Significance
Nitrogen availability influences morphogenesis, antibiotic production, and virulence/pathogenicity in actinomycetes. This work provides novel insights into the molecular mechanisms underlying nitrogen metabolism regulation. Glutamine synthetases (GlnA) were acetylated by the acetyltransferase AcuA, which was regulated by the nitrogen regulator GlnR. Acetylation inactivated GlnA4 (GSII). Acetylated GSI-β (GlnA1) exhibited a remarkable ability to directly interact with GlnR and enhance GlnR–DNA binding, thereby modulating target gene transcription in response to nitrogen availability. The results indicated that GSI-β plays a central and essential role in nitrogen metabolism through not only controlling glutamine synthesis but also serving as the nitrogen signal for GlnR activation. These findings substantively extend our understanding of the regulatory mechanisms underlying nitrogen metabolism in actinobacteria, which include antibiotic-producing actinomycetes and pathogenic mycobacteria.
Keywords: actinomycetes, protein acetylation, glutamine synthetase, nitrogen metabolism, chaperone
Abstract
In cells of all domains of life, reversible lysine acetylation modulates the function of proteins involved in central cellular processes such as metabolism. In this study, we demonstrate that the nitrogen regulator GlnR of the actinomycete Saccharopolyspora erythraea directly regulates transcription of the acuA gene (SACE_5148), which encodes a Gcn5-type lysine acetyltransferase. We found that AcuA acetylates two glutamine synthetases (GlnA1 and GlnA4) and that this lysine acetylation inactivated GlnA4 (GSII) but had no significant effect on GlnA1 (GSI-β) activity under the conditions tested. Instead, acetylation of GlnA1 led to a gain-of-function that modulated its interaction with the GlnR regulator and enhanced GlnR–DNA binding. It was observed that this regulatory function of acetylated GSI-β enzymes is highly conserved across actinomycetes. In turn, GlnR controls the catalytic and regulatory activities (intracellular acetylation levels) of glutamine synthetases at the transcriptional and posttranslational levels, indicating an autofeedback loop that regulates nitrogen metabolism in response to environmental change. Thus, this GlnR-mediated acetylation pathway provides a signaling cascade that acts from nutrient sensing to acetylation of proteins to feedback regulation. This work presents significant new insights at the molecular level into the mechanisms underlying the regulation of protein acetylation and nitrogen metabolism in actinomycetes.
Protein lysine acetylation was first discovered in eukaryotic histones in the 1960s as a posttranslation modification (PTM) (1, 2), and the chemotaxis protein CheY was the first acetylated protein identified in bacteria (3). Furthermore, in Salmonella enterica, the activity of the metabolic enzyme acetyl-CoA (AcCoA) synthetase was shown to be regulated by a reversible lysine acetylation (RLA) system consisting of acetyltransferase (4) and deacetylase (5). Recently, it was found that protein lysine acetylation can occur via either enzymatic or nonenzymatic acetylation, such as chemical acetylation, wherein the intracellular acetyl-phosphate (AcP) plays a critical role (6, 7). In particular, RLA is being increasingly recognized as an important metabolic regulatory mechanism in bacteria. In the last decade, advancements in mass spectrometry (MS) and high-affinity purification of acetylated peptides have accelerated the identification of lysine acetylation sites, and the acetylproteome of several microorganisms has been reported (6–16). The picture emerging from these studies indicates that lysine acetylation is a conserved regulatory mechanism of metabolism. However, at present, our understanding of how the regulation of the acetylation system is integrated with the regulation of genes encoding proteins whose function is controlled by acetylation is limited. More recently, insights into the complexities of such regulatory integration have been reported for Escherichia coli and S. enterica (13, 17). Results from studies by Yu et al. and Schilling et al. suggest that the levels of acetylation are regulated in response to environmental changes (8, 13). Nevertheless, the means by which signals dictate the level of protein acetylation and lysine acylation remain poorly understood.
In Bacillus subtilis, the regulation of expression of the acuA gene, which encodes an acetyltransferase, is under the control of CcpA, a global regulatory protein that is affected by the quality of the carbon source available to the cell (18). In E. coli, two cAMP-mediated acetylation signaling pathways have been identified (13, 19). The carbon regulator cAMP receptor protein (CRP)–cAMP complex induces the expression of yfiQ, which encodes the Gcn5-like acetyltransferase (GNAT) YfiQ (PatZ, Pka) and thus increases the acetylation level of proteins in response to the intracellular cAMP signal (19). Furthermore, cAMP–CRP also regulates glucose-induced AcP-dependent protein acetylation in response to carbon overflow (13). Allosteric effects also control the activity of protein acetyltransferases. In Mycobacterium tuberculosis and Mycobacterium smegmatis, cAMP directly activates the protein acetyltransferases MtKat (Rv0998) and MsKat (MSMEG_5458) by binding to a cyclic nucleotide-binding domain that is fused to the N terminus of the catalytic GNAT domain (20). Recently, we found that an amino acid-binding domain (ACT domain) fused to the GNAT acetyltransferase of Micromonospora aurantica is used to exert amino acid-induced allosteric regulation of the enzyme (21). Thus, it is likely that the protein acetyltransferase enzymes are carefully regulated at the transcriptional and posttranslational levels in response to changes of the intracellular signals that control the acetylation of specific proteins, which in turn mold the metabolic network. Understanding this signaling pathway is critical for scrutinizing how the cellular environment influences protein acetylation.
