Abstract
Spectraplakins are crucially important communicators, linking cytoskeletal components to each other and cellular junctions. Microtubule actin crosslinking factor 1 (MACF1), also known as actin crosslinking family 7 (ACF7), is a member of the spectraplakin family. It is expressed in numerous tissues and cells as one extensively studied spectraplakin. MACF1 has several isoforms with unique structures and well-known function to be able to crosslink F-actin and microtubules. MACF1 is one versatile spectraplakin with various functions in cell processes, embryo development, tissue-specific functions, and human diseases. The importance of MACF1 has become more apparent in recent years. Here, we summarize the current knowledge on the presence and function of MACF1 and provide perspectives on future research of MACF1 based on our studies and others. [BMB Reports 2016; 49(1): 37-44]
Keywords: Function, Isoform, MACF1, Structure, Spectraplakin
INTRODUCTION
Cytoskeleton in most multicellular organisms is a highly organized and interconnected network of filaments composing of microfilaments (F-actin), microtubules (MTs), and intermediate filaments (IFs). Cytoskeletal network acts as a cytoplasmic scaffold, defining the cell shape, cell mechanical properties, and functions of many other cellular events, including cell polarization, migration, division, adhesion, intracellular trafficking, and organelle locomotion. Thus, cytoskeleton is both static and adaptive, providing a dynamic cellular architecture that can remodel itself to respond to cellular changes. The tight organization and dynamics of these filaments strongly depends on their associated proteins. It has been demonstrated that spectraplakins are cytoskeletal crosslinkers with ability of interacting with all three types of cytoskeletal filaments, i.e., F-actin, MTs, and IFs (1).
Spectraplakins are exceptionally long and gigantic with molecular weight of >500 kD. They are multi-domain cytoskeletal proteins and master orchestrators for coordinating cytoskeletal elements by binding to F-actin, MTs, and IFs (1, 2). Spectraplakins are evolutionarily conserved. They belong to both spectrin and plakin superfamilies (3). “Spectraplakins” are named as combinations of “spectrin” and “plakin” because they share features of both spectrins and plakins (3, 4). So far, mammalian spectraplakins have two members: microtubule actin crosslinking factor 1 (MACF1) and bullous pemphigoid antigen 1 (BPAG1). The spectraplakin family also consists of Shot/Kakapo (in Drosophila), Magellan (in zebrafish), and Vab-10 (in Caenorhabditis elegans).
MACF1, also known as actin crosslinking family 7 (ACF7), MACF, macrophin, and trabeculin-α, is a widely expressed critical spectraplakin (1). By crosslinking both MTs and F-actin and by controlling the cytoskeletal dynamics, MACF1 plays key roles in the regulation of cell migration (5-8) and cell proliferation and the maintenance of tissue integrity (9, 10). In addition, MACF1 mediates signal transduction pivotal for embryo development (11). More recently, a human disease caused by MACF1 mutation has been reported (12).
In this paper, basic molecular characteristics of MACF1 are introduced and current knowledge on MACF1 is reviewed by highlighting its versatile functions. In addition, perspectives on future research of MACF1 are provided based on studies by us and others to provide researchers the current status of MACF1 research.
MACF1: GENE, ISOFORM, AND TISSUE DISTRIBUTION
Gene and isoforms of MACF1
MACF1, a large protein with molecular weight of ∼600 kD, was first discovered by Byers et al. in the effort to screen for additional members of the actin crosslinker superfamily (13). They isolated a partial cDNA of MACF1 using degenerate primer-mediated PCR and named it ACF7. Subsequently, murine ACF7 was further characterized (14) and its full-length cDNA encoding a 608-kD protein was cloned (15). Due to the association of murine ACF7 with both actin and MTs, ACF7 was renamed MACF to represent microtubule actin cross-linking factor. Meanwhile, human cDNA was cloned independently by two groups and named macrophin and trabeculin-α, respectively (16, 17).
