Significance
Optogenetics is a rapidly growing field in which light is used to control biological systems. We show that Oscillatoria acuminata photoactivated adenylate cyclase (OaPAC) protein produces the fundamental second messenger cyclic-AMP (cAMP) in response to blue light, is stable and functional in different mammalian cell types, and can be used to trigger events by raising cAMP level. OaPAC consists of a catalytic domain controlled by a photosensitive blue light using flavin (BLUF) domain. We have solved the crystal structure to show how activity is triggered by light, and guide mutagenesis experiments. Although the catalytic domain resembles known cyclases, the BLUF domains form an unusual intertwined structure. The protein activity is the same in solution as in the crystal, showing that the activation mechanism involves only small molecular movements.
Keywords: optogenetics, X-ray crystallography, blue light, allostery
Abstract
Cyclic-AMP is one of the most important second messengers, regulating many crucial cellular events in both prokaryotes and eukaryotes, and precise spatial and temporal control of cAMP levels by light shows great promise as a simple means of manipulating and studying numerous cell pathways and processes. The photoactivated adenylate cyclase (PAC) from the photosynthetic cyanobacterium Oscillatoria acuminata (OaPAC) is a small homodimer eminently suitable for this task, requiring only a simple flavin chromophore within a blue light using flavin (BLUF) domain. These domains, one of the most studied types of biological photoreceptor, respond to blue light and either regulate the activity of an attached enzyme domain or change its affinity for a repressor protein. BLUF domains were discovered through studies of photo-induced movements of Euglena gracilis, a unicellular flagellate, and gene expression in the purple bacterium Rhodobacter sphaeroides, but the precise details of light activation remain unknown. Here, we describe crystal structures and the light regulation mechanism of the previously undescribed OaPAC, showing a central coiled coil transmits changes from the light-sensing domains to the active sites with minimal structural rearrangement. Site-directed mutants show residues essential for signal transduction over 45 Å across the protein. The use of the protein in living human cells is demonstrated with cAMP-dependent luciferase, showing a rapid and stable response to light over many hours and activation cycles. The structures determined in this study will assist future efforts to create artificial light-regulated control modules as part of a general optogenetic toolkit.
Naturally occurring light sensor domains are able to control many biological processes such as plant development and the behavior of microbes by using the photochemical response of prosthetic flavins, and in recent years, there has been growing interest in understanding and exploiting these proteins for synthetic biology (1). One of the most studied photoreceptor families contains the blue light using flavin (BLUF) domain (2), which responds to blue light and either regulates the activity of an attached enzyme domain or changes its affinity for a repressor protein. BLUF domains were discovered through studies of photo-induced movements of Euglena gracilis (3), a unicellular flagellate, and gene expression in the purple bacterium Rhodobacter sphaeroides (4). OaPAC, a previously undescribed photoactivated adenylate cyclase (PAC) from Oscillatoria acuminata, is a homodimer of a 366-aa residue protein carrying an N-terminal BLUF domain and a C-terminal class III adenylate cyclase (AC) domain. Its small size and substantial activation by light (up to 20-fold more than basal levels in the dark) make it of particular interest for biotechnology. OaPAC shows 57% sequence identity with a PAC from the soil bacterium Beggiatoa, bPAC, but neither structure has so far been solved by crystallography. The OaPAC structures determined in this study will assist future efforts to create artificial light-regulated control modules as part of a general optogenetic toolkit.
BLUF domains have been identified in a number of proteins including YcgF in Escherichia coli (5) and blue-light–regulated phosphodiesterase (BlrP1) in Klebsiella pneumonia (6). Both of these proteins are homodimers with a single EAL domain (7) attached to an N-terminal BLUF domain. BlrP1 is a light-regulated cyclic nucleotide phosphodiesterase; the crystal structure of BlrP1 was the first experimental model showing how the flavin controls enzyme activity (8). The two copies of the protein associate in an antiparallel fashion through conserved residues of the EAL domains, so that the BLUF domains are held apart and act independently, each interacting with the EAL domain of the partner chain. Changes in the hydrogen bonding pattern around the flavin on exposure to light change the coordination of essential metal ions at the active site, thus triggering a fourfold rise in activity under suitable conditions. We show here that OaPAC cyclase activity is more strongly stimulated by light than BlrP1 and its dimer structure is completely different.
We have crystallized OaPAC in two different space groups (SI Appendix, Table S1), showing the two BLUF domains sandwiching a pair of helices (α3 and its symmetry mate) that hold the AC domains distant from the flavins. The two flavin mononucleotide (FMN) binding sites are on opposite sides of the dimer, more than 45 Å apart, yet they apparently act in concert on the active sites, which are themselves a similar distance from the light-sensing prosthetic groups (Fig. 1A). The homologous protein bPAC responds differently to mutations of conserved residues around the FMN binding site compared with other BLUF domains and maintains an activated state for time-scales on the order of 1–10 s (9, 10). We have carried out functional studies of a number of OaPAC mutants, designed on the basis of our model, to determine how the different spatial arrangement and connector helices trigger enzyme activity in the light-activated state.
