Significance
Type I IFNs are key cytokines involved in antiviral immunity. A number of innate sensing pathways regulate the induction of type I IFNs. These pathways converge at the activation of the transcription factor IRF-3 (IFN regulatory factor 3). Three different adaptors mediate the recruitment of IRF-3 using a conserved structural motif. In this study, we determined the molecular mechanisms by which these adaptors recruit IRF-3 upon phosphorylation, the mechanism of IRF-3 activation, and how rotavirus subverts these signaling mechanisms to evade innate immune surveillance. These results provide critical insights into the molecular basis of innate immunity against microbial and viral infections.
Keywords: innate immunity, signaling, type I interferon, transcription factor, crystal structure
Abstract
Type I IFNs are key cytokines mediating innate antiviral immunity. cGMP-AMP synthase, ritinoic acid-inducible protein 1 (RIG-I)–like receptors, and Toll-like receptors recognize microbial double-stranded (ds)DNA, dsRNA, and LPS to induce the expression of type I IFNs. These signaling pathways converge at the recruitment and activation of the transcription factor IRF-3 (IFN regulatory factor 3). The adaptor proteins STING (stimulator of IFN genes), MAVS (mitochondrial antiviral signaling), and TRIF (TIR domain-containing adaptor inducing IFN-β) mediate the recruitment of IRF-3 through a conserved pLxIS motif. Here we show that the pLxIS motif of phosphorylated STING, MAVS, and TRIF binds to IRF-3 in a similar manner, whereas residues upstream of the motif confer specificity. The structure of the IRF-3 phosphomimetic mutant S386/396E bound to the cAMP response element binding protein (CREB)-binding protein reveals that the pLxIS motif also mediates IRF-3 dimerization and activation. Moreover, rotavirus NSP1 (nonstructural protein 1) employs a pLxIS motif to target IRF-3 for degradation, but phosphorylation of NSP1 is not required for its activity. These results suggest a concerted mechanism for the recruitment and activation of IRF-3 that can be subverted by viral proteins to evade innate immune responses.
The innate immune system of metazoans detects molecules from viral or bacterial pathogens using pattern recognition receptors to initiate antimicrobial responses (1–4). Type I IFNs, such as IFN-α and -β, are a major family of cytokines mediating antiviral immunity (5, 6). A number of innate sensing pathways stimulate the induction of type I IFNs (1, 3, 7, 8). For example, microbial dsDNA in the cytosol binds to and activates the enzyme cGAS (cGMP–AMP synthase), which catalyzes the synthesis of a cyclic dinucleotide, cGAMP (cyclic [G(2′,5′)pA(3′,5′)p]) (9–13). As a second messenger, cGAMP binds to the adaptor protein STING (stimulator of IFN genes) located on the endoplasmic reticulum (ER) membrane and directs the activation of transcription factor IRF-3 (IFN regulatory factor 3) through the protein kinase TBK1 (TANK-binding kinase 1) (7, 11, 14–16). Phosphorylated IRF-3 (pIRF-3) dimerizes and translocates to the nucleus to initiate the transcription of the IFN-β gene (17). In contrast, viral dsRNA in the cytosol is sensed by the RLRs [ritinoic acid-inducible protein 1 (RIG-I)–like receptors] to activate IRF-3 via the adaptor protein MAVS (mitochondrial antiviral signaling) (3, 18, 19). Moreover, the TLRs (Toll-like receptors) TLR3 and TLR4, which recognize viral dsRNA in the endosome and bacterial cell wall component LPS, respectively, also mediate the induction of type I IFNs and inflammatory cytokines (1). These two TLRs use the adaptor protein TRIF (TIR domain-containing adaptor inducing IFN-β) to mediate the recruitment and activation of IRF-3 (20–22). Strikingly, the signaling pathways of these three families of innate immune sensors converge at the activation of TBK1 and IRF-3. Mechanistically, the adaptor proteins STING, MAVS, and TRIF contain a conserved motif, pLxIS (in which p represents the hydrophilic residue, x represents any residue, and S represents the phosphorylation site), that is phosphorylated by TBK1 or IKKε and mediates the recruitment of IRF-3 to the signaling complexes (23). The induced proximity between TBK1 and IRF-3 results in IRF-3 phosphorylation and activation (23). Moreover, IRF-3 itself also contains a pLxIS motif that is crucial for phosphorylation-induced dimerization and activation of IRF-3 (23). Mutations of the phosphorylation site serine in the pLxIS motif of STING, MAVS, and TRIF abolish the induction of type I IFNs in their respective signaling pathways (23). However, the exact molecular mechanisms of IRF-3 recruitment and activation remain unknown. To elucidate the structural bases of IRF-3 recruitment by phosphorylated STING (pSTING), MAVS (pMAVS), and TRIF (pTRIF), we expressed peptides containing the pLxIS motif from the three adaptor proteins, phosphorylated them in vitro with TBK1, and determined the crystal structures of their complexes with the IRF-3 C-terminal domain (CTD) (hereafter referred to as “IRF-3”). To investigate how phosphorylation activates IRF-3, we determined the structure of a phosphomimetic mutant S386/396E of IRF-3 bound to a fragment of cAMP response element binding protein (CREB)-binding protein (CBP). These structures provide critical insight into the mechanisms of IRF-3 recruitment and activation in innate immunity.
Rotavirus is the most common cause of severe diarrhea among infants and young children. A BLAST homology search revealed that rotavirus nonstructural protein 1 (NSP1), a RING family E3 ubiquitin ligase, also contains a pLxIS motif within its C-terminal 17 residues. Because the pLxIS motif plays a critical role in mediating the recruitment of IRF-3 in the cGAS-, RLR-, and TLR-signaling pathways, this motif is likely used by rotavirus to evade innate immune surveillance. Previous studies showed that NSP1 mediates the ubiquitination and degradation of IRF-3 to suppress type I IFN induction and that its C-terminal 17 residues are essential for IRF-3 targeting (24, 25). The CTD of IRF-3 is necessary and sufficient for NSP1-mediated degradation of IRF-3 (26). However, the exact mechanism by which NSP1 targets IRF-3 is still not established. In this study, we observed that NSP1 mediates the degradation of IRF-3 without phosphorylation of its pLxIS motif. To elucidate the structural basis of IRF-3 targeting by NSP1, we determined the structures of phosphorylated and unphosphorylated NSP1 C-terminal peptide bound to IRF-3. These studies reveal the molecular mechanism by which rotavirus evades the innate immune surveillance by targeting IRF-3 for degradation.
Results
The Structural Basis of IRF-3 Recruitment by pSTING.
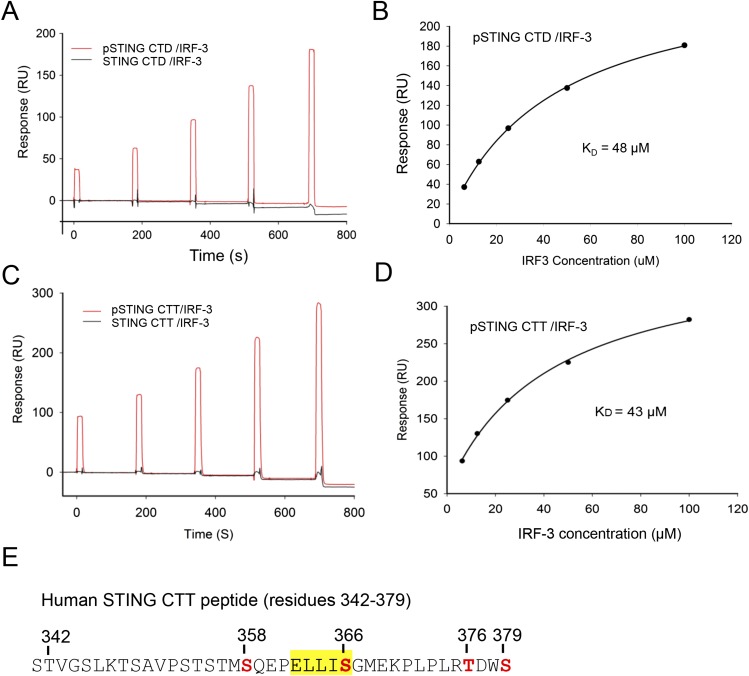
To determine how the phosphorylated STING mediates the recruitment of IRF-3, we expressed small ubiquitin-related modifier (SUMO) fusions of the human STING CTD (residues 155–379) and C-terminal tail (CTT) (residues 342–379) and phosphorylated them in vitro with TBK1. Surface plasmon resonance (SPR) binding studies show that IRF-3 binds only to the pSTING CTD or CTT (Fig. S1 A and C). The affinity of the pSTING CTD for IRF-3 (Kd ∼48 μM) (Fig. S1B) is comparable to that of the pSTING CTT (Kd ∼43 μM) (Fig. S1D). These results show that STING binds to IRF-3 via its C-terminal pLxIS motif upon phosphorylation.
Fig. S1.
SPR-binding studies of the pSTING CTD and CTT with IRF-3. (A) SPR binding studies of the phosphorylated and unphosphorylated STING CTD with IRF-3. (B) Binding affinity between the pSTING CTD and IRF-3. The Kd was determined by fitting the binding data to a one-site binding model. (C) SPR binding studies of the phosphorylated and unphosphorylated STING CTT (residues 342–379, F378W) with IRF-3. The F378W mutation was introduced into human STING CTT for quantification purposes. This mutant peptide also was used for structural analysis. (D) Binding affinity of the pSTING CTT (residues 342–379, F378W) and IRF-3. The Kd was determined by fitting the binding data to a one-site binding model. (E) Potential phosphorylation sites (red) within the pSTING CTT (residues 342–379, F378W). The phosphorylation sites were identified by MS analysis of the pSTING CTT. The pLxIS motif is highlighted in yellow.
