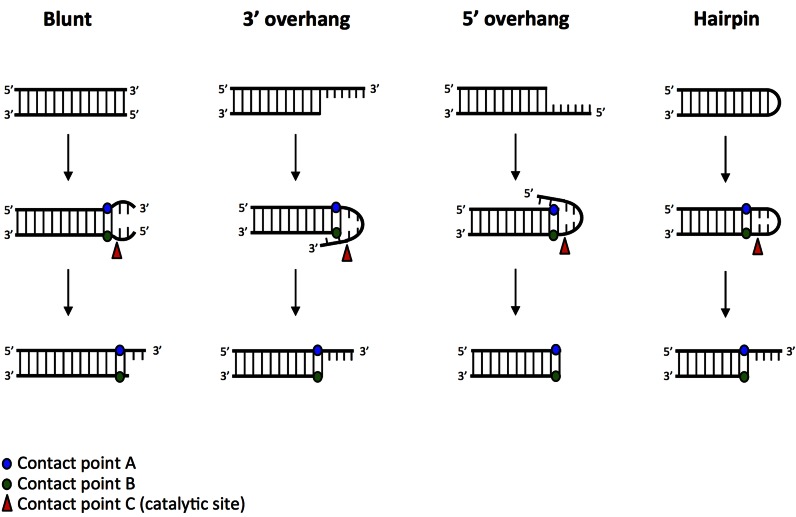
We have now been able to develop a physical model that describes key contact points common to all of the known substrates of Artemis:DNA-PKcs (Figures 1 and 2) (13). This model proposes two contact points (A and B), which are located on the two strands within the duplex portion, directly adjacent to the double- to single-stranded DNA boundary. A third contact point (C, red arrowhead), which is also the catalytic site, is located 1-nt (or an equivalent distance) on the 5′ side of Contact point B (green dot) (Figures 1 and 2); this catalytic site does not have to be on the same DNA strand as Contact point B (see Figure 1, 3′ overhang substrate). In all cases, Artemis:DNA-PKcs is able to distort the single-stranded portion of the substrate into a structure resembling key features of the DNA hairpin substrate. Without these three contact points, Artemis:DNA-PKcs activity is negligible.
Figure 1.
Unifying model describing Artemis activity on common physiological DNA substrates. Functional data on Artemis activity suggests that it has three critical contact points with its DNA substrates that are structurally similar (blunt ends, 3′ and 5′ overhangs, and hairpin structures). Artemis may be able to distort the DNA end to create a ‘hairpin-like’ structure in all cases. Contact point A (blue dot) is located on the 5′→3′ (top) strand at the ss/dsDNA boundary. Contact point B (green dot) is located on the 3′→5′ (bottom) strand directly across from Contact point A. Contact point C (catalytic site, red arrowhead) is located 1-nt (or an equivalent distance) on the 5′ side of Contact point B (green dot), as shown in the figure. Contact point C (catalytic site, red arrowhead) does not need to be on the same DNA strand as Contact point B, but merely an equivalent distance and direction from Contact point B.
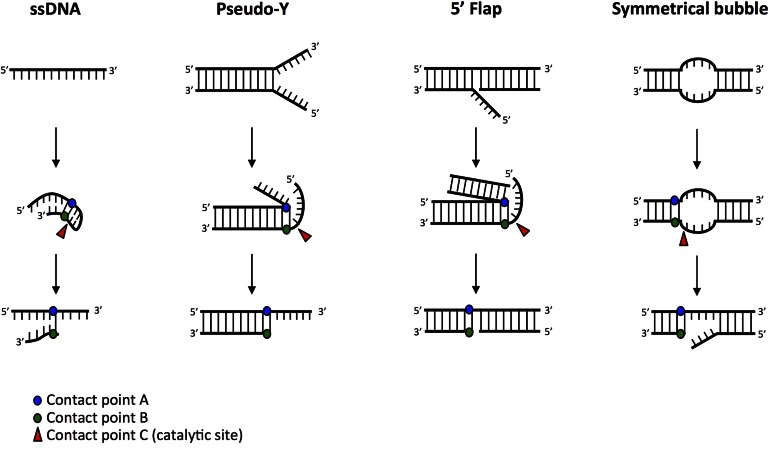
Figure 2.