In this study, we have characterized the acetylation of the glutamine synthetases (GSs) GlnA1 and GlnA4 and the protein acetyltransferase AcuA in Saccharopolyspora erythraea, and found that the global nitrogen-regulator GlnR directly controls these enzymes at the transcriptional level. Lysine acetylation inactivated GlnA4 and revealed no significant effect on activity of GlnA1. Instead, acetylation of GlnA1 modulated its interaction with the GlnR regulator and enhanced GlnR–DNA binding as a chaperone. Nitrogen-mediated signal transduction of protein acetylation provides a possible regulatory mechanism of feedback loop (GlnR–AcuA–GS) at multiple levels controlling nitrogen metabolism. Furthermore, it was also demonstrated that the acetylation levels of GSs are modulated by extracellular nutrient availability. Our results present an example of a previously unidentified cyclic signal-transduction mechanism for regulating protein acetylation through a nutrient-sensing pleiotropic regulator in response to environmental changes.
Results
The Nitrogen-Sensing Regulator GlnR Directly Activates Transcription of acuA.
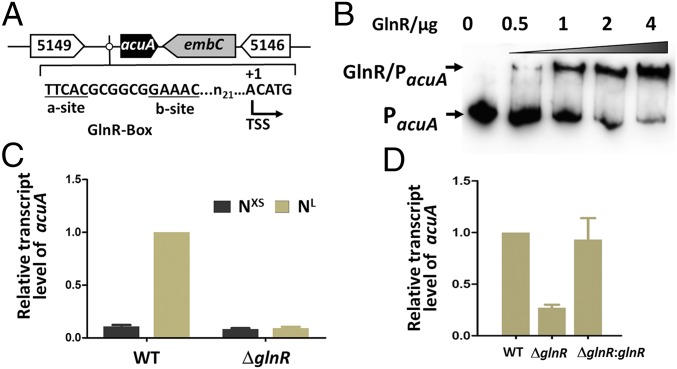
We previously showed that the acetyltransferase AcuA of S. erythraea (SACE_5148) acetylated the AMP-forming acetyl CoA synthetase (AcsA) of this bacterium (22). Herein, we found that a putative GlnR binding site was identified within the acuA promoter (PacuA) (Fig. 1A). Electrophoretic mobility shift assay (EMSA) analysis using purified GlnR suggested that GlnR can bind to the acuA promoter region. Increasing amounts of GlnR protein titrated against a fixed amount of PacuA probe yielded increased amounts of the GlnR–PacuA complex (Fig. 1B). Furthermore, mutations of the GlnR-binding box attenuated or abrogated interaction of the promoter region of acuA gene with GlnR (SI Appendix, Fig. S1). The effect of GlnR on acuA expression was next investigated in vivo. We quantified the amount of acuA transcript in wild-type (WT) and ΔglnR strains in cells grown in Evans medium supplemented with excess (XS) or limited (L) nitrogen (NXS/NL). As shown in Fig. 1C, results of quantitative RT-PCR experiments showed that the level of acuA transcript was increased ∼10-fold in cells grown under nitrogen-limited conditions, whereas no obvious changes in acuA expression were observed in the ΔglnR strain. These results further implicated GlnR in the regulation of acuA in response to nitrogen availability in S. erythraea. The deletion of glnR resulted in a fivefold decrease in acuA expression, lending further support to the idea that GlnR regulates acuA expression in the bacterium. Conversely, the ΔglnR strain harboring the plasmid p7101 encoding the WT glnR+ allele restored acuA transcription to WT levels (Fig. 1D).
Fig. 1.
GlnR directly regulates transcription of acuA, which encodes a protein acetyltransferase in S. erythraea. (A) Genetic organization of the acuA gene of S. erythraea and its putative GlnR-binding site. The predicted transcription start site (TSS) is shown. (B) EMSAs of purified His–GlnR binding toward the predicted motifs. The DNA probes (PacuA) containing the predicted motifs were incubated with GlnR and a 200-fold excess of nonspecific competitor DNA (sperm DNA). (C) Transcriptional analysis of acuA gene response to nitrogen availability (NXS, excess nitrogen; NL, low nitrogen) in S. erythraea WT and glnR-deleted mutant (ΔglnR) strains. (D) Analysis of acuA transcription in S. erythraea WT, ΔglnR mutant, and glnR complementary (ΔglnR::glnR) strains. Error bars show the SD from three independent experiments.
AcuA Acetyltransferase Acetylates Two GSs (GlnA1 and GlnA4).
