Significance
We describe dynamics in assortative mating and fertility patterns by polygenic scores associated with anthropometric traits, depression, and educational attainment across birth cohorts from 1920 to 1955. We find that, for example, increases in assortative mating at the phenotypic level for education are not matched at the genotypic level. We also show that genes related to height are positively associated with fertility and that, despite a widening gap between the more and less educated with respect to fertility, there is no evidence that this trend is associated with genes. These findings are important to our understanding of the roots of shifting distributions of health and behavior across generations in US society.
Keywords: assortative mating, fertility, polygenic scores, cohort trends
Abstract
This study asks two related questions about the shifting landscape of marriage and reproduction in US society over the course of the last century with respect to a range of health and behavioral phenotypes and their associated genetic architecture: (i) Has assortment on measured genetic factors influencing reproductive and social fitness traits changed over the course of the 20th century? (ii) Has the genetic covariance between fitness (as measured by total fertility) and other traits changed over time? The answers to these questions inform our understanding of how the genetic landscape of American society has changed over the past century and have implications for population trends. We show that husbands and wives carry similar loadings for genetic factors related to education and height. However, the magnitude of this similarity is modest and has been fairly consistent over the course of the 20th century. This consistency is particularly notable in the case of education, for which phenotypic similarity among spouses has increased in recent years. Likewise, changing patterns of the number of children ever born by phenotype are not matched by shifts in genotype–fertility relationships over time. Taken together, these trends provide no evidence that social sorting is becoming increasingly genetic in nature or that dysgenic dynamics have accelerated.
The traditional view of evolutionary dynamics in humans was that the history of modern humans was too short for the species to have experienced substantive change in its genetic makeup (1, 2). However, findings from recent population genetics studies suggest the possibility that selective fertility, nonrandom mating, drift, and other violations of the Hardy–Weinberg equilibrium accelerated genetic divergence between modern human populations, particularly since humans began farming and civilization developed (3–6). Extending this logic, rapid economic development and the corresponding demographic transition over the past two centuries may have led to a further shift in the dynamics of reproduction and selection. The present paper uses genetic and phenotypic data from a nationally representative sample of US older adults to test whether the societal changes in the United States during the 20th century were reflected in (i) changes in patterns of genetic assortment in marriage, and (ii) changes in genetic influences on fertility.
Understanding trends with respect to specific deviations from random mating and differential fertility is critical to both social and evolutionary scientists. For example, recent research in sociology has suggested that taking a prospective view on social stratification that incorporates differential fertility yields disparate results for estimands such as levels of intergenerational educational mobility (7). Likewise, genetic research on human populations often assumes that mating in a population is random with respect to genotypes (8, 9). Recent empirical evidence suggests otherwise; married couples tend to be more genotypically similar than would be expected by chance (10–12), although questions remain as to how much of this similarity arises from intraethnic marriage (13, 14). The presence of such “assortative mating” on genotypes has implications for the statistical models used in genetic research because genotype distributions will change across generations (15), causing, by extension, changes in phenotypic trait distributions. Here, we evaluate the implications of genotypic assortative mating for traits of interest in population and health sciences.
To investigate trait-related genotypic assortative mating, we studied polygenic scores (PGSs) derived from genome-wide association studies (GWAS) of educational attainment, height, body mass index (BMI), and major depressive disorder (16–19). PGSs are genome-wide summaries of genetic variation associated with a phenotype (20). They are continuous and typically normally distributed, consistent with biometrical estimates of the genetic architecture of complex traits (21). They are also robust predictors of small amounts of phenotypic variance (22).
