Significance
We show here that binding of the thyroid hormone triiodothyronine to the thyroid hormone receptors (TRs) antagonizes TGF-β/SMAD (mothers against decapentaplegic)-dependent transcription. Transcriptionally inactive TR mutants that do not bind coactivators retained most of the capacity of suppressing transactivation by TGF-β/SMAD, whereas selective mutations in the DNA binding domain abolished this action. TGF-β is a major profibrogenic cytokine, and through this transcriptional mechanism, the hormone-bound TRs act as an endogenous barrier to moderate liver and skin fibrosis. These antagonistic actions on TGF-β/SMAD transcription suggest that TR ligands might be used to block the progression of fibrotic diseases. The natural hormone cannot be used clinically because of severe adverse effects, but novel synthetic ligands with fewer effects might be potentially developed and used.
Keywords: thyroid hormone receptors, TGF-β, SMADs, fibrosis
Abstract
TGF-β, the most potent profibrogenic factor, acts by activating SMAD (mothers against decapentaplegic) transcription factors, which bind to SMAD-binding elements in target genes. Here, we show that the thyroid hormone triiodothyronine (T3), through binding to its nuclear receptors (TRs), is able to antagonize transcriptional activation by TGF-β/SMAD. This antagonism involves reduced phosphorylation of SMADs and a direct interaction of the receptors with SMAD3 and SMAD4 that is independent of T3-mediated transcriptional activity but requires residues in the receptor DNA binding domain. T3 reduces occupancy of SMAD-binding elements in response to TGF-β, reducing histone acetylation and inhibiting transcription. In agreement with this transcriptional cross-talk, T3 is able to antagonize fibrotic processes in vivo. Liver fibrosis induced by carbon tetrachloride is attenuated by thyroid hormone administration to mice, whereas aged TR knockout mice spontaneously accumulate collagen. Furthermore, skin fibrosis induced by bleomycin administration is also reduced by the thyroid hormones. These findings define an important function of the thyroid hormone receptors and suggest TR ligands could have beneficial effects to block the progression of fibrotic diseases.
TGF-β is a cytokine with pleiotropic effects in tissue homeostasis that plays a key role in pathological processes such as cancer and fibrosis (1). Fibrotic diseases, a leading cause of morbility and mortality, are characterized by excessive deposition of extracellular matrix, particularly collagen (2–4). TGF-β1, one of the TGF-β isoforms, is the most potent profibrogenic factor. TGF-β signaling promotes fibroblast differentiation into α-smooth muscle actin (α-SMA) expressing myofibroblasts and stimulates collagen deposition after a variety of injuries in tissues, including, among others, the liver (5–8) and the skin (9). Furthermore, myofibroblast-like cells respond to TGF-β by expressing increased amounts of this factor, thus contributing to fibrosis development through autocrine and paracrine loops of TGF-β‐stimulated collagen production (10).
TGF-β signals via type I and type II serine/threonine kinase receptors. The main signaling pathway involves phosphorylation of regulatory SMADs (mothers against decapentaplegic), such as SMAD2 and SMAD3, which then associate with the coregulatory SMAD4 and translocate to the nucleus (11, 12), where in cooperation with other transcription factors (13), they stimulate transcription of target genes that contain SMAD binding elements (SBEs). Recruitment of coactivators with histone acetyl transferase activity leads to promoter acetylation and transcriptional activation (14). Negative feedback is provided by SMAD-induced expression of the inhibitory SMAD7, which leads to polyubiquitylation and degradation of the TGF-β receptors (15). SMAD factors have been shown to be essential mediators of the fibrogenic actions of TGF-β (16–20).
The actions of the thyroid hormone triiodothyronine (T3) are mediated by binding to nuclear receptors TRα and TRβ. TRα1 and TRβ1 are the two main thyroid hormone-binding isoforms and regulate transcription either by direct binding to thyroid hormone response elements (TREs) in target genes or by modulating the activity of other transcription factors and signaling pathways (21, 22). TRβ mediates the majority of the actions of T3 in the liver (23), playing an important role in regulating metabolism (24, 25), regeneration (26), senescence (27), and carcinogenesis (28). Recent findings suggest that nonalcoholic fatty liver disease, which can progress to liver fibrosis, cirrhosis, and hepatocellular carcinoma, is associated with impaired thyroid action in the liver (29–32). We have shown that expression of TRβ1 in liver cancer cells represses TGF-β-mediated proliferation and induction of metalloproteases (33), suggesting that the receptor could antagonize the response to the growth factor and, therefore, influence carcinogenesis. In this work, we demonstrate that T3 binding to TRs antagonizes TGF-β/SMAD-dependent transcription by a mechanism that involves a direct interaction between the receptors and SMAD factors, reduced SMAD phosphorylation, and decreased recruitment of SMADs to the SBEs in target genes. The importance of this transcriptional antagonism is demonstrated by the finding that the thyroid hormones attenuate fibrotic processes in vivo in the mouse.
Results
T3 Antagonizes TGF-β/SMAD-Dependent Transcription.
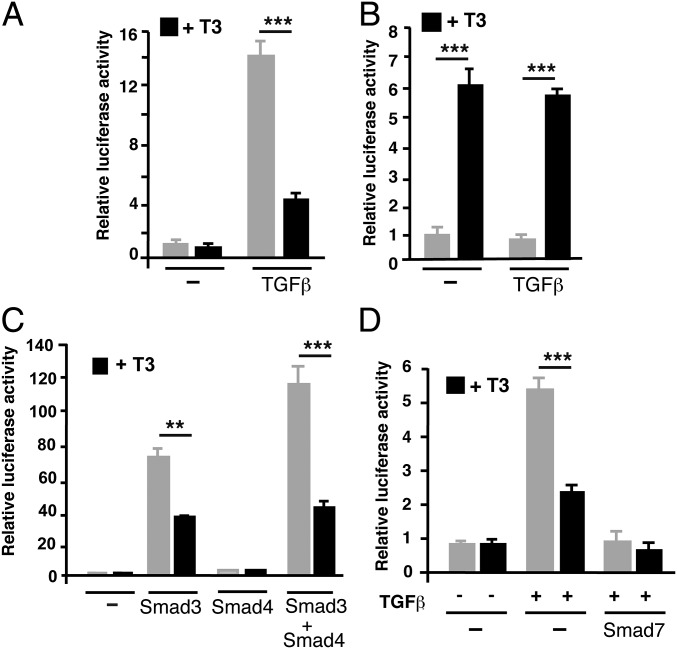
To analyze whether thyroid hormone signaling could regulate TGF-β-mediated transcription, we performed transient transfection assays in rat pituitary GH4C1 cells, which are highly responsive to these hormones, using the p3TP-lux reporter plasmid (34), bearing SBEs. TGF-β markedly stimulated the activity of the reporter construct in these cells, and a physiological T3 concentration strongly antagonized this response (Fig. 1A). The antagonism is not reciprocal, as transactivation of a TRE-containing plasmid by T3 was not altered in TGF-β-treated cells (Fig. 1B). To confirm that the antagonistic effect of T3 was a result of inhibition of SMAD-dependent transcription, and not to changes in TGF-β receptor expression or activation, cells were transfected with expression vectors for SMAD3 and SMAD4 and then incubated in the presence and absence of T3. SMAD3 overexpression increased reporter activity significantly, and this activity was further enhanced in the cells transfected with the coregulatory SMAD4, which by itself lacks transcriptional activity (12). T3 was able to markedly reduce stimulation by these factors, showing that the hormone inhibits TGF-β-dependent transactivation via regulation of SMAD activity (Fig. 1C). This was further demonstrated by the finding that TGF-β-dependent stimulation, as well as T3 antagonism, was abolished after expression of the inhibitory SMAD7 (Fig. 1D). Furthermore, a GAL-SMAD3 fusion plasmid was cotransfected with a luciferase reporter plasmid containing binding motifs for GAL4. GAL-SMAD3 consists of the DNA binding domain (DBD) of GAL4 fused to SMAD3. In parallel with the observed changes in transcriptional activity, incubation with TGF-β strongly increased SMAD3-dependent gene expression, and this response was reduced in T3-treated cells. In contrast, neither TGF-β nor T3 affected the activity of a plasmid containing the GAL4-DBD alone (SI Appendix, Fig. S1). Therefore, T3 appears to directly repress SMAD-dependent transcription.
Fig. 1.
T3 inhibits TGF-β-dependent transactivation. (A) Transient transfection assays in GH4C1 cells with the reporter plasmid p3TP-lux bearing the SBE-containing fragment of the PAI-1 (plasminogen activator inhibitor-1) promoter. Luciferase activity was determined in cells incubated for 36 h with 5 nM T3 and 10 ng/mL TGF-β for the last 5 h. (B) Parallel experiments in cells transfected with a reporter plasmid containing a consensus TRE. (C) Cells were transfected with expression vectors for SMAD3 and/or SMAD4 or with an empty vector (−), and luciferase activity was determined after 36 h incubation in the absence and presence of T3. (D) Luciferase activity in cells transfected with the empty vector or a SMAD7 vector and treated with T3 and/or TGF-β, as indicated. All data (mean ± SD; n = 3) are expressed as fold induction over the values obtained in the untreated cells and are representative of three independent experiments. Significance of ANOVA posttest (Bonferroni) between T3-treated and untreated cells is indicated.
