Significance
Genetic variants in human β-cardiac myosin, which causes muscle contraction in the heart, can lead to hypertrophic cardiomyopathy (HCM), an inherited heart disease that can cause sudden death. New technologies have generated sequence data for large numbers of patients with HCM and unaffected individuals. In this study, we compare the protein structural locations of genetic variants of patients with HCM and the general population to identify spatial regions of the myosin that have a higher than expected proportion of genetic variants associated with HCM and earlier age at diagnosis. In addition, we develop new methods to interrogate the localization of genetic changes in protein structures. Our study demonstrates the power of combining clinical, genetic, and structural data to gain insight into Mendelian disease.
Keywords: hypertrophic cardiomyopathy, myosin, rare disease genetics, genetic burden
Abstract
Myosin motors are the fundamental force-generating elements of muscle contraction. Variation in the human β-cardiac myosin heavy chain gene (MYH7) can lead to hypertrophic cardiomyopathy (HCM), a heritable disease characterized by cardiac hypertrophy, heart failure, and sudden cardiac death. How specific myosin variants alter motor function or clinical expression of disease remains incompletely understood. Here, we combine structural models of myosin from multiple stages of its chemomechanical cycle, exome sequencing data from two population cohorts of 60,706 and 42,930 individuals, and genetic and phenotypic data from 2,913 patients with HCM to identify regions of disease enrichment within β-cardiac myosin. We first developed computational models of the human β-cardiac myosin protein before and after the myosin power stroke. Then, using a spatial scan statistic modified to analyze genetic variation in protein 3D space, we found significant enrichment of disease-associated variants in the converter, a kinetic domain that transduces force from the catalytic domain to the lever arm to accomplish the power stroke. Focusing our analysis on surface-exposed residues, we identified a larger region significantly enriched for disease-associated variants that contains both the converter domain and residues on a single flat surface on the myosin head described as the myosin mesa. Notably, patients with HCM with variants in the enriched regions have earlier disease onset than patients who have HCM with variants elsewhere. Our study provides a model for integrating protein structure, large-scale genetic sequencing, and detailed phenotypic data to reveal insight into time-shifted protein structures and genetic disease.
Myosin motors are molecular machines responsible for converting chemical energy into the mechanical force necessary for cell division, directed cell migration, vesicle trafficking, and muscle contraction (1). Variants in the human β-cardiac myosin heavy chain gene (MYH7), cause hypertrophic cardiomyopathy (HCM), a genetic disease of the heart muscle characterized by an asymmetrical thickening of the ventricular walls and a decrease in the ventricular chamber size. HCM is the most common heritable heart disease, with a prevalence of one in 500 individuals (2). Clinically, the course of HCM is variable, with some patients experiencing minimal symptoms and others developing arrhythmia, heart failure, or sudden death (3). Relationships between genotype and disease expression in HCM have been challenging to establish due to the absence of large-scale genetic population data and lack of multicenter sharing of patient genetic and clinical data (4, 5).
The MYH7 gene is highly constrained for genetic variation (6). Few loss-of-function variants are observed in population cohorts, and identified pathogenic variants are mainly missense. However, although many missense genetic variants in MYH7 cause HCM, not all genetic changes within the gene lead to disease. There are competing hypotheses regarding the localization of pathogenic missense variants within MYH7, analysis of which can offer insight into the underlying mechanism of HCM. Investigators have suggested enrichment of pathogenic variants in many of the functional domains of β-cardiac myosin, including the converter domain, actin-binding site, and ATP-binding domain (7–9). However, others have suggested there is no regional enrichment for HCM variation within MYH7 (4, 10). These inconsistencies could be due to limited sample sizes or to a lack of reference cohorts for comparison. Without information about the natural distribution of rare variants within MYH7, it is impossible to distinguish regions of disease-variant enrichment from regions of increased genetic tolerance (11). In addition, some putatively pathogenic variants are later found at higher than expected allele frequencies in large, ethnically diverse population reference cohorts (12). The study suggesting enriched domains lacked reference cohorts; when a reference cohort was compared against genetic variants from a small sample of patients with HCM, the study failed to detect any significant enrichment for disease-associated variants (10). In addition, a focus on the linear sequence of the gene or previously discovered functional domains could overlook novel functional regions or enrichments spanning multiple domains. These discrepancies point to the need to test for regional HCM variant enrichment within MYH7 using both a large patient population and a large reference cohort while also accounting for the 3D structure of β-cardiac myosin.
Recent advances in next-generation sequencing technology have enabled the assembly of large datasets of human genetic variation in both unselected and disease-affected populations. Comparative analysis of these cohorts enables within-gene inference of disease burden and constraint, a measure of population tolerance to variation that can reveal insight into critical functional residues and disease etiology. The Exome Aggregation Consortium (ExAC) (13) and the DiscovEHR (14) cohort are exome sequencing cohorts of 60,706 and 42,930 individuals, respectively, that provide detailed information regarding the rates and types of genetic variation seen within disease genes. In addition, the Sarcomeric Human Cardiomyopathy Registry (SHaRe) was established as an international consortium of HCM investigators and currently contains detailed longitudinal clinical data on 2,913 patients with HCM who have undergone genetic testing. As large clinical and population sequencing projects such as these become more prevalent, novel methods for statistical analysis of variant burden and constraint will be essential for gaining insight into disease and identifying intragene regions enriched from disease-associated variation.
We hypothesized that assessing regional genetic tolerance in the context of time-shifted 3D structures would reveal novel insights into MYH7 and possible hot spots in the myosin structure of pathogenic variants within HCM. Here, we compare genetic data from the SHaRe, an international HCM registry (15), with variants identified in large-scale exome sequencing projects (13) to identify regions of enrichment for HCM-associated variation within pre- and poststroke structures of β-cardiac myosin. In addition, we develop a general statistical framework based on a modified version of a spatial scan statistic to search for regions of increased disease-associated variation in 3D protein structures and surfaces. Finally, we take advantage of the clinical and phenotypic data in the SHaRe to examine clinical differences between groups of patients based upon variant location within β-cardiac myosin. We demonstrate the power of combining clinical, genetic, and structural data to make inferences regarding disease etiology and 3D structural hotspots for HCM variants.
Results and Discussion
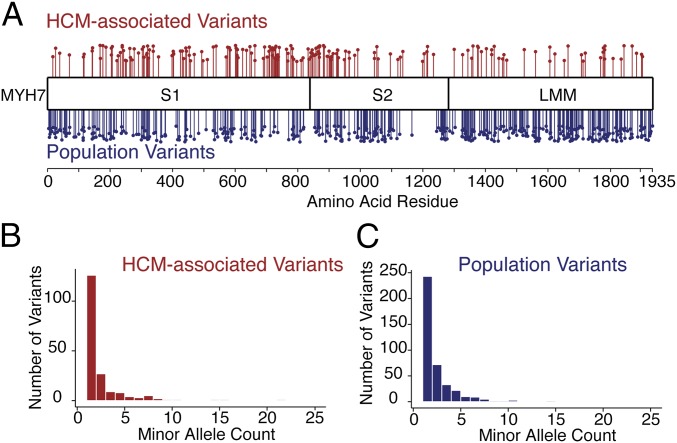
We first compared the linear distribution of missense variants in patients with HCM (SHaRe) with missense variants in a population reference cohort (ExAC). The ExAC cohort contains sequencing information from 60,706 individuals who were part of disease-specific (non-HCM) and population genetic studies, and the SHaRe database contains 2,913 individuals with HCM sequenced for MYH7. Although the ExAC reference contains some pathogenic sarcomere variants and likely some individuals with HCM, it is not enriched for individuals with the disease. We found 192 unique missense variants (in 474 patients with MYH7 variants) in the HCM cohort and 421 unique missense variants in the ExAC database (Dataset S1). In both cases, observed missense variants were very rare and the majority were observed only once (Fig. 1), consistent with previous reports of constraint within the MYH7 gene (6). Further, the vast majority of SHaRe patients with a rare variant in MYH7 carry only one such variant (457 of 474 patients), whereas 16 carry two rare missense variants and a single patient carries three different variants.
Fig. 1.
Differences in the position of missense variants between HCM and population reference cohorts in human β-cardiac myosin. (A) Missense variants identified in SHaRe HCM patients are shown in red, and missense variants identified in ExAC individuals are shown in blue. The height of each point is offset for visibility. (B) Minor allele count of MYH7 missense variants observed in SHaRe HCM probands. (C) Minor allele count of MYH7 missense variants in the ExAC database. Fifteen missense variants with a frequency above 0.005 are not shown.