Assortative genetic mating tells only part of the story with respect to changes in the genetic variance of a population. Differential fertility by genotype also influences the mean levels of a genotype and, consequently, the phenotype. Just as observations have been made regarding the changing patterns of spousal assortment, demographers have documented declines in fertility rates during the 20th century (23). Economic models suggest that the most powerful social (distal) correlates of fertility are child survival and female education, both of which are negatively related to fertility in developed countries (24–28). Meanwhile, twin and molecular genetics studies find that fertility is also influenced by genetic factors (29), although somewhat less so than most complex traits (30, 31). The combination of changes in phenotypic associations with number of children (specifically with respect to years of schooling) along with the documented heritability of fertility suggests that examining the dynamics of the association between specific genetic measures (i.e., PGSs) and number of offspring may be a fruitful avenue of inquiry. One disconcerting possibility is that recent trends in health and illness may be partially driven by negative selection on the genotypic level. This “dysgenic” theory has been framed primarily with respect to cognitive ability (32). To address this question, we test for associations between the same PGSs and number of children ever born and whether such associations are changing over the same time period as our marriage analysis.
Results
We studied pairs of married non-Hispanic white adults participating in the US Health and Retirement Survey (HRS) with DNA samples (n = 4,686 for spousal analysis and n = 8,855 for fertility analysis). We computed PGSs for educational attainment, height, and BMI, and major depressive disorder from genome-wide SNP data using the PLINK software (Materials and Methods). Scores were standardized to have a mean of 0 and a SD of 1 for analysis. Analyses were adjusted for the first 10 principal components estimated from the genome-wide SNP data to account for any residual population structure in the sample.
Deviation from Random Mating.
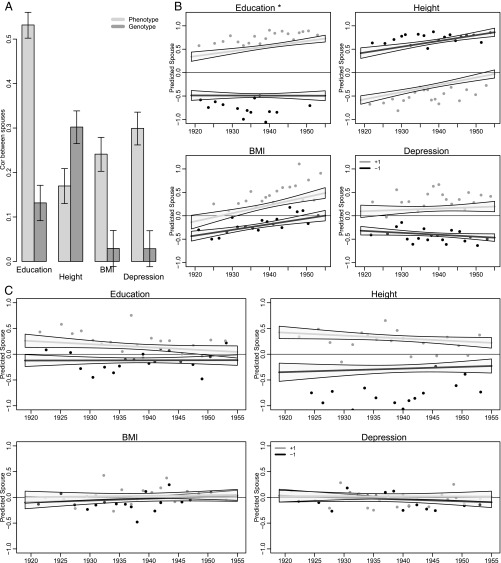
As observed by others, geographic, ethnic, and cultural determinants of spousal assortment declined over the course of the 20th century while assortment on individual attributes increased. In line with these observations, we hypothesized that genetic assortment on the social, mental, and physical characteristics we studied would have increased in tandem with the rising phenotypic assortative mating observed by others (33, 34). We found that husbands and wives in the HRS were similar to each other, both phenotypically and genetically. Fig. 1A shows correlations between husbands and wives across all HRS birth cohorts. These generally correspond to prior estimates (35–37). Fig. 1A also shows a parallel set of results for the PGSs. Although results were directionally similar, only spousal correlations on PGSs for height and educational attainment were statistically different from 0 (for height PGS, r = 0.302 [0.265, 0.339]; for educational attainment PGS, r = 0.132 [0.0917, 0.171]).
Fig. 1.
Spousal associations for both standardized phenotypes and standardized polygenic risk scores among spousal pairs in the HRS, 2012 (n = 4,686; restricted to respondents in their first marriage who have genotypic data and valid phenotypic responses). (A) All birth cohorts pooled. (B and C) Trends in spousal correspondence across birth cohorts. The horizontal axis depicts birth cohort, whereas the vertical axis is the predicted value for the spouse of a focal individual conditional on the focal individual’s birth year and either phenotype or PGS (Eq. 1). The lines show fitted values for those at 1 SD above (gray) and below (black) the mean. Points are based on binned means for two groups of respondents (standardized value below −1, black; standardized value above 1, dark gray). For each group, the distribution of birth years is divided into 20 subgroups with approximately equal numbers. Plotted points are the mean birth year and response for these subgroups. B considers standardized phenotypes. Education demonstrates a change in spousal correlation across birth cohorts. Consider education in B: an individual with relatively low education is predicted to have a spouse of consistently low education across all birth cohorts. In contrast, a high-education individual will have, on average, a spouse with higher education in later birth cohorts compared with earlier birth cohorts. For height, the fact that relatively short individuals are predicted to marry relatively tall individuals is a consequence of the fact that we are looking at opposite sex pairs. C considers standardized PGSs. In contrast to results for phenotypes, spousal correlations in education PGS display reductions across 20th century birth cohorts as do those for height, although these results do not appear significant at conventional α levels.