Artemis activity on other DNA substrates. We propose that Artemis maintains the three contact points on other substrates (ssDNA, pseudo-Y, 5′ flaps and symmetrical bubble structures) by remodeling the end into ‘hairpin-like’ structures. While less apparent, ssDNA has the ability to fold back and base pair with itself to create the ‘hairpin-like’ structures. Contact point A (blue dot) is located on the 5′→3′ (top) strand at the ss/dsDNA boundary. Contact point B (green dot) is located on the 3′→5′ (bottom) strand directly across from Contact point A. Contact point C (catalytic site, red arrowhead) is located 1-nt on the 5′ side of Contact point B as shown in the figure.
With these three contact points, the cutting pattern observed for all of the major substrates can be explained. Among the major substrates, blunt ends are the only ones requiring a DNA end breathing step prior to action by Artemis:DNA-PKcs (13). The requirement for the breathing step explains some key features of the Artemis:DNA-PKcs action at blunt ends (13). First, it explains why Artemis:DNA-PKcs acts on blunt ends less efficiently than DNA termini that have an obvious double- to single-strand boundary (12,13). This is because the breathed state is only very short-lived. Second, it explains why AT-rich DNA termini are cut much faster than the more stable GC-rich DNA-termini (13). Third, once the blunt end breathes open, the transient 5′ single-strand is cut faster than the 3′ single-strand (12,13). This may be explained by steric factors, since removal of the transient 5′ single-strand allows the 3′ single-strand to more readily assume a hairpin conformation (Figure 1).
This model also explains why the resulting product of Artemis:DNA-PKcs activity differs on 5′ and 3′ overhangs (2). The nts on a 5′ overhang substrate are able to fold back toward the duplex to create a hairpin-like configuration. The Artemis:DNA-PKcs complex is able to recognize this hairpin-like structure and cut 1-nt 5′ of Contact point B, resulting in a perfectly blunt-ended product (Figure 1, 5′ overhang substrate). Similarly, the nts on a 3′ overhang substrate also fold back toward the duplex to form this hairpin-like structure. However, in this case, the 4th nt of this overhang is spatially located a distance equivalent to 1-nt 5′ of Contact point B (∼3 to 5 angstroms). Thus, Artemis:DNA-PKcs activity on this substrate results in a 4-nt overhang at the 3′ end (Figure 1, 3′ overhang substrate) (13). This model requires a polarity in the Artemis:DNA-PKcs complex in order to recognize the helical pitch of the double-stranded DNA duplex.
In addition to explaining the mechanism for cleaving the primary substrates of Artemis:DNA-PKcs, this model also explains how less commonly encountered double- to single-stranded boundaries are cut (Figure 2). Among these, only the single-stranded DNA substrate must anneal to itself to form a transient double-stranded substrate, and the short-lived nature of this annealing explains why single-stranded DNA is cut much less efficiently by Artemis or Artemis:DNA-PKcs (4). Various other substrates can also be explained by our model, and, are cleaved at the predicted locations (11). The pseudo-Y, flap and bubble structures may all be encountered during different stages of DNA replication or repair. Importantly, a free DNA terminus (blunt-end, overhang or hairpin) is required to activate the serine/threonine kinase activity of DNA-PKcs (14). Therefore, if a flap or a bubble structure is present internally and far removed from a DNA terminus, DNA-PKcs will not be activated, and thus neither Artemis nor Artemis:DNA-PKcs will cut (11).
CONTRIBUTION OF DNA-PKCS TO SUBSTRATE RECOGNITION
As discussed below, Artemis and DNA-PKcs form a tight complex. DNA-PKcs is only active as a protein kinase when DNA termini, ATP and Mg2+ are present to permit autophosphorylation of DNA-PKcs, which then can stimulate Artemis nuclease activity (2–4). However, using purified proteins, we and others have shown that the divalent cation, Mn2+, enables Artemis to function independent of DNA-PKcs (4,15). All of the substrate structure recognition that we have described above applies not only to Artemis with DNA-PKcs, but also to Artemis in the presence of Mn2+ without DNA-PKcs. This means that the substrate distortion into a hairpin-like configuration is likely due to Artemis alone. It is thought that the C-terminus of Artemis acts as a regulatory region since C-terminal truncation mutants allow Artemis to be constitutively active independent of DNA-PKcs (16). Thus, Mn2+ most likely interacts with Artemis to permit Artemis to be constitutively active. In addition, the three key contact points of Artemis with the DNA substrate are independent of DNA-PKcs.