Both disease (SHaRe) and population/reference (ExAC) variants are nonuniformly distributed throughout the gene, and we find a significant difference in the linear distribution of rare variants between these cohorts [Kolmogorov–Smirnov (KS) test: P = 5.0 × 10−11; Fig. 1]. Disease-associated missense variants are concentrated in the catalytic globular domain and the coiled-coil S2, consistent with some previous results (4) but in contrast to other recent comparisons (10). Even within these domains, however, distributions of disease and population variants are not the same (KS test: P = 0.003). In addition, missense variants in MYH7 in both the SHaRe and ExAC cohorts are extremely rare; the majority in both cohorts are observed only once (Fig. 1 B and C). These results suggest that the likelihood of MYH7 variants causing disease is due, in part, to their location within the gene.
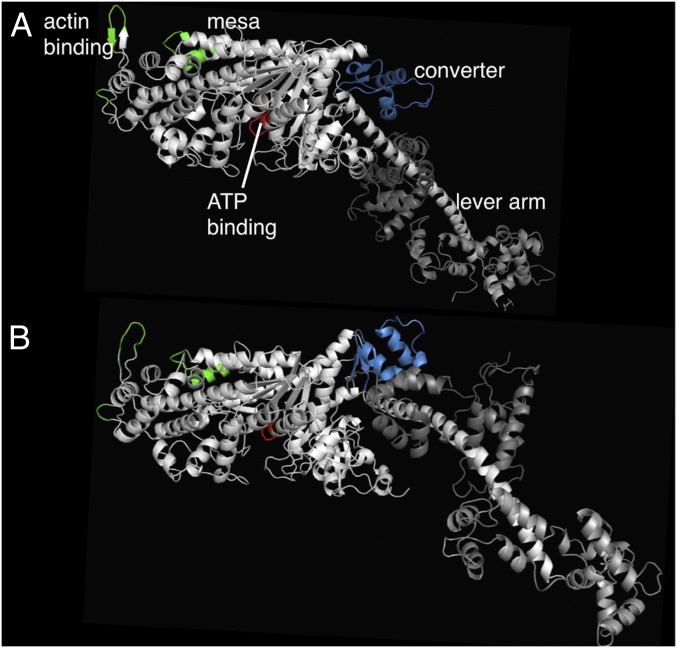
Because molecular motors act in 3D space, we sought a method to investigate patterns of genetic tolerance in the folded structure of human β-cardiac myosin protein. We used multitemplate homology modeling of other myosin proteins in the pre- and poststroke states to build 3D models of human β-cardiac myosin containing the human ventricular light chains (Fig. 2 and Materials and Methods). These models represent two distinct phases of the actin-activated myosin chemomechanical cycle. Four fundamental regions of the myosin motor domain are included: the actin-binding site (Fig. 2, green residues), the ATP-binding pocket (red), the converter domain (blue), and the light chain-binding region or lever arm. In the prestroke state, the converter aligns with a relatively flat surface of the myosin head described as the myosin mesa. Based on its size (>20 nm2), flat topology, and high degree of evolutionary conservation, this feature has been proposed as an interaction site for intra- or intermolecular binding (16). Following the force-producing lever arm stroke of a myosin head, the motor is in its poststroke state (Fig. 2A) and the mesa falls out of alignment with the converter domain.
Fig. 2.
Structural models of the human β-cardiac post- and prestroke obtained by integrating data from solved crystal structures of homologous models. (A) Side view of myosin S1, with the relatively flat mesa at the top, in the poststroke state with important functional domains labeled: the actin-binding site (green residues), the ATP-binding pocket (red), the converter (blue), and the light chain-binding region or lever arm. The converter and its associated lever arm are behind the plane of the figure and below the level of the mesa. (B) Myosin S1 in the prestroke state. Small changes within the globular head region with ADP and Pi in the nucleotide pocket result in a large ∼70° rotation of the converter and lever arm. The converter is moved forward and up compared with the poststroke structure, and the lever arm is projecting forward out of the plane of the image. The distance traversed by the C-terminal end of the lever arm is ∼10 nm, the stroke size of the motor.
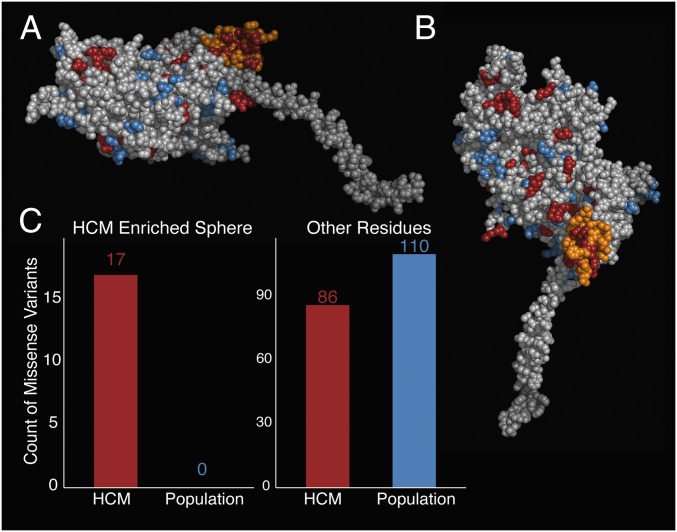
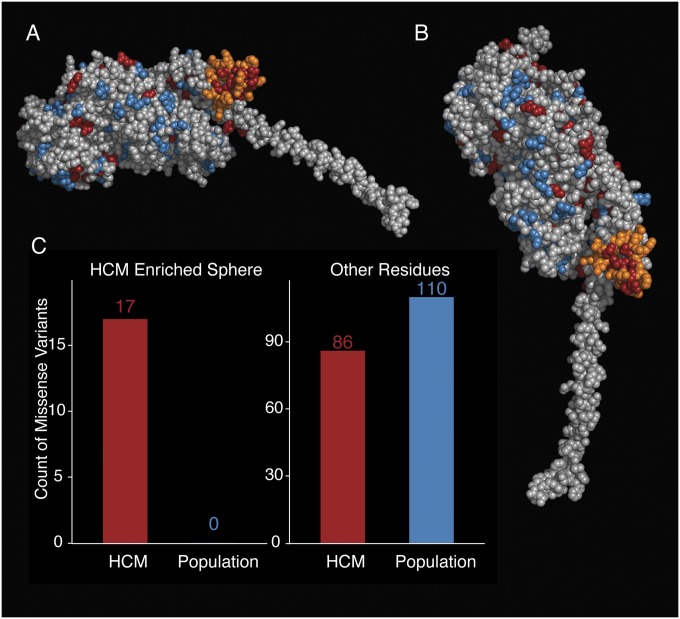
To identify 3D structural regions of interest, we applied a modified version of the spatial scan statistic (17, 18) to the prestroke and poststroke models of β-cardiac myosin S1. This statistic searches for spherical regions with an increased proportion of genetic variants in disease compared with reference cohorts. We defined any rare missense variants observed in patients with HCM in the MYH7 head as disease-associated (n = 103) and any variants seen only in the ExAC cohort as reference (or population) variants (n = 110). Twenty-two variants observed in both the SHaRe and ExAC cohorts were classified as disease-associated. In the myosin prestroke model, we find a striking increase in the proportion of disease-associated missense variation in a 15-Å sphere centered on residue 736 (P = 0.001) (Fig. 3A and Dataset S2). This region, covering a subset of the converter domain, contains 17 missense variants observed in disease and no missense variants observed in reference data (Fig. 3C). Using the poststroke model of β-cardiac myosin, we again observed enrichment of disease-associated variants in a portion of the converter domain (P < 0.001) centered on residue 733 (Fig. S1).
Fig. 3.
Spatial scan statistic identifies a spherical region of the converter domain with an increased proportion of HCM-associated variants. (A) Same side view of the prestroke S1 motor domain as shown in Fig. 1B, without the light chains attached to the α-helical light chain-binding region of the S1. The orange residues define a sphere of residues in the motor domain, which is the only region significantly enriched for HCM variants. The S1 residues are colored as follows: orange, region enriched for HCM variants; blue, missense variants seen only in the ExAC; red, missense HCM variants seen in the SHaRe; light gray, all other residues. (B) Enriched region in the prestroke model of myosin from a different perspective. The view is directly down onto the surface. Coloring is as in A. (C) Number of HCM-variants (SHaRe) and non–disease-associated (ExAC) variants identified in the spherical enriched region (Left) and in the sum of all other parts of the myosin (Right).
Fig. S1.
Poststroke model spherical spatial scan statistic identifies a region of the converter domain as enriched for HCM variants. (A) Side view of the poststroke S1 motor domain as shown in Fig. 2A, without the light chains attached to the S1. The orange residues define a sphere of residues in the motor domain, which is the only region significantly enriched for HCM variants. The S1 residues are colored as follows: orange, enriched region; blue, missense variants seen only in the ExAC; red, missense HCM variants seen in the SHaRe; light gray, all other residues. (B) Enriched region in the poststroke model of myosin, from a different perspective; coloring is as in A. (C) Number of HCM-variants (SHaRe) and non–disease-associated (ExAC) variants identified in the spherical enriched region (Left) and in the sum of all other parts of the myosin (Right).
