Significance
We present a draft genome of the peanut A-genome progenitor, Arachis duranensis, providing details on total genes present in the genome. Genome analysis suggests that the peanut lineage was affected by at least three polyploidizations since the origin of eudicots. Resequencing of synthetic Arachis tetraploids reveals extensive gene conversion since their formation by human hands. The A. duranensis genome provides a major source of candidate genes for fructification, oil biosynthesis, and allergens, expanding knowledge of understudied areas of plant biology and human health impacts of plants. This study also provides millions of structural variations that can be used as genetic markers for the development of improved peanut varieties through genomics-assisted breeding.
Keywords: Arachis duranensis, genome sequence, gene models, polyploidizations, gene duplication
Abstract
Peanut or groundnut (Arachis hypogaea L.), a legume of South American origin, has high seed oil content (45–56%) and is a staple crop in semiarid tropical and subtropical regions, partially because of drought tolerance conferred by its geocarpic reproductive strategy. We present a draft genome of the peanut A-genome progenitor, Arachis duranensis, and 50,324 protein-coding gene models. Patterns of gene duplication suggest the peanut lineage has been affected by at least three polyploidizations since the origin of eudicots. Resequencing of synthetic Arachis tetraploids reveals extensive gene conversion in only three seed-to-seed generations since their formation by human hands, indicating that this process begins virtually immediately following polyploid formation. Expansion of some specific gene families suggests roles in the unusual subterranean fructification of Arachis. For example, the S1Fa-like transcription factor family has 126 Arachis members, in contrast to no more than five members in other examined plant species, and is more highly expressed in roots and etiolated seedlings than green leaves. The A. duranensis genome provides a major source of candidate genes for fructification, oil biosynthesis, and allergens, expanding knowledge of understudied areas of plant biology and human health impacts of plants, informing peanut genetic improvement and aiding deeper sequencing of Arachis diversity.
Peanut (Arachis hypogaea L.) is one of the most economically important crops as an important source of edible oil and protein. With yields averaging 2,898 pounds per acre in the United States in recent years, and with seed comprised of a remarkable 45∼56% oil, peanut typically produces more than twice the amount of oil per unit area than soybean (1,059 vs. 446 L/ha). Rudolf Diesel’s revolutionary engine showcased to the world in Paris in 1900 ran on peanut oil.
Rich in oleic and linoleic acids, peanut oil is associated with several human health benefits. Peanut is a good source of protein (24% by weight), resveratrol implicated in improved cardiovascular health, fiber that reduces the risk of certain cancers and controls blood sugar levels, folic acid, which helps prevent neural tube defects, and contains nearly half of the 13 essential vitamins and 35% of the essential minerals. A hindrance to realizing these benefits is the prevalence of peanut allergy, for example affecting 0.8% of children and 0.6% of adults in the United States, totaling more than 1 million Americans (1).
The genus Arachis is distinguished from most Fabaceae taxa in that all members have geocarpic reproductive habit, with “aerial flower, subterranean fruit” (2). Following fertilization, the peanut gynophore elongates to form a special geotropic “peg” (2) harboring the embryo, which continues to grow and push the developing pod into the soil. Pod formation and embryo differentiation occur and a seed is produced underground (3). Thought to be an adaptation to particularly harsh environments, geocarpy has been reported in only 24 families and 57 genera of flowering plants (4).
Arachis originated in South America and its ∼80 species have been divided taxonomically into nine sections (5). A. hypogaea, cultivated peanut or groundnut, was domesticated (6) and is now grown on 25 million ha between latitudes 40° N and 40° S, with global production of 36 million tons (faostat.fao.org). Peanut is allotetraploid (2n = 4x = 40), with an AABB genomic constitution and genome size of 2,800 Mb (7); its suspected recent origin is thought to account for the paucity of genetic diversity among naturally occurring peanut genotypes. However, the genetic vulnerability of natural tetraploids is in contrast to rich diversity among diploid Arachis.
To gain insight into peanut evolution and opportunities for mitigating its genetic vulnerability, we sequenced the suspected peanut A-genome progenitor, Arachis duranensis, as well as four synthetic tetraploids and their six diploid parents [two A-genome and four B-genome, including the suspected B-genome progenitor, Arachis ipaensis (8, 9)]. Complementing an independent effort to sequence tetraploid peanut, this approach provides new insight into Arachis biology and evolution and into the nature and rate of genomic change following polyploid formation.
Results and Discussion
Genome Assembly and Annotation.