The antagonistic effect of T3 was observed in cells transfected with other SBE-bearing plasmids. Thus, SMADs stimulated the activity of the Smad7 and p15 promoters, and T3 also antagonized this activation (SI Appendix, Fig. S2A). To analyze whether the antagonistic effect of T3 was observed in other cell types, the p3TP–lux construct was transfected into human hepatocarcinoma HepG2-TRβ cells that express ΤRβ1 in a stable manner. Incubation with TGF-β or transfection of SMAD3 and SMAD4 activated the promoter, and T3 attenuated this activation, which was again abolished by SMAD7, whereas T3-dependent transactivation was not affected in TGF-β-treated cells, reproducing the results obtained in GH4C1 cells (SI Appendix, Fig. S2B). Furthermore, T3 caused a strong reduction of SMAD-dependent transactivation in murine neuroblastoma N2aβ cells expressing ΤRβ1, and a much weaker but detectable inhibition in human HeLa cells that express low endogenous levels of TRs (SI Appendix, Fig. S2C).
Both TRα and TRβ Can Mediate the Antagonistic Effect of T3 on TGF-β/SMAD Signaling.
To test whether or not TRα could also antagonize TGF-β/SMAD transcriptional activity, we used mink Mv1Lu epithelial cells, a cell line widely used to study TGF-β signaling. In these cells, T3 was unable to repress stimulation of the SBE-bearing promoter by the TGF or SMADs. However, transfection of either TRα or ΤRβ caused some ligand-independent SBE activation, and T3 repressed very potently both basal activity and transactivation by TGF-β or SMADs (SI Appendix, Fig. S3). Therefore, both receptor isoforms appear to mediate hormone-dependent repression of TGF-β/SMAD activity while inducing activity in the absence of T3.
TR Domains Involved in Repression of TGF-β-Dependent Transcription by T3.
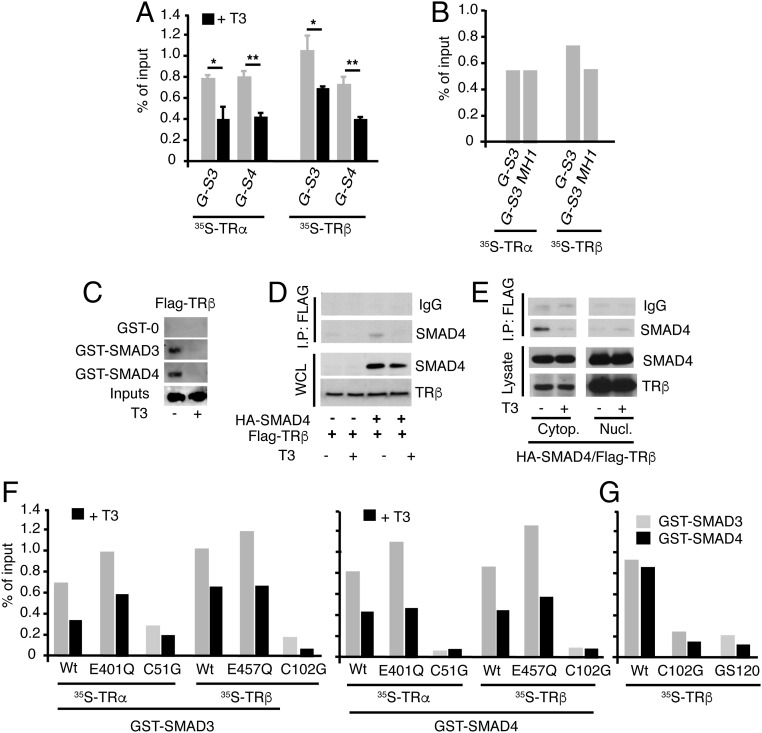
To identify the TR regions involved in this inhibition, we cotransfected Mv1Lu cells with a SBE-bearing plasmid and expression vectors for the native receptors and different TRα and ΤRβ mutants. Endogenous TR expression was not detected in these cells, and the WT and mutant receptors were expressed at comparable levels in the presence and absence of T3 (Fig. 2 C and F). The mutations K288I and E457Q in ΤRβ and the equivalent mutation E401Q in TRα are located in the ligand-binding domain (LBD) and abolish the recruitment of coactivators and T3-dependent transcription, although they still bind T3 (35). However, these mutants maintain the ligand-independent activity and mediate significant T3-dependent repression of TGF-β-dependent transactivation, although with less potency than the WT receptors. In contrast, the TRα LBD alone was unable to antagonize stimulation by TGF-β (Fig. 2A). The involvement of the DBD in the antagonism, was proved using DBD mutants (Fig. 2 B and E). Mutations C51G in TRα or C102G in ΤRβ affect a conserved residue in the first zinc finger that participates in the coordination of the zinc ion (36), and in the TRβ-GS120 mutant, two residues in the P-box sequence responsible for TRE recognition were changed to those present in several steroid receptors, thus altering the specificity of DNA binding. In the absence of T3, expression of the DBD mutants did not increase reporter activity in response to TGF-β and was unable to mediate a significant ligand-dependent inhibition of TGF-β-mediated transactivation. This suggests that residues in the DBD appear to be required for T3-dependent antagonism of TGF-β-induced transcription (Fig. 2 A and D). Other possible interpretation was that the DBD mutants had enhanced inhibition of TGF-β signaling that was not influenced by ligand. To sort out the possible explanations for these functional results, we performed chromatin immunoprecipitation (ChIP) assays in GH4C1 cells to determine the interaction of the wild-type and mutant receptors with a SBE region. Because these cells express predominantly TRβ, we found that endogenous TRβ, but not TRα, was associated with the SBE region, and that this association increased after overexpression of the WT receptor and the LBD mutants. In contrast, binding of the DBD mutants was not observed, suggesting that residues in the DBD are important for TR tethering to the SBEs (Fig. 2G).
Fig. 2.
TRα and TRβ domains involved in repression of TGF-β induced transcription by T3. (A) Reporter assays in Mv1Lu cells transfected with a noncoding vector or with vectors encoding WT and mutant TRα. Luciferase activity was measured in cells incubated with T3 for 36 h and with TGF-β for the last 5 h. Results are expressed relative to the untreated cells transfected with an empty vector. (B) Scheme of TRα showing the position of the mutations. AF-2, ligand-dependent transcriptional activation function. (C) Western blot of TRα in untreated and T3-treated cells transfected with the different mutants. The arrow indicates the mobility of the TRα LBD. ERK2 was used as a loading control. (D) Luciferase activity in cells transfected with the TRβ mutants indicated and treated as in A. (E) Scheme of the mutations of TRβ used. (F) Expression of TRβ mutants in transfected cells. All data are mean ± SD (n = 3). Significance of ANOVA posttest (Bonferroni) of cells treated in the absence and presence of T3 is indicated. (G) ChIP assays performed with control IgG and anti-TRα or anti-TRβ antibodies in GH4C1 cells transfected with an empty plasmid (−) or with expression vectors for the indicated receptors. Precipitated DNA was examined by quantitative RT-PCR with primers specific for the SBE-containing region (−1,300/−1,042) of the Id1 promoter.
TRs Interact with SMADs.
To explore the mechanism underlying T3-dependent inhibition of SMAD signaling, we analyzed the possibility that TRs could interact with SMADs. GST pull-down assays showed that SMAD3 and SMAD4 interacted similarly with 35S-labeled TRα and ΤRβ, and that this interaction was reduced by T3 (Fig. 3A). Furthermore, the Mad Homology 1 (MH1) domain of SMAD3 was sufficient to associate with the receptors (Fig. 3B). Interaction of unliganded TR with SMAD3 and SMAD4 was confirmed in extracts of Cos-1 cells transfected with Flag-ΤRβ and incubated in the absence and presence of T3 (Fig. 3C) and in coimmunoprecipitation experiments of HeLa cells transfected with HA-SMAD4 and Flag-ΤRβ that showed association of the receptor with SMAD4, which was also reversed in T3-treated cells (Fig. 3D). Although both the transfected receptor and SMAD4 were more abundant in the nuclei, the interaction was detected predominantly in the cytoplasmic fraction (Fig. 3E). Furthermore, in GST pull-down assays, the LBD mutants TRα E401Q and ΤRβ E457Q, which were able to mediate T3-dependent inhibition of TGF-β transcriptional activity, showed a strong association with SMAD3 and SMAD4, whereas the DBD mutants that showed little inhibition of TGF-β signaling also showed a markedly reduced association with SMADs (Fig. 3 F and G). There was the possibility that interaction of TRs with SMADs could disrupt the formation of the complex between SMAD2/3 and the coregulatory SMAD4. However, this interaction was not affected in T3-treated cells (SI Appendix, Fig. S4).
Fig. 3.