What then is the role of DNA-PKcs in the Artemis:DNA-PKcs complex? Under physiological conditions (with Mg2+ but not Mn2+), Artemis is inactive without an autophosphoryated DNA-PKcs (3). Therefore, DNA-PKcs is essential for Artemis activity in the presence of Mg2+. Somehow, the Mn2+ must change the conformation of Artemis so as to mimic the effect of an autophophorylated DNA-PKcs. DNA-PKcs requires a broken DNA end for it to bind (usually with Ku) (17,18), and this is necessary for Artemis to be active in cutting the phosphodiester backbone at that DNA end. Therefore, the reliance of Artemis on an activated DNA-PKcs makes the Artemis:DNA-PKcs complex responsive to broken DNA ends.
ROLE OF ARTEMIS: DNA-PKCS IN V(D)J RECOMBINATION
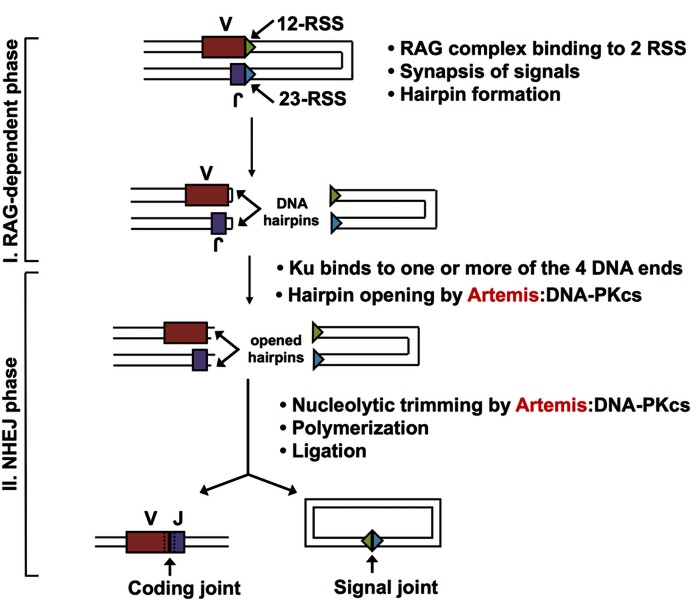
V(D)J recombination relies on the RAG complex to bind and cut at recombination signal sequences (RSS) adjacent to V, D and J segments, resulting in coding ends that are hairpinned and signal ends that are blunt (Figure 3). The hairpin at the coding end must be opened to be joined to form a coding joint. Based on genetic and biochemical evidence, the hairpin opening at coding ends of the V, D and J segments during V(D)J recombination is completely dependent on the Artemis:DNA-PKcs complex (Figure 3) (2,6). Since the Artemis:DNA-PKcs complex is located at each coding end at the time of hairpin opening, the coding end resection is almost certainly due to Artemis:DNA-PKcs. Indeed, the effect of coding end sequence on the extent of resection in vivo matches quite well with the known DNA sequence effects by purified Artemis:DNA-PKcs in biochemical studies (19,20). In addition, the signal ends, which are the DNA fragments cut out and circularized during deletional V(D)J recombination, occasionally suffer a small amount of end resection, which is also due to Artemis:DNA-PKcs activity (21). Thus, the evidence is very strong that the Artemis:DNA-PKcs complex is the nuclease involved in all of the resection that occurs during V(D)J recombination.
Figure 3.
Artemis opens the hairpins generated in V(D)J recombination. V(D)J recombination occurs at sequences called 12-RSS and 23-RSS (triangles in the figure), where RSS designates recombination signal sequence. An RSS contains conserved heptamer and nonamer sequence elements, separated by either 12 or 23 non-conserved base pairs, and hence the designation 12-RSS and 23-RSS. One recombination event requires one 12-RSS and one 23-RSS, and this is called the 12/23 rule. In early lymphoid cells, the RAG complex (RAG1 and 2 along with the constitutively expressed HMGB1 protein) nicks and then hairpins the coding ends at the V and J segments in the figure. Ku can bind to any of the four DNA ends. The Artemis:DNA-PKcs complex then binds to the V and J hairpin ends and nicks the hairpins in a manner that usually results in a 3′ overhang. The ends can be processed further by the Artemis:DNA-PKcs complex and a DNA polymerase to introduce diversity. The NHEJ ligase complex then ligates the ends together. Some antigen receptor loci have not only V and J segments, but also D segments; hence, the name V(D)J recombination.