Fig. 1 B and C represents the test of our main research question: whether phenotypic (B) and genetic (C) correlations among spouses changed from birth cohorts born earlier in the 20th century to those born toward the middle of the 20th century (Eq. 1). We examine the birth cohort of the nonfocal spouse—i.e., the spouse on the right-hand side of Eq. 1 (38). Using this assignment, the similarity of spouse’s education to one’s own education was higher in the later-born compared with the earlier-born cohorts. No similar change was observed for PGS correlations. For education and height, there is some sign of decrease in spousal correlation, although these results are not statistically significant. We suspect that this failure to obtain statistically significant results is largely due to a lack of power related to the measurement error in the PGSs as adjustments to correct for this problem suggest even more pronounced declines (SI Appendix, Text S5).
We also conducted additional sensitivity analyses. First, we tested whether mortality selection might bias the distribution of PGSs in our sample of married couples from the earliest-born cohorts. We compared PGS distributions between the married couples in our analysis sample and birth cohort-matched HRS respondents whose partners had died. Distributions were similar between the married couples we studied and birth cohort-matched HRS respondents whose spouses had died (SI Appendix, Text S2). Next, we tested for changes over the 20th century in genome-wide SNP-level assortative mating—i.e., using a method parallel to previous genotypic assortative mating analyses (12). We found no evidence for such a change. Thus, our results from the PGS analysis are not likely to be confounded by changes in broader patterns of genotypic assortment unrelated to the traits we studied (SI Appendix, Text S3). Additionally, we tested for population stratification bias resulting from changes in coethnic marriage. The original spousal relatedness analysis attempted to account for ancestry differences among the non-Hispanic white husbands and wives in the sample by adjusting for the first 10 principal components estimated from the genome-wide SNP data. These principal components are thought to approximate ancestry differences that have genome-wide effects on allele frequencies. Such differences would bias PGSs and might inflate spousal correlations in the case of coethnic marriage. We tested for such a possibility by repeating our analysis without adjustment for the principal components. Results were unchanged (SI Appendix, Text S4). Finally, analyses based on standardizing height within gender produce the same pattern.
Differential Fertility.
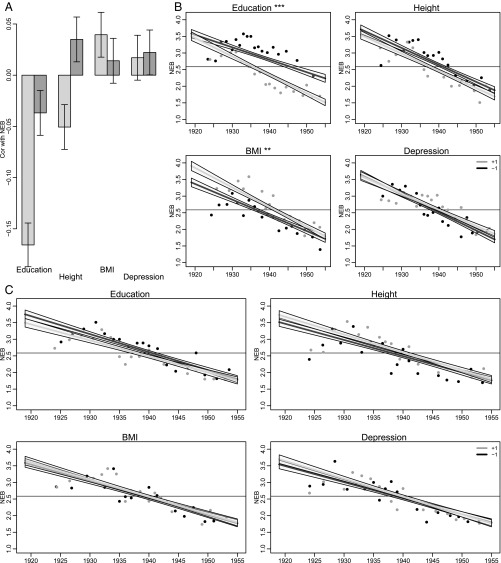
Fig. 2A shows that, of the four studied phenotypes, education, height, and BMI show an association with number of children that is significant at the P < 0.05 level (all of which are also significant at a Bonferroni-corrected α level of P < 0.0125). When we examine the genotypic associations, we find that education and height show statistically significant associations that are below Bonferroni-corrected P value thresholds: the education PGS shows a very small but statistically significant association with fewer children, whereas a PGS predicting higher stature is positively associated with number of children. (The depression PGS is positively associated with fertility at a P value that is conventionally significant but does not survive Bonferroni adjustment.) With respect to trends in the genetics–fertility relationship, we hypothesized that physical phenotypes—such as height and BMI—and their associated PGSs would display a declining association with number of children ever born, whereas behavioral traits would evince stronger associations in younger birth cohorts at both the phenotypic and genotypic levels. We make this prediction based on the shifting nature of the economy from an industrial, manual one to a knowledge-based, postindustrial one over the course of the mid- to late-20th century (39). That is, what traits confer advantage in the reproductive market may have changed to match what characteristics are increasingly rewarded by the labor market (i.e., cognitive ability over physical attributes).