Interaction of TRs with SMADs. (A) Quantifications of pull-down assays with GST-SMAD-3 (G-S3) and GST-SMAD4 (G-S4) and in vitro translated TRα or TRβ in the presence and absence of T3. Data are expressed as percentage of the inputs and are mean ± SD from three independent assays. Significant differences in the absence and presence of T3 are indicated. (B) GST pull-downs of TRα and TRβ with SMAD3 or its N-terminal MH1 fragment. Data were quantitated from two independent assays. (C) GST or GST-SMADs were incubated with 700 μg whole extracts of Cos-1 cells transfected with a Flag-tagged TRβ and treated in the presence and absence of 50 nM T3 for 1 h. The bound receptor was analyzed by Western blot. The input represents 10% of the proteins used in the binding assay. (D) HeLa cells were transfected with HA-SMAD4 and Flag-tagged TRβ, and 36 h later, they were treated with 50 nM T3 for 1 h. Immunoprecipitates (700 µg) with 3 µg anti-Flag antiserum or with a control serum (IgG) were subjected to Western analysis, together with 10% of the cell extract (input) with SMAD4 or TRβ antibodies. (E) Cells were transfected with HA-SMAD4 and Flag-tagged TRβ, treated with T3, and cytoplasmic and nuclear fractions were prepared and immunoprecipitations were performed as in D. (F) Quantification of in vitro GST pull-down assays of SMAD3 and SMAD4 with the indicated TRα and TRβ point mutants in the LBD and the DBD. Assays were performed in the presence and absence of T3 (G) Comparison of interaction of the wild-type receptor with the indicated DBD mutants in GST pull-down assays. Results of F and G represent the mean values obtained in two independent assays.
T3 Reduces Stimulation of SMAD Phosphorylation in Response to TGF-β.
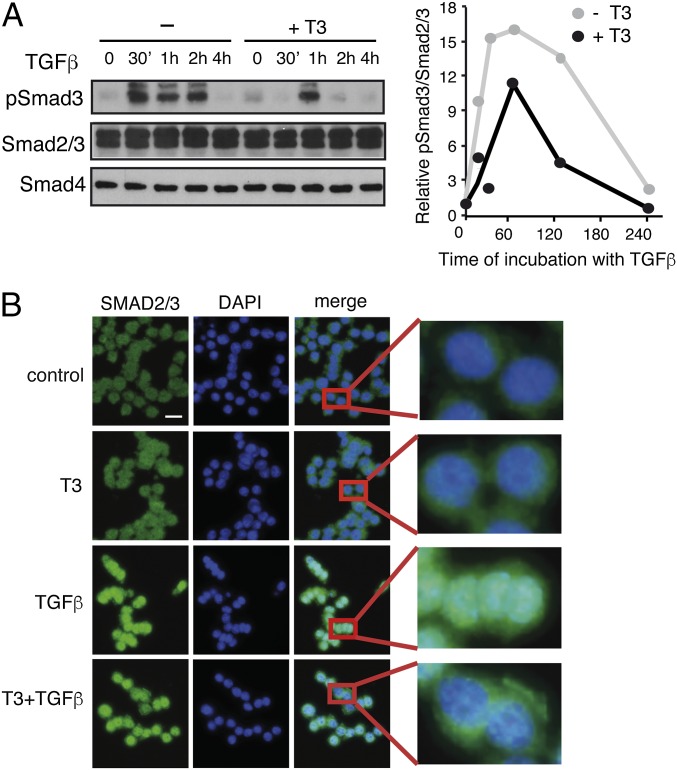
We next tested the possibility that T3 could alter SMAD phosphorylation. As shown in Fig. 4A, TGF-β induced a rapid and strong increase of SMAD3 phosphorylation in GH4C1 cells that was reduced in T3-treated cells. SMAD2/3 and SMAD4 levels were not altered by the treatments, and quantification of the ratio pSMAD/total SMAD confirmed that induction of SMAD3 phosphorylation by TGF-β was attenuated by the hormone. In addition, immunofluorescence staining revealed that T3 caused a moderate but detectable decrease of TGF-β-induced nuclear translocation of SMAD2/3 (Fig. 4B). Reduced SMAD phosphorylation by T3 was also observed in ΤRβ-expressing HepG2 cells, but not in the parental cells (SI Appendix, Fig. S5A), and a significant decrease of nuclear pSMAD2 and SMAD2/3 accumulation was also observed by Western blotting in these cells (SI Appendix, Fig. S5B).
Fig. 4.
Effect of T3 on SMAD phosphorylation and translocation. (A) Western blot analysis of GH4C1 cell extracts with phospho-SMAD3 (pSMAD3) and total SMAD2/3 and SMAD4 antibodies after treatment with T3 for 36 h and with TGF-β for the indicated periods. (Right) Quantification of the pSMAD3/SMAD2,3 ratio from two independent experiments. Data are expressed as fold-induction over the values obtained at time 0 in the untreated cells. (B) The cells were incubated with T3 for 36 h and/or TGF-β during 60 min, fixed, and analyzed by indirect immunofluorescence with SMAD2/3 antibody. Representative images of SMAD2/3, DAPI staining of nuclei, and merge images are shown. (Right) Detail of the indicated areas. (Scale bar, 50 µm.)
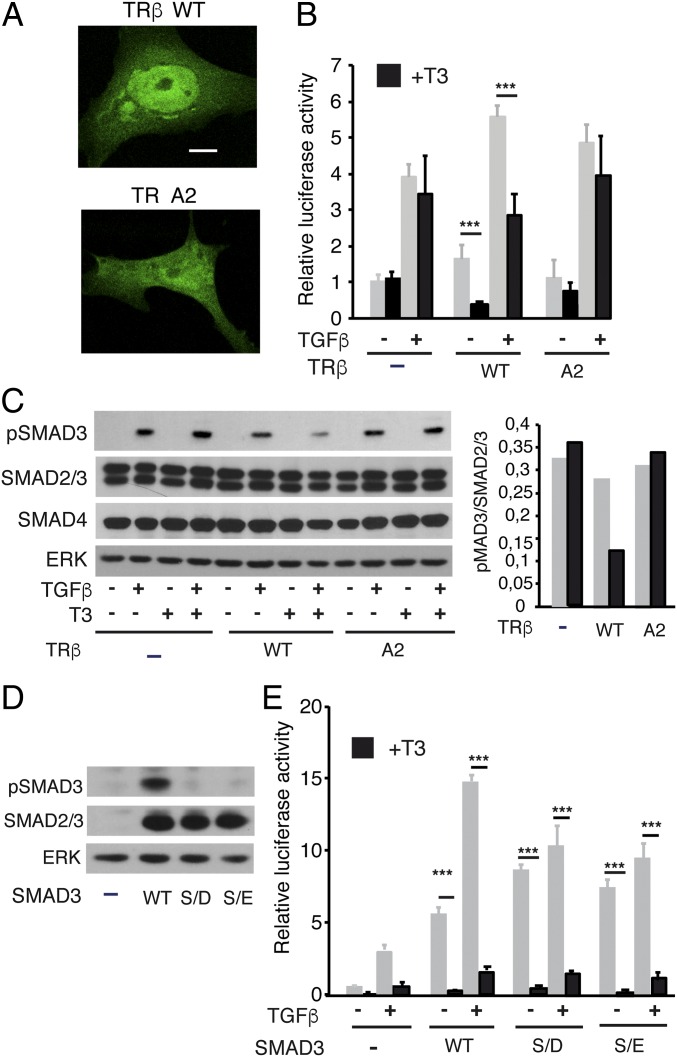
T3 could reduce TGF-β stimulation of transcription by decreasing TR binding to SMADs, cytoplasmic SMAD3 phosphorylation, and nuclear transport or a combination of these events. However, it was also possible that TR binding to SMAD3 could regulate dephosphorylation in the nucleus, as dephosphorylation of SMAD3 by the PPM1A phosphatase leads to its nuclear export (37). To address these issues, we first compared the effect of T3 on the TGF-β transcriptional response in Mv1Lu cells transfected with GFP fusions of the native receptor or a TRβ mutant (2A mutant) in the nuclear localization signal (38). We confirmed that the transfected WT receptor was predominantly, but not exclusively, nuclear, whereas the predominant nuclear localization was lost in the 2A mutant (Fig. 5A). Interestingly, T3-dependent antagonism of TGF-β transcriptional activity was lost in the mutant (Fig. 5B), indicating that nuclear translocation of the receptor is required for this effect, whereas some stimulation by the unliganded receptor was still observed. In addition, analysis of total pSMAD3 levels by Western blot showed that T3 reduced SMAD phosphorylation in cells transfected with WT TRβ, but not in cells expressing an empty vector or the TR2A mutant, and no changes in total levels of SMAD2/3 and SMAD4 were observed (Fig. 5C). pSMAD levels did not increase in cells transfected with the unliganded receptors, but transfection efficiency was low (less than 15%), and therefore the effect of TR and T3 could be underestimated. Thus, we next analyzed the effect of WT and 2A receptors in SMAD3-transfected Cos-1 cells, which have a better transfection efficiency. In Cos-1 cells, the main cytoplasmic distribution of the 2A receptor was also observed (SI Appendix, Fig. S6A). The unoccupied receptors increased pSMAD3 levels mainly in the cytoplasmic compartment, although the WT receptor had a stronger effect. This confirmed that, at least when overexpressed, TRs can induce a ligand-independent effect (SI Appendix, Fig. S6A). In contrast, T3 reduced SMAD3 phosphorylation both in nuclei and cytoplasm of cells expressing the native receptor, although this was not observed with the 2A mutant (SI Appendix, Fig. S6B).
Fig. 5.