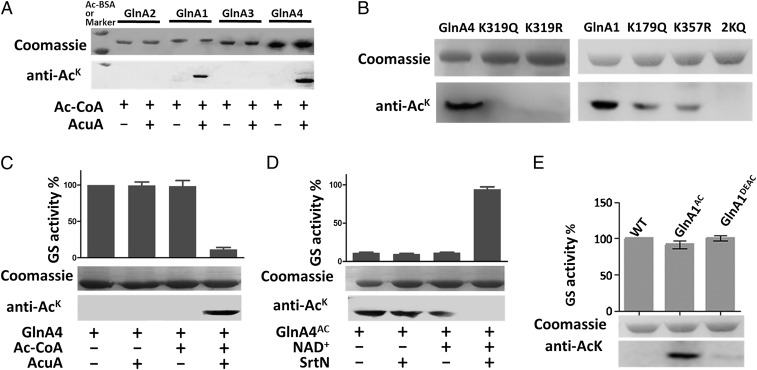
To investigate the effect of AcuA-catalyzed acetylation on nitrogen metabolism, we examined whether the AcuA acetyltransferase acetylated GlnR or other key enzymes involved in nitrogen metabolism. Under the conditions tested, AcuA did not acetylate GlnR, suggesting that AcuA does not exert any direct effect on GlnR. GSs (EC 6.3.1.2) are ubiquitous enzymes that catalyze the ATP-dependent synthesis of glutamine from glutamate and ammonium and play essential roles in nitrogen metabolism. The S. erythraea genome contains four genes encoding putative GSs, including one GSI-β enzyme (glnA1, locus SACE_1623) and three GSII enzymes (glnA2, locus SACE_1613; glnA3, locus SACE_3095; and glnA4, locus SACE_5355) (SI Appendix, Fig. S2; ref. 23), and GlnR has been shown to regulate their expression (24). We found that two GS enzymes, GSI-β GlnA1 and GSII GlnA4, were directly acetylated in vitro after incubation with AcuA and AcCoA; AcuA did not acetylate GlnA2 or GlnA3 (Fig. 2A). Two acetylated peptides MK(179)GGYFPVSPYDHFADLR and IPITGSNPK(357)AK of GlnA1 and one acetylated peptide FAK(319)GSFAPTAVAWGR of GlnA4 were identified. To confirm the sites of acetylation and investigate the effects of the residues on the acetylation levels of GS enzymes, we introduced substitutions at the acetylated sites to generate variants GlnA1K179Q, GlnA1K357Q, GlnA1K179,357Q, GlnA4K319Q, and GlnA4K319R. These substitutions were chosen because, in some cases, glutamine (Q) can serve as a structural mimic for acetyl-lysine and arginine (R) serves as the genetic mimic of unacetylated lysine (25). These GS mutants and native GS enzyme were incubated with AcuA and AcCoA. Western blotting was performed to detect the acetylation level of these GS variants by using an anti-AcK antibody. As shown in Fig. 2B, no acetylation of GlnA1K179,357Q (2KQ), GlnA4K319R, or GlnA4K319Q was detected; however, GlnA1K179Q and GlnA1K357Q mutants exhibited similar acetylation levels that were slightly lower than that of native GlnA1, indicating that two lysine residues of GlnA1, but only one lysine residue of GlnA4, were reversibly acetylated, in agreement with our MS results.
Fig. 2.
Acetyltransferase AcuA acetylates two GSs (GlnA1 and GlnA4) in S. erythraea. (A) The four purified GS enzymes (GlnA2, GlnA1, GlnA3, and GlnA4) were incubated in vitro with or without AcuA and AcCoA at 37 °C for 2 h. After incubation, samples were collected and analyzed by SDS polyacrylamide gel electrophoresis, and the acetylation levels were determined by Western blotting using a specific anti-AcK antibody. (B) GlnA1, GlnA4, and their variants (GlnA1K179Q, GlnA1K357Q, GlnA1K179,357Q, GlnA4K319Q, and GlnA4K319R) were acetylated by AcuA. (C and D) In vitro acetylation/deacetylation affected the activity of GS. GlnA4 was incubated with AcuA and AcCoA at 37 °C for 2 h. Acetylated GlnA4 was incubated with SrtN and NAD+ at 37 °C for 2 h. Error bars show the SD from three independent experiments. (E) GS activity of native GlnA1 (WT), GlnA1AC, and deacetylated GlnA1 (GlnA1DEAC).
Lysine Acetylation Inactivates the GS GlnA4 (GSII) but Not GlnA1 (GSI-β).
To investigate the effect of acetylation on GS enzyme activity, GlnA1 and GlnA4 were incubated with AcuA in the presence or absence of AcCoA for 2 h. The activities of native (nonacetylated), acetylated, and deacetylated GS enzymes were determined. In the presence of both AcCoA and AcuA, GlnA4 was acetylated, and its activity was reduced by ∼88%, indicating that lysine acetylation effectively decreased GlnA4 activity (Fig. 2C). Furthermore, acetylated GlnA4 was deacetylated by the sirtuin-type deacetylase SrtN (encoded by SACE_3798). After deacetylation, GlnA4 activity was restored to a level comparable with that of the nonacetylated GlnA4 control (Fig. 2D). However, acetylation had no significant effect on the catalytic activity of GlnA1 (Fig. 2E). To further examine the effects of acetylation lysine residues on GS activity, the activities of GlnA1, GlnA4, and their variants were also determined. As shown in SI Appendix, Fig. S3, the GlnA4K319Q and GlnA4K319R variants retained only 10% of the WT GlnA4 activity, indicating that residue K319 was critical for catalytic activity because substitution of K319 effectively abolished the activity of GlnA4. Conversely, the K179Q and K357Q GlnA1 mutants showed activity levels that were similar to that of the native protein.
These results suggest that Lys acetylation is not critical for the catalytic activity of GlnA1. However, the far-UV circular dichroism spectra (190–260 nm) (SI Appendix, Fig. S4A) showed decreases in ellipticity at 208 and 222 nm, indicating an increase in the α-helical content of acetylated GlnA1 (GlnA1AC) and suggesting that K179 and K357 acetylation altered the secondary structure of the protein. GlnA1AC showed significantly higher α-helicity and a concomitant reduction in antiparallel and random coil structures compared with native GlnA1 (SI Appendix, Table S1). The effects of acetylation on the tertiary structure, hydrodynamic radius, surface hydrophobicity, and structural stability of GlnA1 were also investigated. Hydrophobicity (as determined by the fluorescence intensity of 6P-toluidinyl-naphthalene-2-sulfonate) increased slightly (SI Appendix, Fig. S4B) as a result of acetylation, whereas the hydrodynamic radius decreased from 8.1 to 7.6 nm (SI Appendix, Fig. S4C), which also altered the tertiary structure of the protein, as evidenced by a 17% increase in Trp fluorescence (SI Appendix, Fig. S4D). Thus, lysine acetylation resulted in a structurally more compact and hydrophobic GlnA1 protein.
Acetylation of GS Enzymes Is Influenced by Extracellular Nitrogen Availability or Lack of Enzymes.