Fig. 1.
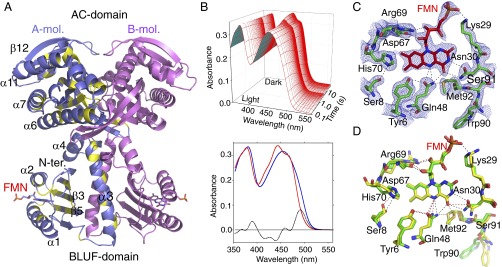
Overall structural properties and solution absorption spectra of OaPAC. (A) The chains are shown as ribbons, with the dyad axis vertical and the BLUF domains at the bottom. One monomer is shown in pink, and the other in blue and yellow, with yellow indicating conserved residues among OaPAC, bPAC (Beggiatoa sp.; ref. 32), and PACαC, the C-terminal region of the α chain of PAC from E. gracilis (3). The β5 strand of the β-sheet is connected to the N terminus of the central α3 helix by a short loop region. The FMN chromophores are shown as stick models. Both domains contribute to the dimer interface; the BLUF domains do not directly contact the AC domain, but the C-terminal end of each α3 helix makes contact with the AC domain of the partner subunit. (B) Absorption spectra of OaPAC in solution. (Upper) Rapid scan spectrophotometry of the switch from light-adapted to dark-adapted conditions, with time shown on a logarithmic scale. (Lower) The visible spectrum of the dark-adapted state (red) is shifted roughly 10 nm to longer wavelength in the light-adapted state (blue). The difference is indicated as a dotted black line. (C) The FMN binding site (hexagonal, open form). The 1.8-Å 2mFo-DFc electron density map, contoured in blue at 1 σ level, of the hexagonal form covering the flavin of one subunit. Nitrogen atoms are colored blue and oxygen red. Carbon atoms of the proteins are colored green, and of the FMN brown. Hydrogen bonds are shown as black dotted lines. Gln-48 adopts a common rotamer, placing both the side-chain nitrogen and oxygen atoms within hydrogen bonding distance of the FMN N5 atom; the nitrogen atom is 3.1 Å from the FMN C4=O carbonyl oxygen, and 2.6 Å from the sulfur atom of Met-92. (D) The FMN binding site (overlay of both forms). Superposition of the different structures shows the similarity of the two models, carbon atoms being colored green and yellow for the open and closed forms of OaPAC, respectively. The differences are close to the experimental error, and the largest change (the side chain of Trp-90) is not well represented in the electron density of the orthorhombic form.
Results and Discussion
Structural Analysis of OaPAC.
Two crystal forms (orthorhombic and hexagonal) were grown in the dark, and have an almost identical monomer structure (backbone rmsd 0.89 Å over 345 residues), although the hexagonal form has much better resolution, 1.8 Å versus 2.9 Å (SI Appendix, Table S1). A ribbon model is shown in Fig. 1A. A nonhydrolyzable ATP analog (ApCpp) was included in the mother liquor of the hexagonal crystal described here but was not found in the final electron density map. The response of OaPAC to light is shown in Fig. 1B. Both crystal forms show similar visible spectra to OaPAC in solution and an equivalent jump in absorption at 492 nm on light exposure that decays with a half-life of a few seconds (SI Appendix, Figs. S1 and S2). The crystal packing does not therefore prevent stimulation by light, or reversion to the dark state, although the hexagonal form shows a more open active site (discussed below) and a slightly longer half-life of the excited state (SI Appendix, Fig. S2 B and D). Attempts to grow crystals of OaPAC under light were unsuccessful, possibly due to a mixture of ground and excited states being present.
BLUF domains consist of a five-stranded β-sheet flanked by helices (11, 12) and use conserved tyrosine and glutamine residues adjacent to the bound FMN to sense light. The simplest proposed sensing mechanism is a rotamer shift of the glutamine, so that after stimulation, this side chain donates a hydrogen bond to the C4=O carbonyl and accepts one from the tyrosine, whereas in the dark state, the glutamine donates to the tyrosine (11, 13, 14). In BlrP1, these changes are accompanied by movement of a nearby methionine on the β5-strand of the BLUF domain, and both “Metin” and “Metout” arrangements have been described (15). The arrangement of Tyr-6, Gln-48, and Met-92 in the two forms of OaPAC is similar to that of the equivalent residues in BlrP1 (Fig. 1 C and D), but the dimer arrangement is different. Light exposure shifts the principal visible FMN absorption bands to longer wavelength, and the carbonyl oxygen at C4 becomes a stronger hydrogen bond acceptor (16). The maximum absorption difference (light state minus dark state) in OaPAC is seen at a wavelength of 492 nm (Fig. 1B).
The main monomer differences between the OaPAC crystal structures are the His-70 side chain moving slightly away from Tyr-6 in the hexagonal form, and Asn-30 breaking a hydrogen bond with N3 of the FMN, leading to a small shift in Trp-90 (Fig. 1D). Superposition of the dimer in the two-crystal forms fails to reveal dramatic conformational changes between domains, but the buried surface area of the orthorhombic form is larger (SI Appendix, Fig. S3 A and B). The hexagonal form shows a movement apart of the AC domains, opening up the active sites formed between the two dimers (SI Appendix, Fig. S3C).