Importantly, Artemis null mice and DNA-PKcs mutant mice have similar phenotypes that are consistent with our biochemical model (22,23). Cells and animals from both mutants show failure of hairpin opening and failure of coding joint formation, despite normal RAG cutting and completion of signal joint formation. Both also show sensitivity to ionizing radiation. Notably, human Artemis mutant patients and the one known human DNA-PKcs mutant patient show similar phenotypes to these mice (6,7,22,23). We note that some level of leakiness (a low level of successful coding joint formation) was observed in some strains of mice, depending on the amount of 129/SvJ strain background (22). The amount of leakiness drops to nearly zero in strains with a pure C57BL6 background (23). The C57BL6 Artemis mutant response to bone marrow transplants is more comparable to children with Artemis SCID than the ‘leaky’ 129/SvJ strain (23). Even in leaky strains of mice, the vast majority of coding joint formation is blocked. In human Artemis null patients, the level of leakiness is nearly zero (6) [i.e. only a few B cell clones survive (reflecting exceedingly rare coding joint formation) out of hundreds of billions of B cells that failed to survive (reflecting a >99.99% failure of coding joint formation)].
ROLES OF ARTEMIS: DNA-PKCS IN NHEJ
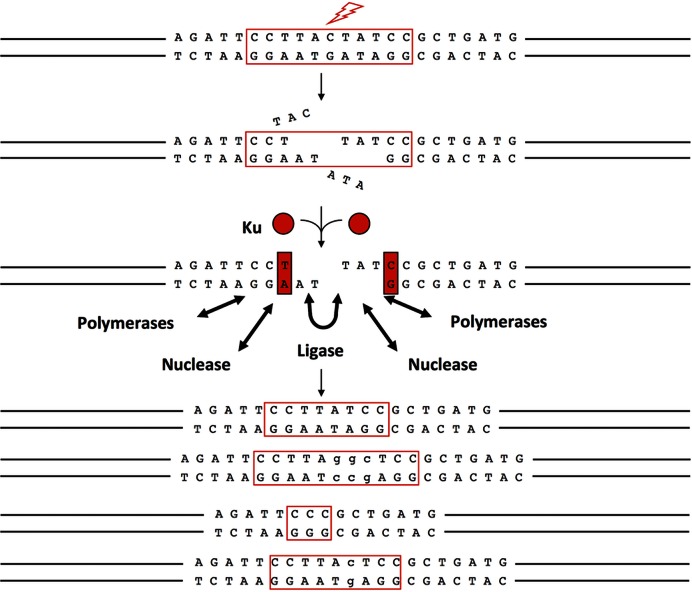
NHEJ is required for the joining phase in V(D)J recombination, and NHEJ relies heavily on Artemis:DNA-PKcs. Based on these two points, we have assumed that Artemis:DNA-PKcs is the major nuclease for all NHEJ processes. We have summarized elsewhere a list of other possible nucleases that might function in NHEJ (e.g., MRN, CtIP, ExoI, WRN and FEN-1) (24). In NHEJ, Ku is the protein that recognizes DSBs and recruits the other NHEJ factors (Figure 4) (17,25). It is one of the most abundant non-histone proteins in mammalian cells (∼400 000 molecules per cell) and tightly binds to DNA ends (KD = 6 × 10−10 M) (26,27). DNA-PKcs is also abundant (50 000 to 100 000 molecules per cell in humans (28,29)), and binds DNA termini well on its own (KD = 3 × 10−9 M) but 100-fold tighter when Ku is present (KD = 3.5 × 10−11 M) (30). Artemis and DNA-PKcs form a tight complex that is stable in vitro even at 1 M monovalent salt (2). We have recently used quantitative Western blots to determine that the number of Artemis molecules per human cell (Reh pre-B cells) is ∼70 000. Therefore, the ratio of Artemis to DNA-PKcs to Ku is approximately 1:1:4. The relative abundance and tight binding of Artemis:DNA-PKcs makes it likely the primary nuclease for mammalian NHEJ and explains why activity is increased when Ku is present (13).