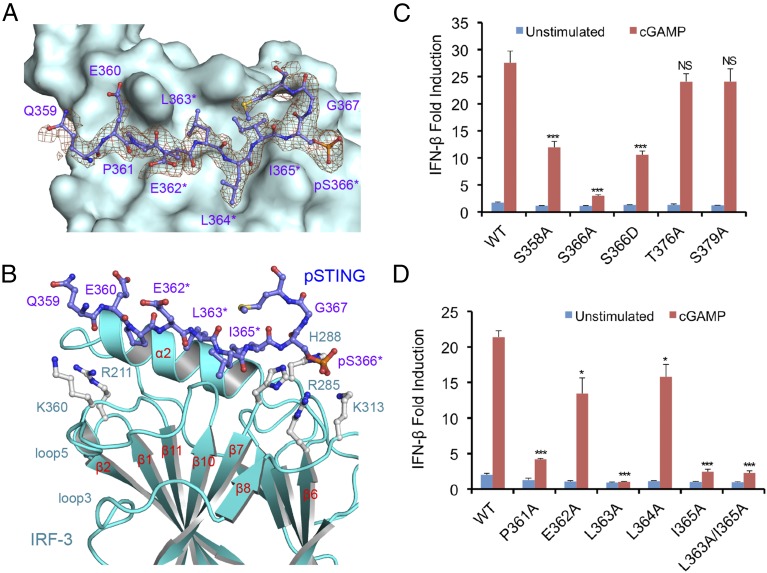
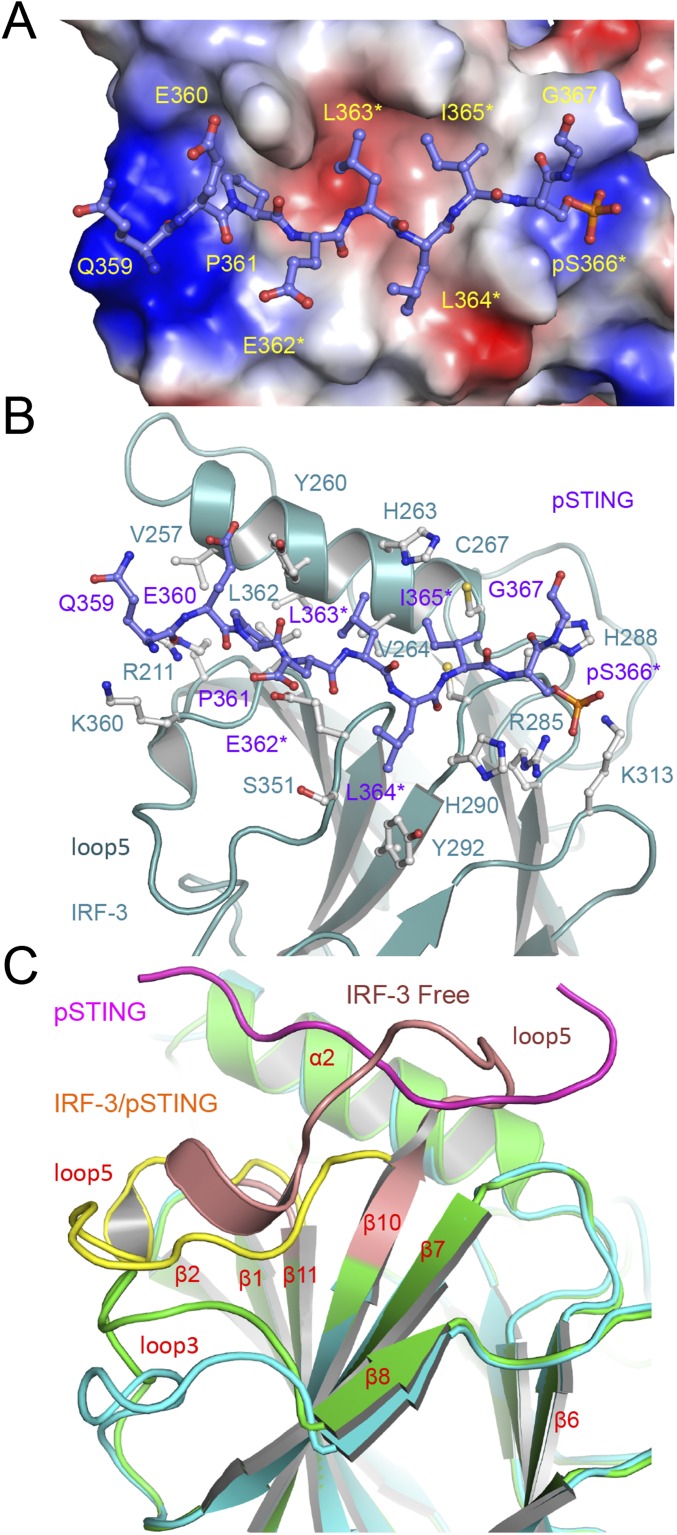
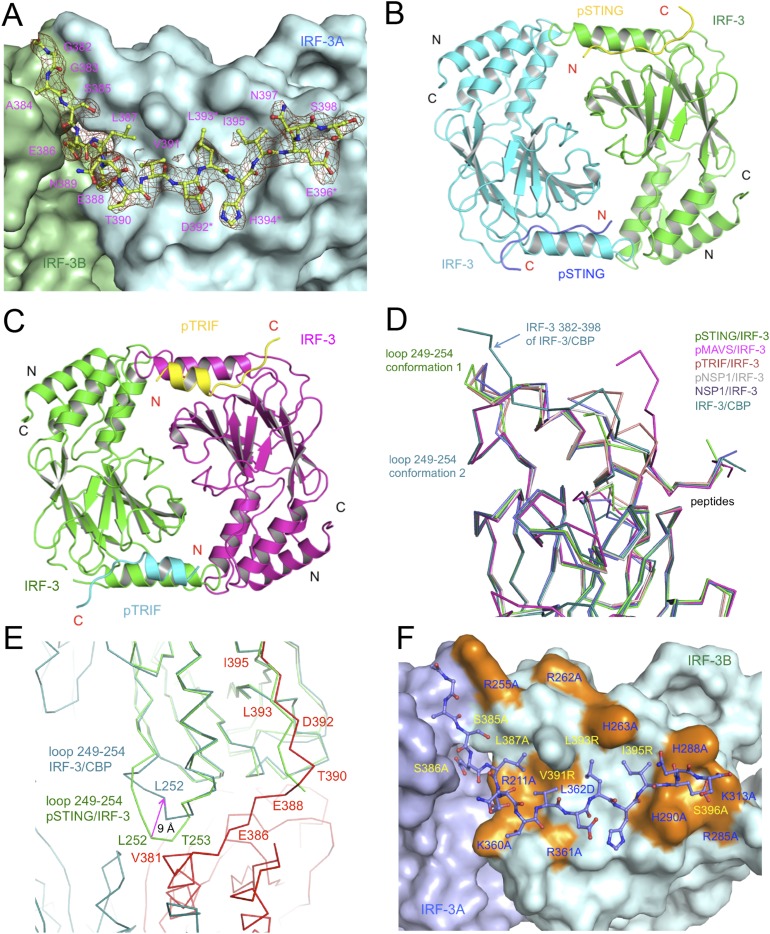
To elucidate the structural basis of IRF-3 recruitment by STING, we determined the structure of the pSTING CTT (referred to as “pSTING” hereafter) bound to IRF-3 (Fig. 1 A and B and Table S1). The binding site for pSTING is located opposite the N and C termini of IRF-3 on a flat surface formed by helix α2, the loop between strands β6 and β7, and the loop between strands β10 and β11 (Fig. 1B). pSTING adopts an extended random coil structure spanning two patches of positively charged residues and a large hydrophobic groove between them (Fig. S2 A and B). Compared with the ligand-free IRF-3 structure (27), restructuring loop5 between strands β10 and 11 exposes the binding surface for the pLxIS motif, allowing IRF-3 to interact with pSTING (Fig. S2C). The pLxIS motif interacts with IRF-3 through electrostatic interactions, hydrogen bonds, and hydrophobic interactions. A cluster of positively charged residues, Arg285, His288, His290, and Lys313, interacts with pSer366 of pSTING through electrostatic interactions (Fig. 1B and Fig. S2B). Residues pSer366, Ile365, and Leu363 form three backbone hydrogen bonds with IRF-3. The side chains of Ile365, Leu363, and Pro361 reach into the long hydrophobic groove of IRF-3 (Fig. S2 A and B). The SPR binding study shows that the S366A mutation abolishes IRF-3 binding by pSTING (Table S2), indicating that phosphorylation of Ser366 is critical for IRF-3 recruitment. The R285S and K313S mutations of IRF-3 also disrupt interactions between pSTING and IRF-3 (Table S2), demonstrating that Arg285 and Lys313 are critical for pSer366 recognition.
Fig. 1.
The structural basis of IRF-3 recruitment by pSTING. (A) Difference map of pSTING bound to IRF-3 contoured at 2.5σ. The σA-weighted Fo-Fc map was calculated with pSTING omitted from the model. pSTING is shown by the purple ball-and-stick model. The background is a surface representation of IRF-3. Residues of the pLxIS motif are indicated by asterisks. (B) Close-up view of the pSTING/IRF-3 complex. IRF-3 is shown by cyan ribbons. The two positively charged clusters of IRF-3 are shown by gray ball-and-stick models. (C) IFN-β reporter assays showing that mutations of potential phosphorylation site residues affect STING-mediated signaling in HEK293T cells. The data (mean ± SEM) are representative of three independent assays. *P < 0.05, ***P < 0.001, NS (P > 0.05) values were calculated based on comparisons of signals between cells transfected with STING mutants and cells transfected with wild-type STING. (D) IFN-β reporter assays showing that mutations of key residues of STING involved in IRF-3 binding affect STING-mediated signaling. The data (mean ± SEM) are representative of three independent assays. *P < 0.05, ***P < 0.001, NS (P > 0.05) values were calculated based on comparisons of signals in cells transfected with STING mutants and in cells transfected with wild-type STING.
Table S1.
Statistics of crystallographic analyses
| pSTING/IRF-3 | pMAVS/IRF-3 | pTRIF/IRF-3 | IRF-3/CBP | pNSP1/IRF-3 | NSP1/IRF-3 | |
| Data collection | ||||||
| Space group | P2 | P21 | P3221 | P21 | P3221 | P212121 |
| Cell dimensions | ||||||
| a, b, c (Å) | 60.08, 56.40, 75.55 | 38.80, 104.91, 66.27 | 72.81, 72.81, 127.60 | 85.23, 88.66, 92.81 | 72.98, 72.98, 127.55 | 67.77, 107.92, 135.65 |
| α, β, γ (°) | 90.0, 104.64, 90.0 | 90.0, 106.13, 90.0 | 90.0, 90.0, 120.0 | 90.0, 113.09, 90.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 |
| Resolution, Å | 2.00 (2.03–2.00) | 2.40 (2.49–2.40) | 1.60 (1.63–1.60) | 2.50 (2.59–2.50) | 1.72 (1.75 to1.72) | 2.90 (2.95–2.90) |
| Unique reflections | 35,085 | 19,995 | 52,282 | 42,426 | 42,593 | 22,409 |
| Molecules per ASU | 2 | 2 | 1 | 4 | 1 | 4 |
| Rmerge (%) | 10.8 (57.0) | 9.9 (136) | 5.6 (>100) | 13.8 (155) | 6.8 (>100) | 14.0 (89.8) |
| Rp.i.m. (%) | 6.6 (35.0) | 4.0 (53.9) | 1.7 (33.4) | 8.5 (94.6) | 2.2 (39.8) | 5.6 (35.6) |
| CC1/2 | (0.817) | (0.697) | (0.818) | (0.384) | (0.823) | (0.781) |
| I/σI | 14.0 (1.8) | 14.3 (1.7) | 45.2 (1.8) | 6.2 (0.8) | 44.5 (1.9) | 15.7 (2.1) |
| Completeness (%) | 98.2 (83.7) | 100 (100) | 99.8 (99.5) | 96.3 (85.9) | 100 (100) | 100 (99.9) |
| Redundancy | 3.6 (3.2) | 7.2 (7.3) | 11.9 (11.6) | 3.5 (3.4) | 11.8 (10.9) | 7.1 (7.3) |
| Refinement | ||||||
| Resolution (Å) | 2.00 | 2.40 | 1.60 | 2.50 | 1.72 | 2.90 |
| No. reflections used | 35,068 | 19,966 | 52,240 | 42,378 | 42,537 | 22,342 |
| Rwork/Rfree (%) | 17.6/23.7 | 18.8/25.1 | 16.8/18.7 | 19.2/24.2 | 17.4/19.0 | 21.0/25.0 |
| No. atoms | 4,341 | 3,968 | 2,228 | 7,876 | 2,207 | 7,718 |
| Protein | 3,669 | 3,678 | 1,851 | 7,650 | 1,885 | 7,386 |
| Ligand/ion | 292 | 220 | 110 | N.A. | 102 | 332 |
| Water | 380 | 70 | 267 | 226 | 220 | 0 |
| B-factors | ||||||
| Protein | 33.6 | 64.9 | 32.7 | 62.8 | 39.4 | 46.3 |
| Ligand/ion | 58.9 | 113.2 | 39.3 | N.A. | 60.4 | 63.9 |
| Water | 37.5 | 52.9 | 42.2 | 52.3 | 46.3 | N.A. |
| Rmsd | ||||||
| Bond lengths (Å) | 0.007 | 0.002 | 0.006 | 0.003 | 0.007 | 0.002 |
| Bond angles (°) | 1.004 | 0.605 | 1.039 | 0.624 | 0.999 | 0.589 |
| PDB ID code | 5JEJ | 5JEK | 5JEL | 5JEM | 5JEO | 5JER |
One crystal was used to collect each of the datasets. Values in parentheses are for the highest-resolution shells.
Fig. S2.
The structural basis of IRF-3 recruitment by pSTING. (A) Electrostatic surface representation of IRF-3 with pSTING (purple ball-and-stick model) bound to it. The positively charged surface is colored blue, and the negatively charged surface is colored red. (B) Interactions between pSTING and IRF-3. pSTING is shown by the purple ball-and-stick model. Residues of IRF-3 involved in pSTING binding are shown by gray ball-and-stick models. (C) Conformational change of IRF-3 upon pSTING binding. IRF-3 bound to pSTING is shown by the cyan and yellow ribbons. Ligand free IRF-3 is shown by the green and pink ribbons. pSTING is shown by the magenta coil.