MACF1/ACF7 is encoded by MACF1 gene located on human chromosome 1p32 or mouse chromosome 4 (13, 14, 18). Human MACF1 gene contains at least 102 exons and spans over 270 kb. Genomic organization analysis of human MACF1 gene has demonstrated that MACF1 and ACF7 are the same gene (18). MACF1 gene is a hybrid of genes encoding plakins and spectrins/dystrophins (15, 18). MACF1 shares similar gene sequences with plectin, a well-characterized plakin. It has the same exon-intron boundaries for N-terminal actin-binding domain (ABD), one large exon encoding plectin repeats, and a similar serine/glycine-rich C-terminus containing GSR repeats. In addition, the spectrin repeats in MACF1 are similar to those of dystrophin gene. This hybrid gene structure of MACF1 reveals that it is related to Drosophila gene shot and Caenorhabditis elegans gene vab-10. Due to the hybrid structure of these genes and their combined features of both spectrin and plakin family members, they were assigned to the spectraplakin family (3).
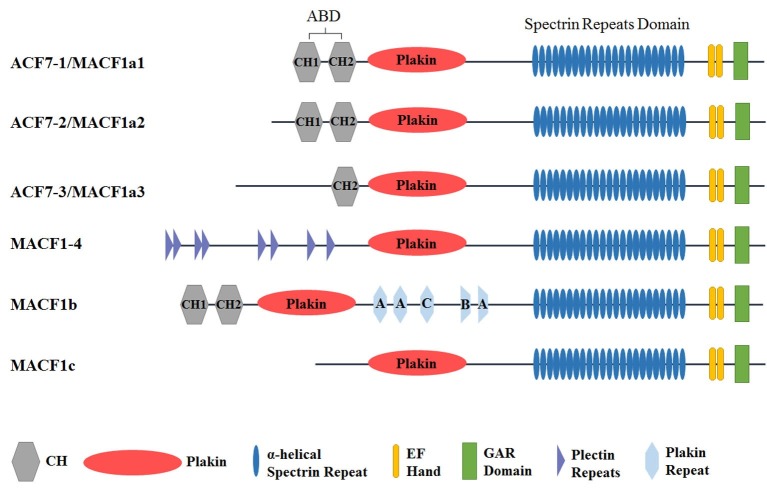
Like dystonin/BPAG1 (19, 20), MACF1 displays isoform diversity. Several MACF1 isoforms (Fig. 1) produced by alternative splicing and different promoter usage have been reported. Three murine ACF7/MACF1 isoforms with a common 3’ sequence but a unique 5’ region were first identified by Bernier et al. (14). Isoforms ACF7-1 and ACF7-2 contain identical actin-binding domains (ABDs) but different 5’UTRs, while isoform ACF7-3 contains a unique 5’UTR and a longer N-terminal sequence with just the second half of the ABD (14). Subsequently, a fourth isoform MACF1-4 was cloned (18). Different from the three MACF1 isoforms mentioned above, MACF1-4 has no ABD at the N-terminus. Instead, it has plectin repeats (Fig. 1). Later, another alternatively spliced murine isoform of MACF1 encoding a gigantic protein of approximately 800-kD was revealed (21). It was named MACF1b with the original isoform renamed as MACF1a. MACF1b harbors extra plakin repeats between the plakin domain and the spectrin repeats of MACF1a (Fig. 1). With different N-terminal domains, the first three isoforms ACF7-1, ACF7-2, and ACF7-3 were renamed as MACF1a1, MACF1a2, and MACF1a3, respectively (21). While studying the role of MACF1 in the nervous system by adopting MACF1a conditional knockout (cKO) mice, a novel isoform MACF1c was found by Goryunov et al. (22). MACF1c is identical to MACF1a except that it has no N-terminal ABD domain (Fig. 1).
Fig. 1. Isoforms of MACF1 with a combination of different unique domain structure. There are seven main types of functional domains, including an actin binding domain (ABD) with CH1 and CH2 fragments, a plakin domain, a spectrin repeats rod domain with 23 α-helical spectrin repeats, two EF hand motifs, a GAR domain, plectin repeats domain, and plakin repeats domain (PRD). A, B, C represents different types.
In addition, a putative MACF/MACF1 isoform MACF2 has been reported by Sun et al. (23). Based on sequence homology analysis, a partial human cDNA clone (KIAA0728) encoding a polypeptide sharing 68% amino acid identities with human MACF was found and called MACF2. However, they found differences in chromosomal locations and nucleotide sequences between MACF and MACF2 transcripts based on unigene database, suggesting that human MACF and MACF2 represent two distinct protein products from two different genes (23). Consequently, MACF2 was demonstrated to be an isoform of BPAG1 and renamed as BPAG1-a (20). Therefore, up to date, a total of six isoforms of MACF1 (MACF1a1, MACF1a2, MACF1a3, MACF1-4, MACF1b, and MACF1c, Fig. 1) have been reported.