The genome of A. duranensis (accession PI475845) (SI Appendix, Fig. S1) was sequenced using a shotgun strategy. About 229.94 Gb of raw data from a Hiseq2500 (SI Appendix, SI Text and Table S1) yielded an optimized assembly of 1.05 Gb with an N50 contig length of 29.6 kb and N50 scaffold length of 649.8 kb (Table 1 and SI Appendix, SI Text, Fig. S2 and Tables S2 and S3). PCR amplification of randomly selected regions, sequence-depth distribution, and expressed sequence tag validation (SI Appendix, Figs. S3–S6 and Tables S4–S6 and Dataset S1) indicated the high quality of the assembled genome. K-mer analysis indicated A. duranensis genome size of 1.38 Gb (SI Appendix, Table S7), consistent with prior data (10). The average heterozygosity rate was estimated to be three SNPs per kilobase (SI Appendix, Fig. S7). The average GC content is 31.79% (Table 1) and its distribution is similar to other legumes, although different from other oilseeds and other plants (SI Appendix, Figs. S8 and S9).
Table 1.
Summary of genome assembly and genome annotation of diploid A-genome progenitor
| Genome features | Measures |
| Genome assembly | |
| No. Scaffolds | 8,173 |
| Total span | 1,051,523,805 bp |
| N50, scaffold | 649,840 bp |
| Longest scaffold | 5,342,956 bp |
| No. contigs | 90,568 |
| Longest contigs | 285,529 |
| N50, contig | 29,584 bp |
| GC content | 31.79% |
| Gene models | |
| No. gene models | 50,324 |
| Mean gene length | 3,057 bp |
| Mean exon length | 312 bp |
| Mean intron length | 709 bp |
| Mean number of exons per gene | 3.37 |
| Mean gene density | 21.4 kb |
| Nonprotein coding genes/elements | |
| No. miRNA genes | 801 |
| Mean length of miRNA genes | 107 bp |
| miRNA genes share in genome | 0.0080% |
| No. rRNA fragment | 115 |
| Mean length of rRNA fragment | 1077 bp |
| rRNA fragment share in genome | 0.011% |
| No. tRNA genes | 913 |
| Mean length of tRNA genes | 73 bp |
| tRNA gene share in genome | 0.0062% |
| No. snRNA genes | 202 |
| Mean length of snRNA genes | 127 bp |
| snRNA genes share in genome | 0.0024% |
| Total transposable elements, bp | 643,886,314 |
| Transposable element percent in genome | 59.77% |
We predicted 50,324 protein-coding gene models (Table 1), with 99% (50,281) supported by transcriptome sequences (SI Appendix, Fig. S6). Compared with the gene sets of legumes, oilseeds, and other plant species (SI Appendix, Table S8), A. duranensis genes showed highest similarity to legumes (SI Appendix, Table S9), with gene number comparable to Medicago truncatula (50,894), lower than soybean (tetraploid Glycine max, 56,044), and higher than other legumes (SI Appendix, Table S10). Of the 50,324 gene models, ∼90% matched entries in publically available databases (SI Appendix, Fig. S10 and Table S11). Approximately 10.9% (5,494) of gene models with no homology to known proteins were supported by transcriptome data and may be peanut-specific. Comparison of gene characteristics indicated that distribution of A. duranensis gene features were similar to legumes, but obviously different from distantly related plant species (SI Appendix, Fig. S11). A. duranensis has an average of one gene per 21.4 kb (Fig. 1 and Table 1), half the gene density of Lotus (10.2 kb) (11) and one-fifth that of Arabidopsis (4.5 kb) (12). The average gene length of 3,057 bp, protein length of 368 aa, coding sequence length of 312 bp with 3.37 exons, and intron length of 709 bp were relatively long among plant species (Table 1 and SI Appendix, Table S10). Gene Ontology (GO) enrichment analysis revealed genes overrepresented in processes such as gravitropism under reaction to stimulus (SI Appendix, Figs. S12–S14 and Datasets S2–S4).