Comparison with Other Proteins.
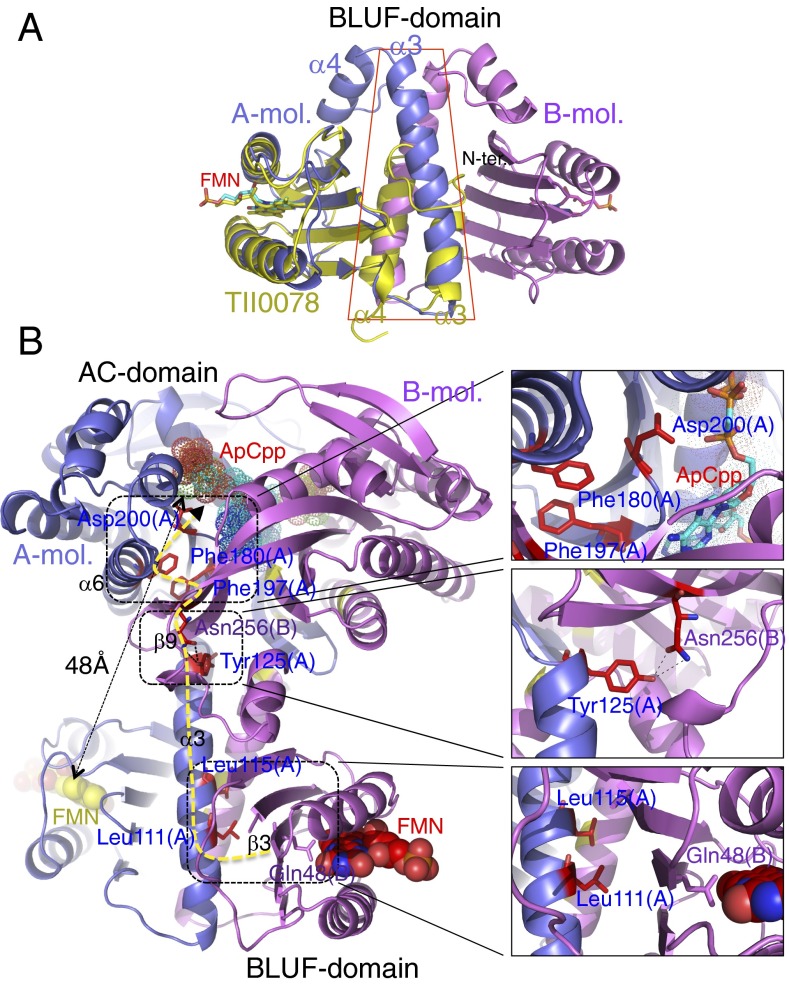
In BlrP1, the two FMN chromophores of the dimer lie more than 54 Å apart, and the two BLUF domains make no contact (8). In OaPAC, the BLUF domains form a large mutual interface by providing one helix each to a central coiled coil so that the two FMN moieties come within 34 Å of each other (Fig. 1A). Overlay of the OaPAC dimer with the BLUF domain of Tll0078 protein from Thermosynechecoccus elongates (12) shows how the C-terminal helices of Tll0078 mimic the central helix pair of OaPAC, but are relatively short, because this protein has no C-terminal domain (Fig. 2A). OaPAC is the first known example to our knowledge of a BLUF protein with this architecture that also possesses a functional output domain.
Fig. 2.
Superposition of OaPAC BLUF domain and positions of mutated residues in the OaPAC structure. (A) Superposition of the BLUF domain of OaPAC (monomers shown in blue and pink) with the monomer BLUF domain of Tll0078 (12) (yellow). The FMN molecule is shown as a stick model, with carbon atoms yellow for Tll0078. The N-terminal residues of the α3 helix pair of OaPAC overlay α3 and α4 of the monomeric Tll0078, so that α3 helix pair of OaPAC and α4 of Tll0078 have opposite direction, but the Tll0078 helices are much shorter. (B) The positions of mutated residues in the structure. The yellow dotted line shows the most direct path that information may travel through the protein once light is detected by the chromophore. ApCpp fitted into the binding site is shown as dotted van der Waals spheres. Insets show more detailed views; Phe-197 fits into an apolar pocket with Phe-180 (Top); Tyr-125 makes hydrogen bonds with Asn-256 of the partner chain (Middle); Leu-111 and Leu-115 lie close to Gln-48 of the partner chain, on the opposite side of strand β3 (Bottom).
Superposition of OaPAC and CyaC, a soluble AC from Spirulina platensis (17), shows the active site residues are conserved (SI Appendix, Fig. S4). Activity of CyaC is promoted by bicarbonate, which triggers relative motion between the active sites on a comparable scale (up to approximately 2 Å) to that suggested by the two crystal forms of OaPAC; bicarbonate ions could not be located directly by X-ray crystallography, but sequence comparisons indicate a binding site close to the substrate ATP (17). OaPAC remains a dimer under all conditions tested (SI Appendix, Fig. S5 A and B). A fundamental issue with OaPAC and the related bPAC is therefore how the signal is transmitted a substantial distance without major conformational switching or change in oligomeric state.