Enrichment of disease-associated variants in both the pre- and poststroke states persists when including only variants formally classified as pathogenic or likely pathogenic (26 disease-associated of 103 total variants in the SHaRe; Supporting Information). Burden tests can also be sensitive to differences in the population prevalence of disease or differences in the population frequency of variants. To ensure that our results were robust to these potential biases, we performed the same analysis limited to individuals of European descent, the largest population in both the SHaRe and ExAC cohorts (2,519 of 2,913 patients in the SHaRe and 89 of 103 disease-associated variants; Supporting Information). We identified very similar enriched regions in both the pre- and poststroke models, indicating that our analysis is not being significantly affected by population stratification. However, large cohorts from other populations may help identify other regions of the myosin enriched for disease variation.
During systolic contraction of the heart, the converter domain serves the critical function of transducing force by swinging about 70° from its prestroke position (Fig. 3B, lever arm projecting outward). Variants in the converter domain have been shown to alter muscle power output and kinetics (19, 20) and have been associated with worse outcomes in HCM (21, 22). We find that the converter domain is the only spherical region significantly enriched for disease variation in contrast to previous reports, which hypothesized that there were many regions of enrichment throughout the myosin head (7–9) or others that found no regions of significant enrichment (10). Although location in an enriched region is not a necessary condition for pathogenicity, novel variants in regions strongly enriched for disease variation should be viewed with increased suspicion. In regions where nearby variants do not have similar effects, there will not be an enrichment of disease-associated variation. Our data provide a complementary line of evidence that variants in part of the β-cardiac myosin converter domain are poorly tolerated and individuals carrying variants in this region are prone to development of HCM.
To replicate these results, we sought an independent source of disease-associated and population genetic variation. We curated publications from medical centers not yet affiliated with the SHaRe registry (Dataset S3 and Supporting Information). We compared this set of 231 unique missense variants with 430 unique missense variants found in 42,930 exomes from unselected individuals in the Geisinger Health System sequenced by the Regeneron Genetics Center (DiscovEHR cohort) (14) (Dataset S4). The converter domain regions identified in both the prestroke and poststroke states showed enrichment of disease-associated variants in the replication dataset (prestroke: P = 0.0079, poststroke: P = 4.1 × 10−4).
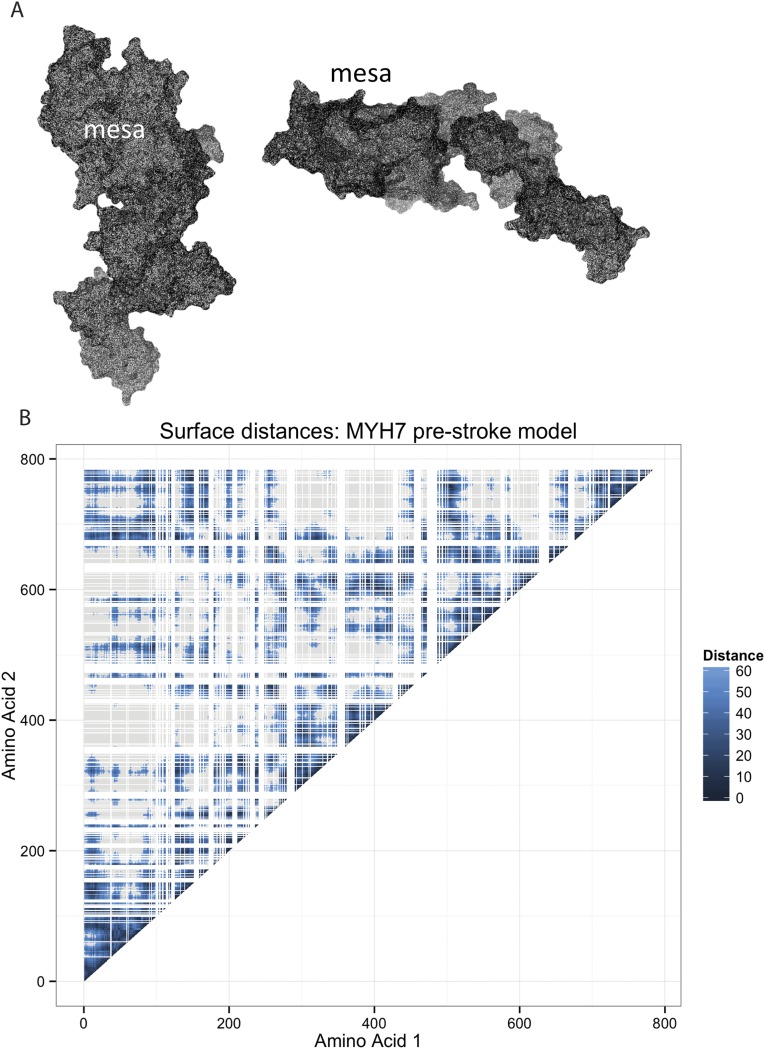
Surface regions of the β-cardiac myosin have been suggested to be important functional domains implicated in HCM (16). We extended our analysis to examine surface regions of β-cardiac myosin and search for regions enriched for HCM-associated variants. We first defined surface-exposed amino acids by their accessibility to a spherical probe with a radius of 2.5 Å (the approximate size of an amino acid side chain) and approximated the surface distance between any two residues (23) (Supporting Information). As expected, surface distances tended to be shorter between amino acids nearby in the primary sequence (Fig. S2B, diagonal). In addition, there were many regions where amino acids far apart in the primary sequence were close together on the surface of the β-cardiac myosin (e.g., amino acids near residues 110 and 684).
Fig. S2.
Solvent-excluded surface mesh of myosin with the labeled mesa region and amino acid surface distances calculated for the myosin head. (A) Solvent-excluded surface is represented by a network of points in space. This weighted network is used to calculate the surface distance between any two points. The mesh was calculated using the MSMS program with a 2.5-Å surface probe. The mesa region is labeled. (B) Surface distance in angstroms between any two amino acid residues in the prestroke model of the myosin head, with darker shading indicating residues that are closer together. White lines indicate amino acids not located on the surface. Gray regions indicate distances of greater than 60 Å. Residues near each other in the linear sequence of the protein are often located near each other (dark shading along diagonal). In addition, there are many instances where residues that are far apart in the linear sequence are close together in 3D space (dark shading off the diagonal).
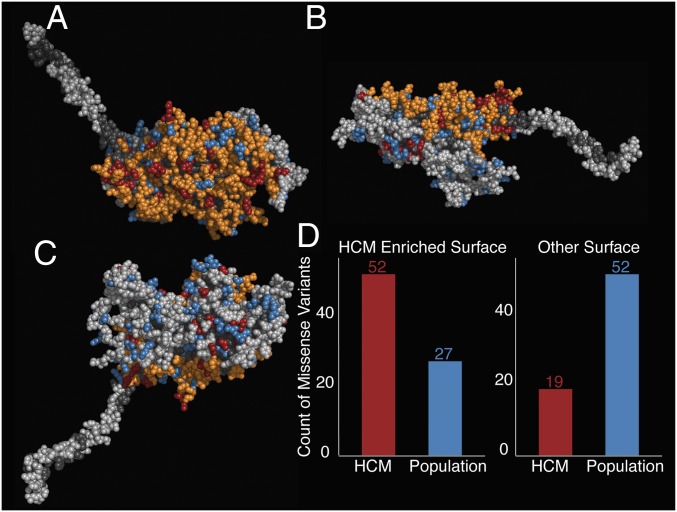
The surface of β-cardiac myosin contains 568 of the 765 residues in the S1 domain (74%). Of these residues, 71 are associated with HCM (72% of all HCM variants) and 79 are found in the ExAC population (71% of all reference variants), suggesting that variants in both cohorts are evenly distributed between the surface and core of the protein (χ2: P = 0.51). We then applied our spatial scan statistic to the surface of β-cardiac myosin. Using the myosin prestroke model, we identified a region of the surface covering 277 of the 568 surface amino acid residues (P = 0.002; Fig. 4 A and B), including the converter domain and the mesa, which is highly enriched for disease-associated variation. Strikingly, the region contains 52 of the 71 surface HCM-associated missense variants and only 27 of the 79 surface non–disease-associated missense variants (Fig. 4D), whereas the remainder of the myosin surface (Fig. 4C) covers 291 residues and contains only 19 disease-associated variants compared with 52 non–disease-associated variants (Fig. 4D). The identified converter/mesa region was also enriched in the replication dataset (P = 4.3 × 10−5).
Fig. 4.
Surface spatial scan analysis identifies a larger surface region enriched for HCM-associated missense variation. (A) Similar view of the prestroke model to the view in Fig. 2B, looking directly down onto the mesa. The residues are colored as follows: orange, surface region enriched for HCM variants; blue, missense variants seen only in the ExAC; red, missense variants seen in the SHaRe; light gray, residues considered to be on the surface; dark gray, residues not considered to be on the surface. The HCM-enriched surface region identified covers the entire mesa plus the adjoining converter domain. (B) Same side view of the prestroke S1 motor domain as shown in Figs. 1 and 2A. (C) View of the prestroke model viewing the side opposite the mesa. This surface is not enriched for HCM variants. (D) Number of HCM variants (SHaRe) and non–disease-associated (ExAC) variants identified in the surface enriched region (Left) and in the sum of all other parts of the myosin surface (Right).