MACF1’s tissue distribution
As a cytoskeletal linker protein, MACF1 is ubiquitously expressed in different tissues with some isoforms having different distributions (Table 1).
Table 1. The isoforms of MACF1.
| Isoform | Tissue distribution | Domains | References |
|---|---|---|---|
|
| |||
| ACF7-1/MACF1a1 | Broadly expressed with predominance in skin, kidney, and stomach | ABD (CH1, CH2), Plakin, Spectrin repeats, EF hand, and GAR. | (14,21,24) |
| ACF7-2/MACF1a2 | Broadly expressed with high level in brain, spinal cord, and lung | ABD (CH1, CH2), Plakin, Spectrin repeats, EF hand, and GAR. Different 5’ UTR with MACF1a1. | (14,16,21,24) |
| ACF7-3/MACF1a3 | Predominant in brain and spinal cord | Half ABD (CH2), Plakin, Spectrin repeats, EF hand, and GAR. | (14,21) |
| MACF1-4 | Broadly expressed with high level in heart, lung, pituitary gland and placenta | Plectin repeats, Plakin, Spectrin repeats, EF hand, and GAR. | (18) |
| MACF1b | Broadly expressed | ABD (CH1, CH2), Plakin, Plakin repeats, Spectrin repeats, EF hand, and GAR. | (21) |
| MACF1c | Nervous system | Plakin, Spectrin repeats, EF hand, and GAR. | (22) |
Bernier et al. were the first to show that MACF1 was widely expressed in postnatal mouse tissues, including brain, spinal cord, spleen, liver, heart, skeletal muscle, stomach, lung, kidney, and skin, with the strongest expression in the lung, followed by brain, spinal cord, cardiac/skeletal muscle, and skin (14). They further demonstrated a predominant expression of MACF1 in neural, muscle, and lung tissue at the beginning of embryonic development which was continued into adulthoods (24). A striking difference in tissue distribution for different MACF1 transcripts has been observed. Relatively higher levels of MACF1a2 have been detected in the brain, spinal cord, and lung, while relatively lower levels of MACF1a2 have been found in the kidney, heart, and skeletal muscles without any detection in the skin, liver, stomach, or the spleen. In contrast, MACF1a1 is predominantly found in the skin, kidney, and stomach. Similarly, MACF1a1 mRNA is detected in embryos from day 7.5 to day 10.5, whereas MACF1a2 mRNA becomes detectable only at day 10.5. MACF1a3 is predominantly detected in the brain and spinal cord. Moderate levels of MACF1a3 has been found in the skin, lung, and kidney without expression in the heart, skeletal muscle, or the liver (24). MACF1b is expressed in all tissues and throughout the development of mouse embryo (21).
MACF1 is widely expressed in human tissues, including pituitary, adrenal, thyroid, salivary gland, mammary glands, pancreas, heart, and skeletal muscle at different levels (13, 16, 17). MACF1a2 is highly expressed in the brain, heart, lung placenta, liver, kidney, and pancreas (16), while the hybridization signals of MACF1-4 are visible in all tissues, with the strongest signals in the heart, lung, pituitary gland, and placenta (18).
UNIQUE DOMAIN STRUCTURE OF MACF1
Generally, the structure of MACF1 contains three main domains: an N-terminal domain containing an ABD and a plakin, a rod domain composed of spectrin repeats, and a C-terminal domain consisting of EF-hand calcium-binding domain and a GAS2-related protein (GAR) domain (23). Seven types of functional domains presenting different combination in different isoforms (Fig. 1) have been found in MACF1.