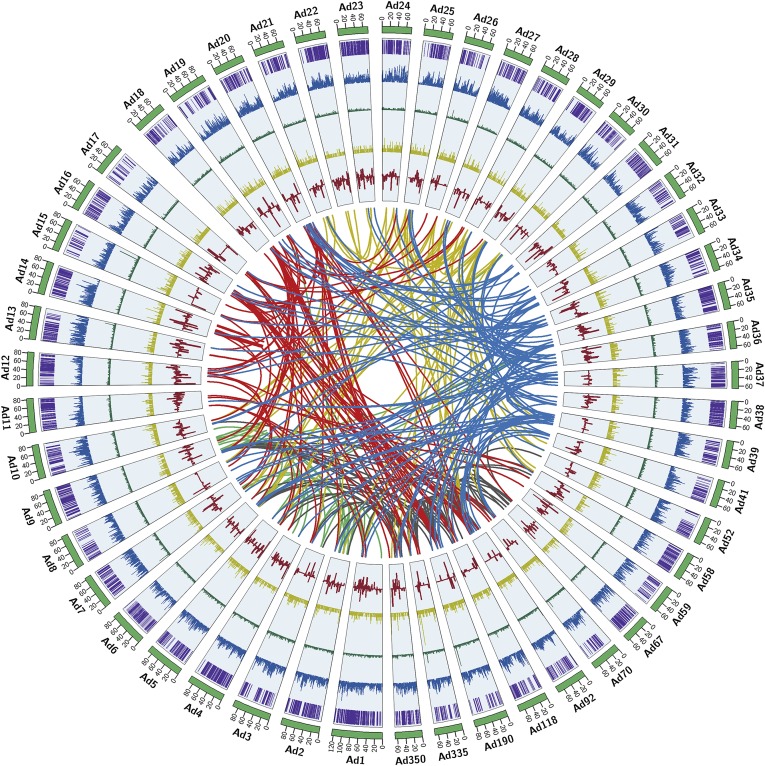
Fig. 1.
A. duranensis genome overview. From the outer edge inward, circles represent the 50 largest DNA sequence scaffolds (green), the genes on each scaffold (purple), SNP density at 100-kb intervals (blue), repeat density at 100 kb (green), transposable element density at 100 kb (yellow), and the fold-change values of transcripts (red). Links in the core connect duplicated sets of genes (E-value threshold of <1e-10 and 85% similarity).
Gene Content and Repeated Sequence Annotation.
In 16 diverse plant species with 54,384 gene families identified using OrthoMCL (13) (SI Appendix, Figs. S15–S20 and Tables S12 and S13), 1,423 families were specific to A. duranensis. Comparison with soybean and Medicago suggested that A. duranensis family-size variations were different from those of other legumes (SI Appendix, Table S13). A total of 9,968 single-copy orthologs and 16,472 unique paralogs were identified in A. duranensis (SI Appendix, Fig. S21 and Table S14).
We identified 5,251 putative A. duranensis transcription factor (TF) genes in 57 families, 10.4% of the predicted A. duranensis genes, slightly higher than soybean, and much higher than most plant species analyzed (Dataset S5). Although distributed among families in a manner generally similar to soybean (SI Appendix, Fig. S22), a few TF families were dominant in A. duranensis, such as B3, E2F/DP, FAR1, GeBP, HSF, NAC, S1Fa-like, and STAT (Fig. 2A and Dataset S5). Families such as ARR-B, CAMTA, DBB, MIKC, and NF-YA, were sparser in A. duranensis than in most plants. Expansion and contraction of TF families may reflect regulatory differences in biological functions of A. duranensis.
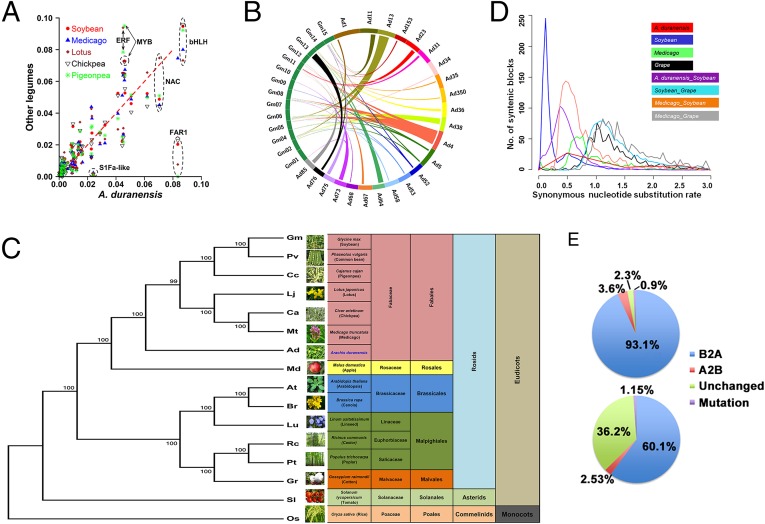
Fig. 2.
Comparative genomic and evolutionary analysis. (A) Scatter plot of percentage of A. duranensis transcription factors in relation to soybean, Medicago, Lotus, chickpea, and pigeonpea. (B) Syntenic relationship between A. duranensis scaffolds and soybean chromosomes. (C) Phylogenetic tree for 16 plant species based on single copy orthologous genes. (D) Distribution of synonymous nucleotide substitutions (Ks) for A. duranensis, soybean, Medicago, and grape. (E) Allelic changes between the A and B subgenomes of synthetic tetraploid peanut lines, ISATGR 184 (Upper) and ISATGR 1212 (Lower).