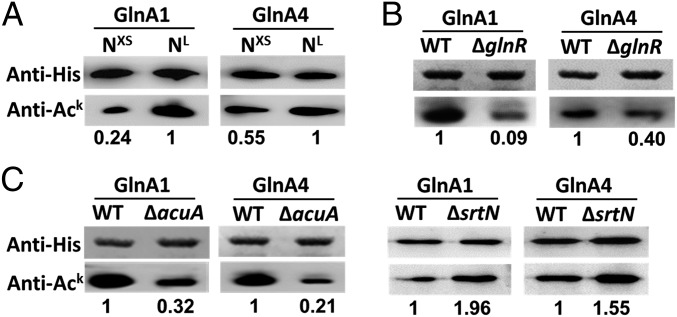
Next, we investigated the acetylation status of S. erythraea GlnA1 in vivo by immunoprecipitation (IP) and GlnA1 overexpression. The results demonstrate that GlnA1 in S. erythraea is acetylated in vivo (SI Appendix, Fig. S5). To investigate whether the intracellular acetylation levels of GS enzymes were influenced by nitrogen availability or by the lack of AcuA, SrtN, or GlnR, we performed Western blotting analyses to assess the acetylation state of GlnA1 and GlnA4 in S. erythraea cells (WT, ΔacuA, ΔsrtN, and ΔglnR strains) on nitrogen-limited (NL) or nitrogen-rich (NXS) media.
The results showed that nitrogen limitation led to an increase in the acetylation levels of GS enzymes (Fig. 3A) and of glnR transcript levels (approximately fourfold) (SI Appendix, Fig. S6). The antibody exhibited weak reactivity against two GS enzymes isolated from ΔglnR strains compared with those from WT strains (Fig. 3B). Conversely, the acetylation levels of overproduced endogenous GlnA1 and GlnA4 in the ΔacuA or ΔsrtN strain were markedly lower or higher than those in WT strains (Fig. 3C), indicating that the two endogenous GS enzymes were acetylated under physiological conditions and confirming that the acetyltransferase AcuA and the deacetylase SrtN were involved in their reversible acetylation. These results suggested that acetylation of the two GS enzymes was positively regulated by GlnR. The overproduced endogenous GlnA1 and GlnA4 in WT and ΔacuA strains were analyzed by MS for identification of the acetylated sites; the data are shown in SI Appendix, Table S2.
Fig. 3.
Acetylation levels of GS enzymes are influenced by extracellular nitrogen availability or the absence of AcuA, SrtN, or GlnR. (A) Acetylation levels of GlnA1 and GlnA4 of S. erythraea grown in minimal medium with NXS/NL nitrogen. (B) Acetylation levels of GlnA1/GlnA4 in S. erythraea cells (WT and ΔglnR strains). (C) Acetylation levels of GlnA1/GlnA4 in S. erythraea cells (WT, ΔacuA, and ΔsrtN strains).
The nonenzymatic acetylation of GlnA1 and GlnA4 was further investigated by using AcP and AcCoA. Western immunoblot analysis demonstrated that AcP could acetylate GlnA1 and GlnA4, whereas AcCoA was unable to acetylate the two GS proteins in vitro (SI Appendix, Fig. S7). The acetylated sites of GlnA1 and GlnA4 acetylated by AcP were also identified, including K246, K390, and K421 of GlnA1; and K162, K207, K235, K394, and K412 of GlnA4 (SI Appendix, Table S2). The results revealed the existence of two distinct mechanisms for the acetylation of GlnA1 and GlnA4: a nonenzymatic, AcP-dependent mechanism as reported recently by the teams headed by Choudhary and Wolfe (6, 7), as well as the AcuA-dependent enzymatic mechanism. As shown in SI Appendix, Table S2, K357 of GlnA1 and K319 of GlnA4 were acetylated in the WT strain, whereas no acetylation of these two sites was observed in the ΔacuA strain. Most of the sites acetylated by AcP in vitro were found to be acetylated in vivo in both WT and ΔacuA strains. These results further confirmed that AcuA was involved in the acetylation of K357 of GlnA1 and K319 of GlnA4 in vivo and in the regulation of the function of these two GS enzymes and suggested that AcP is responsible for acetylation of some of the other residues in vivo.
GlnA1AC (GSI-β) Serves as a Chaperone for GlnR–DNA Binding.