Table S2.
Binding affinities between IRF-3 and STING, MAVS, TRIF, and NSP1 determined by SPR
| Proteins/peptides | IRF-3 | Kd (μM) |
| Human STING 155–379 | ||
| Avi-sumo-STING 155–379 | IRF-3 189–427 | No binding |
| p-Avi-sumo-STING 155–379 | IRF-3 189–427 | 48 |
| p-Avi-sumo-STING 155–379, S366A | IRF-3 189–427 | No binding |
| p-Avi-sumo-STING 155–379, S366D | IRF-3 189–427 | 47.6 |
| Human STING 342–379, F378W | ||
| p-Avi-sumo-STING 342–379, F378W | IRF-3 189–427 | 43.2 |
| p-Avi-sumo-STING 342–379, F378W | IRF-3 189–427, R285S | No binding |
| p-Avi-sumo-STING 342–379, F378W | IRF-3 189–427, K313S | No binding |
| p-Avi-sumo-STING 342–379, F378W | IRF-3 189–427, H288S | 43.6 |
| p-Avi-sumo-STING 342–379, F378W | IRF-3 189–427, H290S | 45.2 |
| Human MAVS 433–450, M449W | ||
| Avi-sumo-MAVS 433–450, M449W | IRF-3 189–427 | No binding |
| p-Avi-sumo-MAVS 433–450, M449W | IRF-3 189–427 | 104 |
| p-Avi-sumo-MAVS 433–450, M449W | IRF-3 189–427, R285S | No binding |
| p-Avi-sumo-MAVS 433–450, M449W | IRF-3 189–427, K313S | No binding |
| p-Avi-sumo-MAVS 433–450, M449W | IRF-3 189–427, H288S | ∼330 |
| p-Avi-sumo-MAVS 433–450, M449W | IRF-3 189–427, H290S | ∼400 |
| p-Avi-sumo-MAVS 433–450, M449W and S442A | IRF-3 189–427 | No binding |
| Human TRIF 199–219, L218W | ||
| Avi-sumo-TRIF 199–219, L218W | IRF-3 189–427 | No binding |
| p-Avi-sumo-TRIF 199–219, L218W | IRF-3 189–427 | 72.4 |
| p-Avi-sumo-TRIF 199–219, L218W | IRF-3 189–427, R285S | No binding |
| p-Avi-sumo-TRIF 199–219, L218W | IRF-3 189–427, K313S | No binding |
| p-Avi-sumo-TRIF 199–219, L218W | IRF-3 189–427, H288S | ∼150 |
| p-Avi-sumo-TRIF 199–219, L218W | IRF-3 189–427, H290S | ∼260 |
| p-Avi-sumo-TRIF 199–219, L218W and S210A | IRF-3 189–427 | No binding |
| p-Avi-sumo-TRIF 199–219, L218W and S202A | IRF-3 189–427 | ∼120 |
| NSP1 479–496, N495W | ||
| Avi-sumo-NSP1 479–496, N495W | IRF-3 189–427 | ∼200 |
| p-Avi-sumo-NSP1 479–496, N495W | IRF-3 189–427 | 15.8 |
| Avi-sumo-NSP1 479–496, N495W and E482A | IRF-3 189–427 | ∼360 |
| Avi-sumo-NSP1 479–496, N495W and F484A | IRF-3 189–427 | No binding |
| Avi-sumo-NSP1 479–496, N495W and L486A | IRF-3 189–427 | No binding |
| Avi-sumo-NSP1 479–496, N495W and I488A | IRF-3 189–427 | No binding |
| Avi-sumo-NSP1 479–496, N495W and S489A | IRF-3 189–427 | ∼290 |
| Avi-sumo-NSP1 479–496, N495W and S489D | IRF-3 189–427 | 27.3 |
To identify the phosphorylation site of STING that is essential for IRF-3 recruitment, we mutated each of the four potential phosphorylated residues (Fig. S1E) to alanine and conducted IFN-β luciferase reporter assays in HEK293T cells, which do not express STING (28). As shown in Fig. 1C, the S366A mutation abolishes IFN-β reporter activation, consistent with previous studies (23). In addition, the S358A mutation partially impairs IFN-β reporter activation (Fig. 1C). However, the T376A and S379A mutations do not affect IFN-β reporter activation (Fig. 1C). These results demonstrate that phosphorylation of Ser366 is crucial for IRF-3 recruitment. Next, we tested how other residues within and upstream of the pLxIS motif contribute to STING-mediated signaling. The L363A and I365A mutations individually or in combination strongly abolish IFN-β reporter activation (Fig. 1D). In contrast, the E362A and L364A mutations have only minor effects on STING-mediated signaling (Fig. 1D). The P361A mutation upstream of the pLxIS motif also dramatically reduces IFN-β reporter activation (Fig. 1D). These results show that the hydrophobic interactions between IRF-3 and pSTING residues Pro361, Leu363, and I365 are also critical for IFN-β induction. Taken together, these structural and functional studies reveal the structural basis of IRF-3 recruitment by pSTING.
Mechanisms of IRF-3 Recruitment by pMAVS and pTRIF.
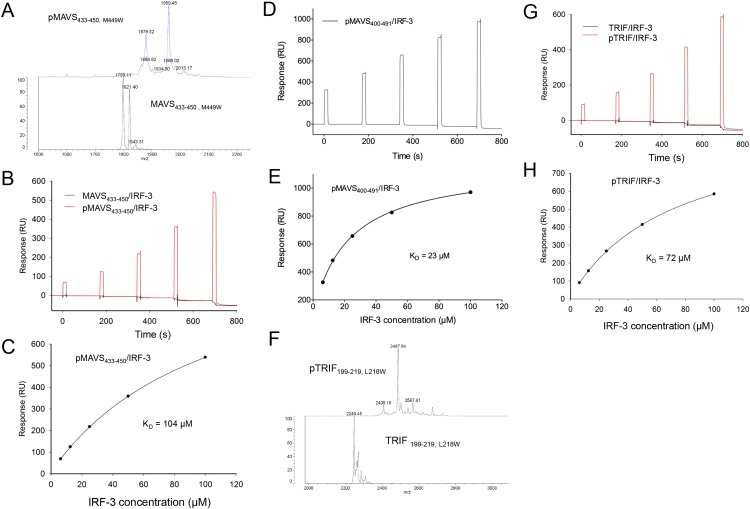
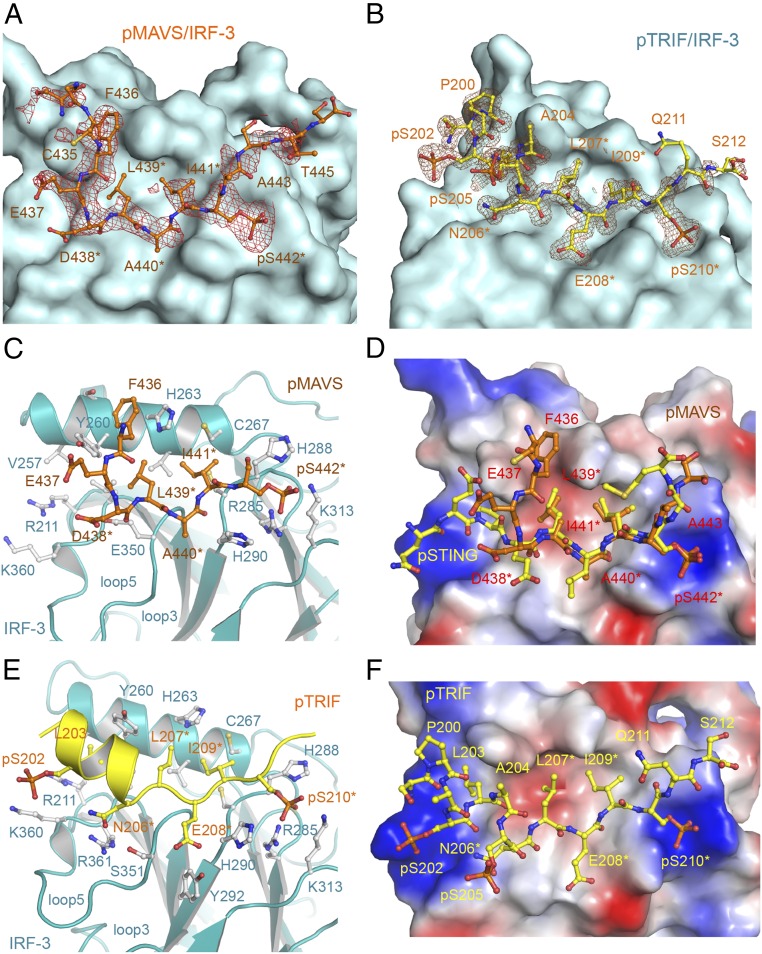
In contrast to dsDNA sensing through the cGAS–STING pathway, the RLRs sense dsRNA in the cytosol and activate IRF-3 via the adaptor MAVS (3, 18, 19), and TLR3 and TLR4 use the adaptor TRIF to recruit IRF-3 (20–22). Phosphorylation of the pLxIS motif of MAVS or TRIF is required for the recruitment and activation of IRF-3 (23). To determine how MAVS and TRIF recruit IRF-3, we phosphorylated SUMO fusions of peptides containing the pLxIS motif derived from human MAVS (residues 433–450) and TRIF (residues 199–219) using TBK1 (Fig. S3 A and F). SPR binding studies show that pMAVS and pTRIF bind IRF-3 with affinities of ∼104 and 72 μM, respectively (Fig. S3 B, C, G, and H). However, unphosphorylated MAVS and TRIF peptides do not bind IRF-3 (Fig. S3 B and G). A longer fragment of MAVS containing residues 400–491 binds IRF-3 at a higher affinity of 23 μM when phosphorylated (Fig. S3 D and E). To elucidate the molecular bases of IRF-3 recruitment by MAVS and TRIF, we determined the structures of IRF-3 bound to pMAVS and pTRIF, respectively (Fig. 2 A and B and Table S1). The pLxIS motifs of pMAVS and pTRIF interact with the same ligand-binding surface on IRF-3 as pSTING (Fig. 2). However, the interaction of IRF-3 with residues upstream of the pLxIS motif in pMAVS and pTRIF is different from that in pSTING (Fig. 2 D and E). pMAVS bends toward helix α2 of IRF-3 at the first residue of the pLxIS motif (Fig. 2 C and D). The side chain of Phe436 sandwiches between the side chains of Tyr260 and His263 of IRF-3 (Fig. 2C). In contrast, residues 199–206 of pTRIF form a short α-helix extending toward the positively charged patch around Arg211 and Lys360 (Fig. 2 E and F). Ser202 and Ser205 in this helix are also phosphorylated, and pSer202 is in close proximity to Lys360 (4.9 Å) (Fig. 2E). In addition, Leu203 upstream of the pLxIS motif reaches into a hydrophobic pocket of IRF-3 (Fig. 2 E and F). Mutating the serine residues in the pLxIS motif to alanine abolishes IRF-3 binding by both pMAVS and pTRIF (Table S2). Conversely, the R285S and K313S mutations in IRF-3 also disrupt pMAVS and pTRIF binding (Table S2). In contrast, the H288S and H290S mutations of IRF-3 or the S202A mutation of TRIF reduce the binding affinities by only two- to fourfold (Table S2). Taken together, these structural and functional analyses reveal the molecular bases of IRF-3 recruitment by MAVS and TRIF upon phosphorylation.
Fig. S3.