Nuclear translocation is required for T3 antagonism. (A) Representative confocal images in Mv1Lu cells transfected with WT TRβ fused to GFP and the 2A mutant in the nuclear localization signal. (Scale bar, 10 µM.) (B) Cells were transfected with an empty vector or with vectors encoding WT and 2A TRβ. Luciferase activity was measured in cells incubated with T3 for 36 h and with TGF-β for the last 5 h. (C) Western blot analysis of pSMAD3, total SMAD2/3, total SMAD4, and ERK in cells transfected with the WT and mutant receptors and treated with T3 for 36 h in the presence and absence of TGF-β for the last 30 min. (D) Western blot analysis of pSMAD3 and SMAD2/3 in Mv1Lu cells transfected with expression vectors for WT SMAD3 or with SMAD3 mutants in the SXS motif (S/E and S/D). (E) Luciferase assays in Mv1Lu cells cotransfected with TRβ and WT and mutant SMADs. After transfection, cells were incubated in the presence and absence of T3 for 24 h and/or with TGF-β for the last 5 h. Luciferase results (mean ± SD) in the different panels are expressed relative to the values obtained in untreated cells transfected with empty vectors. Statistically significant differences between cells treated with and without T3 are indicated.
To analyze whether or not the hormone could regulate dephosphorylation in the nucleus, we next compared the effect of T3 in Mv1Lu cells cotransfected with TRβ and WT SMAD3 or with two mutants in the C-terminal SXS motif (S/D and S/E). These mutations should generate a constitutively active factor, as they mimic phosphorylation and cannot be targeted by the phosphatase. Both the native and mutant SMADs were expressed at similar levels, but as expected, pSMAD3 was only detected in cells transfected with WT SMAD (Fig. 5D). The S/D and S/E mutants indeed acted constitutively, as they strongly increased SBE-dependent transcription in the absence of phosphorylation. In addition, TGF-β further increased transcriptional activation in cells transfected with WT SMAD, although this factor had little if any effect in cells transfected with the mutant SMADs. Interestingly, T3 was still able to strongly reduce transcriptional activity of both mutants, indicating that SMAD3 dephosphorylation by PPM1A or other phosphatase/s does not play a major role in the hormonal effect (Fig. 5E).
T3 Inhibits Recruitment of SMADs to TGF-β Target Gene Promoters.
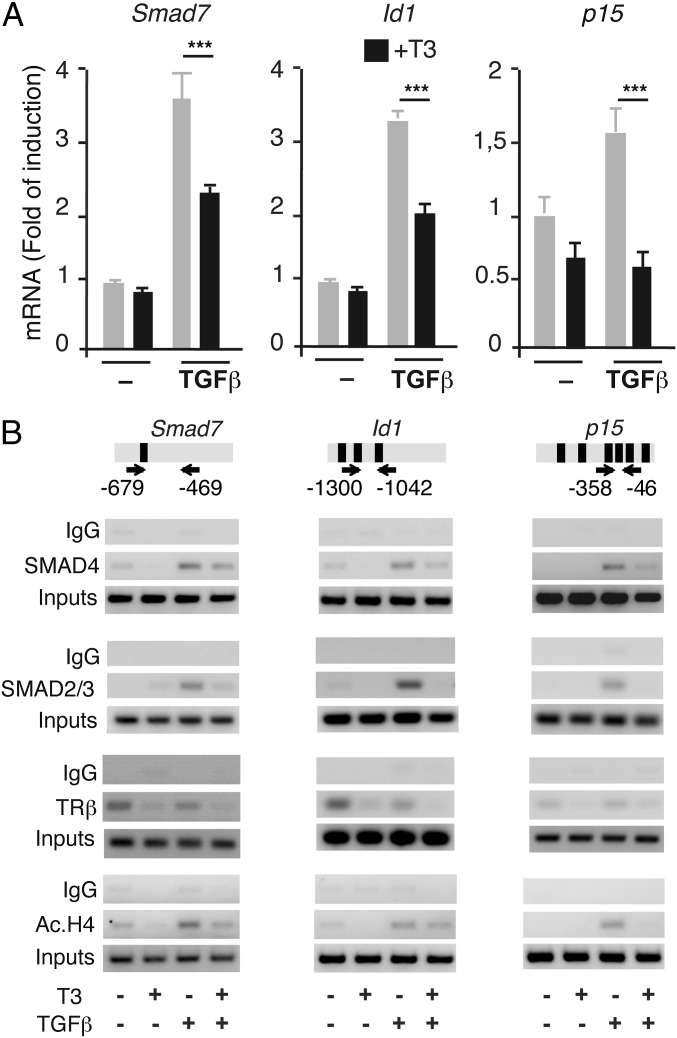
Next, we confirmed that T3 repressed endogenous expression of TGF-β target genes in GH4C1 cells. Transcripts for three well-known TGF-β responsive genes, Smad7, Id1, and p15, were induced by TGF-β, and this response was blunted in cells cotreated with T3 (Fig. 6A). We then analyzed the possibility that T3 could inhibit recruitment of SMADs to the promoters. ChIP assays with the SBE-containing promoter regions (39–41) demonstrated that the recruitment of SMAD2/3 and SMAD4 to these regions was enhanced by TGF-β and that this interaction was significantly attenuated in the presence of T3 (Fig. 6B). TRβ was present in the TGF-β-target promoters in control and TGF-β-treated cells and was released by the hormone. Because recruitment of histone acetyltransferases plays a crucial role in TGF-β/SMAD transcriptional responses (42), we also examined histone H4 acetylation in the TGF-β target promoters in cells treated with T3, TGF-β, or both. As expected, TGF-β increased the abundance of acetylated histone H4 at the promoters, and this induction was also reversed by T3 (Fig. 6B).
Fig. 6.
T3 represses stimulation of endogenous TGF-β target genes and inhibits in vivo occupancy of the promoters by SMADs. (A) Level of transcripts for the Smad7 and p15 genes were determined by quantitative PCR in GH4C1 cells treated with T3 for 36 h and TGF-β for the last 24 h. The early induction of Id1 mRNA was analyzed after 1 h treatment with TGF-β. Data are mean ± SD. Significance of Bonferroni post hoc test (n = 3) is indicated. (B) ChIP assays were performed with the indicated fragments of the TGF-β target promoters that contain SBEs. Location of the primers used is shown with arrows, and the SBEs are depicted as black boxes. Cells were treated for 1 h with T3 and/or TGF-β and analyzed by ChIP with noninmmune IgGs, SMAD2/3, SMAD4, TRβ, and acetylated histone H4 antibodies. One representative experiment of two is shown. The input for each assay is also shown.
Thyroid Hormone Administration Alleviates Fibrotic Responses in Vivo.