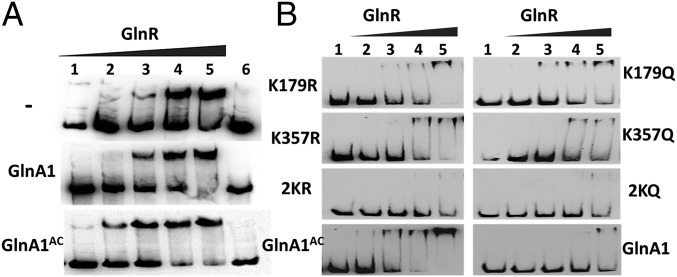
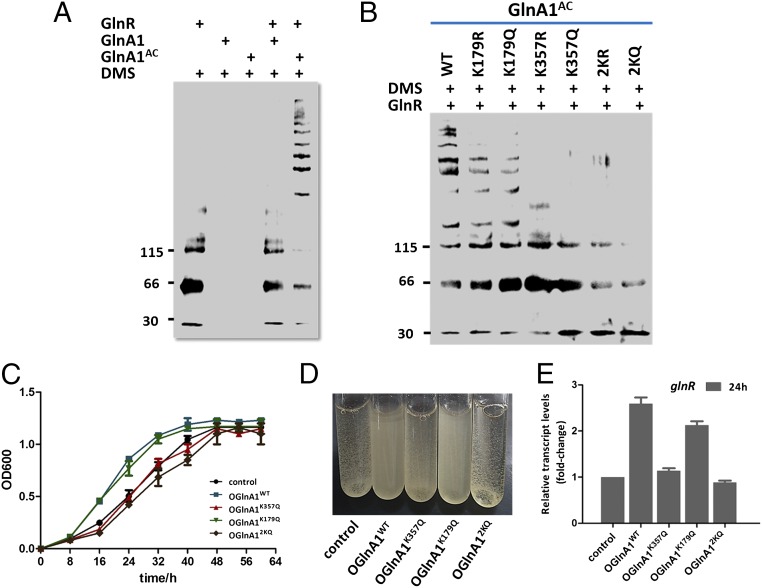
Previous studies have shown that B. subtilis GSI-α also acts as a chaperone [i.e., the feedback-inhibited (FBI) form of GS, or FBI-GS] to improve the DNA binding by MerR-type GlnR and TnrA via protein–protein interaction and thus regulates the expression of genes involved in nitrogen metabolism (26, 27). However, to date there has been no chaperone activity ascribed to GSI-β. Given that acetylation can modulate interactions between proteins by neutralizing the positive charge of the ε-amino group of lysine, we speculated that GlnA1AC (GSI-β) could improve the DNA-binding activity of GlnR as a chaperone through protein–protein interaction in a manner similar to that of FBI-GS in B. subtilis. We tested this possibility by evaluating the effect of GlnA1AC on the binding of GlnR to the glnA1 promoter by EMSA. In the absence of GlnA1, GlnR formed a relatively stable complex with DNA fragments at a concentration of 1.25 μM, as evidenced by the clear mobility shift (Fig. 4A, Top); the DNA-binding activity of GlnR was enhanced in the presence of GlnA1AC, such that GlnR formed stable complexes with DNA fragments at low protein concentrations (125 nM) (Fig. 4A, Bottom). In contrast, under the same conditions, there was no significant change in the GlnR DNA-binding affinity in the presence of native GlnA1 (Fig. 4A, Middle). These results indicate that GlnA1 acetylation enhances the DNA-binding activity of GlnR. This conclusion was further evaluated in the presence or absence of GSs by biolayer interferometry (Octet QKe; ForteBio). When GlnA1AC was present in the reaction, an ∼10-fold increase was observed in the DNA-binding affinity of GlnR (Kd from 8.6 ± 1.2 to 0.6 ± 0.05 nM), whereas no change was detected after adding native GlnA1 (Kd = 5.1 ± 1.6 nM). We next investigated which acetylation site(s) (K179 and/or K357) was involved in these interactions (Fig. 4B). Double mutants (2KR and 2KQ) lost the chaperone function of GlnA1 for enhancing the DNA-binding activity of GlnR, which was also attenuated in single mutants (K179R, K179Q, K357R, or K357Q) compared with GlnA1AC. Notably, K357R/Q mutants exhibited weaker chaperone activity than K179R/Q mutants, indicating that K357 of GlnA1 is critical for its chaperone function.
Fig. 4.
Effect of acetylation on GlnR DNA-binding activity. (A) Binding of GlnR to the glnA promoter, as determined by EMSA. GlnR (1–6 lines: 0.125, 0.625, 1.25, 2.5, 5, and 0 μM) and a 300-bp DNA fragment from the glnA promoter were incubated in the absence of GlnA1 (Top), with 1 μM native GlnA1 (Middle), or 1 μM GlnA1AC (Bottom). (B) GlnR (1–5 lines: 0, 0.125, 0.625, 1.25, and 2.5 μM) and the 300-bp DNA fragment were incubated in the presence of 1 μM GlnA1 or 1 μM mutated GlnA1 (K179R, K179Q, K357R, K357Q, 2KR, or 2KQ). GlnA1 mutants were acetylated before use in the EMSA assay. Representative images are shown from three independent experiments.
The glnA1 gene encoding an essential GS (GSI-β) is required for cell growth in several actinomycetes species. The direct interaction between GlnA1AC (Ac-GSI-β) and GlnR was further examined by using chemical cross-linking and the microscale thermophoresis assay (NanoTemper). Multiple bands with low mobility were observed in chemical cross-linking reactions with dimethyl suberimidate in the presence of GlnA1AC (Fig. 5A, lane 5), demonstrating that more GlnR became cross-linked to GlnA1AC. In contrast, there were no cross-linking bands observed for reactions containing native GlnA1 and GlnR (Fig. 5A, lane 4), suggesting that GlnR preferentially interacts with the acetylated form of GlnA1. The ability of the different GlnA1 mutants to interact with GlnR was also investigated. We found that acetylated K179R/Q single mutants were cross-linked with GlnR, showing equivalent or similar intensities and numbers of cross-linking bands (Fig. 5B, lanes 2 and 3) comparable to those obtained for acetylated GlnA1. However, K357R/Q mutants displayed significantly reduced interaction with GlnR (Fig. 5B, lanes 4 and 5), and the double mutants 2KR and 2KQ lost all GlnR interaction ability (Fig. 5B, lanes 6 and 7). These results revealed that acetylation of the K357 residue was required for its interaction with GlnR, which was consistent with the data shown in Fig. 4B. The GlnA1AC–GlnR interaction had a binding affinity of Kd = 2.86 μM as determined by microscale thermophoresis, whereas no binding was observed between native GlnA1 and GlnR (SI Appendix, Fig. S8).
Fig. 5.