Characterization of pMAVS and pTRIF peptides. (A) MALDI-TOF MS analysis of phosphorylated human MAVS (residues 433–450, M449W) peptide. The M449W mutation was introduced in this peptide for quantification purposes. (B) SPR binding studies of phosphorylated and unphosphorylated MAVS (residues 433–450, M449W) peptide with IRF-3. (C) Binding affinity between pMAVS (residues 433–450, M449W) peptide and IRF-3. (D) SPR binding studies of pMAVS (residues 400–491) peptide with IRF-3. (E) Binding affinity between pMAVS (residues 400–491) peptide and IRF-3. (F) MALDI-TOF MS analysis of phosphorylated human TRIF (residues 199–219, L218W) peptide. (G) SPR binding studies of phosphorylated and unphosphorylated TRIF (residues 199–219, L218W) peptide with IRF-3. (H) Binding affinity between pTRIF (residues 199–219, L218W) peptide and IRF-3.
Fig. 2.
Structures of pMAVS and pTRIF peptides bound to IRF-3. (A) Difference map showing pMAVS bound to IRF-3 contoured at 2.5σ. The σA-weighted Fo-Fc map was calculated with pMAVS omitted from the model. pMAVS is shown by the orange ball-and-stick model, and IRF-3 is shown by the cyan surface. Residues of the pLxIS motif are indicated by asterisks. (B) Difference map showing pTRIF bound to IRF-3 contoured at 2.5σ. The σA-weighted Fo-Fc map was calculated with pTRIF omitted from the model. pTRIF is shown by the yellow ball-and-stick model, and IRF-3 is shown by the cyan surface. (C) Interactions between pMAVS and IRF-3. pMAVS is shown by the orange ball-and-stick model. IRF-3 is shown by cyan ribbons. Residues of IRF-3 involved in pMAVS binding are shown by gray ball-and-stick models. Residues of the pLxIS motif are indicated by asterisks. (D) Superposition of the structures of pMAVS and pSTING bound to IRF-3. pSTING is shown by the yellow ball-and-stick model. IRF-3 is shown by the surface representation colored according to surface electrostatic potential. The positively charged surface is in blue, and the negatively charged surface is in red. (E) Interactions between pTRIF and IRF-3. pTRIF is shown by the yellow ribbon with key residues in ball-and-stick models. IRF-3 is shown by cyan ribbons with residues involved in pTRIF binding shown by gray ball-and-stick models. (F) Structure of pTRIF bound to IRF-3. The ligand-binding surface of IRF-3 is colored according to surface electrostatic potential as in D. pTRIF is shown by the yellow ball-and-stick model.
The pLxIS Motif of IRF-3 Mediates Its Dimerization and Activation.
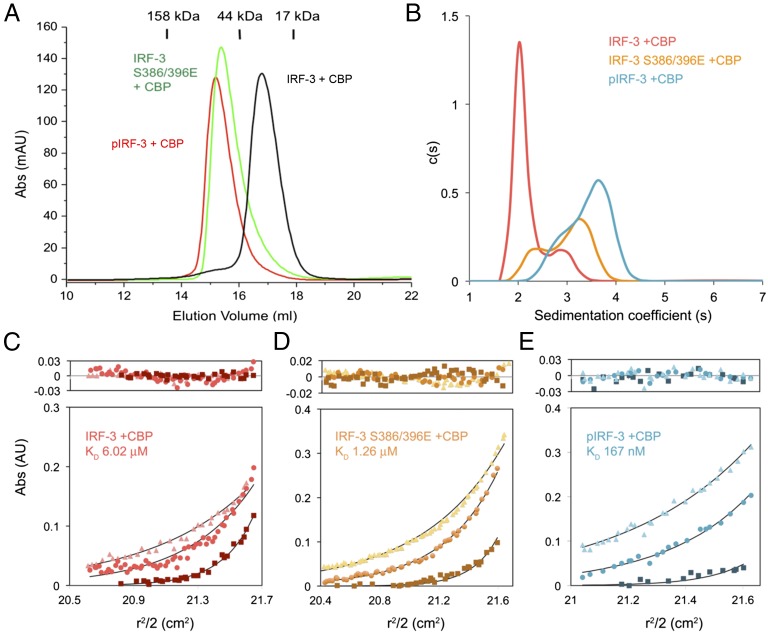
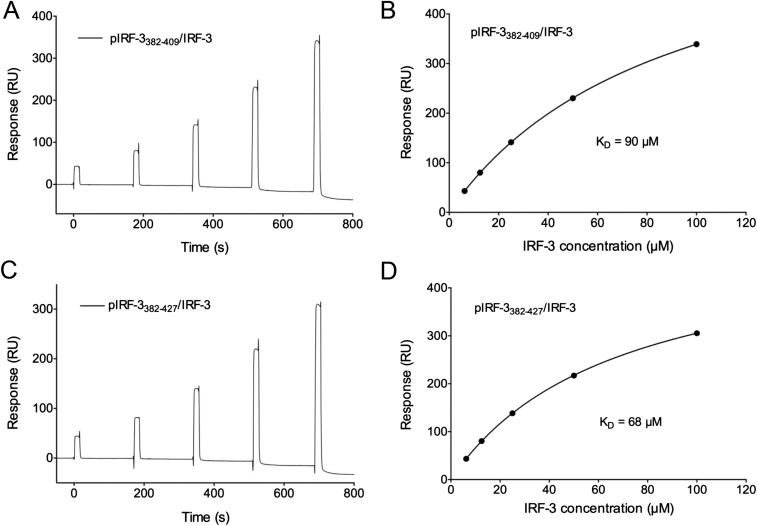
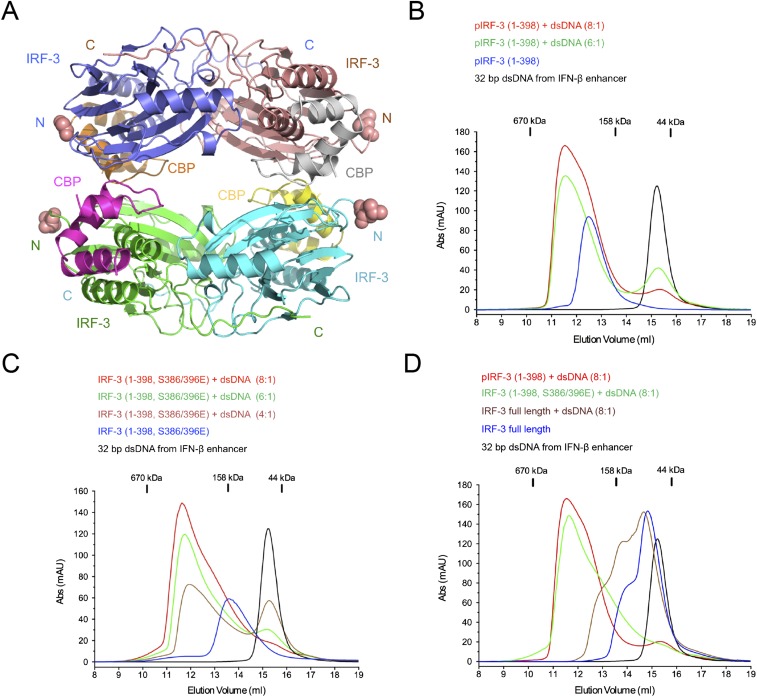
IRF-3 contains not only the binding surface for the pLxIS motif to mediate its recruitment by the adaptors but also a pLxIS motif of its own to mediate its activation (23). Previous studies showed that Ser386 and 396 are critical phosphorylation sites for IRF-3 activation (29, 30). However, the exact mechanism of IRF-3 activation remains elusive. To determine this mechanism, a double phosphomimetic mutant of IRF-3, S386/396E, was coexpressed with the IRF-3–binding domain of CBP and was crystallized for structural analysis (Table S1). Gel-filtration chromatography and sedimentation velocity analytical ultracentrifugation analyses show that a majority of the IRF-3 mutant forms a dimer in solution, whereas the unphosphorylated IRF-3/CBP complex is mostly monomeric (Fig. 3 A and B). Consistent with these results, sedimentation equilibrium analyses show that unphosphorylated IRF-3 in complex with CBP is mostly monomeric and dimerizes weakly with an affinity of ∼6 μM (Fig. 3C). Because the C-terminal tail (CTT) of IRF-3 after Ser398 is truncated, the exposed pLxIS motif likely mediates the low-affinity binding between two IRF-3/CBP complexes. The S386/396E mutations increase the binding affinity between IRF-3 molecules to 1.26 μM (Fig. 3D), whereas phosphorylation of IRF-3 by TBK1 results in a stable dimer with a Kd of 167 nM (Fig. 3E). To test if the internal pLxIS motif of IRF-3 is responsible for the high-affinity interaction between two pIRF-3 molecules, we expressed SUMO fusions of two IRF-3 C-terminal peptides containing residues 382–409 and 382–427 and phosphorylated them with TBK1. SPR binding studies show that these two peptides bind IRF-3 with affinities of ∼90 and 68 μM, respectively (Fig. S4), indicating that the intrinsic affinity of IRF-3 for its phosphorylated tail is comparable to its affinity for phosphorylated adaptor proteins and that phosphorylation-induced dimerization is critical for the high stability of the pIRF-3 dimer. The higher affinity between pIRF-3 molecules compared with the S386/396E mutant is likely a result of the phosphomimetic mutations being unable to recapitulate fully the interactions between two pIRF-3 molecules.
Fig. 3.
The IRF-3 phosphomimetic mutant S386/396E in complex with CBP dimerizes in solution. (A) Gel-filtration chromatography analyses of IRF-3/CBP complexes. The black curve is for unphosphorylated IRF-3 bound to CBP. The green chromatogram is for the IRF-3 S386/396E mutant bound to CBP. The red curve is for the TBK1 pIRF-3/CBP complex. (B) Sedimentation velocity AUC analysis of the samples described in A. The data are consistent with a shift from primarily monomeric IRF-3 in the unphosphorylated state toward the dimeric state upon phosphorylation. (C–E) Sedimentation equilibrium AUC analyses of the three IRF-3/CBP complex samples. Protein samples were spun at 18,000, 22,000, and 32,000 rpm in a Beckman An-60 Ti rotor at loading concentrations of 1, 3.5, and 14 μM. Nine datasets for each sample were analyzed globally; for clarity, three representative data curves for each sample are shown. The data are best described by an equilibrium between 1:1 and 2:2 IRF-3:CBP complexes, with experimental Kd values listed on each plot. In each panel, data from 18,000, 22,000, and 32,000 rpm rotor speeds are shown by squares, circles, and triangles, respectively.
Fig. S4.
SPR binding studies of pIRF-3 C-terminal peptides containing the pLxIS motif with the IRF-3 CTD (residues 189–427). (A) SPR binding study of the pIRF-3 peptide (residues 382–409, Y408W) with IRF-3 CTD. The Y408W mutation was introduced to the IRF-3 sequence for quantification purposes. (B) Binding affinity between the pIRF-3 peptide (residues 382–409, Y408W) with IRF-3 CTD. (C) SPR binding study of the pIRF-3 peptide (residues 382–427, E426W) with IRF-3 CTD. The E426W mutation was introduced to the IRF-3 sequence for quantification. (D) Binding affinity between the pIRF-3 peptide (residues 382–427, E426W) with IRF-3 CTD.