Figure 4.
Artemis is involved in DNA end repair via the NHEJ pathway. Natural causes of pathologic double-strand breaks (DSBs) are expected to generate heterogeneous, incompatible DNA ends with little or no terminal microhomology. Ku (red circle and red rectangle) is the most abundant DNA end binding protein in eukaryotic cells and can slide onto DNA ends that have diverse configurations (38). Once Ku is bound to the DNA end, it can improve the binding equilibrium of the nuclease, polymerases and ligase of NHEJ. The nuclease, polymerases and ligase appear capable of binding to and functioning at a DNA end without Ku, but the binding to the DNA end is tighter when Ku is present. The most clearly identified nuclease thus far is the Artemis:DNA-PKcs complex. The polymerases for NHEJ include the POL X polymerases, pol μ and pol λ (39). The ligase complex of NHEJ consists of XLF, PAXX, XRCC4 and DNA ligase IV (40–42). The bottom portion of the diagram shows four equally plausible outcomes for the joining (the junction is highlighted in a red box). There are hundreds of other possible joining outcomes even for one pair of starting DNA ends with the same end configuration as that shown. This heterogeneity in outcome is in addition to the heterogeneity in the end configuration generated by the original breakage process at that very same set of phosphodiester bonds within the DNA duplex. Thus, there is heterogeneity in the generation of the broken DNA ends and heterogeneity in how these ends are repaired.
In vivo experiments have shown that approximately 20% of DSBs caused by ionizing radiation require Artemis for repair (31). It is important to note that NHEJ is an iterative process (20,32,33). When Artemis is present, it may participate in nearly all NHEJ joining events. But when it is absent as in the SCID T−B−NK+ patients (6), only a small subset of ionizing radiation-induced DNA ends may require Artemis, and thus be unresolvable in its absence. Therefore, the 20% failure to join should be considered a conservative estimate of joining events that would require Artemis in wild type cells. In vivo studies have shown that Artemis is involved in the repair of a subset of DSBs generated by various forms of ionizing radiation (X-ray, γ-ray and α-particles) and reactive oxygen species (ROS) (31,34). These DSBs result in heterogeneous end structures, which make it difficult to determine the specific subset of ends that Artemis is processing. More specific DSB-inducing agents such as neocarzinostatin (NCS) and bleomycin have been shown to generate DSBs that are repaired by Artemis (35,36). NCS generates DSBs with a 5′-phosphate and either a 3′-phosphate or 3′-phosphoglycolate terminus with 1- to 2-nt 3′ overhangs. Bleomycin generates a mixture of blunt-ended or 1-nt 5′-overhang substrates with 5′-phosphates and 3′-phosphglycolate termini (35). It will be interesting to utilize other forms of DSB generating agents to determine the specific subset of DNA ends that require Artemis for repair.
CONCLUSION
We now know the essential structural features for DNA substrates of Artemis. These substrates have a stable or transient double- to single-strand boundary. We have determined how the structural features determine where Artemis will hydrolyze the phosphodiester backbone of each substrate. This model also explains why some DNA substrates are cleaved more efficiently than others. This knowledge will aid in the co-crystallization of Artemis with DNA and with the understanding of Artemis action on its substrates, once a crystal structure is determined (37).
Acknowledgments
The authors thank Dr Raymond D. Mosteller for comments on the manuscript and Dr Nick Pannunzio and Dr Go Watanabe for discussions.