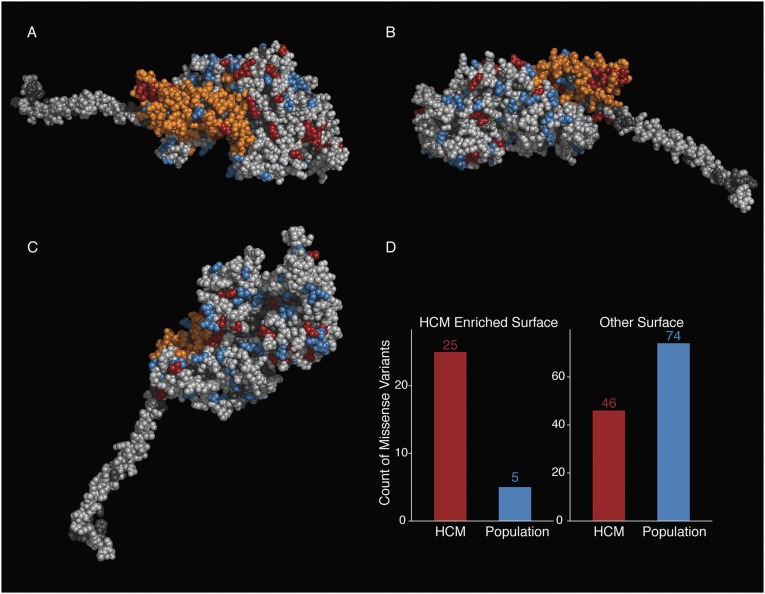
Using the same procedure to search the surface regions of the myosin poststroke structure, we detected a smaller enriched region of 122 amino acid residues again covering the converter but with a reduced portion of the myosin mesa (Fig. S3), a domain that has been proposed to be a binding site for another protein structural element (16). During the myosin power stroke, the converter moves away from the mesa (compare Figs. 2 A and B, 2B, and 4 and Fig. S3), so the enriched converter/mesa region is no longer contiguous and available for intra- or intermolecular interactions in the poststroke state (Fig. 2B and Fig. 4 A and B). The enriched amino acid residues in the poststroke model move significantly more in 3D space between the pre- and poststroke models (Wilcoxon test: P < 2 × 10−16) than other amino acid residues on the surface of the protein. This result suggests that myosin conformational changes during the actin-activated chemomechanical cycle may be important not only for transducing force but also for modulating the size and shape of this surface region and altering its availability for binding to other protein structural elements. The functional importance of the converter and the mesa region is further supported by the presence of the binding site for omecamtiv mecarbil, a recently described small-molecule modulator of cardiac myosin currently in clinical trials for the treatment of heart failure (24, 25).
Fig. S3.
Surface spatial scan statistic identifies a smaller surface region as enriched in the myosin poststroke model. (A) Spatial scan statistic identifies an enriched surface region in the poststroke myosin model that contains many of the residues in the enriched region in the prestroke myosin model. In this view, the orange residues define the surface of residues in the motor domain, which is the region significantly enriched for HCM variants. The S1 residues are colored as follows: orange, enriched region; blue, missense variants seen only in the ExAC; red, missense HCM variants seen in the SHaRe; light gray, all other surface residues; dark gray, nonsurface residues. (B) Side view of the enriched region; coloring is as in A. (C) Nonenriched side of the poststroke myosin; coloring is as in A. (D) Number of HCM variants (SHaRe) and non–disease-associated (ExAC) variants identified in the surface enriched region (Left) and in the remaining regions of the myosin surface (Right).
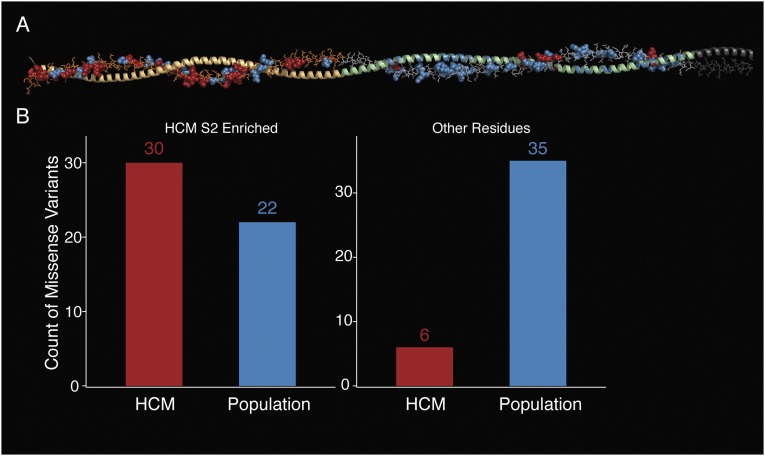
Next, we tested the myosin S2 fragment for regions enriched for disease-associated variation. The spatial scan analysis revealed that the first half of the S2 fragment is enriched for disease variants (P = 0.003; Fig. S4). This proximal part of S2 has been shown to bind to the aminoterminal part of myosin-binding protein C (MyBP-C) (26), a sarcomere protein that is also frequently mutated in patients with HCM (27). The enrichment of disease-associated variants in this region suggests that binding between myosin S2 and MyBP-C (and potentially other partners) is important for development of HCM.
Fig. S4.
Enrichment of HCM residues in the proximal part of myosin S2. (A) Molecular structure of the coiled-coil S2 fragment, modeled as described in Materials and Methods. Only one α-helix of the coiled-coil is shown in detailed structure; the second α-helix is symmetrical and shown only in cartoon mode. HCM variants are labeled in red, whereas ExAC variants are labeled in blue. Orange shading indicates the identified enriched region. (B) Number of HCM variants (SHaRe) and non–disease-associated (ExAC) variants identified in the enriched region of the S2 compared with the remainder of the S2 model.
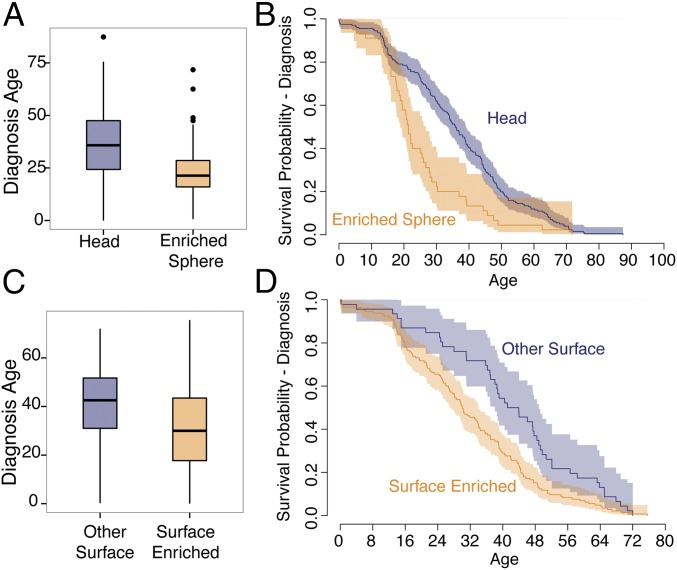
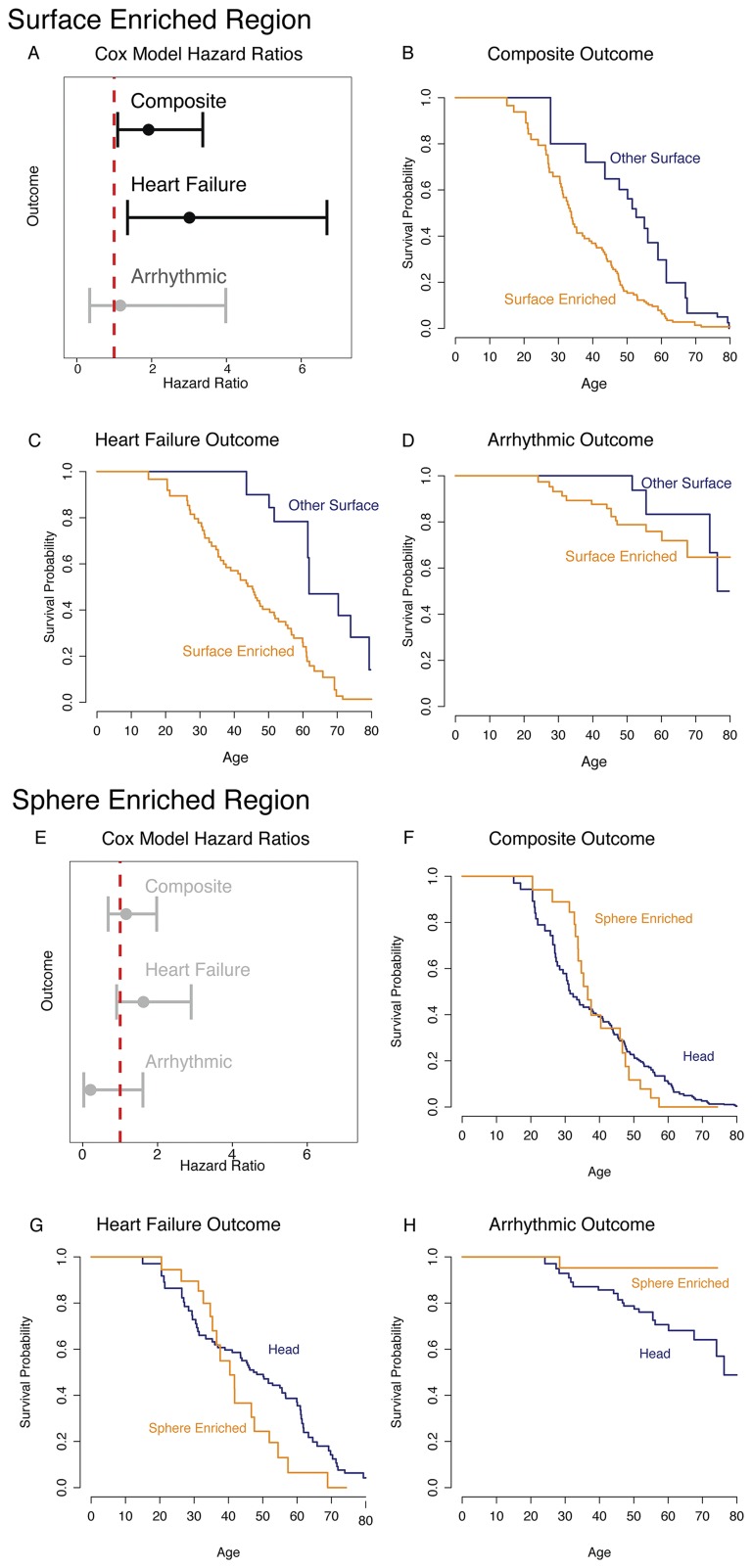
To investigate the contribution of the genetically constrained regions to disease further, we compared the clinical features of patients with variants in these regions with the clinical features of patients with variants elsewhere in MYH7. The clinical profile of HCM is highly variable, with some patients living a normal lifespan with minimal symptoms and others dying suddenly or requiring cardiac transplantation at a young age (28). Similarly, age at presentation of HCM varies widely among patients, and earlier onset is correlated with a more severe phenotype (29). We find that HCM patients with a variant inside the spherical enriched region are 11.2 y younger at diagnosis [24.9 (SE = 1.0) vs. 36.1 (SE = 1.2) y old; P = 5.6 × 10−5; Fig. 5 A and B] than patients harboring other variants in the myosin head. The presence of a variant in the HCM-enriched surface region is associated with a 10.0-y younger age at diagnosis [31.5 (SE = 2.4) vs. 41.5 (SE = 2.6) y old; P = 1.6 × 10−4] than in those patients with other surface variants (Fig. 5 C and D). In addition, we find an increased hazard for clinical outcomes in the surface enriched region after adjusting for differences in age at diagnosis (hazard ratio = 1.918, P = 0.023), although not in the spherical enriched region (Fig. S5 and Supporting Information). These findings demonstrate that analysis of genetic constraint in protein space can reveal domains with both increased disease burden and pathogenicity.
Fig. 5.