We annotated 816 Arachis microRNAs (miRNAs), 913 transfer RNAs (tRNAs), 115 ribosomal RNAs (rRNAs), and 202 small nucleolar RNAs (snRNAs) (Table 1, SI Appendix, Table S15, and Dataset S6). A total of 64 target genes were predicted after aligning 15 new miRNAs to our gene models (SI Appendix, Table S16). These target genes were annotated in diverse GO categories (SI Appendix, Fig. S23 and Table S17).
About ∼59.77% of the A. duranensis genome appeared to be transposable elements (Fig. 1 and Table 2), with ∼40% long terminal repeat retrotransposons, and with one major burst of amplification ∼2 Mya (SI Appendix, Fig. S24). Most A. duranensis transposable element sequences had a >10% divergence rate (SI Appendix, Fig. S25). In addition, we identified 105,003 simple sequence repeats (SSRs) in A. duranensis, and resequencing of two other A-genome genotypes and four B-genome genotypes (SI Appendix, Tables S18 and S19 and Datasets S7–S10) allowed the discovery of ∼8 million SNPs and other structural variations, useful as molecular markers (SI Appendix, Tables S20 and S21 and Dataset S11).
Table 2.
Organization of repetitive sequences in the diploid A-genome progenitor
| Repetitive elements | Repeat number | Length, bp | Repeat, % | Genome, % |
| Retrotransposons | 386,961 | 445,273,366 | 69.15 | 41.34 |
| LINE retrontransposons | 16,479 | 13,561,787 | 2.11 | 1.26 |
| SINE retrontransposons | 783 | 95,460 | 0.01 | 0.01 |
| LTR retrotransposons | 369,699 | 431,616,119 | 67.03 | 40.07 |
| Gypsy | 332,073 | 397,492,954 | 61.73 | 36.90 |
| Copia | 28,832 | 28,817,768 | 4.48 | 2.68 |
| Other | 8,794 | 5,305,397 | 0.82 | 0.49 |
| DNA transposons | 114,098 | 55,942,274 | 8.69 | 5.19 |
| Unclassified elements | 527,605 | 142,670,674 | 22.16 | 13.24 |
| Total transposon elements | 1,028,664 | 643,886,314 | — | 59.77 |
LINE, long interspersed nuclear element; LTR, long terminal repeat; SINE, short interspersed nuclear element.
Phylogenetic Analysis.
Synteny analysis based on corresponding genes between genomes revealed high numbers of syntenic blocks between A. duranensis and soybean and common bean, and low numbers with Arabidopsis and rice (Fig. 2B, SI Appendix, Fig. S26, and Dataset S12). A species tree based on single-copy orthologous genes indicated A. duranensis to be in an individual clade, not including any other legume species and consistent with its phylogenetic placement (Fig. 2C).
Speciation of Peanut A and B Subgenomes.
By performing a trio comparison of the synthetic tetraploid ISATGR 184 and its parents, ICG 8123 and ICG 8206, we studied the divergence between the A and B subgenomes. A total of 847,676 high-quality single nucleotide variants were identified between the two parental lines, meaning a mutation rate of ∼4.5 × 10−4 in each line. Upon mapping the reads of ISATGR 184 onto the reference genome, we identified 748,802 single nucleotide variants sites between the two parental lines.
Recursive Peanut-Specific Genome Duplications.
Peanut shares a genome duplication with Medicago and soybean. Collinear genes from Medicago, soybean, and grape were used to locate related evolutionary events. The synonymous nucleotide substitution (Ks) distribution of peanut homologs shows a prominent peak around Ks = 0.5 (Fig. 2D and SI Appendix, Fig. S27), overlapping the peak of soybean duplicated genes resulting from a pan-legume tetraploidization previously inferred to be ∼60 Mya (14) (SI Appendix, Fig. S28). Adding the pan-eudicot γ-hexaploidy (∼130 Mya) and polyploidy-producing tetraploid peanut by joining the Arachis A and B subgenomes, estimated to have diverged 3.5 Mya (15), the Arachis lineage has been affected by at least three polyploidizations since the origin of eudicots, with a collective 12× paleo-duplication depth.
Gene Conversion has Already Occurred in Synthetic Polyploids.