Fig. 2.
Association of selected phenotypes and corresponding PGSs with fertility. (A) Overall association with number of children ever born for all birth cohorts. (B and C) Birth cohort differences in associations between number of children ever born between both standardized phenotypes and standardized polygenic risk scores among non-Hispanic whites in the HRS, 2012 (n = 8,855; restricted to respondents who have genotypic data and valid phenotypic responses). The horizontal axis depicts birth cohort, whereas the vertical axis is the predicted number of offspring conditional on an individual’s birth year and either phenotype or PGS (Eq. 2). The lines show fitted values for those at 1 SD above (gray) and below (black) the mean. The horizontal line shows the mean number of offspring in the sample. Points are based on binned means for two groups of respondents (standardized value below −1, black; standardized value above 1, dark gray). For each group, the distribution of birth years is divided into 20 subgroups with approximately equal numbers. Plotted points are the mean birth year and response for these subgroups. B considers standardized phenotypes. The number of predicted offspring is lower for later birth cohorts. One important observation is that this decrease in the number of offspring is driven by the more educated. C considers standardized PGSs. The number of offspring does not appear to be changing as a function of PGSs over the birth cohorts.
The most marked feature of the fertility data are a secular decline in the number of offspring over the observed period such that those born in 1919 are predicted to have 3.6 children, whereas those born in 1955 are predicted to have 1.7 children (the overall mean is 2.6 children; SI Appendix, Table S1). This decline matches patterns documented by prior researchers (40). These trends are reflected in Fig. 2B by the generally negative widening gaps in the number of predicted offspring for those with high and low values on the associated phenotypes in more recent cohorts (with the exception of BMI, which appears to have a lessening effect on number of children in more recent cohorts).
For example, although the less educated respondents in the population have a fairly stable number of offspring over the birth cohorts, those with greater observed (i.e., phenotypic) education levels have fewer children over time. A similar pattern can be observed for height where only in more recent birth cohorts do we see those with higher stature having fewer children. Both of these phenotypic trends would seem to imply dynamics of emergent or strengthening dysgenic reproductive patterns. However, when we look at the relevant genetic scores in Fig. 2C, we find that the dysgenic trends inferred from phenotypic associations between education and height on the one hand, and fertility on the other, are not present with respect to the genotypic data. [We show in SI Appendix, Text S5, that this pattern of results is not likely due to the following: measurement error of the PGS on the right-hand side, misspecification of number of children as a normal rather than count (i.e., Poisson-distributed) variable, or the missing data present in this sample (SI Appendix, Table S14).] Although we may recall from Fig. 2A that the PGSs for height and education predict number of children ever born in the overall sample, these associations are consistent across all birth cohorts in this study. Thus, although there may be positive selection on height and slight negative selection on additive measures of the genetic architecture of education, these are not accelerating (32).