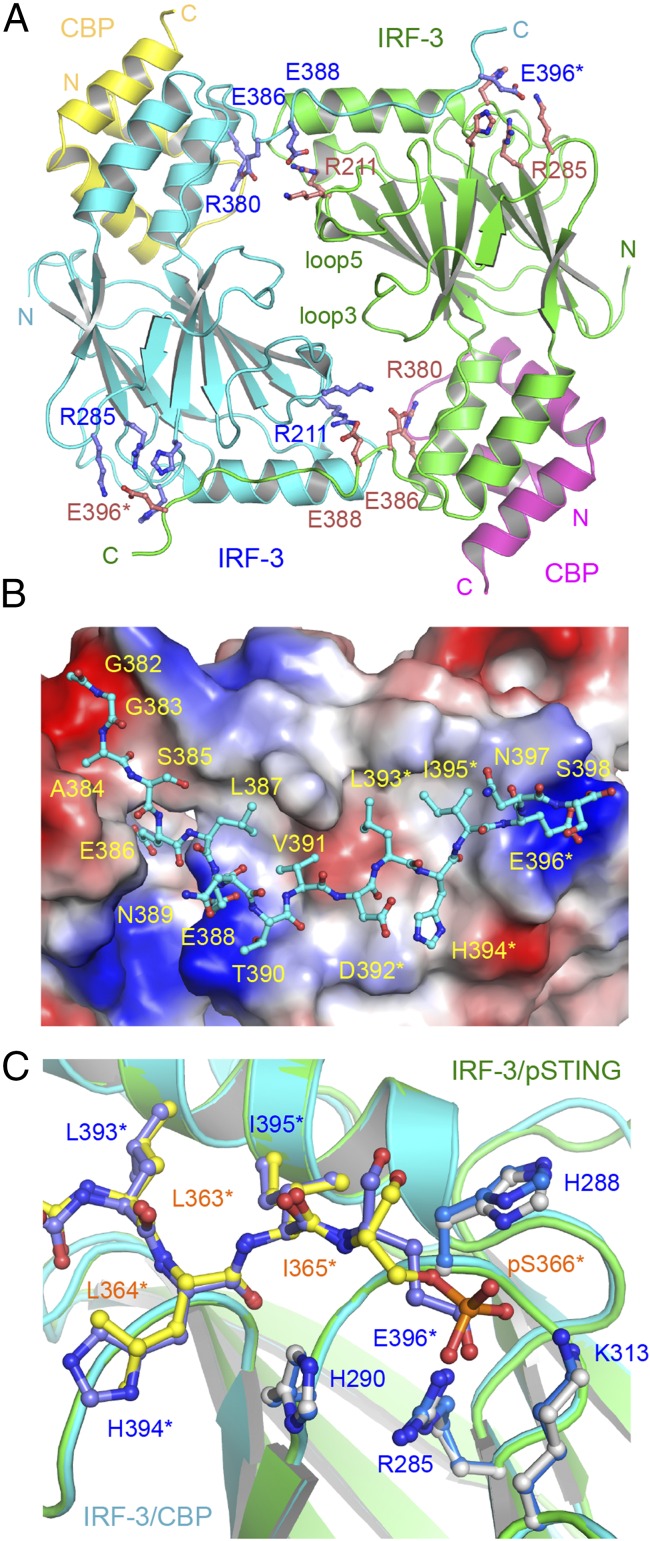
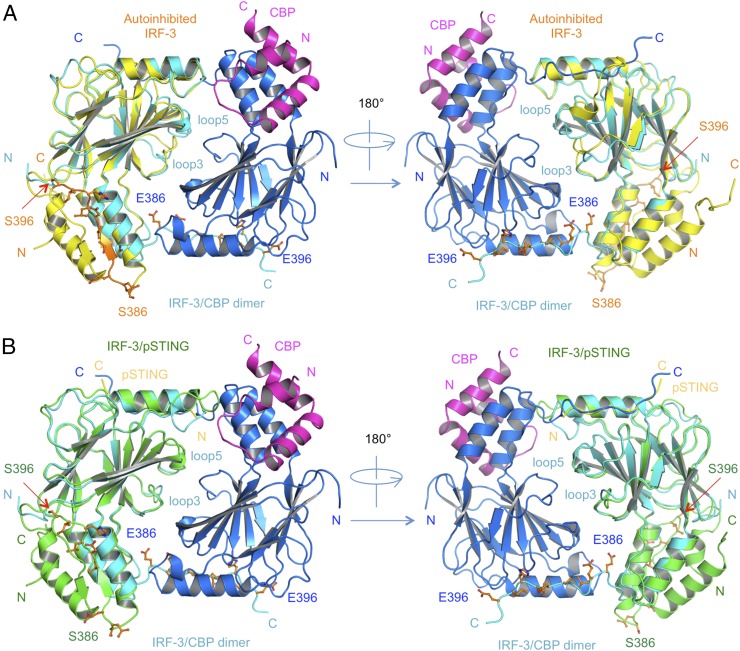
The structure of the IRF-3/CBP complex reveals that the CTT of IRF-3 containing the pLxIS motif undergoes a dramatic conformational change from a buried autoinhibitory configuration (27) to an extended coil that mediates the formation of a domain-swapped dimer (Fig. 4A and Fig. S5A). The IRF-3 dimer interface can be subdivided into a smaller central region and tail-mediated interactions (Fig. 4A). Hydrophobic interactions among Leu299, Leu300, and Trp358 in loop3 and loop5 form the core of the central dimer interface, surrounded by Pro357, Thr359, Glu297, Asn302, Arg213, and Lys360, which forms a salt bridge with Glu297 of the opposite protomer. The C-terminal tail contributes to more than two-thirds of the total buried surface area (∼3,800 Å2) at the IRF-3 dimer interface (Fig. 4B and Fig. S5A). Leu387, Val391, Leu393, and Ile395 reach into a long hydrophobic groove on the surface of IRF-3 (Fig. 4B). Glu386, the phosphomimetic of pSer386, interacts with Arg380 upstream of it (Fig. 4A), introducing a bend to the CTT of IRF-3 so that it can reach into the pLxIS motif-binding surface of a neighboring IRF-3 molecule. In addition, the electrostatic interaction between Glu388 and Arg211 also contributes to the dimerization of IRF-3 (Fig. 4A). Glu396, the phosphomimetic of pSer396, interacts with the positively charged cluster around Arg285 (Fig. 4 A and C). Of note, the phosphomimetic pLxIS motif interacts with a neighboring IRF-3 in a manner similar to that of the phosphorylated pLxIS motif of STING (Fig. 4C). Because the affinity between the S386/396E mutant of IRF-3 is ∼7.5-fold lower than that of pIRF-3, it is likely that other phosphorylated residues also contribute to the higher stability of the pIRF-3 dimer. Indeed, we observed that the phosphorylation site residue Thr253 is at the IRF-3 dimer interface. It may interact with Arg380 of a neighboring IRF-3 molecule upon phosphorylation. Sequence alignment of IRF-3 and IRF-7, which also regulates IFN-β induction, reveals that key residues that mediate IRF-3 dimerization are conserved in IRF-7, suggesting that a similar mechanism is likely involved in IRF-7 dimerization or IRF-3/7 heterodimerization.
Fig. 4.
Structure of the IRF-3 phosphomimetic mutant S386/396E bound to CBP. (A) The domain-swapped dimer of the IRF-3 S386/396E mutant bound to CBP. Key residues mediating IRF-3 dimerization are shown by blue and pink ball-and-stick models. Residues of the pLxIS motif are indicated by asterisks. (B) Structure of the C-terminal tail of IRF-3 (cyan ball-and-stick model) that mediates IRF-3 dimerization. The IRF-3 dimer with the C-terminal tail omitted is shown by the surface representation colored according to surface electrostatic potential. The positively charged surface is in blue, and the negatively charged surface is in red. (C) Superposition of the pLxIS motifs of IRF-3 and pSTING. The pLxIS motif of IRF-3 with the S396E mutation is shown by the blue ball-and-stick model. The pLxIS motif of pSTING is shown by the yellow ball-and-stick model.
Fig. S5.
Structure of the IRF-3/CBP complex, crystallographic dimers of IRF-3 bound to pSTING and pTRIF, and comparison of IRF-3 structures. (A) Difference map showing the CTT (residues 382–398) of IRF-3 in the IRF-3/CBP complex contoured at 2.5σ. The σA-weighted Fo-Fc map was calculated with these residues omitted from the model. The omitted residues are shown by the light green ball-and-stick models, and the rest of the model is shown by the green and cyan surface. Residues of the pLxIS motif are indicated by asterisks. (B) Structure of the pSTING/IRF-3 dimer in the crystallographic asymmetric unit. (C) Structure of the crystallographic symmetry-related pTRIF/IRF-3 dimer. (D) Superposition of IRF-3 structures from the pSTING/IRF-3, pMAVS/IRF-3, pTRIF/IRF-3, pNSP1/IRF-3, NSP1/IRF-3, and IRF-3/CBP complexes. (E) Superposition of IRF-3 structures from the IRF-3/CBP complex (cyan and red) and the pSTING/IRF-3 complex (green). The conformational change in loop 249–254 is indicated by the magenta arrow. (F) Mutations of IRF-3 affecting its dimerization and activation mapped onto the structure of the IRF-3/CBP complex. Mutations of residues at the IRF-3 CTT binding surface are highlighted in orange and are labeled in blue. Mutations of IRF-3 C-terminal residues (purple ball-and-stick models) are labeled in yellow.
Comparison of the structures of autoinhibited IRF-3, IRF-3/adaptor complexes, and the phosphomimetic IRF-3/CBP dimer reveal the mechanism of IRF-3 activation by the adaptors (Fig. 5 A and B). Binding of pSTING, pTRIF, and pMAVS induces a major reorganization of loop5 of IRF-3 necessary to create the binding surface for the pLxIS motif (Figs. 1B and 2 C and E and Fig. S2C). Binding of the adaptor proteins induces a sharp bend at Gly349 in loop5 that moves the backbone by as much as 16 Å (Fig. S2C), forming a solvent-exposed cluster of previously buried residues, Pro357, Trp358, and Thr359. This reorganization of loop5 induces a shift in loop3 (Fig. 5 and Fig. S2C), forming a continuous surface at the central dimer interface. Interestingly, the central dimer interface is conserved in crystallographic dimers observed in the pSTING/IRF-3 and pTRIF/IRF-3 complexes (Fig. S5 B and C), suggesting that this dimer likely represents a “predimer” configuration that then can be locked into the domain-swapped active dimer upon phosphorylation of the IRF-3 tail by TBK1. Comparisons of the structure of the IRF-3/CBP dimer with the structures of the pSTING/IRF-3 and pTRIF/IRF-3 complexes reveal that a conformational change in the loop between β4 and α2 containing residues Gly249 to Asp254 is also required for the formation of the IRF-3/CBP dimer (Fig. S5 D and E). This conformational change moves the loop away from the IRF-3 dimer interface, avoiding clashes between residues Leu252 and Val381 and residues Thr253 and Arg380 (Fig. S5E). Once the highly stable IRF-3 dimer forms, it will be free from the adaptors and can translocate to the nucleus and initiate IFN-β transcription. In summary, these structural analyses of the IRF-3/adaptor and IRF-3/CBP complexes provide critical insights into the mechanisms of IRF-3 activation in the three innate sensing pathways.
Fig. 5.
Comparison of structures of autoinhibited IRF-3, the pSTING/IRF-3 complex, and the IRF-3/CBP dimer. (A) Comparison of the autoinhibited IRF-3 and IRF-3/CBP dimer structures. Autoinhibited IRF-3 is shown by yellow ribbons. The key residues at the CTT of IRF-3 are shown by orange ball-and-stick models. Blue and cyan ribbons show IRF-3 in the IRF-3/CBP dimer structure. CBP bound to the blue IRF-3 is shown by a magenta ribbon. CBP bound to the cyan IRF-3 is deleted for clarity. (B) Comparison of the pSTING/IRF-3 complex and IRF-3/CBP dimer structures. The pSTING/IRF-3 complex is shown by yellow (pSTING) and green (IRF-3) ribbons. The key residues of the CTT of IRF-3 are shown by orange ball-and-stick models. The blue and cyan ribbons show IRF-3 in the IRF-3/CBP dimer structure. CBP bound to the blue IRF-3 is shown by a magenta ribbon. CBP bound to the cyan IRF-3 molecule is deleted for clarity.
Interestingly, a causal relationship between the IRF-3 R285Q mutation and herpes simplex encephalitis has been reported recently (31). The R285Q mutation of IRF-3 impairs innate immune responses to herpes simplex virus infection. Consistent with this finding, the IRF-3 mutation R285D disrupts IFN-β reporter activation by Newcastle disease virus in IRF-3−/− fibroblasts (32). Crystal structures of IRF-3 bound to the adaptors and the IRF-3/CBP dimer complex suggest that R285Q or R285D mutations impair IRF-3 recruitment and activation by disrupting the electrostatic interactions between the pLxIS motif and IRF-3. The IRF-3/CBP dimer structure also explains how dozens of previously reported mutations affect IRF-3 activation (Fig. S5F) (27, 29, 32–34). Most of these mutations are located at the tail-mediated IRF-3 dimer interface and play crucial roles in mediating IRF-3 dimerization (Fig. S5F). Taken together, these observations unanimously confirm the functional relevance of the active IRF-3 dimer structure.