Effect of acetylation on GlnA–GlnR interaction and cell growth. Cross-linking reactions containing the indicated components were separated by SDS polyacrylamide gel electrophoresis and evaluated by Western blotting with anti-GlnR antiserum. GlnR and GlnA1 were used at concentrations of 1 and 20 μM, respectively. (A) Cross-linking with native GlnA1 or GlnA1AC. (B) Cross-linking with GlnA1AC or mutated GlnA1 (K179R, K179Q, K357R, K357Q, 2KR, or 2KQ). Molecular size markers are shown to the left (in kDa). Representative images are shown. (C) Growth curves of S. erythraea strains overexpressing WT or mutant glnA1 cultured with 2 mM NH4Cl as the sole nitrogen source. S. erythraea without glnA1 overexpression was used as a control. (D) Images were captured at 24 h (exponential growth phase). (E) Transcriptional profiles of the glnR gene at 24 h. The relative expression of the glnR gene in the WT strain (control) was set to 1.0. Error bars show the SDs of three independent experiments.
To test the effects of acetylation at the two different Lys residues in GlnA1 in vivo, alleles encoding WT and variant GlnA1 proteins (GlnA1K357Q, GlnA1K179Q, and GlnA12KQ) were introduced into S. erythraea. The resulting strains were grown at 30 °C in minimal Evans medium (2 mM NH4Cl as the sole nitrogen source), and growth was monitored in triplicate at an optical density of 600 nm. Differences were observed in the growth behavior of the strains (Fig. 5 C and D). For example, the strains overexpressing GlnA1WT or GlnA1K179Q showed increased growth rates during the exponential growth phase (10–40 h), whereas those overexpressing GlnA1K357Q or GlnA12KQ showed growth rates similar to that of the WT strain. The levels of glnR transcript were also investigated in the S. erythraea strains at 24 h. As expected, the expression of glnR was up-regulated (more than twofold) in strains overexpressing GlnA1WT or GlnA1K179Q (Fig. 5E). These in vivo data are consistent with the activity measured in the GlnA1 variants in vitro (Figs. 4 and 5 A and B) and furthermore suggest that GlnA1 K357 acetylation is required for its chaperone function. Thus, the results demonstrated that acetylation of the GSI-β enzyme plays an important physiological role for nitrogen metabolism in living cells.
Chaperone Activity of Acetylated GSI-β Is Conserved in Actinomycetes.
GS GlnA1 is present and essential across actinomycetes, exhibiting high sequence similarity (>60%) among homologs (23). The S. erythraea GlnA1 (Sen_GlnA1) sequence was aligned with GSI-β homologs in other species, including SCO2198 from Streptomyces coelicolor, MSMEG_4290 from M. smegmatis, and RV2220 from M. tuberculosis (SI Appendix, Fig. S9). Notably, two lysine residues and their flanking sequences (KACGGYFPV and NPKACAKR) were highly conserved in all actinomycetes GSI-β–type enzymes (SI Appendix, Fig. S10A), whereas this conservation was not observed in GSII enzymes (GlnA2, GlnA3, and GlnA4) (SI Appendix, Fig. S2). K179GGYFPV was located in the long Tyr-179 loop containing 36 residues, whereas KGGYF was also found in the short Tyr-179 loop (only 11 residues, residues 148–158) of the B. subtilis GSI-α enzyme, which contained no Lys residue corresponding to K357. In particular, K179 was at the right side of a specific 25-amino acid insertion distinguishing GSI-α and -β (SI Appendix, Fig. S9), although the significance of this feature is unknown.
The Sen_GlnA1 structure was reconstructed by using Modeler (Version 9.14) based on the structure of M. tuberculosis Mtb_GlnA1 (Protein Data Bank ID code 1HTQ), which is 77% identical to Sen_GlnA1. We inferred that residue K179 (K183 in Mtb_GlnA1) lies within the loop extending from the active site to the central dodecamer channel, suggesting that its acetylation would have no effect on the interaction between GlnA1 and GlnR. The structural analysis of actinomycetes GSI-β enzymes revealed that residue K357 (K361 in Mtb_GlnA1) is localized at the edge of the dodecamer; i.e., the GlnR interaction interface. Upon modeling of an acetylated lysine residue at position 357, we found that the introduction of the acetyl moiety increased the hydrophobicity of the protein surface (SI Appendix, Fig. S10B).
Sequences flanking the two acetylation sites K179 and K357 were conserved in the β-subgroup of GSI-type enzymes, suggesting that similar regulatory mechanisms for nitrogen metabolism might exist in all actinomycetes. To test this hypothesis, we investigated the chaperone function of S. coelicolor and M. smegmatis GSI-β enzymes for enhancing GlnR–DNA binding. Acetylated Sco_GlnA1 and Msm_GlnA1 were found to interact with their corresponding GlnRs, which showed increased DNA binding (SI Appendix, Figs. S11A and S12A). Notably, GlnA1AC enzymes were able to interact with the GlnR of a different species (Sen_GlnA1AC with Sco_GlnR in SI Appendix, Fig. S11B and Msm_GlnA1AC with Sco_GlnR in SI Appendix, Fig. S12B), indicating that the GlnA1AC–GlnR–DNA interaction is highly conserved across actinomycetes. OmpR-type GlnR proteins have a conserved winged helix–turn–helix motif characterized by three α-helices that recognize and bind similar sequence motifs (23). However, the C- and N-terminal regions of GlnR proteins have low sequence conservation (SI Appendix, Fig. S13). The molecular mechanism underlying the interaction between GlnA1AC and GlnR that enhances GlnR–DNA binding activity remains to be investigated. Acetylation of GlnA1 (GSI-β) resulted in increased chaperone activity that enhanced GlnR-mediated transcription of nitrogen-related genes in response to nitrogen starvation. We therefore speculated that the chaperone role of acetylated GlnA1 might be necessary during nitrogen limitation and that the chaperone activity of acetylated GSI-β enzymes would be highly conserved across actinomycetes.