Mutational Analysis.
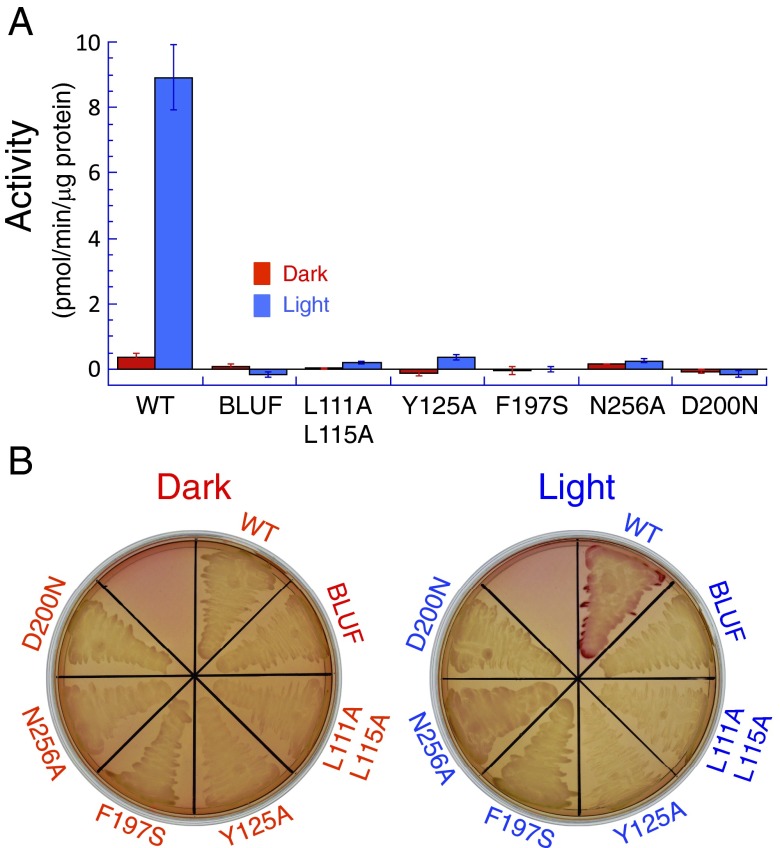
To detail the path that information travels from the FMN to the active site, a number of mutants were constructed and tested for light-stimulated cyclase activity (Figs. 2B and 3). Both in vitro and in vivo assays show that the wild-type protein shows low activity in the dark state and significant activity under light (Fig. 3). The in vivo assay tested restoration of lactose metabolism in an AC-deficient strain of E. coli that also expressed OaPAC. The BLUF domain alone shows no activity in either assay under any conditions (Fig. 3). OaPAC activity (∼9 pmol⋅min−1⋅μg−1 protein) was as high as that of PAC from Euglena measured under the same conditions (18). Adenylate cyclases require a catalytic metal ion coordinated by an aspartate residue (19, 20), and sequence comparisons show the equivalent residue in OaPAC is Asp-200. Replacing this residue with asparagine abolishes any activity under all conditions (Fig. 3). Other mutations between the FMN and active sites were also found to block activation, and are difficult to reconcile with a simple, mechanical conformational control mechanism. The α3 helices are tightly associated by hydrophobic faces (SI Appendix, Fig. S6 A and B), with Leu-111 and Leu-115 facing their symmetry partners across the dimer twofold symmetry axis (Fig. 2B). Replacing either of these leucines with alanine destabilized the protein too strongly to allow significant expression of the mutant, but the double mutant L111A/L115A could be purified. Although these two residues appear to be simply part of a relatively immobile dimer interface, the double mutant showed no activation on light exposure (Fig. 3). This loss of function indicates the important role of hydrophobic interactions between the α3 helices, which has not been noted for other BLUF proteins including BlrP1. At the junction between the α3 helices and the AC domains, the conserved Tyr-125 (at the C-terminal end of the α3 helix) and Asn-256 share an intersubunit hydrogen bond through their side chains (Fig. 2B); both are required for normal protein function although these surface residues play no apparent part in determining protein fold. Phe-197 lies at the dimer interface where one AC domain packs against the other, forming π–π interactions with Phe-180, and close to the hairpin loop carrying Asp-200 (Fig. 2B). Replacing Phe-197 with serine also blocked activity stimulation (Fig. 3). Both polar and apolar contacts across the dimer interface are therefore necessary for signal transmission to the active site.
Fig. 3.
Enzyme assays and photoactivation of OaPAC. (A) Adenylate cyclase activity assay. Activity of wild-type (WT) and mutant OaPAC was determined as described in Methods. Red bars indicate cyclase activity under dark conditions; blue bars indicate activity with light exposure. The BLUF domain alone and every mutant indicated showed no significant activity under any conditions. Error bars indicate the SD for three independent experiments. (B) Restoration of cyclase activity in an adenylate cyclase deficient strain of E. coli (33) by OaPAC. WT and mutant OaPAC were tested for their ability to rescue lactose metabolism, shown by a red color on MacConkey agar. Only the WT protein showed a significant effect under conditions of light exposure.