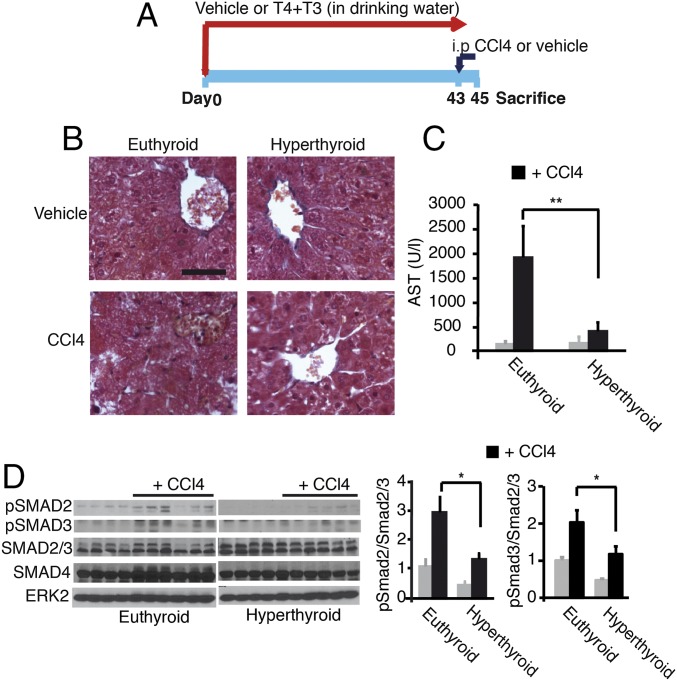
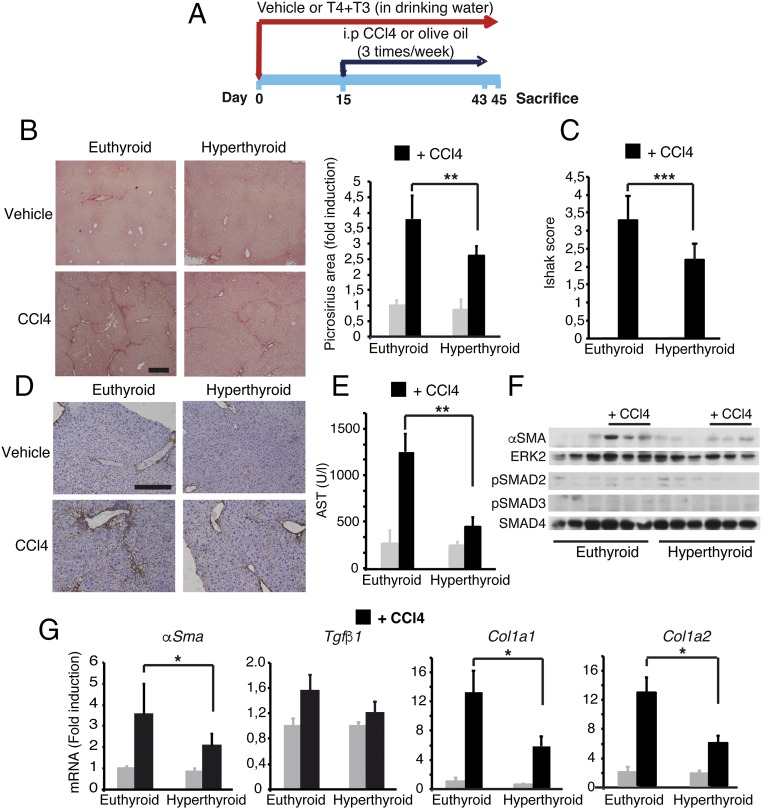
In the light of the finding that T3 can antagonize TGF-β/SMAD-dependent transcription, we hypothesized that thyroid hormone treatment should also lead to a reduced susceptibility of mice to developing fibrosis. We thus treated mice with T3 and T4 in the drinking water before administration of carbon tetrachloride (CCl4), a widely used hepatotoxic agent (Fig. 7A). Oral thyroid hormone treatment strongly increased circulating T3 levels. Because T3 treatment depletes pituitary TSH and blocks endogenous thyroid secretion (43), mice were also given T4 to maintain near-normal circulating levels of this hormone, although they were still below the normal range. The increased circulating levels of the active hormone T3, as well as the increased hepatic Dio1 gene expression, an accurate functional marker of thyroid hormone status, shows that the oral treatment induced functional hyperthyroidism in the mice (SI Appendix, Fig. S7A). As shown in Fig. 7B, acute treatment (48 h) with CCl4 caused significant hepatocyte necrosis with nuclei loss in cells surrounding centrilobular veins in normal mice, whereas this was not observed in hyperthyroid mice. Decreased liver damage in thyroid hormone-treated mice was confirmed by the markedly reduced induction of serum aspartate aminotransferase (AST) activity by CCl4 (Fig. 7C). After acute liver injury, TGF-β enhances SMAD2/3 phosphorylation, which is involved in extracellular matrix deposition and the development of liver fibrosis after chronic damage (44). Thyroid hormone treatment decreased pSMAD2 and pSMAD3 levels both before and after CCl4 injection, and quantification of the pSMAD/SMAD2,3 ratio confirmed the reduced SMAD2 and SMAD3 activation in hyperthyroid livers (Fig. 7D). In addition, the induction of Tgfβ1 and Egr-1 mRNA that occurs after acute CCl4-induced liver injury in the normal mice (45) was abrogated in thyroid hormone-treated mice (SI Appendix, Fig. S7B), showing an early reduction of the response to CCl4 in these animals compared with their control littermates. Suppression of the initial liver response suggested that the thyroid hormones could prevent subsequent development of liver fibrosis at later stages. Indeed, hyperthyroid mice developed less severe liver fibrosis than control mice after chronic exposure to CCl4 (three times per week for 30 d) (Fig. 8A). This difference was evidenced by significantly decreased Picrosirius red-positive area in histology (Fig. 8B), a lower Ishak fibrotic score (Fig. 8C), reduced immunostaining of collagen 1 (Fig. 8D), and diminished AST activity (Fig. 8E). In accordance with the reduced fibrosis, induction of α-SMA, the most reliable myofibroblast marker, was significantly reduced in CCl4-treated hyperthyroid mice (Fig. 8F). This finding was confirmed by quantitative RT-PCR analysis that showed that hepatic α-Sma mRNA levels were also lower in the hyperthyroid mice. Furthermore, thyroid hormone administration down-regulated hepatic expression of the key fibrotic marker genes Col1a1 and Col1a2 in response to CCl4, whereas at this time, it did not significantly decrease Tgfβ1 mRNA levels (Fig. 8G).
Fig. 7.
Administration of thyroid hormones attenuates the acute hepatic response to CCl4 treatment in mice. (A) Scheme of the treatments (vehicle, n = 4; CCl4, n = 6). (B) Representative Masson’s trichrome staining of livers of the different experimental groups after acute CCl4 treatment, showing extensive damage with nuclei loss only in euthyroid animals. (Scale bar, 50 µm.) (C) AST activity in serum. Data are mean ± SEM. Significance of ANOVA posttest (Bonferroni) between euthyroid and hyperthyroid CCl4-treated mice is indicated. (D) Western blot of pSMAD2, pSMAD3, SMAD2/3, and SMAD4 in the livers after acute CCl4 treatment. ERK2 was used as a loading control. (Right) pSMAD2/SMAD2,3 and pSMAD3/SMAD2,3 ratios (mean ± SEM) were quantitated and are shown.
Fig. 8.
Administration of thyroid hormones attenuates liver fibrosis induced by CCl4 treatment in mice. (A) Scheme of the treatments (vehicle, n = 4; CCl4, n = 6). (B) Representative Picrosirius red staining of livers of euthyroid and hyperthyroid mice after chronic treatment with vehicle or CCl4. (Scale bar, 400 µm.) Fibrosis was quantified from the stained area. (Right) Results (mean ± SEM) are shown. (C) Fibrosis grading with the Ishak score obtained with the same samples. (D) Immunohistochemistry of Collagen1 expression. (Scale bar, 200 µm.) (E) Serum AST activity (mean ± SEM). (F) Western blot of α-SMA, pSMAD2, and pSMAD3 in livers after chronic CCl4 treatment. ERK2 and SMAD4 were used as loading controls. (G) Relative mRNA levels of the fibrogenic genes α-Sma, Tgfβ1, Col1a1, and Col1a2. Results (mean ± SEM) are expressed relative to the values obtained in control euthyroid mice treated with vehicle. Statistically significant ANOVA posttest differences (Bonferroni) between CCl4-treated euthyroid and hyperthyroid groups are shown.
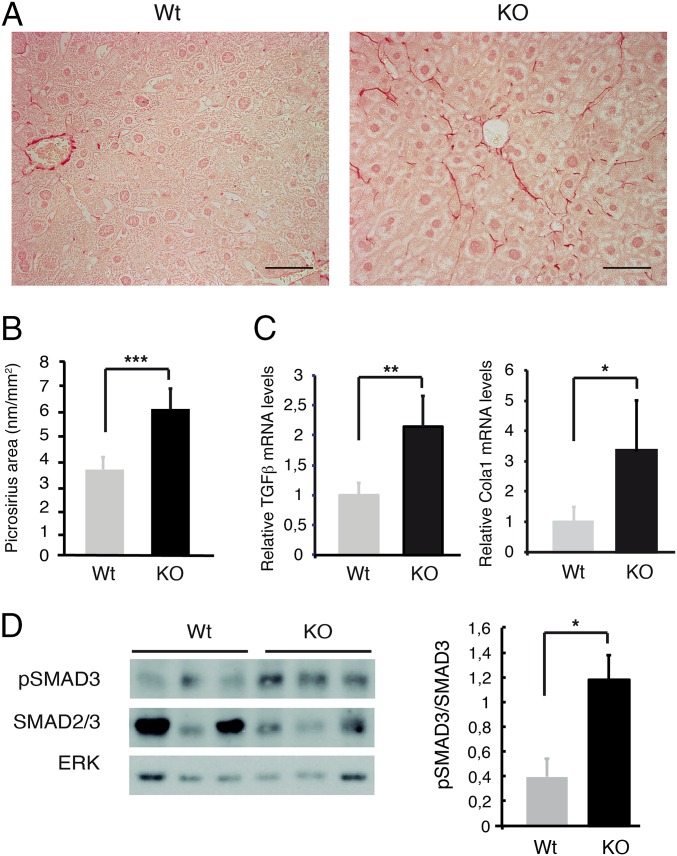
The finding that thyroid hormone administration prevents liver fibrosis led us to examine whether lack of thyroid hormone signaling could promote fibrogenesis. For this purpose, we compared livers of 18-mo-old wild-type mice and mice lacking both TRα1 and TRβ, the main thyroid hormone binding isoforms. Indeed, all TR KO mice exhibited spontaneously increased collagen deposition assessed by Picrosirius staining, whereas no fibers were essentially found in the livers of wild-type mice (Fig. 9 A and B). Collagen fibers were present in the perisinusoidal Disse space, a pattern characteristic of the initial stages of hepatic cirrhosis in human liver pathologies. Histological findings were confirmed by the increased hepatic expression of the profibrotic genes Tgfβ1 and Col1a1 in TR-deficient mice (Fig. 9C). In addition, pSMAD3/SMAD2,3 ratio was increased in the livers of double TR KO mice (Fig. 9D), reinforcing the idea that regulation of SMAD activity by thyroid hormone signaling could play a role in the fibrotic phenotype found in TR KO mice.
Fig. 9.