Following polyploid formation in plants, extensive and highly structured unidirectional homeologous exchanges between genes from different subgenomes can overwrite one progenitor allele with additional copies of the other, often called gene conversion (16, 17). Implicated as a possible contributor to the transgressive properties of polyploids relative to their progenitors (16), extensive gene conversion has occurred as recently as the past 7,500–12,500 y since formation of the Neolithic species Brassica napus (18).
By performing a three-way comparison of the synthetic tetraploid ISATGR 184 and its progenitor lines, ICG 8123 and ICG 8206, we find evidence of extensive gene conversion between the At and Bt subgenomes in the ∼three seed-to-seed generations since its formation by human hands. The vast majority (∼93%) of alleles have been converted to homozygosity for the A genome allele in ISATGR 184 (Fig. 2E), an asymmetry resembling those found in cotton and canola (16, 18). Further analysis with only the singleton genes in the present peanut dataset revealed a similar conclusion (not shown). ISATGR 1212, a reciprocal cross between the same parental lines as ISATGR 184, shared Bt to At bias of conversion (Fig. 2E and Dataset S13) but had far fewer converted sites than ISATGR 184 (χ2 << 0.001), perhaps indicating a contribution of germ-line types to genomic variation in the offspring.
Aerial Flower but Subterranean Fruit.
A characteristic feature of Arachis is the gynophore (“peg”), a specialized organ that transitions from upward growth habit to downward outgrowth upon fertilization, driving the developing pod into the soil. Arachis pod formation, embryo differentiation, and seed production occur in subterranean darkness (Fig. 3A).
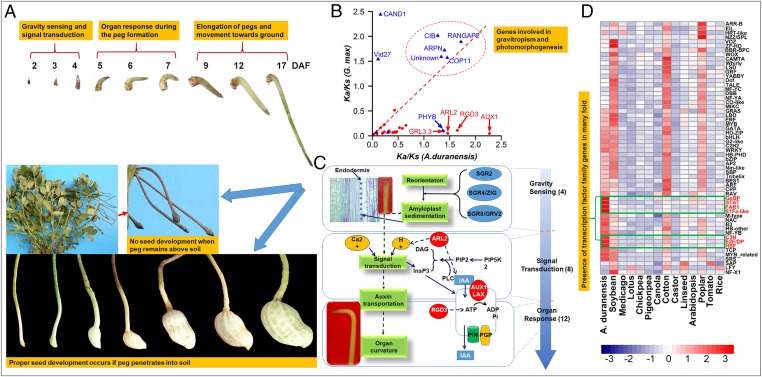
Fig. 3.
Genes, TFs, and pathways related to the biological process “Aerial flower, subterranean fruit.” (A) Peanut fruit development (2, 3, 4, 5, 6, 7, 9, 12, and 17 indicate days after fertilization, DAF). Upon fertilization the gynophore reverts from upward growth after sensing gravity followed by signal transduction and organ response. Seed development is triggered only if the elongated peg penetrates soil. (B) Comparison of ω (Ka/Ks) of gravitropic orthologs in Arabidopsis thaliana-A. duranensis and A. thaliana-G. max for genes involved in photomorphogenesis (blue) or gravitropism (red). Circled genes show evidence of positive selection in both A. duranensis and G. max. (C) Gravitropism phases (gravity sensing, signal transduction and organ response) where three functionally identified genes (ARL2, AUX1, and RGD3) were found positively selected. (D) Numbers of TF family members identified in A. duranensis compared with other plant species (blue indicating low and red indicating high numbers). TFs in brackets have been found in large numbers in A. duranensis.
We searched our gene set against genes in the GO category “gravitropism” (GO:0009630) and experimentally identified in Arabidopsis using an E-value cut-off of 1E-10, identifying 151 gravitropism-related genes expressed during pod development. Using the branch-site likelihood ratio test, eight A. duranensis genes showed evidence of positive selection, (Fig. 3B, SI Appendix, Fig. S29, and Dataset S14). Three of these genes were identified in Arabidopsis by several lines of experimental evidence (19–21) to be involved in signal transduction and organ response (Fig. 3C). Further analysis based on previous functional studies (19, 21–25) identified 24 A. duranensis genes likely to be gravitropic, including 4 involved in gravity perception, 8 in signal transduction, and 12 in organ response (Fig. 3C and Dataset S15).