FUNDING
National Institutes of Health (NIH). Funding for open access charge: National Institutes of Health (NIH) [CA100504 to M.R.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA Repair and Mutagenesis. 2nd edn. Washington D.C: ASM Press; 2006. [Google Scholar]
- 2.Ma Y., Pannicke U., Schwarz K., Lieber M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 3.Goodarzi A.A., Yu Y., Riballo E., Douglas P., Walker S.A., Ye R., Harer C., Marchetti C., Morrice N., Jeggo P.A., et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu J., Li S., Zhang X., Wang L.C., Niewolik D., Schwarz K., Legerski R.J., Zandi E., Lieber M.R. DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst) 2010;9:429–437. doi: 10.1016/j.dnarep.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen F.S., Cooper M.D., Wedgwood R.J. The primary immunodeficiencies. N. Engl. J. Med. 1995;333:431–440. doi: 10.1056/NEJM199508173330707. [DOI] [PubMed] [Google Scholar]
- 6.Li L., Moshous D., Zhou Y., Wang J., Xie G., Salido E., Hu D., de Villartay J.P., Cowan M.J. A founder mutation in Artemis, an SNM1-like protein, causes SCID in Athabascan-speaking Native Americans. J. Immunol. 2002;168:6323–6329. doi: 10.4049/jimmunol.168.12.6323. [DOI] [PubMed] [Google Scholar]
- 7.van der Burg M., van Dongen J.J., van Gent D.C. DNA-PKcs deficiency in human: long predicted, finally found. Curr. Opin. Allergy Clin. Immunol. 2009;9:503–509. doi: 10.1097/ACI.0b013e3283327e41. [DOI] [PubMed] [Google Scholar]
- 8.Jones J.F., Ritenbaugh C.K., Spence M.A., Hayward A. Severe combined immunodeficiency among the Navajo. I. Characterization of phenotypes, epidemiology, and population genetics. Hum. Biol. 1991;63:669–682. [PubMed] [Google Scholar]
- 9.Buckley R.H., Schiff R.I., Schiff S.E., Markert M.L., Williams L.W., Harville T.O., Roberts J.L., Puck J.M. Human severe combined immunodeficiency: genetic, phenotypic, and functional diversity in one hundred eight infants. J. Pediatr. 1997;130:378–387. doi: 10.1016/s0022-3476(97)70199-9. [DOI] [PubMed] [Google Scholar]
- 10.Kwong P.C., O'Marcaigh A.S., Howard R., Cowan M.J., Frieden I.J. Oral and genital ulceration: a unique presentation of immunodeficiency in Athabascan-speaking American Indian children with severe combined immunodeficiency. Arch. Dermatol. 1999;135:927–931. doi: 10.1001/archderm.135.8.927. [DOI] [PubMed] [Google Scholar]
- 11.Ma Y., Schwarz K., Lieber M.R. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Yannone S.M., Khan I.S., Zhou R.Z., Zhou T., Valerie K., Povirk L.F. Coordinate 5' and 3' endonucleolytic trimming of terminally blocked blunt DNA double-strand break ends by Artemis nuclease and DNA-dependent protein kinase. Nucleic Acids Res. 2008;36:3354–3365. doi: 10.1093/nar/gkn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang H.H., Watanabe G., Lieber M.R. Unifying the DNA end-processing roles of the artemis nuclease: Ku-dependent artemis resection at blunt DNA ends. J. Biol. Chem. 2015;290:24036–24050. doi: 10.1074/jbc.M115.680900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meek K., Dang V., Lees-Miller S.P. DNA-PK: the means to justify the ends? Adv. Immunol. 2008;99:33–58. doi: 10.1016/S0065-2776(08)00602-0. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y., Giblin W., Kubec M., Westfield G., St Charles J., Chadde L., Kraftson S., Sekiguchi J. Impact of a hypomorphic Artemis disease allele on lymphocyte development, DNA end processing, and genome stability. J. Exp. Med. 2009;206:893–908. doi: 10.1084/jem.20082396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niewolik D., Pannicke U., Lu H., Ma Y., Wang L.C., Kulesza P., Zandi E., Lieber M.R., Schwarz K. DNA-PKcs dependence of Artemis endonucleolytic activity, differences between hairpins and 5' or 3' overhangs. J. Biol. Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- 17.Dvir A., Peterson S.R., Knuth M.W., Lu H., Dynan W.S. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovanovic M., Dynan W.S. Terminal DNA structure and ATP influence binding parameters of the DNA-dependent protein kinase at an early step prior to DNA synapsis. Nucleic Acids Res. 2006;34:1112–1120. doi: 10.1093/nar/gkj504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gauss G.H., Lieber M.R. Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell. Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y., Lu H., Tippin B., Goodman M.F., Shimazaki N., Koiwai O., Hsieh C.L., Schwarz K., Lieber M.R. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004;16:701–713. doi: 10.1016/j.molcel.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Touvrey C., Couedel C., Soulas P., Couderc R., Jasin M., de Villartay J.P., Marche P.N., Jouvin-Marche E., Candeias S.M. Distinct effects of DNA-PKcs and Artemis inactivation on signal joint formation in vivo. Mol. Immunol. 2008;45:3383–3391. doi: 10.1016/j.molimm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Rooney S., Sekiguchi J., Zhu C., Cheng H.L., Manis J., Whitlow S., DeVido J., Foy D., Chaudhuri J., Lombard D., et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell. 2002;10:1379–1390. doi: 10.1016/s1097-2765(02)00755-4. [DOI] [PubMed] [Google Scholar]