Genetic deletion of TRs causes spontaneous liver fibrosis in aged mice. (A) Picrosirius red staining in representative liver sections of WT mice (n = 9) and KO mice (n = 5) lacking the thyroid hormone binding isoforms TRα1 and TRβ. (Scale bar, 50 µm.) (B) Quantification (mean ± SEM) of the Picrosirius red stained area. (C) Relative levels of Tgfβ1 and Col1a1 mRNA in both groups. (D) Western blot of pSMAD3, total SMAD2/3, and ERK in livers from WT and KO mice. (Right) pSMAD3/SMAD2,3 ratio (mean ± SEM) obtained in four WT and six KO mice. Statistically significant unpaired two-tail Student´s t test differences between WT and KO groups are shown.
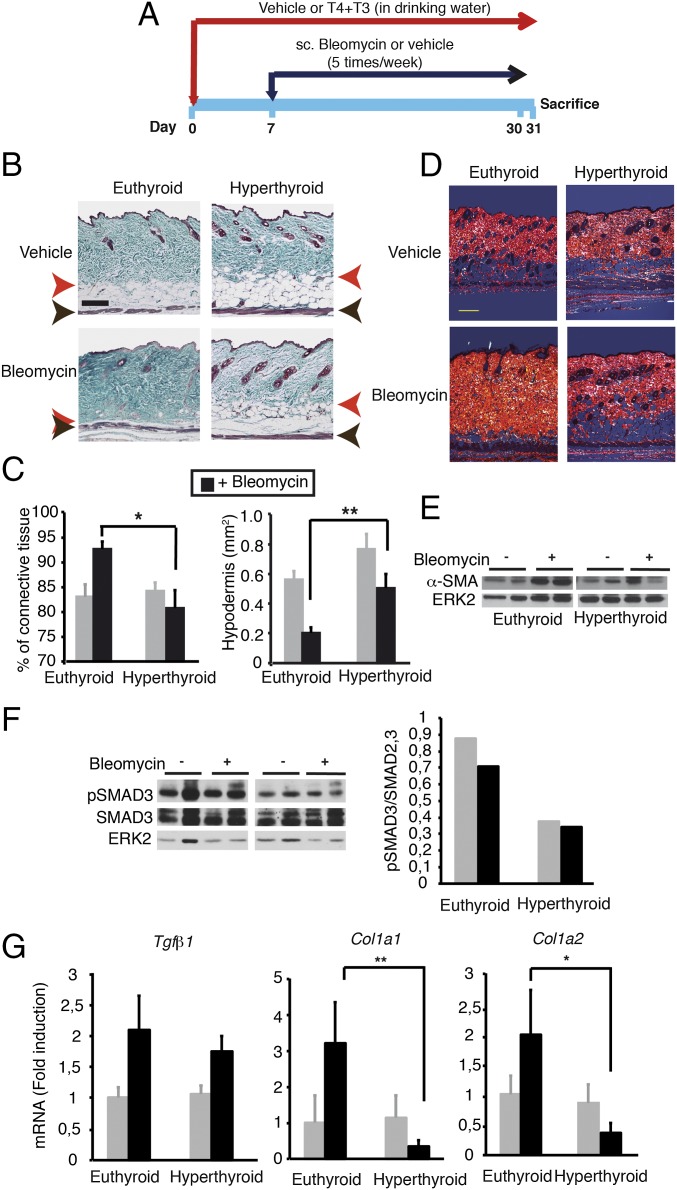
We also tested the effect of thyroid hormone administration on fibrosis, using a mice model of cutaneous scleroderma (Fig. 10A) induced by repeated local injections of bleomycin in the dorsal skin (46), in which SMAD-dependent signaling plays a crucial role (9, 17, 19). Thyroid hormone treatment caused the expected increase of circulating T3, reduction of T4, and induction of hepatic Dio1 mRNA levels (SI Appendix, Fig. S8). In the bleomycin-injected areas of euthyroid mice, we observed a significant increase of dermal thickness that was much less evident in thyroid hormone-treated mice. In the control mice, these areas showed a high accumulation of collagen fiber bundles in the dermis that extends to the hypodermis, destroying the adipose tissue (Fig. 10B). Quantification of the images indicated a significant increase of the dermal connective tissue concomitant with an important reduction of the area of the hypodermis in euthyroid mice, which was not observed in the hyperthyroid mice (Fig. 10C). Furthermore, the polarization of Picrosirius red staining showed a high packaging of collagen fibers in the dermis of bleomycin-treated controls. This polarized pattern is characteristic of increased collagen presence in fibrotic areas. The collagen fibers were very thick and compact in these animals, whereas samples from hyperthyroid mice treated with bleomycin showed a decrease of thickness of bundle collagen fibers and extracellular matrix associated with low collagenization of the hypodermis (Fig. 10D). Reduced skin fibrosis in bleomycin-treated hyperthyroid mice was associated with reduced induction of α-SMA expression (Fig. 10E), with decreased pSMAD3 levels (Fig. 10F), and with a significantly weaker induction of profibrotic genes (Fig. 10G). Therefore, the thyroid hormones alleviate fibrotic responses in vivo in two different models of TGF-β-dependent fibrosis.
Fig. 10.
Thyroid hormone treatment alleviates skin fibrosis induced by bleomycin administration. (A) Scheme of the experimental model (n = 6). (B) Masson’s trichrome staining of skins. The red arrow shows the junction of the dermis with the s.c. fat layer (hypodermis), and the black arrow marks the muscle. (Scale bar, 200 µm.) (C) The percentage of the dermal connective tissue and the area of the hypodermis were calculated from the stained slides (mean ± SEM). ANOVA differences using the Bonferroni post hoc test between vehicle and bleomycin-treated mice are indicated. (D) Representative Picrosirius red stained slides of the skin of the different experimental groups observed under polarized light to detect collagen. (Scale bar, 200 µm.) (E) Western blot of α-SMA from skins of the different groups. ERK2 was used as a loading control. (F) Western blot of pSMAD3, SMAD2/3, and ERK. (Right) Ratio pSMAD3/total SMAD in euthyroid and hyperthyroid animals. (G) Transcript levels of profibrotic genes (mean ± SEM) in the skin of the different groups. Statistically significant ANOVA differences between euthyroid and hyperthyroid mice treated with bleomycin are shown.
Because fibroblast-like cells are the primary effectors for fibrosis development, we examined the possibility that T3 could directly inhibit TGF-β responses in primary cultures of fibroblasts. α-Sma transcripts were strongly stimulated by TGF-β, and this response was significantly attenuated in T3-treated cells (SI Appendix, Fig. S9A). Furthermore, T3 also reduced induction of Tgfβ1 and Col1a1 by TGF-β, suggesting that the hormone could directly regulate myofibroblast activation. As expected, this correlated with a reduced response to TGF-β and SMAD in transactivation assays and a reduced induction of SMAD2 phosphorylation in the presence of T3 (SI Appendix, Fig. S9 B and C).
Discussion
In this article, we show that the thyroid hormone-bound TRs can attenuate TGF-β-mediated responses. Transcriptional activity of TGF-β is mainly mediated by SMAD transcription factors and in TR-expressing cells, T3 was able to repress activation of TGF-β target promoters containing binding sites for these transcription factors. This antagonism occurred irrespective of the promoter and in rat, mouse, mink, and human cells. Therefore, the ability of T3 to repress SMAD transcriptional activity is likely conserved across mammalian species. One of the genes repressed by T3 was SMAD7, which acts as an attenuator of SMAD-dependent transcription. Because of negative feedback, inhibition of SMAD7 expression could counteract the T3 suppressing effect of TGF-β signaling. However, both transactivation experiments and measure of TGF-β-stimulated transcripts show that the inhibitory effect of T3 is predominant, despite lower SMAD7 induction.
Transcriptional antagonism between nuclear receptors and other transcription factors can involve direct protein-to-protein interactions. We observed that the receptors associate with SMAD3 and SMAD4 and that the MH1 domain of SMAD3 that contains a DNA binding motif is involved in the interaction. However, this association was significantly inhibited by T3 both in vitro and in T3-treated cells. Therefore, a mechanism by which hormone binding could promote TR/SMAD interactions and formation of transcriptionally inactive complexes can be dismissed. Rather, the unoccupied receptors might act by enhancing the activity or stability of transcriptional complexes by interaction with SMADs, other transcription factors, or coregulators, and this would be reversed in the presence of T3. This is suggested by the finding that in ChIP assays, unoccupied TRs are tethered to the SBE and are released by T3.
Mechanistically, we found that mutant receptors that are unable to recruit classical coactivators and to mediate T3-dependent transcriptional activation in transfection studies (35) still showed significant repression of TGF-β/SMAD transcriptional activity, although with less potency than the wild-type receptors. In addition, analysis of DBD mutants revealed that residues in this domain are required for repression of TGF-β-dependent transcription. DNA binding is essential for many functions of TRβ in vivo, although others can occur in its absence (47). The finding that mutations C51G or C102G, which alter the structure of the DBD, do not show repressive activity could be a consequence of its inability to bind DNA. However, other explanations are possible, as these mutations also display a strongly reduced interaction with SMADs and cannot associate with the SBE-containing promoters. In addition, the GS120 mutant (47) also shows reduced interaction with SMADs, suggesting the DBD could be involved in mediating antagonism independent of its capacity to bind a TRE. Therefore, the inhibition of TFGβ transcriptional activity by the hormone involves TR domains distinct from those responsible for classical transcriptional activation that requires both binding to a TRE and recruitment of coactivators. Furthermore, the parallelism of TR mutants to mediate T3-dependent repression of SMAD transcriptional activity, to associate with SMADs, and to be tethered to the SBE suggests this interaction might be functionally important for the antagonism observed.