Allosteric Mechanism of OaPAC.
Overall, the combined structural and functional studies presented here show that light exposure of OaPAC leads directly to minor structural rearrangements around the chromophore, inducing a relatively small relative motion of the cyclase domains (SI Appendix, Table S2 and Movie S1). A recent mutational study of bPAC highlighted the importance of the flavin pocket in the allosteric mechanism (9), but, without an atomic model of the protein to guide the analysis, changes were confined to this region, and did not explore residues that are shown here to connect the BLUF and AC domains. The β5-strand methionine and tryptophan residues (Trp-90 and Met-92 in OaPAC) were, however, shown to have strong effects on dark state recovery and enzyme activity; replacing the methionine with alanine, or the tryptophan with phenylalanine, gave pseudolit proteins with much increased dark-state activity and weak amplification under illumination (9). These two residues are much discussed in regard to signaling mechanism of BLUF domains (8, 13), but are not conserved in PAC from Euglena (SI Appendix, Fig. S7), although this protein shares in common with OaPAC and bPAC a number of resides in the α3 helix, including Pro107, which lies close to Trp-90 of OaPAC and causes a kink after the first turn of the helix. Sequence comparisons therefore suggest that OaPAC, bPAC, and PAC share the same allosteric mechanism, but PAC differs in detail. Mutant forms of bPAC created by Stierl et al. (9) highlight the role of certain conserved residues in communication between chromophore and active site. The mutant bPAC-Ser27Ala (based on a BLUF protein from Naegleria gruberi) showed a 15-nm red shift, which was hypothesized to arise from loss of a hydrogen bond to the flavin. The crystal structures of OaPAC show this serine side chain to sit directly over the isoalloxazine ring, making no hydrogen bond to the flavin or protein.
An important feature of OaPAC is the fact that it shows lower dark-state activity and slower stimulation by blue light than BlrP1. This tighter control possibly arises from the fact the two partner subunits are closely associated and act through the same coiled coil. The process of OaPAC activation is notably different from classical, domain-level conformational changes (such as those observed in hemoglobin) because it occurs rapidly and without substantial domain motions (SI Appendix, Table S2 and Movie S1). A simple mechanical model of coiled-coil proteins has shown that sliding, bending, and twisting modes can yield an allosteric free energy of roughly 2kBT; changes in the Young’s modulus of the α-helices also give significant allosteric effects (21). These vibrational responses to stimulation allow rapid signal transduction through a protein structure with minimal structural change. In fact, BLUF domains are known to photoactivate on a subnanosecond time-scale, faster than protein structural reorganization (16). Comparison of the open and closed structures described here suggests that these are both dark-adjusted forms, although the longer relaxation time of the open (hexagonal) form indicates it may be closer to the fully activated protein (SI Appendix, Fig. S2 B and D).
Examples are already known of proteins switching between states by helix rotation. The complex formed by sensory rhodopsin II and its cognate transducer is a light-activated protein operating by rotation of α-helices about an axis nearly parallel the helical axis. The crystal structure of the transmembrane protein complex has been solved in both the ground state and an intermediate “M” state (22, 23), but molecular dynamics simulations suggest that these atomic models do not demonstrate the full range of conformation change due to restraints of crystal packing (24). The helix movement occurs on a timescale of tens to hundreds of nanoseconds, weakening the binding between the rhodopsin and transducer by 25- to 50-fold (25). In the case of OaPAC, the activity stimulation by light is almost as large, but the fact the protein behavior is almost unaltered in a crystal environment is strong evidence that the allosteric mechanism involves only subtle structural changes, which is consistent with the rapid photoresponse. Adopting the language of the Monod–Wyman–Changeux model (26), the dark state of OaPAC is “tense” and the illuminated state “relaxed,” but there is no thermodynamic reason for supposing these states must have dramatically different atomic coordinates (27).
Application in Optogenetics of OaPAC.