Comparison of clinical phenotypes between enriched regions and other regions in β-cardiac myosin. (A) Age of diagnosis of patients with HCM variants in the enriched spherical converter region (orange, n = 45) compared with patients with variants in other parts of the myosin head (blue, n = 201) (Wilcoxon test: P = 6.7 × 10−5). Box plots show the median surrounded by the interquartile range (IQR), with whiskers extending to 1.5-fold the interquartile range (IQR). (B) Kaplan–Meier curves of age at diagnosis compared between HCM variants in the enriched spherical converter region (orange) and patients with variants in other parts of the myosin head (blue). Shading indicates 95% confidence intervals for the survival curve. (C) Age of diagnosis of patients with HCM variants in the enriched surface region (orange, n = 145) compared with patients with variants in other parts of the myosin head surface (blue, n = 46) (Wilcoxon test: P = 1.6 × 10−4). Box plots show the median and IQR, with whiskers extending to 1.5-fold the IQR. (D) Kaplan–Meier curves of age at diagnosis compared between HCM variants in the enriched surface region (orange) and patients with variants in other parts of the myosin head surface (blue). Shading indicates 95% confidence intervals for the survival curve.
Fig. S5.
Patients with HCN who carry a missense variant in the surface enriched region of MYH7 have an increased hazard for clinical events. (A) Hazard ratios and 95% confidence intervals for the surface enriched region for each of the three outcome classes. (B) Kaplan–Meier survival curve for the composite outcome comparing the two regions. (C) Kaplan–Meier survival curve for the heart failure outcome comparing the two regions. (D) Kaplan–Meier survival curve for the arrhythmic outcome comparing the two regions. (E) Hazard ratios and 95% confidence intervals for the spherical enriched region for each of the three outcome classes. (F) Kaplan–Meier survival curve for the composite outcome comparing the two regions. (G) Kaplan–Meier survival curve for the heart failure outcome comparing the two regions. (H) Kaplan–Meier survival curve for the arrhythmic outcome comparing the two regions.
Our study demonstrates the power of integrating detailed structural information with large clinical and genetic databases to identify regions associated with functional importance and disease severity in Mendelian diseases. We find that variants associated with HCM are enriched in the β-cardiac myosin converter domain, where they lead to more severe outcomes. We provide the first evidence, to our knowledge, that similar clustering and pathogenicity are present in a surface spanning the converter domain and the relatively flat surface of the myosin catalytic domain called the myosin mesa (16). Because amino acid residues forming the mesa come from disparate locations in the nucleotide sequence, discovery of this region depends on integration of protein structural information. The pronounced shift of the converter/mesa surface during the power stroke raises the mechanistic hypothesis that these variants exert their deleterious effect selectively in the prestroke state, perhaps by disrupting dynamic binding interactions. In summary, these findings highlight the importance of considering data from human genetics in the context of the dynamic 3D protein structure, and illustrate an approach to structure/function analysis in genetic diseases.
Materials and Methods
SHaRe Database.
The SHaRe is a multicenter database that pools deidentified patient-level data from established institutional datasets at participating sites. At the time of analysis, the registry contained clinical and genetic testing data on 2,913 patients with HCM, 514 with variants in MYH7. This database contains individuals from nine inherited disease centers throughout the world and includes demographic data, medical history, echocardiogram data, genetic testing results, and many other data relating to cardiac health and clinical outcomes.
ExAC.
The ExAC (13) released data from 60,706 exomes from multiple sequenced cohorts that are not enriched for rare diseases such as HCM. We downloaded ExAC data for the canonical MYH7 transcript ENST00000355349 on August 27, 2015.
Variant Filtering and Inclusion Criteria.
Variants in MYH7 from the SHaRe database were filtered for quality purposes. Only exonic missense variants in MYH7 were included in the analysis. We included all exonic missense variants seen in HCM patients in clinical genetic testing. For comparison, we downloaded data from the MYH7 gene from the ExAC on August 27, 2015.
For the spatial scan and surface scan analyses, variants were considered to be one of two categories: disease-associated and reference. Any rare missense variants identified in HCM patients in the SHaRe cohort were considered to be disease-associated, regardless of their presence in the ExAC population. Rare missense variants identified only in the ExAC population were considered to be non–disease-associated, or reference, variants. Any variants identified in both the SHaRe and ExAC cohorts were considered to be disease-associated. Within the MYH7 S1 head, we identified 103 disease-associated variants (seen in the SHaRe) and 110 reference (or population) variants seen only in the ExAC cohort. Twenty-two variants observed in both the SHaRe and ExAC cohorts were classified as disease-associated.
Enrichment testing was performed by searching for regions of the myosin protein that had a significantly higher proportion of disease-associated variants than non–disease-associated variants. The ExAC population likely contains some individuals with cardiomyopathy induced by changes in MYH7; therefore, we cannot assume variants identified in the ExAC are not pathogenic. For example, there are multiple individuals carrying a well-known pathogenic variant for HCM in the MYBPC3 gene, p.Arg502Trp, in the ExAC cohort. However, the incidence of cardiomyopathies in the ExAC population is not expected to be greater than the incidence in the general population. A large proportion of the rare variants identified in MYH7 in the ExAC cohort are likely not associated with cardiomyopathy. In contrast, rare variants identified in SHaRe HCM patients are likely enriched for disease-causing variation.