Discussion
The change from an agrarian society to an industrial and postindustrial one has been well-noted (41). This change, along with others, resulted in dramatic shifts in the environments encountered by humans during the course of the 20th century. Expansion of schooling (42), medical improvements (43), increased longevity (44), and caloric abundance are just some of the changes that may influence not only relationships between important phenotypes but the underlying salience of their associated genotypes as well. In the present paper, we demonstrate that observed changes in mating preferences or fertility associations do not always correspond to shifts in the underlying genetic architecture. For example, whereas higher educational attainment has come to predict having fewer children in more recent birth cohorts, the same is not true of genetics correlated with educational attainment—i.e., the association between a person’s PGS for educational attainment and their fertility has remained constant across birth cohorts. Likewise, the spousal correlation in educational phenotype has been increasing even as the sorting on measured polygenic predictors of education has been flat (if not decreasing).
Contrary to worries about negative selection and/or increased polarization on education-related genetic measures, we predict—based on the results here—that any dysgenic dynamics with respect to education are not, in fact, increasing even as phenotypic associations strengthen. Although the environmental landscape of US society is certainly changing, the genetic makeup of the population may also be shifting along with it—although possibly in conflicting directions. Future researchers examining cohort trends in genetic influences—under the assumption that these are entirely driven by changing environmental conditions while genetic variance is constant—would be wise to reexamine that assumption (45–48).
In addition to informing our knowledge about the changing genetic and phenotypic landscape of marriage and reproduction, knowing the degree to which spouses are correlated on phenotypically predictive genetic measures as opposed to merely observed phenotype has important implications for models of additive heritability that rely on an assumption of random mating (49). Namely, most heritability models—notably the classic twin or extended twin design—assume that siblings (i.e., fraternal twins) share, on average, 50% of the relevant genetic markers that are associated with the phenotype of interest. If there is significant positive assortative mating on the relevant, underlying genetic measures, then heritability is underestimated (even if overall genome-wide assortative mating is nil). Prior attempts to relax that assumption have operationalized assortative mating through the phenotypic correlation among parents, but this assumes that phenotypic correlations can act as accurate proxies for genotypic correlations, the true parameter of interest. Our results suggest the opposite: stable or potentially decreased genetic assortment on genetic measures linked to education in the face of increased phenotypic assortment. Likewise, differential fertility by genotype affects the likelihood of parents being included in any studies of intergenerational transmission—genetic or social—due to the practice of sampling on offspring in most retrospective studies or the exclusion of childless individuals from prospective studies of parent–child pairs. That is, if a given genetic measure is pleiotropic for affecting an outcome of interest—say education—as well as fertility, then estimating its effects based on a sample of living offspring (as is typically done) will yield substantially different results than sampling on the parental generation and allowing for nonfertile members of that generation to remain in the analysis and modeling offspring education conditional on being born at all.
Materials and Methods
We tested changes during the 20th century in genetic influences on assortative mating and fertility. We used data from spousal pairs in the HRS, a nationally representative survey of adults born during the first half of the 20th century, 1919–1955, and their partners. We analyzed data from non-Hispanic whites (due to concerns regarding population stratification) with available genome-wide SNP genotype and phenotype data. We included only spousal pairs in their first marriage. Our sample consisted of n = 2,343 pairs. (Details on sample characteristics and selection analysis are reported in SI Appendix, Text S1.) Phenotype measurements consisted of self-assessments of height and weight, the CES-D scale score, and self-reports of the number of years of education completed (2012 or otherwise most recent wave with available data):
• Number of children ever born (NEB): Maximum number of children reported ever born to or fathered by an individual (waves 3–11). This information was missing for 871 respondents. Despite being a count variable, it displays a normal distribution as shown in SI Appendix, Fig. S1.
• Education: Total years of educational attainment.
• BMI: Mean BMI over all available waves.
• Height: Maximum height over all available waves.
• Depression: Mean CES-D score over all available waves. This variable had a skewed distribution, so it was transformed via the logarithm (after adding 1 to everyone’s score).
Sample descriptives are shown in SI Appendix, Table S1. We calculated PGSs for each participant for height, BMI, major depression, and educational attainment based on published results from GWAS consortia (16–19) using the methods described below. The HRS collected genotype information from consenting subjects in 2006 and 2008 and assayed with the Illumina 2.5 Human Omni Quad Array.