Actin-binding domain
MACF1 has an ABD located at the N-terminus, a conserved structure among spectraplakins inherited from members of the spectrin superfamily (15, 25). ABD consists of two calponin homology (CH) domains CH1 and CH2 that can bind F-actin and enable spectraplakins to interact directly with the actin cytoskeleton. CH1 alone can bind to actin. CH2 exerts a weaker binding affinity for actin. However, CH1-CH2 tandem domain has high binding affinity (26, 27). Not all MACF1 isoforms have CH1 and CH2 domains. MACF1a3 only has CH2 domain while MACF1-4 and MACF1c have no CH domain (Fig. 1). Regardless of the CH domain, the interaction of MACF1 with F-actin is directly affected (15, 25). Although the crystal structure of MACF1 has not been reported yet, structure analysis of ABD in dystrophin and utrophin spectrins has demonstrated an interdomain linker between the CH1 and CH2 domains, with the interdomain linker primarily determining the structural stability of tandem CH domains (28).
Plakin domain
Following the ABD at the N-terminal of MACF1, there is a plakin domain (23). The plakin domain is also a feature of members of the plakin superfamily (3). It is characterized by a high α-helical content (29). Structure analysis of BPAG1 and plectin has revealed a number of spectrin repeats in the plakin domain (30, 31), suggesting that the plakin domain is derived from spectrin repeats. The plakin domain is responsible for binding to various adhesion and signaling molecules (e.g. β4 integrin, BPAG2, and Erbin) (32-35). It plays a role in the mediation of the binding to cell junctions (29). However, the structure and the function of the plakin domain in MACF1 require further investigation.
Spectrin repeats
MACF1 contains both the spectrin repeats and the plakin domain. It is classified as a spectraplakin. Spectrin repeats are the typical architecture of the spectrin family. MACF1 has 23 dystrophin-like spectrin repeats (15). Each repeat contains 110∼120 residues that are folded into three α helices. These helices then form an antiparallel three-helical coiled coil (36, 37). These α-helical spectrin repeats then form an extended rod-like structure and act as a spacer region that separates different functional domains at the N- and C-termini. In addition, these spectrin repeats endow the protein MACF1 with flexibility (1, 3).
EF hand
MACF1 contains two EF-hand calcium-binding motifs located at the C-terminus that are conserved within the spectrin superfamily (23). Sun et al. have indicated that the EF-hand motifs of MACF1 have no effect on the ability of MACF1 to interact with MTs (23).
GAS2-related protein (GAR) domain
Following the EF-hand motifs, there is a GAR domain located at the C-terminal of MACF1 (Fig. 1). The GAR domain is restricted to spectraplakins. It can be used to distinguish spectraplakins from spectrins and plakins (3). The GAR domain is the homology region shared between Gas2 (growth arrest-specific protein 2) and GAR22 (Gas2 related on chromosome 22) proteins. It has the function to bind and stabilize MTs (15, 23).
Plakin repeat domain (PRD)
PRD is unique to members of the plakin family. It only exists in the MACF1b isoform (21). Based on well-characterized A, B, and C type PRDs in desmoplakin (38, 39), Lin et al. have revealed five PRDs in MACF1b, including two A type, one C type, one incomplete B type, and one incomplete A type (21). They have further demonstrated that PRD targets MACF1b to the Golgi complex via its N-terminal portion of two consecutive A-type subdomains.
Plectin repeats
Plectin repeats are unique to the MACF1-4 isoform (18). MACF1-4 contains eight plectin repeats instead of ABD at the N-terminus (Fig. 1). Studies from the plakins that contain plectin repeats have demonstrated the essential role of plectin repeats in binding IFs. Therefore, the same function of plectin repeats may exist in MACF1-4. This requires further study.
MACF1 IN CELL MIGRATION
MACF1 has a well-known function in cell migration by coordinating F-actin and MTs dynamics (5-7). MACF1 binds to both F-actin and MTs with its ABD and MT-binding domains. In endodermal cells, MACF1 can bind along MTs, especially at the plus-ends of those MTs toward the edges of migrating cells (5, 25). Loss of MACF1 causes perturbations in MTs trajectories, dynamic stabilities, and tracking ability along actin cables, which can affect cell migration in response to wounding (5). MACF1 deficiency compromises the targeting of MTs along F-actin to focal adhesions (FAs), stabilizes FA-actin networks, and impairs epidermal migration (6). The underlying mechanisms involve the F-actin binding domains in MACF1 and an intrinsic actin-regulated ATPase domain of MACF1 for the regulation of cell migration. Therefore, there is a new functional domain in MACF1. MACF1 also sustains directional cell migration in stem cells responsible for homeostasis and wound healing (7). It has been demonstrated that GSK3β can phosphorylate and uncouple MACF1 from MTs during the regulation of cell migration by MACF1 (7). Moreover, MACF1 can regulate cell migration in neurons (8) with several partners, including ErbB2 receptor and ELMO (40, 41).