Although the TR DBD appears to be required both for T3-independent induction and T3-dependent inhibition of SMAD-mediated transcriptional activity, a mutant receptor with a main cytoplasmic localization still maintains some ligand-independent effect, although T3-dependent antagonism appears to require a more predominant nuclear receptor localization. Phosphorylation of SMADs that increase nuclear uptake and DNA binding could play a role in stimulation of transcription by the unliganded receptors, and indeed augmented pSMAD3 levels are found in the cytoplasm of cells transfected with unoccupied receptors. However, increased transcriptional activity by unliganded TRs was obtained under conditions of receptor overexpression, and therefore its functional significance in vivo in cells expressing endogenous receptor levels is still unclear. The interaction of T3 with TR might be reflected in an altered accessibility of SMAD complexes for phosphorylation in response to TGF-β signaling. Indeed, we observed that T3 partially blocked SMAD phosphorylation that is required for nuclear translocation. In agreement with the idea that TRs and SMADs shuttle between the nuclear and cytoplasmic compartments (38, 48), we noticed the presence of a detectable fraction of transfected native TRβ in the cytoplasm. Therefore, SMADs and TRs could have the opportunity to form a complex in this cellular compartment. Although we do not have a good explanation for this observation, there was more TRβ/SMAD4 interaction in the cytoplasmic fraction, even if both SMADs and the receptor are more abundant in the nuclei. Furthermore, the A2 mutant receptor in the nuclear localization signal was ineffective in mediating T3-dependent inhibition of SMAD phosphorylation and TGF-β transcriptional activity, showing that TR nuclear translocation is required for these receptor actions. These results suggested that T3-bound TRs could suppress TGF-β/SMAD signal transduction by reducing the association of SMAD transcription factors to TGF-β target genes. This was confirmed in ChIP assays in which T3 significantly antagonized TGF-β-induced recruitment of SMADs to the target promoters, thus inhibiting promoter acetylation. This could be a result of reduced SMAD phosphorylation and nuclear translocation, and/or of a less-efficient binding of SMADs to the SBEs in the presence of T3. There was also the possibility that liganded TR may increase dephosphorylation of pSMAD3 in the nucleus, leading to its nuclear export (37). However, we ruled out the possibility that phosphatases could play a major role in regulation of TGF-β/SMAD signaling by the receptor.
The functional importance of the repression of TGF-β signaling by hormone-bound TR was demonstrated in vivo, using mouse models of fibrosis. During pathological fibrosis, tissue damage-induced TGF-β signaling results in an excessive wound healing response, characterized by synthesis of large amounts of extracellular matrix promoted by the differentiation of fibroblasts, or hepatic stellate cells in the case of the liver, into myofibroblasts (49). Our results show that activation of TR signaling attenuates the development of chemically induced liver and skin fibrosis, reducing collagen deposition and α-SMA expression. Furthermore, fibrosis appears spontaneously in livers of mice in which TRs have been genetically ablated, thus implicating these receptors as physiological negative regulators of the fibrotic response in vivo in this organ. Fibroblast activation by TGF-β1 induces profibrotic gene expression via SMAD binding to the SBEs, as well as chromatin remodeling by histone acetylation. By decreasing accessibility of SMADs to SBEs, liganded TRs could directly antagonize myofibroblast activation and profibrotic gene expression, as observed here both in cultured cells and in vivo. This TR/SMAD transcriptional cross-talk represents a previously unrecognized mechanism for regulating fibrotic processes. However, we cannot exclude the possibility of multiple suppressive effects of TR on fibrogenesis. For instance, inflammation appears to play an important role on the initial stages of fibrosis, sensitizing cells for TGF-β/SMAD activation (4, 7), and we have previously shown that TR deletion increases inflammatory responses in the skin (50). Although the role of TRs in the inflammatory reaction to CCl4 or bleomycin administration remains to be explored, it is possible that a reduced inflammation as a consequence of increased TR signaling could also play a role in the observed antifibrotic effects of the thyroid hormones in vivo.
Previous studies have reported that both a deficiency and an excess of thyroid hormones could inhibit fibrosis. Thus, it has been reported that hypothyroidism induced by antithyroidal drugs can prevent pulmonary and cutaneous fibrosis induced by hypochlorous acid (51) and to improve survival after acute thioacetamide administration in the rat (52). However, hypothyroidism also induces cardiac fibrosis in the rat (53, 54), thyroid hormone administration ameliorates kidney fibrosis (55), and thyroid disfunction and subclinical hypothyroidism have been associated with primary biliary cirrhosis, primary sclerosing cholangitis, and nonalcoholic fatty liver disease in humans (29, 32). These observations reveal the complex role of the thyroid hormones on fibrosis that could depend on the type of insult or tissue examined. In the case of the liver, and in agreement with our results, a TRβ selective analog was effective at reducing hepatic disease and carcinogenesis in rodents (28, 29, 32). Although the natural hormone and TR agonists present adverse effects (21, 24), novel thyromimetic drugs with fewer adverse effects might be potentially developed and used to block the progression of fibrotic diseases. Recently, it has been reported that other ligands of nuclear receptors can also repress fibrosis. In particular, vitamin D receptor signaling prevents CCl4-induced liver fibrosis, inhibiting SMAD3 occupancy and SMAD-dependent transcriptional activity (56). Furthermore, vitamin D receptor agonists without hypercalcemic effects showed antifibrotic activity (57). This serves as a proof of concept that development of nuclear receptor ligands capable of disrupting TGF-β/SMAD activity could constitute a novel pharmacological approach to treat human diseases.
Materials and Methods
Extended materials and methods are provided in SI Appendix. Animal studies were approved by the Ethics Committee of the Consejo Superior de Investigaciones Científicas. Mice were made hyperthyroid by adding thyroxine and T3 to the drinking water. Circulating thyroid hormones were measured by RIA (43). Euthyroid and hyperthyroid animals were subjected to a liver fibrosis protocol by i.p. administration of CCl4 and to a bleomycin model of skin fibrosis, as detailed in the SI Appendix. Double KO mice for TRα1 and TRβ have been described (50). Experimental procedures for transfections, luciferase reporter assays, Western blot, GST pull-down assays, coimmunoprecipitation, mRNA determination by real-time PCR, and chromatin immunoprecipitation assays have been published previously and are described together with the antibodies and primers used in SI Appendix. Histology, immunohistochemistry and morphometric analysis were performed by standard procedures and are described with more detail in SI Appendix. Significance of ANOVA posttest or the Student t test among the experimental groups indicated in the figures is shown as *P < 0.05, **P < 0.01, and ***P < 0.001.
Supplementary Material
Acknowledgments
We thank J. Massagué, J. Seoane, J. Derynck, S.-Y. Cheng, and P. Santisteban for plasmids; B. Vennström for animals used to establish the KO colony; M. Privalsky for HepG2-TRβ cells; and M. J. Obregón for help with the radioimmunoassays. The technical help of M. Sanchez-Prieto and C. Sanchez-Palomo is also acknowledged. This work was supported by Grants BFU2011-28058 and BFU2014-53610P from Ministerio de Economía y Competitividad; S2011/BMD-2328 TIRONET from the Comunidad de Madrid; and RD12/0036/0030 from the Instituto de Salud Carlos III. The cost of this publication has been paid in part by FEDER funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506113113/-/DCSupplemental.