Fruit development, a genetically controlled process under light conditions, occurs in darkness (subterranean fructification) in Arachis, with peg elongation responding conversely to light/dark conditions (26). Five TF families related to photomorphogenesis were identified in very large numbers in A. duranensis, namely S1Fa-like, FAR1, HSF, NAC, and STAT (Fig. 3D). Notably, A. duranensis has 126 S1Fa-like TFs versus no more than 5 in other plants (Fig. 3D and Dataset S5). S1Fa-like TFs containing a small peptide (70 aa) with a nuclear localization and DNA binding domain were more highly expressed in roots and etiolated seedlings than green leaves (27). Also greatly expanded in A. duranensis is the FAR1 TF family, which plays an important role in modulating phyA-signaling homeostasis in higher plants (28). Importantly, phyA localized in the cytosol of dark-grown seedlings acts primarily as a far-red sensor, which regulates the transition from skotomorphogenesis to photomorphogenesis (29). Furthermore, each phytochrome shows strong dark reversion in some species (29). PhyB, exhibiting a fast and strong but incomplete dark conversion in some cases, is the main light receptor responsible for the shade-avoidance response in mature plants (30) and shows evidence of positive selection in A. duranensis (Fig. 3B), suggesting a role in skotomorphogenesis.
Oil Biosynthesis and Allergen-Encoding Genes.
Comparative genome analysis identified 1,671 A. duranensis genes related to acyl lipid synthesis, comparable to soybean (1,695 genes) and more than the nonoilseed plant Arabidopsis (1,291 genes). Significant enrichment of lipid gene-functional groups were found in both A. duranensis and soybean relative to Arabidopsis (SI Appendix, Table S22). A. duranensis and soybean had small differences in gene numbers for five categories of lipid synthesis genes (phospholipase, lipid related, fatty acid synthesis in plastids, lipase, and cuticular wax synthesis), whereas the remaining 17 categories were indistinguishable.
Considering the importance of peanut as an oil crop, we manually investigated the annotations of 67 gene models with similarity to known genes involved in fatty acid biosynthesis and triacylglycerol (TAG) assembly, which in peanut correspond mainly to oleic and linoleic acids (31). Of these 67 gene models, 51 have homologs in the castor genome (32) (Dataset S16), and 62 were supported by RNA-seq data from peanut seeds during seed filling, maturation, and desiccation (SI Appendix, Fig. S30 and Table S23). FAD2 encoding δ-12 oleic acid desaturase, the key enzyme controlling the high oleate trait (33), was expressed highly during seed filling but little during desiccation (Fig. 4A). Genes encoding key enzymes in the TAG pathway were expressed at diverse levels at different developmental stages (Fig. 4A). Multiple copies or isoforms of some key genes were detected in the A. duranensis genome like glycerol-3-phosphate acyltransferase and diacylglycerol acyltransferase, which catalyze the first and final steps in the TAG pathway. Information on copy number and expression diversity of these metabolic genes is important for improvement of oil quality parameters in peanut, such as a high oleic to linoleic acid ratio (O/L).
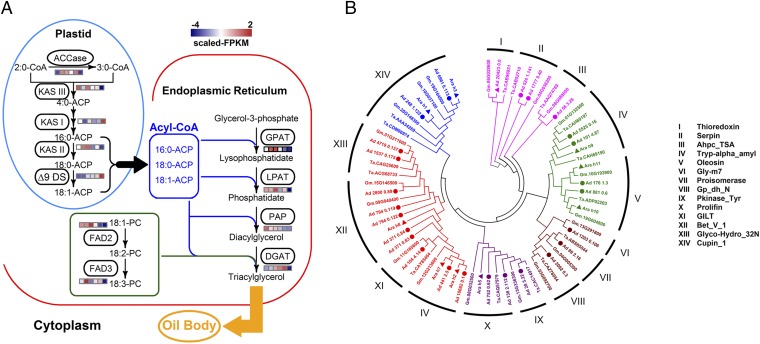
Fig. 4.
Genes involved in oil biosynthesis and encoding allergens. (A) Expression patterns of genes involved in fatty acid synthesis and TAG assembly in peanut seed. Expression levels were estimated by fragments per kilobase of transcript per million mapped fragments (FPKM) for each gene obtained by sequencing RNA samples from peanut seeds at six developmental stages. (B) Phylogenetic tree of allergens and their homologs identified in A. hypogaea and A. duranensis and other allergenic crops (wheat and soybean). A total of 69 allergens or homologous genes from 14 families group into 6 major clusters differentiated with colors. Different families having the same colors are more similar to each other than to other families. “Dots” indicate genes identified in the A. duranensis genome and “triangles” indicate genes previously identified in A. hypogaea. This figure also shows the distribution of 12 newly identified putative allergen genes from the A. duranensis assembly falling into different clusters and families.