MACF1 IN CELL SIGNALING
Given the implication that MACF1−/− mice and Wnt3−/− and Lrp5/6 double-knockout mice have similar phenotype, the role of MACF1 in Wnt/β-catenin signal transduction has been determined by Chen et al. (11). Their results have indicated that MACF1 can interact with the Axin complex, including Axin, GSK-3β, β-catenin, and APC. They have also revealed that MACF1 is responsible for the translocation of the Axin complex from the cytoplasm to the cell membrane where MACF1 also binds to co-receptor LRP5/6. Reduction of MACF1 will impair the translocation of the Axin complex, thus inhibiting Wnt-induced TCF/β-catenin-dependent transcriptional activation. In addition, MACF1 has a phosphorylation site for GSK-3β (7). It can also mediate GSK-3 signaling (8). These findings have demonstrated a role of MACF1 in cell signaling. In addition, MACF1 functions in vesicles transport. In axonal vesicle transportation, MACF1 acts as a medium to transport vesicles from the trans-Golgi network to Kif5A. Malfunction of MACF1 can cause vesicles incapable of being mobilized to the cell periphery (42). Interaction between Trans-Golgi protein p230 and MACF1 can mediate the transport of mAtg9 from the trans-Golgi network to peripheral phagophores in the early step of autophagy (43).
MACF1 IN EMBRYO
As a ubiquitously-expressed protein, MACF1 is present at embryonic day 7.5 (E7.5) in mouse (24), which is quite early, suggesting that MACF1 may act as a critical molecule during embryo development. Chen et al. have confirmed the essential role of MACF1 in embryo development using MACF1−/− mice (11). The MACF1−/− mice died at the gastrulation stage with a “head-without-trunk”. They had developmental retardation at E7.5, lacking primitive streak, node, and mesoderm, similar to the phenotypes of Wnt3−/− embryos (44) and LRP5/6 double-knockout embryos (45). Moreover, a deletion mutation in the magellan gene encoding MACF1 in zebrafish caused oocyte polarization failure (46). Taken together, these data demonstrate the importance of MACF1 in embryo development.
MACF1 IN THE NERVE
High levels of MACF1 and significant physiological function of the homolog of MACF1 Kakapo/Shot in the nervous system (15, 47) indicate that MACF1 might play important role in the nervous system. By generating a nervous system-specific Macf1 knockout (cKO) mouse model, Goryunov et al. (22) have underscored the importance of MACF1 in nervous system development. MACF1 cKO brains have been found to have multiple neuronal developmental defects (22). Defects in neuronal migration in the cKO cortex are also observed, suggesting a critical role of MACF1 in neuronal migration (22). In addition, the MACF1c isoform that lacks an ABD compared to MACF1a cannot compensate for the loss of MACF1a (22). Their findings indicate the necessity to interact with actin for MACF1’s function in the nervous system. Other studies have confirmed the role of MACF1 in the regulation of neuronal growth, survival, and migration (8, 48, 49). These evidences indicate the importance of MACF1 in the nerve.
MACF1 IN SKIN
The physiological function of ACF7/MACF1 in mammalian skin has been uncovered mainly by Elaine Fuchs’ group. Karakesisoglou et al. first showed the expression of MACF1 in the epidermis of mice and identified the functional binding sites of MACF1 for both actin and MTs (25). Subsequently, Wu et al. elucidated the function of MACF1 in the skin by conditionally knocking out (cKO) MACF1 in epidermal cells (6, 7). No gross morphological change in the skin or hair coat was observed in MACF1 cKO mice. However, a significant delay in repairing full-thickness wounds due to defects in epidermal cell migration was detected in the skin of cKO mice in response to injury. Further studies revealed that cell migration defects were rooted in perturbations in epidermal-ECM adhesion caused by a malfunction in focal adhesion dynamics due to MACF1 absence (6). Moreover, a link between MACF1 and GSK3β controlling skin stem cell migration and polarized locomotion upon injury was identified (7). Taken together, these results suggest a key role of MACF1 in maintaining skin integrity and regulating epidermal cell migration through coordinating with F-actin and MTs dynamics. Recently, MACF1 has been linked to cellular processes involved in human epidermal cells in response to extremely-low-frequency electric fields (50).