We calculated the maximum population-specific allele frequency for each variant in the ExAC as the maximum frequency of the variant in any of the ExAC populations (excluding Finnish). To compare the burden of rare variants within each cohort, we removed from both sets (SHaRe and ExAC) any allele with a population-specific allele frequency higher than 1:2,000. This filter removed 16 variants in MYH7 from our ExAC list (of 421 total variants), six of which were also reported in the SHaRe list (of 192 total variants). Within the MYH7 head, only four ExAC variants were found at a higher frequency than 1:2,000 (of 136 total variants), two of which were also seen in the SHaRe cohort (of 105 total variants).
For validation purposes, we also performed a subset of analyses including variants classified as “pathogenic” or “likely pathogenic” according to the American College of Medical Genetics and Genomics guidelines (30), although excluding variants of unknown significance, likely benign, and benign variants (Supporting Information). In addition, we performed a validation analysis using variants found in individuals of European ancestry from the ExAC and individuals with a reported race of white to ensure that global population structure was not confounding our analysis (Supporting Information).
We generated an independent validation dataset combining previously published analyses of HCM variants from other medical centers with 42,930 exomes from the DiscovEHR sequencing project (14). We included only missense variants and removed variants with an allele frequency greater than 1 in 2,000 in the DiscovEHR exomes (Supporting Information).
Development of Human β-Cardiac Myosin Protein Models.
We developed human β-cardiac myosin S1 models based on the known motor domain structural data to represent the human form of the cardiac myosin. We retrieved the protein sequence of human β-cardiac myosin and the human cardiac light chains from the UniProt database (31): myosin heavy chain motor domain (MYH7), P12883; myosin essential light chain (MLC1), P08590; and myosin regulatory light chain (MLC2), P10916. We used a multitemplate homology modeling approach to build the structural coordinates of MYH7 (residues 1–840), MLC1 (residues 1–195), MLC2 (residues 1–166), and S2 (residues 841–1,280). We obtained the 3D structural model of S1 in the pre- and poststroke states by integrating the known structural data from solved crystal structures (details are provided in Supporting Information). Homology modeling of the prestroke structure was performed with template structures of the smooth muscle myosin motor domain (32) [Protein Data Bank (PDB) ID code 1BR1; with MgADP.AlF4 bound at the active site, which is thought to mimic most closely the ADP.Pi-bound state, or prestroke state, of the myosin] and the scallop smooth muscle myosin light chain domain (33) (PDB ID code 3TS5). The templates used for the modeling of the poststroke structure were obtained from the human β-cardiac myosin motor domain (25, 34) (PDB ID code 4P7H; no nucleotide in the active site) and the rigor structure from the squid myosin motor domain (35) (PDB ID code 3I5G; no nucleotide in the active site). Modeling was done using the MODELER package (36). Visualizations were performed using PyMOL version 1.7.4 (www.pymol.org).
Statistical Methods.
Comparisons between the ExAC and SHaRe variant locations in MYH7 were performed using the KS test statistic. All statistical analyses were performed in R version 3.1.2 (37), and many graphs were prepared using ggplot2 (38).
Spatial Scan Statistic.
For the spatial scan analysis, we compared the locations of unique HCM-associated variants with the locations of reference variants. The spatial scan statistic exhaustively searches 3D windows of a predefined set of sizes and shapes throughout the human β-cardiac myosin molecule for regions with an increased proportion of HCM-associated variants. It identifies the 3D region of the protein with the largest binomial likelihood ratio test of enrichment of HCM-associated variants (17, 18) (Supporting Information). The maximum binomial ratio test statistic for the entire set of windows in the model was calculated, and significance was assessed through permutation of variant labels using 1,000 permutations. For validation, we performed the analysis above, removing all missense variants classified as variants of unknown significance using only missense variants observed in the European individuals (Supporting Information).
Surface Analysis.
In addition to the spherical windows defined above, we define windows based upon the exposed surface of the human β-cardiac myosin molecule. For this analysis, we estimate the surface distance between any two amino acids based upon the solvent-excluded surface calculated by the MSMS program (23) (Supporting Information). Based upon this set of pairwise calculated distances, we define planar surface regions of the β-cardiac myosin for analysis as all of the amino acids within a certain distance of any given “center” amino acid. We exclude nonsurface amino acids, based upon their depth. We once again perform the spatial scan statistic as described above to identify surface regions of increased genetic burden.
S2 Fragment Analysis.
We performed the spherical spatial scan statistic as described above to test for enrichment in the S2 fragment of myosin, including the region between amino acids 838 and 1,112 (Supporting Information).
Clinical Phenotype Analysis.
For the analysis of age at diagnosis, only known probands were included (n = 260 myosin head, n = 201 myosin surface). Patients with multiple MYH7 variants were excluded from the age at diagnosis analysis, because the presence of multiple variants causes an earlier onset of HCM. We compared the primary diagnosis ages using a Wilcoxon test. The overall composite outcome combined the arrhythmic and heart failure outcomes, as well as the outcomes of atrial fibrillation, stroke, and all-cause death (Supporting Information). Individuals were considered to enter the study at their diagnosis age and were censored at their last known age. We compared hazard ratios for each region using the Cox proportional hazards model, adjusting for gender.
Spatial Scan Statistic
For the spatial scan analysis, we compared the locations of unique HCM-associated variants with the locations of reference variants in the myosin S1 subfragment. The spatial scan statistic exhaustively searches 3D windows of a predefined set of sizes and shapes throughout the human β-cardiac myosin molecule for regions that have an increased proportion of HCM-associated (SHaRe) variants. It identifies the 3D region of the protein with the largest binomial likelihood ratio test of enrichment of HCM-associated variants (17, 18).
Let pw be the proportion of variation within a window that is HCM-associated, and let qw be the proportion of variation outside the window, which is HCM-associated. For each window, we calculate the binomial likelihood ratio statistic comparing the null model, where pw and qw both equal the overall rate r against the model where pw is not equal to qw. The likelihood ratio statistic (Λw) for each window is as follows:
where yw is the number of HCM-associated variants within the window, zw is the number of other variants in the window, yg is the number of HCM-associated variants outside the window, zg is the number of other variants outside the window, and r is the overall proportion of HCM-associated variants. The test statistic is the maximum of the observed likelihood ratio statistics (maximum value of Λw) for all windows tested over the protein. Significance is assessed through permutation analysis by permuting the variant labels.
For the first analysis, we used spherical windows based upon the 3D locations of amino acids in the human β-cardiac myosin molecule. The myosin S1 models include residues 1–841. We excluded the loop regions between residues 205 and 211 and residues 627 and 640, because the positions of these amino acid residues are not well defined. We generated lists of unique missense variants associated with HCM and reference variants. Variants were assigned the 3D location in the human β-cardiac myosin model of the α-carbon atom of their corresponding residue. For acid residue in the myosin S1, we tested spherical windows with radii of 10, 12.5, 15, 17.5, 20, 22.5, and 25 Å centered on the α-carbon for enrichment of HCM-associated variants.
The maximum binomial ratio test statistic for the entire set of windows in the model was calculated, and significance was assessed through permutation of variant labels using 1,000 permutations.
Myosin S2 Spatial Scan Analysis
To assess enrichment in the S2 fragment of myosin, we performed a similar spatial analysis to the one performed on the S1 fragment. We used spherical window sizes of 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100 Å centered on the α-carbon of each amino acid residue in the S2 model. For this analysis, we compared the region between amino acid residues 838 and 1,112. The analysis was truncated at residue 1,112 due to low sequencing coverage following residue 1,112 in the ExAC data.
Protein Modeling
We used homology modeling to determine the prestroke and poststroke structures of the β-cardiac myosin.
Homology modeling of the prestroke structure was performed with template structures of the smooth muscle myosin motor domain (32) (PDB ID code 1BR1, with MgADP.AlF4 bound at the active site, which is thought to mimic most closely the ADP.Pi-bound state, or prestroke state, of the myosin) and the scallop smooth muscle myosin light chain domain (33) (PDB ID code 3TS5). The templates used for the modeling of the poststroke structure were obtained from the human β-cardiac myosin motor domain (34) (PDB ID code 4P7H, no nucleotide in the active site) and the rigor structure from the squid myosin motor domain (35) (PDB ID code 3I5G, no nucleotide in the active site). Missing regions in the myosin motor domain (loop1, loop2) were each separately built using the ModLoop program (39), and regions in the regulatory light chains that were not solved in the crystal structures were independently modeled using the I-TASSER prediction (40) method, and they were used as individual templates. Sequence alignment between MYH7, MLC1, and MLC2 with their respective structural templates was obtained. The models of pre- and poststroke structures were acquired using the MODELER package (36). We used a multitemplate modeling method: A total of 100 models were obtained, and the best model was selected based on the discrete optimized protein energy score. The structural models were energy-minimized using SYBYL7.2 (Tripos, Inc.) to remove potential short contacts. The final 3D models of the pre- and poststroke structures were validated using RAMPAGE (41), which provides a detailed check on the stereochemistry of the protein structure using the Ramachandran map.