Peanut allergy, one of most serious life-threatening food sensitivities, is becoming increasingly prevalent, particularly among children. Comparison with known allergenic proteins from peanut and other crops identified 21 candidate allergen-encoding genes in A. duranensis (Fig. 4B and Dataset S17), of which 9 have already been reported in peanut (SI Appendix, Fig. S31), whereas the remaining 12 (SI Appendix, Fig. S32) are A. duranensis homologs of genes from other crops. Interestingly, we found three putative paralogous genes for Ara h 8 in A. duranensis, but none for Ara h 3, which had 16 homologous sequences in peanut (34). Sequence information for additional allergen-encoding genes may nurture studies leading to either genetic or medical approaches to allergy mitigation.
Concluding Remarks
This draft A. duranensis genome provides rich new information about an important branch of the legume clade, building on the genomes of Lotus (11), Medicago (14), soybean (35), pigeonpea (36), chickpea (37), and common bean (38). The Arachis genome sequences presented here provide new insights and new resources for evolution and polyploidization research. The biological interpretation of the A. duranensis genome enhances understanding of the unusual fruit development of peanut. Resequencing 10 additional genotypes including four tetraploids provides a backdrop for reference sequencing of cultivated peanut, as well as molecular markers (SSR or SNP) for investigating genetic diversity and aiding breeding programs.
Materials and Methods
The diploid species A. duranensis (AA 2n = 2x = 20) is the most possible progenitor and donor of the A subgenome to cultivated tetraploid (AABB 2n = 4x = 40) peanut (6, 9, 39, 40). Within species A. duranensis, the accession PI 475845 falls into the group that includes 14 of the total 18 accessions (39). Furthermore, South America is the center of origin, the accession PI475845 is a collection from Bolivia. Therefore, being a representative accession, we have sequenced the genome of PI 475845 (A. duranensis). See SI Appendix for the details of sequencing, assembly, annotation, structural variation, evolutionary, synteny, and transcriptomic analysis.
Supplementary Material
Acknowledgments
This project was supported by the National Natural Science Foundation of China (31501246 and 31271767); the Modern Agro-industry Technology Research System (CARS-14); the Science and Technology Planning Project of Guangdong Province (2015B020231006, 2016B020201003, 2012B050700007, and S2013020012647); Pearl River Science and Technology Nova of Guangzhou (2013J2200088), the Southeastern US Region Peanut Research Initiative; the Georgia Peanut Commission; the University System of Georgia Regent’s Professorship funds; and USAID–ICRISAT (International Crops Research Institute for the Semi-Arid Tropics) Linkage Grants. This work has also been undertaken as part of the CGIAR (Consultive Group for International Agricultural Research) Research Program on Grain Legumes. ICRISAT is a member of the CGIAR Consortium.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI database (BioProject ID PRJNA288069; BioSample accession no. SAMN03799425).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600899113/-/DCSupplemental.
References
- 1.Sampson HA. Update on food allergy. J Allergy Clin Immunol. 2004;113(5):805–819, quiz 820. doi: 10.1016/j.jaci.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 2.Smith BW. Arachis hypogaea. Aerial flower and subterranean fruit. Am J Bot. 1950;37(10):802–815. [Google Scholar]
- 3.Feng QL, et al. Arachis hypogaea plant recovery through in vitro culture of peg tips. Peanut Science. 1995;22(2):129–135. [Google Scholar]
- 4.Tan D, Zhang Y, Wang A. A review of geocarpy and amphicarpy in angiosperms, with special reference to their ecological adaptive significance. Chinese Journal of Plant Ecology. 2010;34(1):72–88. [Google Scholar]
- 5.Krapovickas A, Gregory WC. Taxonomia del genero Arachis (Leguminosae) Bonplandia. 1994;8:1–186. [Google Scholar]
- 6.Simpson CE, Krapovickas A, Valls JFM. History of Arachis including evidence of A. hypogaea L. progenitors. Peanut Science. 2001;28(2):78–80. [Google Scholar]
- 7.Temsch EM, Greilhuber J. Genome size variation in Arachis hypogaea and A. monticola re-evaluated. Genome. 2000;43(3):449–451. [PubMed] [Google Scholar]
- 8.Seijo JG, et al. Physical mapping of the 5S and 18S-25S rRNA genes by FISH as evidence that Arachis duranensis and A. ipaensis are the wild diploid progenitors of A. hypogaea (Leguminosae) Am J Bot. 2004;91(9):1294–1303. doi: 10.3732/ajb.91.9.1294. [DOI] [PubMed] [Google Scholar]
- 9.Burow MD, Simpson CE, Faries MW, Starr JL, Paterson AH. Molecular biogeographic study of recently described B- and A-genome Arachis species, also providing new insights into the origins of cultivated peanut. Genome. 2009;52(2):107–119. doi: 10.1139/g08-094. [DOI] [PubMed] [Google Scholar]