Discussion
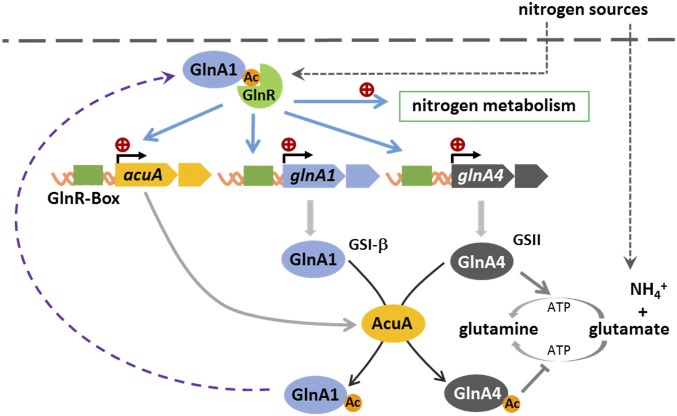
This study reveals the GlnR-mediated acetylation signaling pathway—from nutrient sensing to acetylation of proteins to feedback regulation—and provides new insights into the integration of nitrogen metabolism and the protein lysine acetylation system to construct a forward autofeedback loop mechanism of a GlnR–AcuA–GS circuit controlling nitrogen metabolism in response to environmental change (Fig. 6).
Fig. 6.
GlnR activates expression of the acuA, glnA1, and glnA4 genes in S. erythraea, integrating nitrogen metabolism and lysine acetylation to construct a forward autofeedback loop mechanism of the GlnR–AcuA–GS circuit.
GS is encoded by the glnA gene, and its expression and activity are strictly controlled via transcriptional regulation, allosteric regulation, and PTM. The two major GS families, GSI and GSII, are widely distributed in prokaryotes. GSI is largely absent in eukaryotes, but plays a vital role in many microorganisms, existing as α- and β-isoforms that differ in terms of sequence (35–41% identity), regulatory mechanisms (feedback allosteric regulation or adenylylation regulation), and biological functions (catalytic and chaperone activities) (28). Both the GSI-β and GSII enzymes are found in actinomycetes, and are distinguishable from each other by specific 25-amino acid insertion sequences (23). In this study, we found that AcuA-dependent lysine acetylation exerts different effects on four GS enzymes in S. erythraea: inactivation of one GSII enzyme (GlnA4) by acetylation, gain of chaperone activity of one GSI-β (GlnA1) by acetylation, and no acetylation of the other two GSII enzymes (GlnA2 and GlnA3). Actinomycetes generally contain one GSI-β enzyme (GlnA1) and two or three GSII enzymes. In many species of actinomycetes, such as M. tuberculosis, only the glnA1 gene (encoding a GSI-β) has been shown to be essential for growth, whereas the genes encoding GSII enzymes are not (29). GSI-β plays a central and essential role in nitrogen metabolism in actinomycetes because it not only controls glutamine synthesis, but also serves as the nitrogen signal for GlnR activation.
As one of the largest bacterial genera, actinomycetes are of great importance and are closely associated with human activities. For example, ∼70% of antibiotics are produced by actinomycetes. Conversely, some species such as M. tuberculosis (the causative agent of tuberculosis) are pathogenic. Nitrogen availability and metabolic regulation influence morphogenesis, antibiotic production, and virulence/pathogenicity in these species. The GS GlnA1 (GSI-β) is essential for cell growth in several actinomycetes organisms and plays important roles in nitrogen metabolism and pathogenic mechanisms (30). It has been found that the M. tuberculosis GlnA1 is critical for its pathogenicity and thus represents a potential pharmacological target for tuberculosis drugs. In addition, the level of extracellular GS in different mycobacterial strains has been shown to be correlated with their virulence (30, 31). Inhibiting GS has a strong impact on Mycobacterium growth in many experimental systems, including human macrophages and guinea pigs (32). Our work provides novel insight into the molecular mechanisms underlying the GlnA1 and pathogenicity in actinomycetes and suggests that drugs targeting the GlnR–GlnA1 circuit might be useful for controlling populations of pathogenic mycobacteria.
In summary, we found that acetylation exerted differing effects on the two GS subtypes. Acetylation inactivated GSII enzymes, whereas acetylated GSI-β exhibited a marked ability to directly interact with the GlnR nitrogen regulator and enhance GlnR–DNA binding, thereby modulating the transcription of target genes in response to nitrogen availability. GSI-β acetylation was directly responsible for its chaperone function, and this regulatory function of acetylated GSI-β enzymes was shown to be highly conserved across actinomycetes. Together, these findings substantively extend our understanding of the regulatory mechanisms underlying nitrogen metabolism in Gram-positive actinobacteria, which include antibiotic-producing actinomycetes as well as pathogenic mycobacteria.
Materials and Methods
Detailed methods of protein expression/purification, site-directed mutagenesis, GS-overproducing strains, the GS activity and in vitro acetylation/deacetylation assays, IP and immunoblotting, MS, EMSA, and cross-linking experiments are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by China NSF Grants 21276079, 21421004, and 21335003; Chinese Ministry of Education Grant SRFDP 20120074110009; and 863 Program Grant 2014AA02150.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525654113/-/DCSupplemental.