PGSs.
PGSs were calculated using the PLINK software and published GWAS results (16–19). Briefly, polygenic scoring was done with the PLINK software (50). SNPs in the HRS genetic database were matched to SNPs with reported results in a GWAS. For each SNP, a loading was calculated as the number of phenotype-associated alleles multiplied by the effect-size estimated in the original GWAS. Loadings were summed across SNPs to calculate the PGS. Finally, scores were residualized for the first 10 principal components estimated from the genome-wide SNP data using PLINK (51). Residuals were calculated to eliminate variance attributable to ancestry (although analysis of raw scores does not change the pattern of results; SI Appendix, Text S4). Scores were standardized to have a mean of 0 and SD of 1 for analysis for ease of interpretation. We studied PGSs because they are the best available method to summarize molecular genetic predisposition to a complex trait; however, PGSs have limitations. For example, they do not capture nonadditive combinations of genetic influences. As a result, PGSs capture only a fraction of genetic influences on complex traits. Our analysis is therefore a preliminary observation of population dynamics in distributions of genetic influences on complex traits, not a comprehensive summary of them. As large-scale and more sophisticated GWAS uncover additional components of genetic influence on complex traits, this science will evolve.
Phenotypes.
Phenotypes were computed based on RAND Fat Files, version N (up to wave 11 in 2012).
Statistical Models.
We used two statistical models. The first model tested whether genetic assortative mating has changed over time. We estimated equations of the following form:
| [1] |
where each spouse in pair i is double-entered on both sides of the equation. Three separate versions of this equation were used focusing on spousal phenotype, PGS, and principal component ( and represented standardized phenotypes, PGSs, or principal components in the three respective versions of this equation). We used Huber–White SEs to correct for the nonindependence of spousal pairs.
The second model investigated changes in fertility. We estimated equations of the following form:
| [2] |
where i now indexes individuals, is the number of ever-born children reported by individual i, and the now represents either an individual’s phenotype or PGS. In reporting results, we show conventional α levels; however, we also indicate whether our results achieve statistical significance using Bonferroni-corrected α levels for four independent tests (because we have four phenotypes and associated PGSs). That said, our argument largely rests on the nonsignificance of genetic patterns of change over time in light of evident phenotypic trends during the same time period.
Human Subjects.
The analysis plan was reviewed by the Institutional Review Board of New York University and deemed exempt human subjects research. Genetic data were accessed via the National Center for Biotechnology Information Genotypes and Phenotypes Database Data Access Request system at the National Institutes of Health (Project 2260).
Supplementary Material
Acknowledgments
This work was supported by Russell Sage Foundation Grant “GxE and Health Inequality over the Life Course.” This research uses data from the HRS, which is sponsored by National Institute on Aging Grants NIA U01AG009740, RC2AG036495, and RC4AG039029, and conducted by the University of Michigan. Research was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) of the National Institutes of Health (NIH) under Award R21HD078031. We also acknowledge cofunding from the NICHD and the Office of Behavioral and Social Sciences Research (Grant 1R21HD071884). Further support was provided by the NIH/NICHD-funded University of Colorado Population Center (Grant R24HD066613). D.W.B. is an Early Career Fellow of the Jacobs Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523592113/-/DCSupplemental.