- 10.Temsch EM, Greilhuber J. Genome size in Arachis duranensis: A critical study. Genome. 2001;44(5):826–830. [PubMed] [Google Scholar]
- 11.Sato S, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15(4):227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielen S, et al. Matita, a new retroelement from peanut: Characterization and evolutionary context in the light of the Arachis A-B genome divergence. Mol Genet Genomics. 2012;287(1):21–38. doi: 10.1007/s00438-011-0656-6. [DOI] [PubMed] [Google Scholar]
- 16.Paterson AH, et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature. 2012;492(7429):423–427. doi: 10.1038/nature11798. [DOI] [PubMed] [Google Scholar]
- 17.Wang XY, Paterson AH. Gene conversion in angiosperm genomes with an emphasis on genes duplicated by polyploidization. Genes (Basel) 2011;2(1):1–20. doi: 10.3390/genes2010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalhoub B, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 19.Bennett MJ, et al. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science. 1996;273(5277):948–950. doi: 10.1126/science.273.5277.948. [DOI] [PubMed] [Google Scholar]
- 20.Guan C, Rosen ES, Boonsirichai K, Poff KL, Masson PH. The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway. Plant Physiol. 2003;133(1):100–112. doi: 10.1104/pp.103.023358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison BR, Masson PH. ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. Plant J. 2008;53(2):380–392. doi: 10.1111/j.1365-313X.2007.03351.x. [DOI] [PubMed] [Google Scholar]
- 22.Young LS, et al. Adenosine kinase modulates root gravitropism and cap morphogenesis in Arabidopsis. Plant Physiol. 2006;142(2):564–573. doi: 10.1104/pp.106.084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspar T, Pickard BG. Gravitropism in a starchless mutant of Arabidopsis: Implications for the starch-statolith theory of gravity sensing. Planta. 1989;177(2):185–197. doi: 10.1007/BF00392807. [DOI] [PubMed] [Google Scholar]
- 24.Swarup R, et al. Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell. 2004;16(11):3069–3083. doi: 10.1105/tpc.104.024737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature. 2003;423(6943):999–1002. doi: 10.1038/nature01716. [DOI] [PubMed] [Google Scholar]
- 26.Shlamovitz N, Ziv M, Zamski E. Light, dark and growth regulator involvement in groundnut (Arachis hypogaea L.) pod development. Plant Growth Regul. 1995;16(1):37–42. [Google Scholar]
- 27.Zhou DX, Bisanz-Seyer C, Mache R. Molecular cloning of a small DNA binding protein with specificity for a tissue-specific negative element within the rps1 promoter. Nucleic Acids Res. 1995;23(7):1165–1169. doi: 10.1093/nar/23.7.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin R, et al. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318(5854):1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitelam GC, Halliday KJ. Light and Plant Devlopment. Blackwell Publishing; Oxford: 2007. [Google Scholar]
- 30.Medzihradszky M, et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell. 2013;25(2):535–544. doi: 10.1105/tpc.112.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore KM, Knauft DA. The inheritance of high oleic acid in peanut. J Hered. 1989;80(3):252–253. [Google Scholar]
- 32.Chan AP, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol. 2010;28(9):951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.López Y, et al. Isolation and characterization of the Δ12-fatty acid desaturase in peanut (Arachis hypogaea L.) and search for polymorphisms for the high oleate trait in Spanish market-type lines. Theor Appl Genet. 2000;101(7):1131–1138. [Google Scholar]
- 34.Ratnaparkhe MB, et al. Comparative and evolutionary analysis of major peanut allergen gene families. Genome Biol Evol. 2014;6(9):2468–2488. doi: 10.1093/gbe/evu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmutz J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463(7278):178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 36.Varshney RK, et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol. 2011;30(1):83–89. doi: 10.1038/nbt.2022. [DOI] [PubMed] [Google Scholar]
- 37.Varshney RK, et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol. 2013;31(3):240–246. doi: 10.1038/nbt.2491. [DOI] [PubMed] [Google Scholar]
- 38.Schmutz J, et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 2014;46(7):707–713. doi: 10.1038/ng.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stalker HT, Dhesi JS, Kochert G. Genetic diversity within the species Arachis duranensis Krapov. &W.C. Gregory, a possible progenitor of cultivated peanut. Genome. 1995;38(6):1201–1212. doi: 10.1139/g95-158. [DOI] [PubMed] [Google Scholar]
- 40.Kochert G, et al. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut, Arachis hypogaea (Legumenosae) Am J Bot. 1996;83(10):1282–1291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.