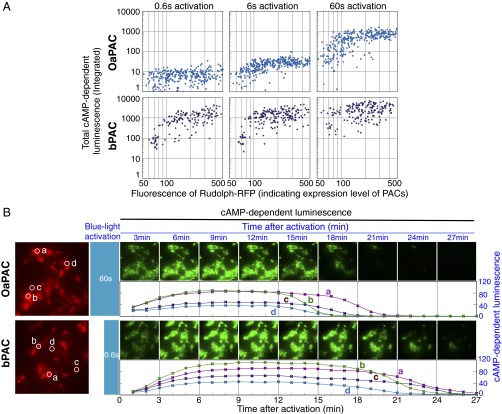
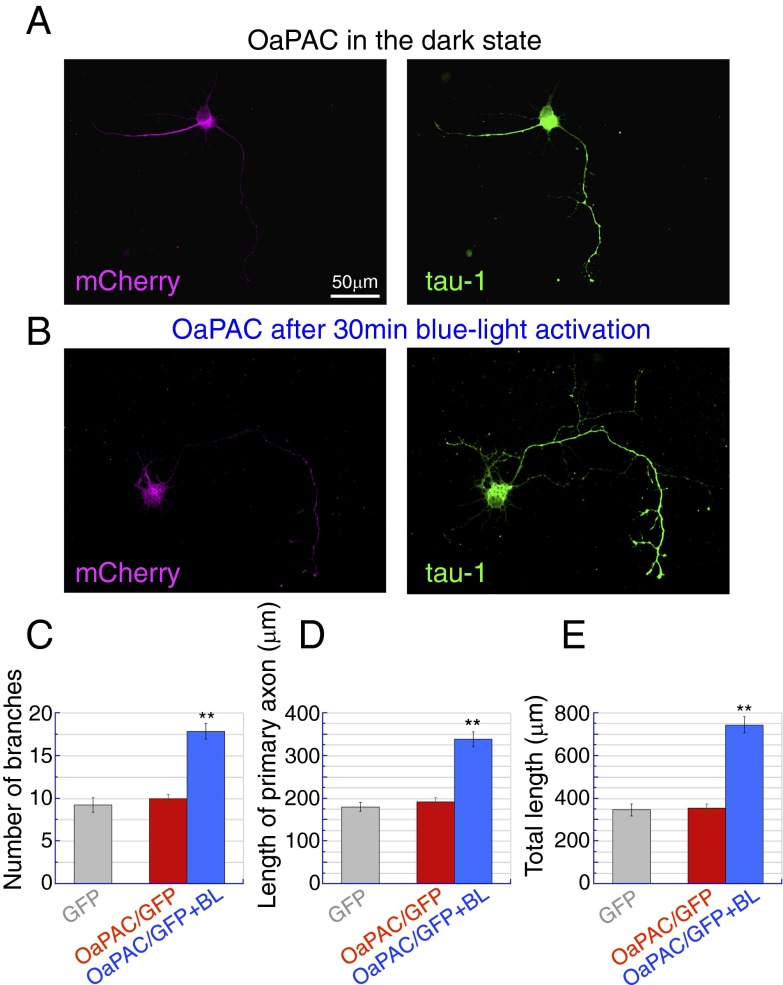
To be of use as an optogenetic tool, a light-stimulated enzyme must show consistent responses over time. This consistency requires stability of the protein itself as well as commensurate activation responses to equal stimuli at different times. HEK293 cells, a cultured cell line derived from human embryonic kidney cells, were used to demonstrate OaPAC function in living tissue. OaPAC was expressed with GloSensor-22F cAMP, a luciferase-based cAMP reporter, allowing OaPAC stimulation to be measured directly by luminescence (SI Appendix, Fig. S8 and Movie S2). In one experiment, emitted light was constantly monitored while stimulating blue light was applied in 30-s pulses every 4 min. Intracellular cAMP concentration showed a highly reproducible response, rising immediately on blue-light exposure and continuing to rise for a further minute afterward, before peaking and returning to basal level over the course of several minutes (SI Appendix, Fig. S8B). In a separate experiment, blue light was applied in 1-min pulses, and the luminescence was measured 2 min after each pulse (SI Appendix, Fig. S8C). Luminescence decayed to background levels within 10 min, allowing the experiment to be repeated over a period of hours, six times per hour. The same luminescence response was observed over a period of 8 h, indicating the system is stable (SI Appendix, Fig. S8 C and D). A comparison of photostimulation of OaPAC and bPAC in HEK293 cells is shown in Fig. 4. Luminescence jumped approximately two orders of magnitude on light exposure of both proteins, but OaPAC shows lower minimum photoactivity (in the dark) and lower maximum photoactivity (when illuminated). OaPAC also requires longer illumination times to give the same rise in cAMP level, allowing finer control of the degree of stimulation. The lower light sensitivity allows much more precise control of cAMP level in human cells than achievable with Euglena PAC proteins (28). The ability of OaPAC to work in different cells types in demonstrated in Fig. 5, which shows rat hippocampal neurons expressing OaPAC together with the fluorescent protein mCherry. After blue light stimulation, the neurons show significant increases in axonal growth (both length and branching) due to the raised cAMP level. A comparison between the effects of OaPAC and PACα on neurons is given in SI Appendix, Fig. S9.
Fig. 4.