References
- 1.Phillips DM. The presence of acetyl groups of histones. Biochem J. 1963;87:258–263. doi: 10.1042/bj0870258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;31(41):10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- 4.Starai VJ, Escalante-Semerena JC. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol. 2004;340(5):1005–1012. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298(5602):2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- 6.Weinert BT, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51(2):265–272. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn ML, et al. Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation. PLoS One. 2014;9(4):e94816. doi: 10.1371/journal.pone.0094816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu BJ, Kim JA, Moon JH, Ryu SE, Pan JG. The diversity of lysine-acetylated proteins in Escherichia coli. J Microbiol Biotechnol. 2008;18(9):1529–1536. [PubMed] [Google Scholar]
- 9.Zhang J, et al. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8(2):215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang K, Zheng S, Yang JS, Chen Y, Cheng Z. Comprehensive profiling of protein lysine acetylation in Escherichia coli. J Proteome Res. 2013;12(2):844–851. doi: 10.1021/pr300912q. [DOI] [PubMed] [Google Scholar]
- 12.Kim D, et al. The acetylproteome of Gram-positive model bacterium Bacillus subtilis. Proteomics. 2013;13(10-11):1726–1736. doi: 10.1002/pmic.201200001. [DOI] [PubMed] [Google Scholar]
- 13.Schilling B, et al. Protein acetylation dynamics in response to carbon overflow in Escherichia coli. Mol Microbiol. 2015;98(5):847–863. doi: 10.1111/mmi.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang D, Li ZH, You D, Zhou Y, Ye BC. Lysine acetylproteome analysis suggests its roles in primary and secondary metabolism in Saccharopolyspora erythraea. Appl Microbiol Biotechnol. 2015;99(3):1399–1413. doi: 10.1007/s00253-014-6144-2. [DOI] [PubMed] [Google Scholar]
- 15.Hentchel KL, Escalante-Semerena JC. Acylation of biomolecules in prokaryotes: A widespread strategy for the control of biological function and metabolic stress. Microbiol Mol Biol Rev. 2015;79(3):321–346. doi: 10.1128/MMBR.00020-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolfe AJ. Bacterial protein acetylation: New discoveries unanswered questions. Curr Genet. 2016;62(2):335–341. doi: 10.1007/s00294-015-0552-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hentchel KL, Thao S, Intile PJ, Escalante-Semerena JC. Deciphering the regulatory circuitry that controls reversible lysine acetylation in Salmonella enterica. MBio. 2015;6(4):e00891–e00815. doi: 10.1128/mBio.00891-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy FJ, Turinsky AJ, Henkin TM. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176(15):4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castaño-Cerezo S, Bernal V, Blanco-Catalá J, Iborra JL, Cánovas M. cAMP-CRP co-ordinates the expression of the protein acetylation pathway with central metabolism in Escherichia coli. Mol Microbiol. 2011;82(5):1110–1128. doi: 10.1111/j.1365-2958.2011.07873.x. [DOI] [PubMed] [Google Scholar]
- 20.Nambi S, Basu N, Visweswariah SS. cAMP-regulated protein lysine acetylases in mycobacteria. J Biol Chem. 2010;285(32):24313–24323. doi: 10.1074/jbc.M110.118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu JY, You D, Leng PQ, Ye BC. Allosteric regulation of a protein acetyltransferase in Micromonospora aurantiaca by the amino acids cysteine and arginine. J Biol Chem. 2014;289(39):27034–27045. doi: 10.1074/jbc.M114.579078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.You D, Yao LL, Huang D, Escalante-Semerena JC, Ye BC. Acetyl coenzyme A synthetase is acetylated on multiple lysine residues by a protein acetyltransferase with a single Gcn5-type N-acetyltransferase (GNAT) domain in Saccharopolyspora erythraea. J Bacteriol. 2014;196(17):3169–3178. doi: 10.1128/JB.01961-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward D, van Helden PD, Wiid IJ. Glutamine synthetase sequence evolution in the mycobacteria and their use as molecular markers for Actinobacteria speciation. BMC Evol Biol. 2009;9:48. doi: 10.1186/1471-2148-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao LL, et al. GlnR-mediated regulation of nitrogen metabolism in the actinomycete Saccharopolyspora erythraea. Appl Microbiol Biotechnol. 2014;98(18):7935–7948. doi: 10.1007/s00253-014-5878-1. [DOI] [PubMed] [Google Scholar]
- 25.Ren Q, Gorovsky MA. Histone H2A.Z acetylation modulates an essential charge patch. Mol Cell. 2001;7(6):1329–1335. doi: 10.1016/s1097-2765(01)00269-6. [DOI] [PubMed] [Google Scholar]
- 26.Fisher SH, Wray LV., Jr Bacillus subtilis glutamine synthetase regulates its own synthesis by acting as a chaperone to stabilize GlnR-DNA complexes. Proc Natl Acad Sci USA. 2008;105(3):1014–1019. doi: 10.1073/pnas.0709949105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wray LV, Jr, Zalieckas JM, Fisher SH. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell. 2001;107(4):427–435. doi: 10.1016/s0092-8674(01)00572-4. [DOI] [PubMed] [Google Scholar]
- 28.Brown JR, Masuchi Y, Robb FT, Doolittle WF. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38(6):566–576. doi: 10.1007/BF00175876. [DOI] [PubMed] [Google Scholar]
- 29.Harth G, Maslesa-Galić S, Tullius MV, Horwitz MA. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol Microbiol. 2005;58(4):1157–1172. doi: 10.1111/j.1365-2958.2005.04899.x. [DOI] [PubMed] [Google Scholar]
- 30.Chandra H, Basir SF, Gupta M, Banerjee N. Glutamine synthetase encoded by glnA-1 is necessary for cell wall resistance and pathogenicity of Mycobacterium bovis. Microbiology. 2010;156(Pt 12):3669–3677. doi: 10.1099/mic.0.043828-0. [DOI] [PubMed] [Google Scholar]
- 31.Tullius MV, Harth G, Horwitz MA. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect Immun. 2003;71(7):3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harth G, Zamecnik PC, Tang JY, Tabatadze D, Horwitz MA. Treatment of Mycobacterium tuberculosis with antisense oligonucleotides to glutamine synthetase mRNA inhibits glutamine synthetase activity, formation of the poly-L-glutamate/glutamine cell wall structure, and bacterial replication. Proc Natl Acad Sci USA. 2000;97(1):418–423. doi: 10.1073/pnas.97.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.