References
- 1.Lewontin R. The apportionment of human diversity. In: Dobzhansky T, Hecht MK, Steere WC, editors. Evolutionary Biology. Springer; New York: 1972. pp. 381–398. [Google Scholar]
- 2.Gould SJ. Biological potential vs. biological determinism. Nat Hist. 1976;85(5):12–22. [PubMed] [Google Scholar]
- 3.Bulayeva KB, et al. Ethnogenomic diversity of Caucasus, Daghestan. Am J Hum Biol. 2006;18(5):610–620. doi: 10.1002/ajhb.20531. [DOI] [PubMed] [Google Scholar]
- 4.Laland KN, Odling-Smee J, Myles S. How culture shaped the human genome: Bringing genetics and the human sciences together. Nat Rev Genet. 2010;11(2):137–148. doi: 10.1038/nrg2734. [DOI] [PubMed] [Google Scholar]
- 5.Shen P, et al. Population genetic implications from DNA polymorphism in random human genomic sequences. Hum Mutat. 2002;20(3):209–217. doi: 10.1002/humu.10117. [DOI] [PubMed] [Google Scholar]
- 6.Mathieson I, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528(7583):499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mare RD, Maralani V. The intergenerational effects of changes in women’s educational attainments. Am Sociol Rev. 2006;71(4):5542–5564. doi: 10.1177/000312240607100402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th Ed Pearson; Essex, UK: 1996. [Google Scholar]
- 9.Neale MC, Cardon LR. Methodology for Genetic Studies of Twins and Families. Kluwer Academic; Dordrecht, The Netherlands: 1992. [Google Scholar]
- 10.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visscher PM, et al. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2006;2(3):e41. doi: 10.1371/journal.pgen.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domingue BW, Fletcher J, Conley D, Boardman JD. Genetic and educational assortative mating among US adults. Proc Natl Acad Sci USA. 2014;111(22):7996–8000. doi: 10.1073/pnas.1321426111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdellaoui A, Verweij KJH, Zietsch BP. No evidence for genetic assortative mating beyond that due to population stratification. Proc Natl Acad Sci USA. 2014;111(40):E4137. doi: 10.1073/pnas.1410781111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domingue BW, Fletcher JM, Conley D, Boardman JD. Reply to Abdellaoui et al.: Interpreting GAM. Proc Natl Acad Sci USA. 2014;111(40):E4138. doi: 10.1073/pnas.1413105111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harpending HC. The population genetics of interactions. Am Nat. 1979;113(4):622–630. [Google Scholar]
- 16.Locke AE, et al. LifeLines Cohort Study ADIPOGen Consortium AGEN-BMI Working Group CARDIOGRAMplusC4D Consortium CKDGen Consortium GLGC ICBP MAGIC Investigators MuTHER Consortium MIGen Consortium PAGE Consortium ReproGen Consortium GENIE Consortium International Endogene Consortium Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood AR, et al. Electronic Medical Records and Genomics (eMEMERGEGE) Consortium MIGen Consortium PAGEGE Consortium LifeLines Cohort Study Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rietveld CA, et al. LifeLines Cohort Study GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340(6139):1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ripke S, et al. Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18(4):497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17(10):1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nat Rev Genet. 2009;10(12):872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 22.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9(3):e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bongaarts J. Household size and composition in the developing world in the 1990s. Popul Stud (Camb) 2001;55(3):263–279. [Google Scholar]
- 24.Lutz W, Kc S. Global human capital: Integrating education and population. Science. 2011;333(6042):587–592. doi: 10.1126/science.1206964. [DOI] [PubMed] [Google Scholar]
- 25.Herzer D, Strulik H, Vollmer S. The long-run determinants of fertility: One century of demographic change 1900-1999. J Econ Growth. 2012;17(4):357–385. [Google Scholar]
- 26.Vogl TS. Differential fertility, human capital, and development. Rev Econ Stud. 2015 doi: 10.1093/restud/rdv026. [DOI] [Google Scholar]