Comparison of adenylate cyclase activities between OaPAC and bPAC expressed in cultured HEK293 cells. HEK293 (human embryonic kidney) cells expressing OaPAC or bPAC were activated by blue light, and the rise in intracellular cAMP was monitored by luminescence imaging of cAMP-dependent luciferase. The cellular expression level of each PAC was also monitored by coexpressed fluorescent protein using fluorescence microscopy. (A) The activities of OaPAC and bPAC after different stimulation times. The total cAMP-dependent-luminescence measured within each cell (indicating PAC activity) is plotted against fluorescence intensity of RFP (indicating PAC expression level) on a log-log scale. Blue-light activation of PACs was carried out for three different time periods (0.6 s, 6 s, and 60 s). Upper shows OaPAC and Lower shows bPAC. For quantitative image analysis, cells were randomly sampled under each set of conditions (for OaPAC: 0.6 s, n = 373; 6 s, n = 339; 60 s, n = 343, for bPAC: 0.6 s, n = 157; 6 s, n = 208; 60 s, n = 193), each result being indicated by a dot. (B) Rise and fall of intracellular cAMP level after photoactivation of OaPAC (Upper, 60 s of blue-light activation) or bPAC (Lower, 0.6 s of blue-light activation). A fluorescence micrograph on the left of each row shows RFP (red), indicating expression level of PAC, and time-sequential images of cAMP-dependent luminescence (60 s per image, green color) are shown. A plot of luminescence intensity (indicating concentration of cAMP) at four independent cells (a–d, shown by white circles in the fluorescence micrographs) is also shown.
Fig. 5.
The effect of OaPAC activation on axonal growth in neurons. Primary cultures of hippocampal neurons were prepared from postnatal day 3-4 rat pups. Membrane-GFP (mGFP), mGFP/OaPAC were transfected on 1 d in vitro (DIV). Blue light stimulation (BL) was applied for 30 min on 4 DIV, and cells were then fixed on 7 DIV and immunostained for GFP, mCherry, and the neurite marker tau-1. (A and B) Representative images of OaPAC-transfected neurons (7 DIV) cultured without (A) or with (B) blue light stimulation. OaPAC expression was confirmed by mCherry (magenta), and the axonal morphology was visualized by tau-1 (green). (Scale bar: 50 μm.) (C) Bar graph indicating the number of axon branches for cells transfected with GFP alone (gray), OaPAC/GFP (dark state, red), or OaPAC/GFP with blue light stimulation (blue). **P < 0.01 vs. OaPAC without light; Steel–Dwass test after Kruskal–Wallis test, n = 30 cells for each group. (D) Bar graph indicating the length of primary axons. Colors represent the same cell groups as (C). **P < 0.01 vs. OaPAC without light, Tukey’s test after one-way ANOVA, n = 30 cells for each group. (E) Bar graph indicating the total axonal length. **P < 0.01 vs. OaPAC without light; Tukey’s test after one-way ANOVA, n = 30 cells for each group.
Recently bPAC has been used to control the motility of transgenic mouse sperm through light-stimulated production of cAMP (29). Here, we have not only shown that OaPAC offers stable control of cAMP levels in mammalian cells over extended periods, but also the structural basis for light stimulation. As a small, blue-light sensitive system of known structure, OaPAC offers the chance to create new photoactivated proteins that can operate alongside near-infrared wavelength sensitive biliverdin IXα-containing bacteriophytochromes (30, 31) with minimal interference, allowing independent control of different artificial light-stimulated systems.
Methods
Recombinant OaPAC protein was expressed in E. coli and purified by using standard protocols. Details of the materials and methods used in this study, including cloning and protein purification, crystallography, in vivo and in vitro activity assays, spectroscopic and optogenetic analysis of OaPAC are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Prof. Masakatsu Watanabe, recently deceased, for assistance and encouragement and the staff of the beam lines (BL-1A and BL-17A) at Photon Factory and for their help with X-ray data collection (Proposal 2014-G014). This research was supported by the Ministry of Education, Culture, Sports, Science and Technology Platform Project for Supporting in Drug Discovery and Life Science Research grant (to S.-Y.P.) and Japan Society for the Promotion of Science (to M.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.L. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4YUS and 4YUT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517520113/-/DCSupplemental.
References
- 1.Gomelsky M. Special issue on synthetic photobiology. ACS Synth Biol. 2014;3(11):780–781. doi: 10.1021/sb500350t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomelsky M, Klug G. BLUF: A novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci. 2002;27(10):497–500. doi: 10.1016/s0968-0004(02)02181-3. [DOI] [PubMed] [Google Scholar]
- 3.Iseki M, et al. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415(6875):1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 4.Masuda S, Bauer CE. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell. 2002;110(5):613–623. doi: 10.1016/s0092-8674(02)00876-0. [DOI] [PubMed] [Google Scholar]
- 5.Rajagopal S, Key JM, Purcell EB, Boerema DJ, Moffat K. Purification and initial characterization of a putative blue light-regulated phosphodiesterase from Escherichia coli. Photochem Photobiol. 2004;80(3):542–547. doi: 10.1562/2004-06-16-RA-203. [DOI] [PubMed] [Google Scholar]
- 6.Tyagi A, et al. Photodynamics of blue-light-regulated phosphodiesterase BlrP1 protein from Klebsiella pneumoniae and its photoreceptor BLUF domain. Chem Phys. 2008;354(1–3):130–141. [Google Scholar]