Previous structural studies show that there are four IRF-3–binding sites on the IFN-β enhancer (35, 36). Interestingly, we observed that the IRF-3/CBP complex forms a tetramer in the crystallographic asymmetric unit (Fig. S6A). All the IRF-3 N termini in the tetramer are exposed (Fig. S6A). The avidity of a similar tetramer of full-length IRF-3 likely facilitates DNA binding by pIRF-3 and plays a critical role in transcription activation. Because residues 106–196 of IRF-3 form a long and flexible linker between its DNA-binding and dimerization domains, it is likely that no conformational change of the IRF-3 dimers or tetramer as observed in the structures is needed for efficient DNA binding by full-length IRF-3. To test this hypothesis, we coexpressed the CBP fragment with IRF-3 or its S386/396E mutant containing residues 1–398, which include both the DNA-binding and dimerization domains. Gel-filtration chromatography shows that the pIRF-3(1–398)/CBP complex forms a tetramer in solution (Fig. S6B), whereas the phosphomimetic S386/396E mutant of IRF-3(1–398) bound to CBP is mostly dimeric (Fig. S6C). DNA-binding studies using a 32-bp dsDNA derived from the IFN-β enhancer, which contains all four IRF-3–binding sites, show that both the pIRF-3(1–398)/CBP tetramer and the IRF-3(1–398, S386/396E)/CBP dimer bind the IFN-β enhancer DNA and form similar large complexes (Fig. S6 B and C). In contrast, unphosphorylated full-length IRF-3 is mostly monomeric and does not form a similar high-molecular-weight complex with the same dsDNA (Fig. S6D). These results indicate the functional relevance of the IRF-3 dimer and tetramer in IFN-β enhancer DNA binding and transcription activation.
Fig. S6.
Structure of the IRF-3/CBP tetramer in the ASU and binding studies of the pIRF-3 (residues 1–398)/CBP complex, IRF-3 (residues 1–398, S386/396E)/CBP complex, and unphosphorylated full-length IRF-3 with a 32-bp dsDNA derived from the IFN-β enhancer. (A) Crystallographic tetramer of IRF-3 (residues 189–398, S386/396E) in complex with CBP. (B) Gel-filtration chromatography DNA-binding studies of pIRF-3 (residues 1–398) in complex with CBP. The sequence of the DNA used in this study is 5′-CATAGGAAAACTGAAAGGGAGAAGTGAAAGTG-3′. (C) Gel-filtration chromatography DNA-binding studies of the IRF-3 (residues 1–398, S386/396E) mutant in complex with CBP. (D) Gel-filtration chromatography DNA-binding study of unphosphorylated full-length IRF-3.
Rotavirus NSP1 Employs a pLxIS Motif to Target IRF-3 for Degradation.
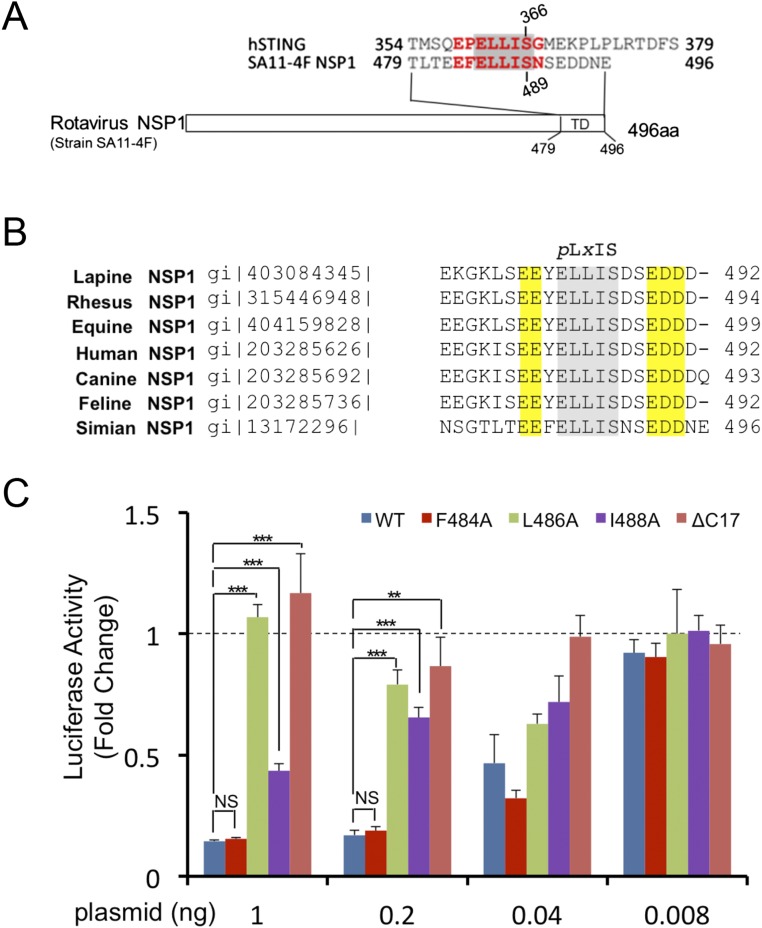
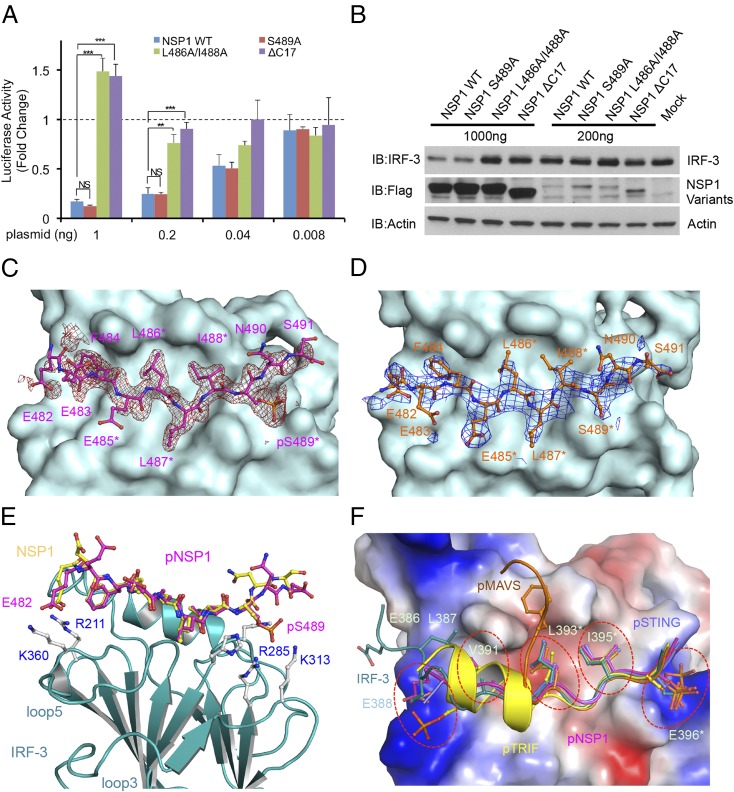
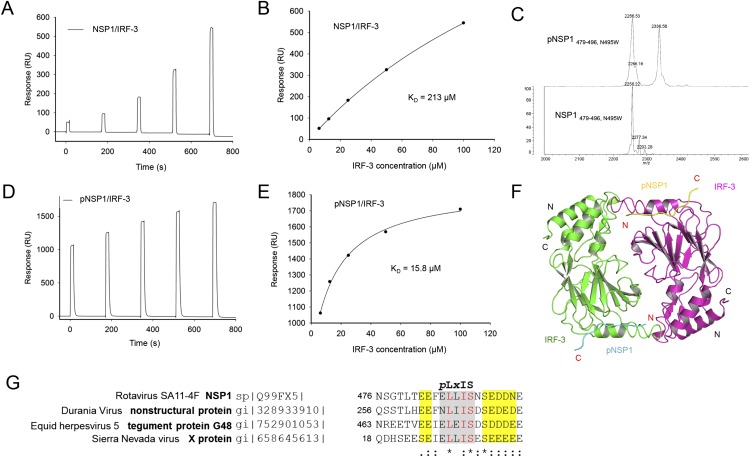
BLAST homology searches revealed that NSP1 of the simian group A rotavirus (RVA) strain SA11-4F also contains a pLxIS motif, which is conserved in NSP1 of rotavirus from other hosts (Fig. S7 A and B). Previous studies showed that NSP1 targets IRF-3 for degradation to evade innate immune surveillance (24–26). The C-terminal 17 residues of NSP1 containing the pLxIS motif are essential for IRF-3 targeting (24). Consistent with this finding, the IFN-β luciferase reporter assay shows that full-length NSP1, but not the ΔC17 truncated form, inhibits IFN-β reporter activation upon cGAMP stimulation (Fig. 6A). Next, we tested whether phosphorylation of the pLxIS motif is required for the activity of NSP1. Strikingly, the S489A mutation within the pLxIS motif of NSP1 does not affect IFN-β reporter suppression (Fig. 6A), indicating that phosphorylation of NSP1 is not required for IRF-3 targeting. In contrast, the L486A mutation abolishes the activity of NSP1, whereas the I488A mutation partially impairs the activity of NSP1 (Fig. S7C). Consistent with these results, Western blot analyses show that overexpression of wild-type NSP1 or its S489A mutant still induces IRF-3 degradation (Fig. 6B). In contrast, either C-terminal truncation or the L486A and I488A mutations impair NSP1-mediated degradation of IRF-3 (Fig. 6B). Consistent with these cell-based analyses, SPR binding studies show that unphosphorylated NSP1 still binds IRF-3 with an affinity of ∼200 μM (Fig. S8 A and B), whereas phosphorylated NSP1 (pNSP1) binds IRF-3 with a moderate affinity of ∼16 μM (Fig. S8 C–E).
Fig. S7.
Sequences of rotavirus NSP1 and luciferase reporter assays of NSP1 mutants. (A) Sequence alignment of the pLxIS motif of rotavirus NSP1 and human STING. (B) Sequence alignment of NSP1 C-terminal peptides from RVA strains in different host species. The pLxIS motifs are shaded. The acidic amino acid clusters flanking the pLxIS motif are highlighted in yellow. (C) Luciferase assays showing the effects of NSP1 mutations in the suppression of IFN-β reporter activation in HEK293T cells transfected with NSP1 and human STING upon cGAMP stimulation. **P < 0.01, ***P < 0.001, NS (P > 0.05) values were calculated based on comparisons of signals in cells transfected with NSP1 mutants and in cells transfected with wild-type NSP1.
Fig. 6.
The pLxIS motif of rotavirus NSP1 targets IRF-3 for degradation and binds to IRF-3 without phosphorylation. (A) Luciferase assays showing that NSP1 suppresses IFN-β reporter activation in HEK293T cells transfected with NSP1 and STING upon stimulation by cGAMP. The S489A mutation does not affect the activity of NSP1. Mutations L486A/I488A within the pLxIS motif or deletion of the C-terminal 17 residues of NSP1 abolish its ability to suppress IFN-β reporter activation. The data (mean ± SEM) are representative of three independent assays. **P < 0.01, ***P < 0.001, NS (P > 0.05) values were calculated based on comparisons of signals in cells transfected with NSP1 mutants and in cells transfected with wild-type NSP1. (B) Western blot showing that wild-type NSP1 and its S489A mutant mediate IRF-3 degradation in HEK293T cells. Mutations within the pLxIS motif or deletion of the C-terminal residues of NSP1 impair its ability to target IRF-3 for degradation. (C) Difference map showing pNSP1 bound to IRF-3 contoured at 2.5σ. The σA-weighted Fo-Fc map was calculated with pNSP1 deleted from the model. pNSP1 is shown by the magenta ball-and-stick model, and IRF-3 is shown by the cyan surface. Residues of the pLxIS motif are indicated by asterisks. (D) Difference map showing unphosphorylated NSP1 bound to IRF-3 contoured at 2.0σ. NSP1 is shown by the orange ball-and-stick model. (E) Superposition of the structures of phosphorylated (magenta) and unphosphorylated (yellow) NSP1 bound to IRF-3 (cyan ribbons). Key residues of IRF-3 involved in NSP1 binding are shown by gray ball-and-stick models. (F) Superposition of structures of pSTING, pMAVS, pTRIF, and pNSP1 bound to IRF-3 and IRF-3 CTT in the IRF-3/CBP dimer. Key residues involved in IRF-3 binding or IRF-3 dimerization are shown by ball-and-stick models. IRF-3 is shown by the surface representation colored by surface electrostatic potential. The positively charged surface is in blue, and the negatively charged surface is in red. The five ligand-binding sites of IRF-3 are indicated by red circles.