- 27.Clark G. Human capital, fertility, and the industrial revolution. J Eur Econ Assoc. 2005;3(2):505–515. [Google Scholar]
- 28.Soares RR. Mortality reductions, educational attainment, and fertility choice. Am Econ Rev. 2005;95(3):580–601. doi: 10.1257/0002828054201486. [DOI] [PubMed] [Google Scholar]
- 29.Kohler HP, Rodgers JL, Christensen K. Between nurture and nature: The shifting determinants of female fertility in Danish twin cohorts. Soc Biol. 2002;49(3-4):218–248. doi: 10.1080/19485565.2002.9989060. [DOI] [PubMed] [Google Scholar]
- 30.Tropf FC, Barban N, Mills MC, Snieder H, Mandemakers JJ. Genetic influence on age at first birth of female twins born in the UK, 1919–68. Popul Stud (Camb) 2015;69(2):129–145. doi: 10.1080/00324728.2015.1056823. [DOI] [PubMed] [Google Scholar]
- 31.Tropf FC, et al. Human fertility, molecular genetics, and natural selection in modern societies. PLoS One. 2015;10(6):e0126821. doi: 10.1371/journal.pone.0126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisenberg G. The reproduction of intelligence. Intelligence. 2010;38(2):220–230. [Google Scholar]
- 33.Schwartz CR. Trends and variation in assortative mating: Causes and consequences. Annu Rev Sociol. 2013;39:451–470. [Google Scholar]
- 34.Schwartz CR. Earnings inequality and the changing association between spouses’ earnings. AJS. 2010;115(5):1524–1557. doi: 10.1086/651373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alford JR, Hatemi PK, Hibbing JR, Martin NG, Eaves LJ. The politics of mate choice. J Polit. 2011;73(2):362–379. [Google Scholar]
- 36.Gualtieri CT. Husband-wife correlations in neurocognitive test performance. Psychology (Irvine) 2013;4(10):771–775. [Google Scholar]
- 37.Watson D, et al. Match makers and deal breakers: Analyses of assortative mating in newlywed couples. J Pers. 2004;72(5):1029–1068. doi: 10.1111/j.0022-3506.2004.00289.x. [DOI] [PubMed] [Google Scholar]
- 38.Heeringa SG, Connor JH. Technical Description of the Health and Retirement Survey Sample Design. Institute for Social Research; Ann Arbor, MI: 1995. [Google Scholar]
- 39.Goldin C, Katz LF. The Race Between Education and Technology. Harvard Univ Press; Cambridge, MA: 2009. [Google Scholar]
- 40.Rindfuss RR, Sweet JA. Postwar Fertility Trends and Differentials in the United States. Elsevier; New York: 2013. [Google Scholar]
- 41.Lobao L, Meyer K. The great agricultural transition: Crisis, change, and social consequences of twentieth century US farming. Annu Rev Sociol. 2001;27(1):103–124. [Google Scholar]
- 42.Lleras-Muney A. The relationship between education and adult mortality in the United States. Rev Econ Stud. 2005;72(1):189–221. [Google Scholar]
- 43.Cutler D, Deaton A, Lleras-Muney A. The determinants of mortality. J Econ Perspect. 2006;20(3):97–120. [Google Scholar]
- 44.Bongaarts J, Feeney G. How long do we live? Popul Dev Rev. 2002;28(1):13–29. [Google Scholar]
- 45.Liu H, Guo G. Lifetime socioeconomic status, historical context, and genetic inheritance in shaping body mass in middle and late adulthood. Am Sociol Rev. 2015;80(4):705–737. doi: 10.1177/0003122415590627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domingue BW, Conley D, Fletcher J, Boardman JD. Cohort effects in the genetic influence on smoking. Behav Genet. 2016;46(1):31–42. doi: 10.1007/s10519-015-9731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenquist JN, et al. Cohort of birth modifies the association between FTO genotype and BMI. Proc Natl Acad Sci USA. 2015;112(2):354–359. doi: 10.1073/pnas.1411893111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boardman JD, Blalock CL, Pampel FC. Trends in the genetic influences on smoking. J Health Soc Behav. 2010;51(1):108–123. doi: 10.1177/0022146509361195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conley D, Rauscher E, Dawes C, Magnusson PKE, Siegal ML. Heritability and the equal environments assumption: Evidence from multiple samples of misclassified twins. Behav Genet. 2013;43(5):415–426. doi: 10.1007/s10519-013-9602-1. [DOI] [PubMed] [Google Scholar]
- 50.Chang CC, et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.