MACF1 IN BONE
MACF1 is a widely expressed protein. However, its role in bone tissue is not well understood. Our group have focused on the role of MACF1 in bone and reported some primary findings in osteoblasts. MACF1 is widely expressed in osteoblastic cells. It partially co-localizes with F-actin and MTs. It is scattered in the cytoplasm and cell periphery. MACF1 also participates in the osteoblast in response to environmental stimuli (51). Under diamagnetic levitation conditions, the distribution of MACF1 in osteoblasts was altered and the co-localization of MACF1 with F-actin and its co-alignment with MTs were also changed (51). Our further studies have demonstrated that MACF1 is crucial for maintaining cell shape and cell proliferation in osteoblasts (52). Knockdown of MACF1 can induce large cells with a binuclear or multinuclear structure, disrupt the organization of F-actin and MTs, and inhibit osteoblast proliferation (52). These data indicate that MACF1 is important in osteoblasts and bone.
MACF1 IN OTHER TISSUES
MACF1 also plays important roles in the heart, colony, and other tissues (53, 54). Fassett et al. (53) have identified a role of MACF1 in regulating cardiomyocyte MTs distribution and adaptation to hemodynamic overload for the first time using inducible cardiac-specific MACF1 knockout (KO) mice. The heart size or function of MACF1 KO mice was not affected under basal conditions. However, exacerbated pressure overload induced left ventricular (LV) hypertrophy, LV dilation, and contractile dysfunction (53). This manifestation has been found to be related to altered MT distribution and several relevant signaling proteins caused by the absence of MACF1 (53). MACF1 is also critical to the maintenance of colonic paracellular permeability because cKO of MACF1 causes significant decrease in colonic mucosal permeability associated with disturbed epithelial arrangement and cytoskeleton dysregulation (54).
MACF1 AND HUMAN DISEASE
In 2014, a human disease caused by a mutation in MACF1 was reported (12). This disease was found in a 12-year-old boy (the proband) who suffered from a skeletal muscle problems with mainly manifestation of generalized hypotonia and pain. Genetic analysis revealed a single duplicated region on chromosome 1p34.3 covering a large part of the MACF1 gene. This affected all 4 major MACF1 isoforms in the proband. Further studies demonstrated that the duplication resulted in a remarkable reduction in MACF1 expression, resulting in reduced motility and internal structural changes of endothelial and satellite cells in the skeletal muscle in the proband. This novel myopathy caused by the duplication in MACF1 was named spectraplakinopathy type 1 (12). The discovery of this MACF1-related genetic human disease has shed light on the importance of MACF1 in human.
Recently, it was found that MACF1 was involved in cancer. A genetic alteration of MACF1 has been detected in numerous cancers, including breast cancer, colorectal cancer, gliomas, and others cancers (55-59). Although the function of MACF1 in cancer requires further investigation, these data suggest that MACF1 might be used as a marker for cancer diagnosis and cancer therapy.
CONCLUSIONS AND FUTURE PERSPECTIVES
In this review, we summarized the current knowledge of MACF1. As an extensively studied mammalian spectraplakin, MACF1 is broadly expressed with versatility. It has differentisoforms, structures, and functions. Similar to other spectraplakins, MACF1 has several isoforms expressed in different tissues. MACF1 participates in many cellular processes and specific-tissue functions. MACF1 is crucial to cell polarization and migration by regulating the dynamics of F-actin and MTs. In addition, MACF1 plays a key role in mediating cellular signaling transduction. The embryonic lethality due to a lack of MACF1 has demonstrated its essential role in embryo development. In addition, MACF1’s ubiquitous expression in all tissue indicates its physiological significance. In fact, MACF1 plays critical roles in controlling the normal development of the nervous system, maintaining skin integrity and colonic paracellular permeability, and adapting cardiomyocyte to hemodynamic overload. Moreover, our findings suggest a novel role of MACF1 in bone tissue. More importantly, mutation in MACF1 has been described in a novel myopathy disease in humans and cancer development. Thus, MACF1 is attracting more attention.