Fig. S8.
Binding studies of unphosphorylated and phosphorylated NSP1 C-terminal peptides with the IRF-3 CTD. (A) SPR binding study of unphosphorylated NSP1 C-terminal peptide (residues 479–496, N495W) with IRF-3. (B) Binding affinity of unphosphorylated NSP1 peptide (residues 479–496, N495W) and IRF-3. (C) MALDI-TOF analysis of the pNSP1 peptide (residues 479–496, N495W). (D) SPR binding study of the pNSP1 peptide (residues 479–496, N495W) with IRF-3. (E) Binding affinity of the pNSP1 peptide (residues 479–496, N495W) and IRF-3. (F) Structure of the crystallographic symmetry-related pNSP1/IRF-3 dimer. Similar dimers also are present in the structure of IRF-3 bound to unphosphorylated NSP1. (G) Amino acid sequences of other viral proteins containing the pLxIS motif. Acidic residues flanking the motif are highlighted in yellow.
To elucidate the structural basis of IRF-3 targeting by NSP1, we determined the structures of phosphorylated and unphosphorylated NSP1 C-terminal peptides bound to IRF-3 (Fig. 6 C–E and Table S1). pNSP1 forms an extended coil structure similar to that of pSTING (Fig. 6C and Fig. S8F). At the N terminus, pNSP1 interacts with IRF-3 through electrostatic interactions between Glu482 and Arg211 (Fig. 6E). In the middle, Phe484, Leu486, and Ile488 reach into the hydrophobic groove of IRF-3 (Fig. 6C). Near the C terminus, pSer489 interacts with the positively charged cluster around Arg285 (Fig. 6E). Strikingly, unphosphorylated NSP1 binds IRF-3 in a manner similar to that of pNSP1 (Fig. 6 D and E). Although the electrostatic interactions mediated by pSer489 are lost, the electrostatic interaction between Glu482 and Arg211, the hydrophobic interactions, and the hydrogen bonds are all preserved, suggesting that these interactions mediate the lower-affinity binding between unphosphorylated NSP1 and IRF-3. Consistent with this observation, mutation of Leu486 or Ile488 within the pLxIS motif to alanine abolished IRF-3 binding (Table S2). The E482A mutation also reduces the binding affinity by about twofold (Table S2). Taken together, these structural and functional studies reveal the molecular basis of IRF-3 targeting by rotavirus NSP1.
Discussion
The cGAS–STING, RLR–MAVS, and TLR–TRIF pathways in innate immunity converge at the recruitment and activation of transcription factor IRF-3. The adaptor proteins STING, MAVS, and TRIF recruit IRF-3 through their phosphorylated pLxIS motifs. Here, we determined a series of crystal structures of IRF-3 bound to pSTING, pMAVS, and pTRIF; the activated IRF-3 dimer bound to CBP; and the complexes of rotavirus NSP1 bound to IRF-3 to elucidate the molecular mechanisms of IRF-3 recruitment and activation and viral evasion of the innate immune response. These comprehensive structural and functional studies reveal that the pLxIS motif not only mediates IRF-3 recruitment but also plays critical roles in IRF-3 activation and in the targeting of IRF-3 by virus. Although phosphorylation of the pLxIS motif is needed for IRF-3 recruitment and activation, it is not required for the targeting of IRF-3 by rotavirus NSP1.
These structural analyses reveal that the pLxIS motifs adopt similar structures and interact with IRF-3 in similar manners (Fig. 6F). However, the pLxIS motif alone is not sufficient to mediate effective binding of IRF-3. Other interactions, such as the hydrophobic and electrostatic interactions upstream of the motif, are also critical for IRF-3 binding (Fig. 6F and Table S2). Based on these structures, we propose a five-site binding model for proteins containing the pLxIS motif (Fig. 6F). The positively charged patch around Arg211 interacts with negatively charged residues or phosphoserine upstream of the pLxIS motif, followed by binding sites for three hydrophobic residues. At the C terminus, the positively charged cluster around Arg285 recognizes the phosphoserine of the pLxIS motif (Fig. 6F). All five sites are used in pNSP1 binding and IRF-3 dimerization, whereas subsets of these five binding sites are involved in pSTING, pMAVS, pTRIF, and NSP1 binding. Because the N terminus of the pMAVS peptide does not interact with site 1 and site 2 of IRF-3, this lack of interaction likely contributing to the lower binding affinity between pMAVS and the higher average B-factor of pMAVS peptides in the crystal structure. It is likely that other proteins containing the pLxIS motif (23) also interact with IRF-3 in similar manners.
The structural data presented here suggest a model for concerted recruitment and activation of IRF-3 by the innate immune adaptors STING, MAVS, and TRIF. In all three pathways, recognition of a pathogen-associated molecular pattern (PAMP) leads to the oligomerization or clustering of the adaptors at membrane surface (e.g., STING at the ER surface, MAVS at the mitochondrial surface, and TRIF at the inner surface of the plasma or endosome membranes). Next, the adaptors are phosphorylated on the pLxIS motif, creating a scaffold with a high local concentration of phosphorylated adaptors capable of recruiting IRF-3. Binding of the phospho-pLxIS motif to IRF-3 induces conformational changes that begin to reorganize key IRF-3 loops and uncover surfaces involved in dimerization, whereas colocalization of IRF-3 with TBK1 induces phosphorylation of the pLxIS motif in the IRF-3 tail. Phosphorylation of IRF-3 by TBK1 can occur in vitro but is relatively inefficient and requires high concentrations (34); colocalization of TBK1 and IRF-3 through recruitment via the adaptors increases local concentrations of both, thus facilitating the phosphorylation and dimerization of IRF-3. The scaffold mechanism for IRF-3 activation would protect effectively against the activation of IRF-3 in the cytosol by TBK1 until appropriate formation of the innate signaling machinery. Because the pLxIS motif-binding surface of IRF-3 plays critical roles in both IRF-3 recruitment and activation, and because the adaptors bind to IRF-3 at relatively low affinities, high-affinity ligands can be designed based on this set of structures to suppress undesired innate immune responses in autoimmune disorders, such as lupus.
Because the pLxIS motif plays a critical role in mediating the recruitment and activation of IRF-3 in the cGAS, RLR, and TLR signaling pathways, rotavirus employs the same motif to evade innate immune surveillance. Our work presented here reveals that this motif is used by rotavirus NSP1 to target IRF-3 for degradation. However, it is surprising that phosphorylation of the pLxIS motif is not essential for NSP1-mediated IRF-3 degradation. Instead, NSP1 binds IRF-3 through hydrophobic residues within and upstream of the pLxIS motif and acidic residues flanking the motif, thus avoiding the triggering of innate signaling pathways to activate protein kinases such as TBK1 to phosphorylate its pLxIS motif. Consistent with this finding, we observed that NSP1 C-terminal peptide is not a good substrate for TBK1 and cannot be fully phosphorylated under the same conditions as STING, MAVS, or TRIF. Extended incubation with TBK1 at higher concentrations also failed to phosphorylate the NSP1 peptide fully. Two clusters of negatively charged residues flank the pLxIS motif of NSP1 (Fig. S7B). These residues are not observed in the sequences of STING, TRIF, and MAVS. It is likely these residues affect NSP1 phosphorylation by TBK1. On the other hand, because NSP1 does not have the binding surface for the pLxIS motif, it is unlikely to be recruited by the adaptors and phosphorylated by TBK1 at the signaling complexes. However, we cannot rule out the possibility that other protein kinases may phosphorylate NSP1 and facilitate IRF-3 degradation. It will be interesting to explore if such an elegant mechanism is unique to rotavirus hijacking of IRF-3. Indeed, a BLAST search of viral proteomes with the pLxIS motif and the flanking acidic amino acid clusters revealed that other viral genomes contain similar sequences (Fig. S8G), suggesting that the strategy used by rotavirus is likely involved in innate immune evasion by other viruses.
Materials and Methods
Protein Expression and Purification.
The cDNA encoding human STING, human MAVS, human TRIF, rotavirus NSP1, and human IRF-3 were cloned into a modified pET28(a) vector with an N-terminal Avi-His6-SUMO tag. For protein quantification, mutations F378W, M449W, L218W, N495W, Y408W, and E426W were introduced into human STING (residues 342–379), human MAVS (residues 433–450), human TRIF (residues 199–219), and NSP1 (residues 479–496), IRF-3 (residues 382–409), and IRF-3 (residues 382–427) sequences, respectively. The cDNA encoding human IRF-3 (residues 189–427, 189–398, 1–398, and full-length) were cloned into a modified pET28(a) vector with an N-terminal His6-SUMO tag. The proteins were expressed in Escherichia coli BL21 (DE3) cells induced with 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) overnight at 16 °C and purified using a Ni2+-NTA column (Qiagen). The purified proteins were cleaved with SUMO protease at 4 °C overnight. The SUMO tag was removed using a Ni2+-NTA column, and the target proteins or peptides in the flow through were further purified by gel-filtration chromatography using a HiLoad 16/60 Superdex 75 column (GE Healthcare) eluted with 20 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl. Human STING CTD (residues 155–379) and mouse TBK1 (residues 1–657) were expressed and purified as described previously (34). SUMO fusion of human CBP (residues 2065–2111) was cloned into the pET22b(+) vector. IRF-3 (residues 189–398 and 1–398) and CBP were coexpressed in E. coli BL21 (DE3) cells and purified using a Ni2+-NTA column. After gel-filtration chromatography on a Superdex 200 column (GE Healthcare), the IRF-3/CBP complex was further purified using a Mono Q 5/50 GL column (GE Healthcare) eluted with a linear gradient of 0–400 mM NaCl in 50 mM Tris buffer at pH 8.5. Biotin-Avi-His6-SUMO peptides were expressed in E. coli BL21 (DE3) cells cotransformed with the pET28(a) plasmids coding for the peptides and the pBirAcm plasmid coding for BirA. Protein expression was induced with 0.4 mM IPTG in the presence of 5 μg/mL biotin at 16 °C overnight. The proteins were first purified using a Ni2+-NTA column and were further purified over a Superdex75 column eluted with 20 mM Tris⋅HCl (pH 7.5) and 150 mM NaCl. Mutants of STING, MAVS, TRIF, IRF-3, and NSP1 were generated using the QuikChange site-directed mutagenesis kit (Agilent) or a PCR-based technique with appropriate primers. Sequences of all of the plasmids were confirmed by DNA sequencing.
Proteins and Peptides Phosphorylation with TBK1.
The purified proteins or peptides were mixed with GST-mTBK1 in a 10:1 (wt/wt) ratio in 20 mM Hepes (pH 7.5), 10 mM MgCl2, 100 mM NaCl, 5 mM ATP, 0.1 mM Na3VO4, 5 mM NaF, 5 mM DTT at 27 °C. The concentration of the proteins or peptides was about 1 mg/mL. After 24 h incubation, the phosphorylated proteins or peptides were concentrated and purified over a Superdex 75 column eluted with 20 mM Tris⋅HCl buffer with 150 mM NaCl at pH 7.5.
MS.