The S2 region is a long coiled-coil structure; hence, we used the template from the Myosinome database (42). Modeling was done using the MODELER package, wherein selection of best model and validation of the model were done as described above. Visualizations were performed using PyMOL version 1.7.4 (www.pymol.org).
Estimation of Surface Distances
We developed the following procedure to estimate the surface distances between any two amino acids. First, we calculated the solvent-excluded surface for the human β-cardiac myosin models using the MSMS program with a 2.5-Å radius sphere as a probe, which approximates the size of amino acid side-chain interactions. For the calculated surface, the MSMS program returns a large net of vertices of the surface, each connected to multiple other points on the surface. We use these vertices and connections to build a weighted graph of the surface of the molecule, with the vertices as nodes and the edges as connections weighted by the Euclidean distance between the two connected vertices. Then, for each amino acid, we assign it to the point on the surface that is the closest point, on average, to all of the nonhydrogen atoms of the amino acid. This average distance is used as an estimate of the depth of the amino acid. Amino acids with average depths of greater than 4 Å for the surface model were considered not to be on the surface. We used the A-star algorithm to calculate the distance on the surface graph between two amino acid residues. In this analysis, we included only amino acids in the “head” of the human β-cardiac myosin molecule, defined as amino acids 1–784. The lever arm was excluded. We excluded the disordered loop regions between residues 205 and 211 and residues 627 and 640.
Replication Dataset for Genetic Analyses
To replicate the associations above, we searched the literature for HCM studies that reported the observed MYH7 variants and that occurred at medical centers not part of the SHaRe consortium. We combined the reported MYH7 missense variants previously reported in the literature into a single set of unique missense variants (43–52). We compared this set against exome sequencing data of MYH7 from the DiscovEHR sequencing project involving the Regeneron Genetics Center and Geisinger Health System using the spatial scan statistic. MYH7 missense variants were extracted from exome sequencing data generated on 42,930 individuals as described. We used the observed set of MYH7 missense variants identified through this exome sequencing set as a general population cohort. Due to the fact that some literature studies only reported amino acid differences, we compared unique amino acid changes from each dataset for the validation analysis.
We tested the enriched regions identified from the SHaRe vs. ExAC analysis for HCM-associated variant enrichment in the validation data. We performed a likelihood ratio test comparing the enrichment of literature HCM-associated variants with the variants found in the DiscovEHR exome population cohort. For each of the enriched regions, we calculated the likelihood ratio test statistic for enrichment for HCM-associated variation. Two times the logarithm of the likelihood ratio statistic follows a χ2 distribution with 1 df, because the alternative model has two parameters, whereas the null model has a single parameter. For the prestroke model, both the spherical enriched region (P = 0.0079) and the surface-enriched region (P = 4.3 × 10−5) validate. For the poststroke model, both regions are enriched in the validation set as well (P = 0.00082 for the surface region and P = 0.00041 for the spherical region).
Age at Event Survival Analysis
We performed an age at event survival analysis comparing individuals with variants in the identified regions vs. individuals with other variants in the head of the human β-cardiac myosin molecule. We focused on multiple types of composite clinical outcomes in SHaRe subjects, including combined measures of heart-failure outcomes and arrhythmic events. For the age at event analyses, the composite outcomes were defined as follows. The arrhythmic outcome consisted of cardiac arrest, implantable cardioverter-defibrillator firing, and sudden cardiac death. The heart failure outcome combined the events of end-stage HCM (defined as the left ventricular ejection fraction falling below 55%), New York Heart Association class III or IV status, transplant operation, and left ventricular assistive device implantation. The overall composite outcome combined the arrhythmic and heart failure outcomes, as well as the outcomes of atrial fibrillation, stroke, and death (all causes).
We used the Kaplan–Meier method to estimate survival curves for two groups of patients: patients carrying variants in the prestroke spherical variant enriched region in the converter domain of β-cardiac myosin and those patients carrying variants located elsewhere in the β-cardiac myosin head. Patients are censored at their last known age and enter the study at their diagnosis age. We also tested for differences in the survival between patients with variants in the enriched region identified on the surface of β-cardiac myosin and patients with variants in other surface amino acids. Using a Cox proportional hazards model, we find no significant differences between individuals with variants in the spherical hit and individuals with variants in other regions of the β-cardiac myosin head. We do find significant differences in the hazard of the heart failure and composite outcomes between individuals with variants in the surface enriched region and individuals with variants in other regions of the myosin surface.
Assessing the Effects of Variants of Unknown Significance and Ancestry on the Spatial Scan Results
To assess whether the observed enrichments of HCM-associated variants are affected by the inclusion of missense variants with less compelling evidence for pathogenicity, we excluded any sites with only variants of unknown significance (VUS) according to the American College of Medical Genetics and Genomics guidelines and again performed the same spatial scan analysis. Within this framework, variants are classified as one of five categories: pathogenic, likely pathogenic, of unknown significance, likely benign, and benign. Within MYH7 in the SHaRe, there were 48 pathogenic, 82 likely pathogenic, 66 of unknown significance, 2 likely benign, and 0 benign variants. Within the MYH7 S1 head domain, there were 33 pathogenic, 48 likely pathogenic, 27 of unknown significance, 1 likely benign variant, and 0 benign variants. This assessment is to ensure that VUS are not driving the results of the analyses. Variant classifications were provided by each site and were made by the site’s clinical team or the clinical genetic testing laboratory that performed the test.
For the prestroke model, we once again identified the same 15-Å region centered at amino acid residue 736 (P = 0.003) as in the analysis that included all missense variants. For the surface analysis, we also find the same surface region is identified when excluding all VUS variants (P < 0.001). For the rigor surface analysis, we also identify the same surface region as in the full analysis (P < 0.001).
Some burden analyses can be strongly confounded by ancestry differences between case and reference groups. To test if our results were confounded by population structure, we performed the spatial scan analysis only including variants seen in individuals with European ancestry in both the SHaRe and ExAC. This analysis identified a similar enriched region centered at amino acid 749 with a 17.5-Å radius (P = 0.003), which included 75% of the residues identified in the previous analyses. For the surface analysis, the same surface region is the most significant hit when including only European variants (P = 0.007). For the rigor surface analysis, we also identify the same surface region as the analysis including all of the variants (P = 0.002). Both the SHaRe and ExAC contain a large majority of individuals with European ancestry. As such, we currently do not have the power to perform this analysis in a subset of individuals from an additional population. As genetic datasets expand further in size, we hope to perform this analysis in other populations to identify additional regions of enrichment or to refine the enriched regions identified here.
Together, these analyses demonstrate that our analyses are not significantly affected by the inclusion of VUS or by ancestry differences between patients with HCM and population references.
Assessing the Effects of Inclusion of Variants of Unknown Significance on the Clinical Phenotype Data
To assess the effects of the exclusion of individuals with VUS on the clinical phenotype analysis, we performed the same clinical comparisons using only individuals with a pathogenic or likely pathogenic variant. Any variants with a frequency above 1:2,000 in the ExAC were excluded from the analysis. For the spherical enriched region, we find similar results as before, with a 9.79-y difference in diagnosis age (Wilcoxon test: P = 2.4 × 10^−4). For the surface enriched region, there is a 7.96 decrease in age at diagnosis (P = 0.0036) when comparing individuals with VUS in the surface enriched region vs. VUS in other surface regions of the myosin molecule.
Supplementary Material
Acknowledgments
We thank the investigators and participants in the SHaRe for their support of this project. We also thank Jonathan Fox for guidance during the early stages of the SHaRe. J.R.H. is supported by a Stanford Graduate Fellowship. R.E.T. is supported by NIH Grant F32 HL123247. This work supported by NIH Grants U01HG007436 (to C.D.B.), 1U01HG007708 (to E.A.A.), and U01HG006382 (to R.P.R.M.), and a grant from SAP (to C.D.B.),
Footnotes
Conflict of interest statement: J.A.S. is a founder of and owns shares in Cytokinetics, Inc. and MyoKardia, Inc., biotechnology companies that are developing therapeutics that target the sarcomere. E.M.G. is an employee and owns shares in MyoKardia, Inc. E.A.A. is a founder of Personalis, Inc. C.D.B. is on the Scientific Advisory Boards of Ancestry.com, Personalis, Liberty Biosecurity, and Etalon DX, and is also a founder and chair of the Scientific Advisory Board of IdentifyGenomics.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606950113/-/DCSupplemental.