- 23.Xiao Z., Dunn E., Singh K., Khan I.S., Yannone S.M., Cowan M.J. A non-leaky Artemis-deficient mouse that accurately models the human severe combined immune deficiency phenotype, including resistance to hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2009;15:1–11. doi: 10.1016/j.bbmt.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pannunzio N.R., Li S., Watanabe G., Lieber M.R. Non-homologous end joining often uses microhomology: implications for alternative end joining. DNA Repair (Amst) 2014;17:74–80. doi: 10.1016/j.dnarep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mimori T., Hardin J.A. Mechanism of interaction between Ku protein and DNA. J. Biol. Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 27.Mimori T., Hardin J.A. Ku polypeptides synthesized in vitro assemble into complex which recognize ends of double-stranded DNA. J. Biol. Chem. 1992;267:331–338. [PubMed] [Google Scholar]
- 28.Anderson C.W., Lees-Miller S.P. The nuclear serine/threonine protein kinase DNA-PK. Crit. Rev. Euk. Gene Exp. 1992;2:283–314. [PubMed] [Google Scholar]
- 29.Anderson C.W., Carter T.H. In: Molecular Analysis of DNA Rearrangements in the Immune System. Jessberger R, Lieber MR, editors. Heidelberg: Springer-Verlag; 1996. pp. 91–112. [Google Scholar]
- 30.West R.B., Yaneva M., Lieber M.R. Productive and nonproductive complexes of Ku and DNA-PK at DNA termini. Mol. Cell. Biol. 1998;18:5908–5920. doi: 10.1128/mcb.18.10.5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riballo E., Kuhne M., Rief N., Doherty A., Smith G.C., Recio M.J., Reis C., Dahm K., Fricke A., Krempler A., et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol. Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 32.Ma Y., Lu H., Schwarz K., Lieber M.R. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 33.Lieber M.R., Lu H., Gu J., Schwarz K. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008;18:125–133. doi: 10.1038/cr.2007.108. [DOI] [PubMed] [Google Scholar]
- 34.Woodbine L., Brunton H., Goodarzi A.A., Shibata A., Jeggo P.A. Endogenously induced DNA double strand breaks arise in heterochromatic DNA regions and require ataxia telangiectasia mutated and Artemis for their repair. Nucleic Acids Res. 2011;39:6986–6997. doi: 10.1093/nar/gkr331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohapatra S., Kawahara M., Khan I.S., Yannone S.M., Povirk L.F. Restoration of G1 chemo/radioresistance and double-strand-break repair proficiency by wild-type but not endonuclease-deficient Artemis. Nucleic Acids Res. 2011;39:6500–6510. doi: 10.1093/nar/gkr257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodarzi A.A., Noon A.T., Deckbar D., Ziv Y., Shiloh Y., Lobrich M., Jeggo P.A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 37.Ochi T., Wu Q., Blundell T.L. The spatial organization of non-homologous end joining: from bridging to end joining. DNA Repair (Amst) 2014;17:98–109. doi: 10.1016/j.dnarep.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falzon M., Fewell J., Kuff E.L. EBP-80, a transcription factor closely resembling the human autoantigen Ku, Recognizes Single- to Double-Strand Transitions in DNA. J. Biol. Chem. 1993;268:10546–10552. [PubMed] [Google Scholar]
- 39.Povirk L.F. Biochemical mechanisms of chromosomal translocations resulting from DNA double-strand breaks. DNA Repair (Amst) 2006;5:1199–1212. doi: 10.1016/j.dnarep.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Buck D., Malivert L., deChasseval R., Barraud A., Fondaneche M.-C., Xanal O., Plebani A., Stephan J.-L., Hufnagel M., LeDiest F., et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 41.Ahnesorg P., Smith P., Jackson S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 42.Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V., et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]