- 7.Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: Enzymatically active and inactive EAL domains. J Bacteriol. 2005;187(14):4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barends TRM, et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature. 2009;459(7249):1015–1018. doi: 10.1038/nature07966. [DOI] [PubMed] [Google Scholar]
- 9.Stierl M, Penzkofer A, Kennis JT, Hegemann P, Mathes T. Key residues for the light regulation of the blue light-activated adenylyl cyclase from Beggiatoa sp. Biochemistry. 2014;53(31):5121–5130. doi: 10.1021/bi500479v. [DOI] [PubMed] [Google Scholar]
- 10.Stierl M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2011;286(2):1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson S, et al. Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry. 2005;44(22):7998–8005. doi: 10.1021/bi0502691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kita A, Okajima K, Morimoto Y, Ikeuchi M, Miki K. Structure of a cyanobacterial BLUF protein, Tll0078, containing a novel FAD-binding blue light sensor domain. J Mol Biol. 2005;349(1):1–9. doi: 10.1016/j.jmb.2005.03.067. [DOI] [PubMed] [Google Scholar]
- 13.Conrad KS, Manahan CC, Crane BR. Photochemistry of flavoprotein light sensors. Nat Chem Biol. 2014;10(10):801–809. doi: 10.1038/nchembio.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathes T, Götze JP. A proposal for a dipole-generated BLUF domain mechanism. Front Mol Biosci. 2015;2(62):62. doi: 10.3389/fmolb.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung A, Reinstein J, Domratcheva T, Shoeman RL, Schlichting I. Crystal structures of the AppA BLUF domain photoreceptor provide insights into blue light-mediated signal transduction. J Mol Biol. 2006;362(4):717–732. doi: 10.1016/j.jmb.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Stelling AL, Ronayne KL, Nappa J, Tonge PJ, Meech SR. Ultrafast structural dynamics in BLUF domains: Transient infrared spectroscopy of AppA and its mutants. J Am Chem Soc. 2007;129(50):15556–15564. doi: 10.1021/ja074074n. [DOI] [PubMed] [Google Scholar]
- 17.Steegborn C, Litvin TN, Levin LR, Buck J, Wu H. Bicarbonate activation of adenylyl cyclase via promotion of catalytic active site closure and metal recruitment. Nat Struct Mol Biol. 2005;12(1):32–37. doi: 10.1038/nsmb880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshikawa S, Suzuki T, Watanabe M, Iseki M. Kinetic analysis of the activation of photoactivated adenylyl cyclase (PAC), a blue-light receptor for photomovements of Euglena. Photochem Photobiol Sci. 2005;4(9):727–731. doi: 10.1039/b417212d. [DOI] [PubMed] [Google Scholar]
- 19.Steegborn C, et al. A novel mechanism for adenylyl cyclase inhibition from the crystal structure of its complex with catechol estrogen. J Biol Chem. 2005;280(36):31754–31759. doi: 10.1074/jbc.M507144200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tesmer JJ, Sunahara RK, Gilman AG, Sprang SR. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsalpha.GTPgammaS. Science. 1997;278(5345):1907–1916. doi: 10.1126/science.278.5345.1907. [DOI] [PubMed] [Google Scholar]
- 21.Hawkins RJ, McLeish TC. Dynamic allostery of protein alpha helical coiled-coils. J R Soc Interface. 2006;3(6):125–138. doi: 10.1098/rsif.2005.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordeliy VI, et al. Molecular basis of transmembrane signalling by sensory rhodopsin II-transducer complex. Nature. 2002;419(6906):484–487. doi: 10.1038/nature01109. [DOI] [PubMed] [Google Scholar]
- 23.Moukhametzianov R, et al. Development of the signal in sensory rhodopsin and its transfer to the cognate transducer. Nature. 2006;440(7080):115–119. doi: 10.1038/nature04520. [DOI] [PubMed] [Google Scholar]
- 24.Nishikata K, Ikeguchi M, Kidera A. Comparative simulations of the ground state and the M-intermediate state of the sensory rhodopsin II-transducer complex with a HAMP domain model. Biochemistry. 2012;51(30):5958–5966. doi: 10.1021/bi300696b. [DOI] [PubMed] [Google Scholar]
- 25.Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc Natl Acad Sci USA. 2006;103(44):16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: A plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 27.Kotani M. 1968. Fluctuation in quaternary structure of proteins and cooperative ligand binding I. Suppl Prog Theor Phys (Extra number):233–241.
- 28.Schröder-Lang S, et al. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods. 2007;4(1):39–42. doi: 10.1038/nmeth975. [DOI] [PubMed] [Google Scholar]
- 29.Jansen V, et al. Controlling fertilization and cAMP signaling in sperm by optogenetics. eLife. 2015;4(4):05161. doi: 10.7554/eLife.05161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu MH, Gomelsky M. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol. 2014;3(11):802–810. doi: 10.1021/sb400182x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu MH, et al. Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci USA. 2014;111(28):10167–10172. doi: 10.1073/pnas.1324301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mussmann M, et al. Insights into the genome of large sulfur bacteria revealed by analysis of single filaments. PLoS Biol. 2007;5(9):e230. doi: 10.1371/journal.pbio.0050230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawamukai M, et al. Nucleotide sequence and characterization of the sfs1 gene: sfs1 is involved in CRP*-dependent mal gene expression in Escherichia coli. J Bacteriol. 1991;173(8):2644–2648. doi: 10.1128/jb.173.8.2644-2648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.