References
- 1.Krendel M, Mooseker MS. Myosins: Tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ, et al. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92(4):785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ, Casey SA, Hauser RG, Aeppli DM. Clinical course of hypertrophic cardiomyopathy with survival to advanced age. J Am Coll Cardiol. 2003;42(5):882–888. doi: 10.1016/s0735-1097(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 4.Walsh R, Rutland C, Thomas R, Loughna S. Cardiomyopathy: A systematic review of disease-causing mutations in myosin heavy chain 7 and their phenotypic manifestations. Cardiology. 2010;115(1):49–60. doi: 10.1159/000252808. [DOI] [PubMed] [Google Scholar]
- 5.Jellis CL, Desai MY. Hypertrophic cardiomyopathy: Still connecting the dots between genotype and phenotype. Cardiovasc Diagn Ther. 2015;5(2):156–159. doi: 10.3978/j.issn.2223-3652.2015.03.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan S, et al. Cardiac structural and sarcomere genes associated with cardiomyopathy exhibit marked intolerance of genetic variation. Circ Cardiovasc Genet. 2012;5(6):602–610. doi: 10.1161/CIRCGENETICS.112.963421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore JR, Leinwand L, Warshaw DM. Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res. 2012;111(3):375–385. doi: 10.1161/CIRCRESAHA.110.223842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buvoli M, Hamady M, Leinwand LA, Knight R. Bioinformatics assessment of beta-myosin mutations reveals myosin’s high sensitivity to mutations. Trends Cardiovasc Med. 2008;18(4):141–149. doi: 10.1016/j.tcm.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayment I, Holden HM, Sellers JR, Fananapazir L, Epstein ND. Structural interpretation of the mutations in the beta-cardiac myosin that have been implicated in familial hypertrophic cardiomyopathy. Proc Natl Acad Sci USA. 1995;92(9):3864–3868. doi: 10.1073/pnas.92.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapplinger JD, et al. Distinguishing hypertrophic cardiomyopathy-associated mutations from background genetic noise. J Cardiovasc Transl Res. 2014;7(3):347–361. doi: 10.1007/s12265-014-9542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minikel EV, et al. Quantifying prion disease penetrance using large population control cohorts. Sci Transl Med. 2016;8(322):322ra9. doi: 10.1126/scitranslmed.aad5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Golbus JR, et al. Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet. 2012;5(4):391–399. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxivorg. October 30, 2015 doi: 10.1101/030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewey FE, et al. Inactivating Variants in ANGPTL4 and Risk of Coronary Artery Disease. N Engl J Med. 2016;374(12):1123–1133. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho CY, et al. Examining Prevailing Genotype-Phenotype Correlations in Hypertrophic Cardiomyopathy Late-Breaking Clinical Trial Abstracts. Circulation. 2015;132(23):2267–2285. doi: 10.1161/CIR.0000000000000334. [DOI] [PubMed] [Google Scholar]
- 16.Spudich JA. The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem Soc Trans. 2015;43(1):64–72. doi: 10.1042/BST20140324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulldorff M, Martin K. A spatial scan statistic. Commun Stat Theory Methods. 1997;26(6):1481–1496. [Google Scholar]
- 18.Ionita-Laza I, Makarov V, Buxbaum JD. ARRA Autism Sequencing Consortium Scan-statistic approach identifies clusters of rare disease variants in LRP2, a gene linked and associated with autism spectrum disorders, in three datasets. Am J Hum Genet. 2012;90(6):1002–1013. doi: 10.1016/j.ajhg.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seebohm B, et al. Cardiomyopathy mutations reveal variable region of myosin converter as major element of cross-bridge compliance. Biophys J. 2009;97(3):806–824. doi: 10.1016/j.bpj.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Köhler J, et al. Mutation of the myosin converter domain alters cross-bridge elasticity. Proc Natl Acad Sci USA. 2002;99(6):3557–3562. doi: 10.1073/pnas.062415899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.García-Giustiniani D, et al. Phenotype and prognostic correlations of the converter region mutations affecting the β myosin heavy chain. Heart. 2015;101(13):1047–1053. doi: 10.1136/heartjnl-2014-307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo A, et al. Mutations of the beta myosin heavy chain gene in hypertrophic cardiomyopathy: Critical functional sites determine prognosis. Heart. 2003;89(10):1179–1185. doi: 10.1136/heart.89.10.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanner MF, Olson AJ, Spehner JC. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers. 1996;38(3):305–320. doi: 10.1002/(SICI)1097-0282(199603)38:3%3C305::AID-BIP4%3E3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Malik FI, et al. Cardiac myosin activation: A potential therapeutic approach for systolic heart failure. Science. 2011;331(6023):1439–1443. doi: 10.1126/science.1200113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winkelmann DA, Forgacs E, Miller MT, Stock AM. Structural basis for drug-induced allosteric changes to human β-cardiac myosin motor activity. Nat Commun. 2015;6:7974. doi: 10.1038/ncomms8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Starr R, Offer G. The interaction of C-protein with heavy meromyosin and subfragment-2. Biochem J. 1978;171(3):813–816. doi: 10.1042/bj1710813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alfares AA, et al. Results of clinical genetic testing of 2, 912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet Med. 2015;17(11):880–888. doi: 10.1038/gim.2014.205. [DOI] [PubMed] [Google Scholar]
- 28.Maron BJ, et al. Clinical course of hypertrophic cardiomyopathy in a regional United States cohort. JAMA. 1999;281(7):650–655. doi: 10.1001/jama.281.7.650. [DOI] [PubMed] [Google Scholar]
- 29.Maron BJ, et al. Hypertrophic Cardiomyopathy in Children, Adolescents, and Young Adults Associated With Low Cardiovascular Mortality With Contemporary Management Strategies. Circulation. 2016;133(1):62–73. doi: 10.1161/CIRCULATIONAHA.115.017633. [DOI] [PubMed] [Google Scholar]
- 30.Richards S, et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.UniProt Consortium Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41(Database issue):D43–D47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 1998;94(5):559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 33.Kumar VSS, et al. Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle myosin. Biophys J. 2011;101(9):2185–2189. doi: 10.1016/j.bpj.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkelmann DA, Miller MT, Stock AM, Liu L. Structure of Human beta-Cardiac Myosin Motor Domain at 3.2 A. Mol Biol Cell. 2011;22(24):4705. [Google Scholar]
- 35.Yang Y, et al. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15(5):553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 37.R Core Team 2014 The R Project for Statistical Computing. Available at www.R-project.org/. Accessed October 31, 2014.
- 38.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer Science & Business Media; New York: 2009. [Google Scholar]
- 39.Fiser A, Sali A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics. 2003;19(18):2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, et al. The I-TASSER Suite: Protein structure and function prediction. Nat Methods. 2015;12(1):7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovell SC, et al. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins. 2003;50(3):437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 42.Syamaladevi DP, et al. Myosinome: A database of myosins from select eukaryotic genomes to facilitate analysis of sequence-structure-function relationships. Bioinform Biol Insights. 2012;6:247–254. doi: 10.4137/BBI.S9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bos JM, et al. Characterization of a phenotype-based genetic test prediction score for unrelated patients with hypertrophic cardiomyopathy. Mayo Clin Proc. 2014;89(6):727–737. doi: 10.1016/j.mayocp.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ingles J, et al. Compound and double mutations in patients with hypertrophic cardiomyopathy: Implications for genetic testing and counselling. J Med Genet. 2005;42(10):e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kubo T, et al. Prevalence, clinical significance, and genetic basis of hypertrophic cardiomyopathy with restrictive phenotype. J Am Coll Cardiol. 2007;49(25):2419–2426. doi: 10.1016/j.jacc.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 46.Millat G, Chanavat V, Créhalet H, Rousson R. Development of a high resolution melting method for the detection of genetic variations in hypertrophic cardiomyopathy. Clin Chim Acta. 2010;411(23-24):1983–1991. doi: 10.1016/j.cca.2010.08.017. [DOI] [PubMed] [Google Scholar]
- 47.Otsuka H, et al. Prevalence and distribution of sarcomeric gene mutations in Japanese patients with familial hypertrophic cardiomyopathy. Circ J. 2012;76(2):453–461. doi: 10.1253/circj.cj-11-0876. [DOI] [PubMed] [Google Scholar]
- 48.Roncarati R, et al. Unexpectedly low mutation rates in beta-myosin heavy chain and cardiac myosin binding protein genes in Italian patients with hypertrophic cardiomyopathy. J Cell Physiol. 2011;226(11):2894–2900. doi: 10.1002/jcp.22636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richard P, et al. EUROGENE Heart Failure Project Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107(17):2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 50.Song L, et al. Mutations profile in Chinese patients with hypertrophic cardiomyopathy. Clin Chim Acta. 2005;351(1-2):209–216. doi: 10.1016/j.cccn.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, et al. Malignant effects of multiple rare variants in sarcomere genes on the prognosis of patients with hypertrophic cardiomyopathy. Eur J Heart Fail. 2014;16(9):950–957. doi: 10.1002/ejhf.144. [DOI] [PubMed] [Google Scholar]
- 52.Yu B, et al. Denaturing high performance liquid chromatography: High throughput mutation screening in familial hypertrophic cardiomyopathy and SNP genotyping in motor neurone disease. J Clin Pathol. 2005;58(5):479–485. doi: 10.1136/jcp.2004.021642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.