References
- 1.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115(2):209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandez-Gea V, Friedman SL. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 4.Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14(3):181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 5.Clouthier DE, Comerford SA, Hammer RE. Hepatic fibrosis, glomerulosclerosis, and a lipodystrophy-like syndrome in PEPCK-TGF-beta1 transgenic mice. J Clin Invest. 1997;100(11):2697–2713. doi: 10.1172/JCI119815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dooley S, ten Dijke P. TGF-β in progression of liver disease. Cell Tissue Res. 2012;347(1):245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seki E, et al. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13(11):1324–1332. doi: 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- 8.Weng HL, et al. The etiology of liver damage imparts cytokines transforming growth factor beta1 or interleukin-13 as driving forces in fibrogenesis. Hepatology. 2009;50(1):230–243. doi: 10.1002/hep.22934. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto T, Takagawa S, Katayama I, Nishioka K. Anti-sclerotic effect of transforming growth factor-beta antibody in a mouse model of bleomycin-induced scleroderma. Clin Immunol. 1999;92(1):6–13. doi: 10.1006/clim.1999.4720. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki Y, Okazaki I. Emerging insights into Transforming growth factor beta Smad signal in hepatic fibrogenesis. Gut. 2007;56(2):284–292. doi: 10.1136/gut.2005.088690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 12.Heldin CH, Moustakas A. Role of Smads in TGFβ signaling. Cell Tissue Res. 2012;347(1):21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- 13.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147(3):565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross S, et al. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. EMBO J. 2006;25(19):4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavsak P, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6(6):1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 16.Breitkopf K, Godoy P, Ciuclan L, Singer MV, Dooley S. TGF-beta/Smad signaling in the injured liver. Z Gastroenterol. 2006;44(1):57–66. doi: 10.1055/s-2005-858989. [DOI] [PubMed] [Google Scholar]
- 17.Lakos G, et al. Targeted disruption of TGF-beta/Smad3 signaling modulates skin fibrosis in a mouse model of scleroderma. Am J Pathol. 2004;165(1):203–217. doi: 10.1016/s0002-9440(10)63289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnabl B, et al. The role of Smad3 in mediating mouse hepatic stellate cell activation. Hepatology. 2001;34(1):89–100. doi: 10.1053/jhep.2001.25349. [DOI] [PubMed] [Google Scholar]
- 19.Takagawa S, et al. Sustained activation of fibroblast transforming growth factor-beta/Smad signaling in a murine model of scleroderma. J Invest Dermatol. 2003;121(1):41–50. doi: 10.1046/j.1523-1747.2003.12308.x. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida K, et al. Transforming growth factor-beta and platelet-derived growth factor signal via c-Jun N-terminal kinase-dependent Smad2/3 phosphorylation in rat hepatic stellate cells after acute liver injury. Am J Pathol. 2005;166(4):1029–1039. doi: 10.1016/s0002-9440(10)62324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aranda A, Alonso-Merino E, Zambrano A. Receptors of thyroid hormones. Pediatr Endocrinol Rev. 2013;11(1):2–13. [PubMed] [Google Scholar]
- 22.Brent GA. Mechanisms of thyroid hormone action. J Clin Invest. 2012;122(9):3035–3043. doi: 10.1172/JCI60047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pascual A, Aranda A. Thyroid hormone receptors, cell growth and differentiation. Biochim Biophys Acta. 2013;1830(7):3908–3916. doi: 10.1016/j.bbagen.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Mullur R, Liu YY, Brent GA. Thyroid hormone regulation of metabolism. Physiol Rev. 2014;94(2):355–382. doi: 10.1152/physrev.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sinha RA, Singh BK, Yen PM. Thyroid hormone regulation of hepatic lipid and carbohydrate metabolism. Trends Endocrinol Metab. 2014;25(10):538–545. doi: 10.1016/j.tem.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 26.López-Fontal R, et al. Mice lacking thyroid hormone receptor Beta show enhanced apoptosis and delayed liver commitment for proliferation after partial hepatectomy. PLoS One. 2010;5(1):e8710. doi: 10.1371/journal.pone.0008710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zambrano A, et al. The thyroid hormone receptor β induces DNA damage and premature senescence. J Cell Biol. 2014;204(1):129–146. doi: 10.1083/jcb.201305084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perra A, Kowalik MA, Pibiri M, Ledda-Columbano GM, Columbano A. Thyroid hormone receptor ligands induce regression of rat preneoplastic liver lesions causing their reversion to a differentiated phenotype. Hepatology. 2009;49(4):1287–1296. doi: 10.1002/hep.22750. [DOI] [PubMed] [Google Scholar]
- 29.Chung GE, et al. Non-alcoholic fatty liver disease across the spectrum of hypothyroidism. J Hepatol. 2012;57(1):150–156. doi: 10.1016/j.jhep.2012.02.027. [DOI] [PubMed] [Google Scholar]
- 30.Perra A, et al. Thyroid hormone (T3) and TRbeta agonist GC-1 inhibit/reverse nonalcoholic fatty liver in rats. FASEB J. 2008;22(8):2981–2989. doi: 10.1096/fj.08-108464. [DOI] [PubMed] [Google Scholar]
- 31.Pihlajamäki J, et al. Thyroid hormone-related regulation of gene expression in human fatty liver. J Clin Endocrinol Metab. 2009;94(9):3521–3529. doi: 10.1210/jc.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silveira MG, Mendes FD, Diehl NN, Enders FT, Lindor KD. 2009. Thyroid dysfunction in primary biliary cirrhosis, primary sclerosing cholangitis and non-alcoholic fatty liver disease. Liver Int 2009;29(7):1094-1100. [DOI] [PubMed]
- 33.Martínez-Iglesias O, et al. Thyroid hormone receptor beta1 acts as a potent suppressor of tumor invasiveness and metastasis. Cancer Res. 2009;69(2):501–509. doi: 10.1158/0008-5472.CAN-08-2198. [DOI] [PubMed] [Google Scholar]
- 34.Wrana JL, et al. TGF beta signals through a heteromeric protein kinase receptor complex. Cell. 1992;71(6):1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 35.García-Silva S, Martínez-Iglesias O, Ruiz-Llorente L, Aranda A. Thyroid hormone receptor β1 domains responsible for the antagonism with the ras oncogene: Role of corepressors. Oncogene. 2011;30(7):854–864. doi: 10.1038/onc.2010.464. [DOI] [PubMed] [Google Scholar]
- 36.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375(6528):203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 37.Lin X, et al. PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell. 2006;125(5):915–928. doi: 10.1016/j.cell.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu XG, Hanover JA, Hager GL, Cheng SY. Hormone-induced translocation of thyroid hormone receptors in living cells visualized using a receptor green fluorescent protein chimera. J Biol Chem. 1998;273(42):27058–27063. doi: 10.1074/jbc.273.42.27058. [DOI] [PubMed] [Google Scholar]
- 39.Feng XH, Lin X, Derynck R. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15(Ink4B) transcription in response to TGF-beta. EMBO J. 2000;19(19):5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Gersdorff G, et al. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor beta. J Biol Chem. 2000;275(15):11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 41.Liang YY, Brunicardi FC, Lin X. Smad3 mediates immediate early induction of Id1 by TGF-beta. Cell Res. 2009;19(1):140–148. doi: 10.1038/cr.2008.321. [DOI] [PubMed] [Google Scholar]
- 42.Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19(23):2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 43.Obregon MJ, Pascual A, de Escobar GM, Escobar del Rey F. Pituitary and plasma thyrotropin, thyroxine, and triiodothyronine after hyperthyroidism. Endocrinology. 1979;104(5):1467–1473. doi: 10.1210/endo-104-5-1467. [DOI] [PubMed] [Google Scholar]
- 44.Yoshida K, Matsuzaki K. Differential Regulation of TGF-β/Smad Signaling in Hepatic Stellate Cells between Acute and Chronic Liver Injuries. Front Physiol. 2012;3:53. doi: 10.3389/fphys.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pritchard MT, Cohen JI, Roychowdhury S, Pratt BT, Nagy LE. Early growth response-1 attenuates liver injury and promotes hepatoprotection after carbon tetrachloride exposure in mice. J Hepatol. 2010;53(4):655–662. doi: 10.1016/j.jhep.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto T, et al. Animal model of sclerotic skin. I: Local injections of bleomycin induce sclerotic skin mimicking scleroderma. J Invest Dermatol. 1999;112(4):456–462. doi: 10.1046/j.1523-1747.1999.00528.x. [DOI] [PubMed] [Google Scholar]
- 47.Shibusawa N, et al. Thyroid hormone action in the absence of thyroid hormone receptor DNA-binding in vivo. J Clin Invest. 2003;112(4):588–597. doi: 10.1172/JCI18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicolás FJ, De Bosscher K, Schmierer B, Hill CS. Analysis of Smad nucleocytoplasmic shuttling in living cells. J Cell Sci. 2004;117(Pt 18):4113–4125. doi: 10.1242/jcs.01289. [DOI] [PubMed] [Google Scholar]
- 49.Rosenbloom J, Castro SV, Jimenez SA. Narrative review: Fibrotic diseases: Cellular and molecular mechanisms and novel therapies. Ann Intern Med. 2010;152(3):159–166. doi: 10.7326/0003-4819-152-3-201002020-00007. [DOI] [PubMed] [Google Scholar]
- 50.Contreras-Jurado C, et al. The thyroid hormone receptors as modulators of skin proliferation and inflammation. J Biol Chem. 2011;286(27):24079–24088. doi: 10.1074/jbc.M111.218487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bagnato G, et al. Propylthiouracil prevents cutaneous and pulmonary fibrosis in the reactive oxygen species murine model of systemic sclerosis. Arthritis Res Ther. 2013;15(5):R120. doi: 10.1186/ar4300. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Bruck R, et al. Hypothyroidism minimizes liver damage and improves survival in rats with thioacetamide induced fulminant hepatic failure. Hepatology. 1998;27(4):1013–1020. doi: 10.1002/hep.510270417. [DOI] [PubMed] [Google Scholar]
- 53.Ghose Roy S, Mishra S, Ghosh G, Bandyopadhyay A. 2007. Thyroid hormone induces myocardial matrix degradation by activating matrix metalloproteinase-1. Matrix Biol 2007;26(4):269-279. [DOI] [PubMed]
- 54.Zhang Y, et al. Both hypothyroidism and hyperthyroidism increase atrial fibrillation inducibility in rats. Circ Arrhythm Electrophysiol. 2013;6(5):952–959. doi: 10.1161/CIRCEP.113.000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Y, Sun Z. Thyroid hormone ameliorates diabetic nephropathy in a mouse model of type II diabetes. J Endocrinol. 2011;209(2):185–191. doi: 10.1530/JOE-10-0340. [DOI] [PubMed] [Google Scholar]
- 56.Ding N, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153(3):601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito I, et al. A nonclassical vitamin D receptor pathway suppresses renal fibrosis. J Clin Invest. 2013;123(11):4579–4594. doi: 10.1172/JCI67804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.