MS analyses of pSTING, pMAVS, pTRIF, pIRF-3, and pNSP1 peptides were performed using a Shimadzu/Kratos Axima CFR MALDI-TOF mass spectrometer. pSTING CTT peptide was analyzed using a Dionex 3000 nRLC coupled to an Orbitrap Fusion mass spectrometer (Thermo Scientific) to identify the phosphorylation sites. The sample was injected onto a trap column for desalting and then was eluted onto an analytical column. A 60,000 resolution data MS scan from 650 to 4,000 was obtained, followed by data-dependent MS2 (second stage of the MS/MS experiment) scans. Both higher-energy collisional dissociation and electron-transfer/higher-energy collision dissociation fragmentation were obtained. MS data were analyzed using the Xcaliber package (Thermo Scientific).
SPR.
The binding affinities between IRF-3 and the adaptor proteins or peptides were determined by SPR using a Biacore X100 SPR instrument (GE Healthcare). Biotin-labeled SUMO-fusion proteins or peptides were coupled on the sensor chip using the Biotin CAPture Kit (GE Healthcare). Dilution series of human IRF-3 (6.25, 12.5, 25, 50, and 100 μM) in 1× HBS-EP+ buffer (GE Healthcare) were injected over the sensor chip at a flow rate of 30 µL/min. The single-cycle kinetic/affinity protocol was used in all binding studies. The sensor chip was regenerated with a buffer containing 6 M guanidine hydrochloride and 0.25 M NaOH. All measurements were duplicated under the same conditions. The equilibrium Kd was determined by fitting the data to a steady-state 1:1 binding model using Biacore X100 Evaluation software version 2.0 (GE Healthcare).
Analytical Ultracentrifugation.
All analytical ultracentrifugation (AUC) experiments were performed at 10 °C in a Beckman XL-I analytical ultracentrifuge with a Beckman An-60 Ti rotor (item no. 361964) using absorbance optics. Sedimentation velocity was performed at 48,000 rpm overnight with 8 μM of each sample, and the data were analyzed with the continuous c(s) distribution model from Sedfit. Partial specific volumes for IRF-3/CBP, S386/396E IRF-3/CBP, and pIRF-3/CBP were calculated using Sednterp to be 0.7425 (representing a weight-averaged value for a 1:1 complex of IRF-3 and CBP), and extinction coefficients were calculated based on amino acid sequence to be 56,950 M/cm. Sedimentation equilibrium data were collected at 18,000 rpm, 22,000 rpm, and 32,000 rpm using samples loaded at 1, 3.5, and 14 μM concentrations. Data for nine curves per sample were analyzed globally using WinNonlin.
Crystallization, Data Collection, and Structure Determination.
Purified IRF-3189–427 was mixed with the peptides in a 1:3 molar ratio with a final concentration of IRF-3 at ∼7.5 mg/mL. Crystals of IRF-3 in complex with pSTING342–379, F378W, pMAVS433–450, M449W, and unphosphorylated NSP1479–496 were grown in 0.1 M Bis-Tris at pH 5.5, ∼0.08–0.2 M MgCl2, ∼20–25% (wt/vol) PEG 3350. Crystals of IRF-3 in complex with pTRIF199–219, L218W were grown in 0.1 M NaAc at pH 4.6 and 2.0 M NaCl. The S386E/S396E mutant of IRF-3 in complex with CBP was crystallized in 0.1 M Hepes buffer at pH 7.5 with 25% (wt/vol) PEG 3350. Crystals of IRF-3 in complex with pNSP1479–496, N495W were grown in 0.1 M Hepes at pH 7.5 plus 0.8 M NaH2PO4 and 0.8 M KH2PO4. All proteins were crystallized by hanging-drop vapor-diffusion method at 4 °C. The crystals were flash-frozen in liquid nitrogen in the reservoir solution containing either 25% (vol/vol) glycerol or 20% (vol/vol) ethylene glycol. Diffraction data were collected at the Advanced Light Source beamlines 5.0.1 using a Quantum 315R CCD detector or a Rigaku R-AXIS IV++ image plate detector at home. The diffraction data for the pSTING/IRF-3, pTRIF/IRF-3, pNSP1/IRF-3, and NSP1/IRF-3 complexes were processed with the HKL2000 package (37). The diffraction data for the pMAVS/IRF-3 and IRF-3/CBP complexes were indexed and integrated with iMosflm and merged with Aimless in the CCP4 package (38). The structure of the pSTING/IRF-3 complex was determined by molecular replacement (MR) using the structural model of IRF-3 CTD [Protein Data Bank (PDB) ID code 1QWT] as the search model using Phaser in the Phenix package (27, 39). The pSTING/IRF-3 complex model was rebuilt with Coot (40). Structures of the pMAVS/IRF-3 and pTRIF/IRF-3 complexes were determined by MR using the IRF-3 structure in the pSTING/IRF-3 complex as the search model. The structure of the IRF-3/CBP complex was determined by MR using the structure of the IRF-3/CBP complex (PDB ID code 1ZOQ) without the C-terminal tail as the search model (41). The structures of the pNSP1/IRF-3 and NSP1/IRF-3 complexes were solved by MR using IRF-3 in the pTRIF/IRF-3 complex structure as the search model. All structures were refined with Phenix (39). Details of data quality and structure refinement are summarized in Table S1. Pairwise structural superposition was carried out using Superpose in the CCP4 package (38). All structural figures were generated with PyMOL (https://www.pymol.org).
Cell Culture.
HEK293T cells (ATCC, CRL-3216) were cultured in DMEM + GlutaMAX (Gibco) supplemented with 10% FBS (Gibco), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37 °C in a humidified atmosphere with 5% CO2.
IFN-β Luciferase Reporter Assays.
The pcDNA3.1 STING carrying a C-terminal Flag tag was a gift from Cheng Kao (Indiana University, Bloomington, IN). Mutants of STING were generated using the QuikChange site-directed mutagenesis kit (Agilent). Sequences of all mutants were confirmed by DNA sequencing. HEK293T cells were plated in CoStar White 96-well plates at 4 × 104 cells per well. When the cells were ∼80% confluent, they were transfected with a mixture of Lipofectamine 2000 reagent (Invitrogen) and a constant amount of IFN-β firefly luciferase reporter plasmids (20 ng per transfection), phRL-TK–Renilla luciferase plasmids (2 ng per transfection) (Promega), and plasmids of either wild-type or mutant STING (0.2 ng per transfection). Empty pcDNA3.1 plasmid was used as transfection control. The cells were incubated for a further 24 h to allow the expression of the genes and then were treated with 30 μg/mL cGAMP. After 16 h incubation, the cells were analyzed using the Dual-Glo Luciferase Reporter Assay Kit (Promega). Luminescence was quantified with the BioTek Synergy HTX Multi-Mode microplate reader. The relative firefly luciferase activity was normalized by the Renilla luciferase activity. The relative IFN-β reporter fold of induction represents the ratio normalized to control plasmid values with the same treatment. The inhibition of IRF-3 activation by NSP1 and its mutants was evaluated by a luciferase-based inhibition assay. The indicated amounts of NSP1 variant plasmids were cotransfected with a constant amount of IFN-β firefly luciferase reporter plasmids (20 ng per transfection), phRL-TK–Renilla luciferase plasmids (2 ng per transfection), and wild-type STING (0.2 ng per transfection). After cGAMP stimulation (30 μg/mL), the luminescence assay and data analysis were performed as described above.
Western Blot.
HEK293T cells were washed and suspended in PBS. The cells were lysed in 200 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 1% Nonidet P-40 supplemented with one complete EDTA-free protease inhibitor mixture tablet (Roche) and one PhosSTOP phosphatase inhibitor mixture tablet (Roche) for each 10 mL of lysis buffer. The proteins were resolved by SDS/PAGE and transferred to PVDF membrane, which then was incubated overnight with primary antibodies. The following antibodies were used in the Western blot analyses: anti–IRF-3 (1:1,000l; sc-9082; Santa Cruz), anti-Flag M2-Peroxidase (1:2,000; A8592; Sigma), and anti-Actin (1:5,000; HHF35; Pierce). Except for flag-tagged proteins probing, the membrane was further incubated with the corresponding HRP-conjugated secondary antibodies. Proteins of interest were visualized using the Western Lightening Plus ECL (PerkinElmer) according to the manufacturer’s protocol.
Statistical Analysis.
For all figures, statistical analyses were carried out using Microsoft Excel. All data are presented as mean ± SEM. A two-tailed Student's t test assuming equal variants was used to compare two groups. In all figures, the statistical significance between the indicated samples and control is designated as *P < 0.05, **P < 0.01, ***P < 0.001, or NS (P > 0.05).
Acknowledgments
This research was supported in part by National Institutes of Health Grant AI 087741 (to P.L.); Welch Foundation Grant A-1816 (to P.L.); and Cancer Prevention and Research Institute of Texas Grant RP150454 (to P.L. and J.-Y.J.). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, the National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structural factors of the pSTING/IRF-3, pMAVS/IRF-3, pTRIF/IRF-3, IRF-3/CBP, pNSP1/IRF-3, and NSP1/IRF-3 complexes have been deposited in the Worldwide Protein Data Bank, www.wwpdb.org (PDB ID codes 5JEJ, 5JEK, 5JEL, 5JEM, 5JEO, and 5JER, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603269113/-/DCSupplemental.
References
- 1.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol. 2014;32:461–488. doi: 10.1146/annurev-immunol-032713-120156. [DOI] [PubMed] [Google Scholar]
- 3.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: Cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243(1):91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 4.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27(3):370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25(3):373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: A complex web of host defenses. Annu Rev Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barber GN. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 2014;35(2):88–93. doi: 10.1016/j.it.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Shu C, Li X, Li P. The mechanism of double-stranded DNA sensing through the cGAS-STING pathway. Cytokine Growth Factor Rev. 2014;25(6):641–648. doi: 10.1016/j.cytogfr.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498(7454):380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153(5):1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, et al. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51(2):226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity. 2013;39(6):1019–1031. doi: 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao P, et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154(4):748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5(214):ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda K, Takaoka A, Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25(3):349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122(5):669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 19.Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, et al. Cutting edge: A novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol. 2002;169(12):6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301(5633):640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 22.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 23.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347(6227):aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 24.Barro M, Patton JT. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci USA. 2005;102(11):4114–4119. doi: 10.1073/pnas.0408376102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arnold MM, Patton JT. Rotavirus antagonism of the innate immune response. Viruses. 2009;1(3):1035–1056. doi: 10.3390/v1031035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold MM, Barro M, Patton JT. Rotavirus NSP1 mediates degradation of interferon regulatory factors through targeting of the dimerization domain. J Virol. 2013;87(17):9813–9821. doi: 10.1128/JVI.01146-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin BY, et al. Crystal structure of IRF-3 reveals mechanism of autoinhibition and virus-induced phosphoactivation. Nat Struct Biol. 2003;10(11):913–921. doi: 10.1038/nsb1002. [DOI] [PubMed] [Google Scholar]
- 28.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478(7370):515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, et al. Contribution of Ser386 and Ser396 to activation of interferon regulatory factor 3. J Mol Biol. 2008;379(2):251–260. doi: 10.1016/j.jmb.2008.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panne D, McWhirter SM, Maniatis T, Harrison SC. Interferon regulatory factor 3 is regulated by a dual phosphorylation-dependent switch. J Biol Chem. 2007;282(31):22816–22822. doi: 10.1074/jbc.M703019200. [DOI] [PubMed] [Google Scholar]
- 31.Andersen LL, et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J Exp Med. 2015;212(9):1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen W, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15(11):1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahasi K, et al. Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change. Genes Cells. 2010;15(8):901–910. doi: 10.1111/j.1365-2443.2010.01427.x. [DOI] [PubMed] [Google Scholar]
- 34.Shu C, et al. Structural insights into the functions of TBK1 in innate antimicrobial immunity. Structure. 2013;21(7):1137–1148. doi: 10.1016/j.str.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129(6):1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Escalante CR, Nistal-Villán E, Shen L, García-Sastre A, Aggarwal AK. Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer. Mol Cell. 2007;26(5):703–716. doi: 10.1016/j.molcel.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology, Vol 276, Macromolecular Crystallography Part A pp 307–326.
- 38.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin BY, et al. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13(9):1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]