Although MACF1’s functions have been extensively studied and uncovered, many aspects of MACF1 have not been well understood. For example, how is the MACF1 expression regulated? What is the role of MACF1 with its ATPase activity? What is the exact role of MACF1 in cancer development? Considering the fact that MACF1 is involved in both osteoblast function and Wnt/β-catenin signal transduction critical for the regulation of osteoblast differentiation and bone formation, it will be interesting to see whether MACF1 can control bone formation via Wnt/β-catenin signaling. Further study of MACF1 is merited to unravel interesting and exciting new functions of MACF1 in both physiological and pathological settings.
Acknowledgments
This study is partly funded by the National Natural Science Foundation of China (31400725, 31570940), Project Funded by China Postdoctoral Science Foundation (2014M562450, 2015T81051), Project Supported by Natural Science Basic Research Plan in Shaanxi Province of China (2015JQ3076), and Fundamental Research Funds for the Central Universities(3102014JKY15007).
References
- 1.Suozzi KC, Wu X, Fuchs E. Spectraplakins: master orchestrators of cytoskeletal dynamics. J Cell Biol. (2012);197:465–475. doi: 10.1083/jcb.201112034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huelsmann S, Brown NH. Spectraplakins. Curr Biol. (2014);24:R307–308. doi: 10.1016/j.cub.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Roper K, Gregory SL, Brown NH. The 'spectraplakins': cytoskeletal giants with characteristics of both spectrin and plakin families. J Cell Sci. (2002);115:4215–4225. doi: 10.1242/jcs.00157. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenberg A, Liem RK. Plakins in development and disease. Exp Cell Res. (2007);313:2189–2203. doi: 10.1016/j.yexcr.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 5.Kodama A, Karakesisoglou I, Wong E, Vaezi A, Fuchs E. ACF7: an essential integrator of microtubule dynamics. Cell. (2003);115:343–354. doi: 10.1016/S0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Kodama A, Fuchs E. ACF7 regulates cytoskeletal-focal adhesion dynamics and migration and has ATPase activity. Cell. (2008);135:137–148. doi: 10.1016/j.cell.2008.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu X, Shen QT, Oristian DS, et al. Skin stem cells orchestrate directional migration by regulating microtubule-ACF7 connections through GSK3β. Cell. (2011);144:341–352. doi: 10.1016/j.cell.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ka M, Jung EM, Mueller U, Kim WY. MACF1 regulates the migration of pyramidal neurons via microtubule dynamics and GSK-3 signaling. Dev Biol. (2014);395:4–18. doi: 10.1016/j.ydbio.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roper K, Brown NH. Maintaining epithelial integrity: a function for gigantic spectraplakin isoforms in adherens junctions. J Cell Biol. (2003);162:1305–1315. doi: 10.1083/jcb.200307089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouameur JE, Favre B, Borradori L. Plakins, a versatile family of cytolinkers: roles in skin integrity and in human diseases. J Invest Dermatol. (2014);134:885–894. doi: 10.1038/jid.2013.498. [DOI] [PubMed] [Google Scholar]
- 11.Chen HJ, Lin CM, Lin CS, Perez-Olle R, Leung CL, Liem RK. The role of microtubule actin cross-linking factor 1 (MACF1) in the Wnt signaling pathway. Genes Dev. (2006);20:1933–1945. doi: 10.1101/gad.1411206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorgensen LH, Mosbech MB, Faergeman NJ, Graakjaer J, Jacobsen SV, Schroder HD. Duplication in the microtubule-actin cross-linking factor 1 gene causes a novel neuromuscular condition. Sci Rep. (2014);4:5180. doi: 10.1038/srep05180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byers TJ, Beggs AH, McNally EM, Kunkel LM. Novel actin crosslinker superfamily member identified by a two step degenerate PCR procedure. FEBS Lett. (1995);368:500–504. doi: 10.1016/0014-5793(95)00722-L. [DOI] [PubMed] [Google Scholar]
- 14.Bernier G, Mathieu M, De Repentigny Y, Vidal SM, Kothary R. Cloning and characterization of mouse ACF7, a novel member of the dystonin subfamily of actin binding proteins. Genomics. (1996);38:19–29. doi: 10.1006/geno.1996.0587. [DOI] [PubMed] [Google Scholar]
- 15.Leung CL, Sun D, Zheng M, Knowles DR, Liem RK. Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J Cell Biol. (1999);147:1275–1286. doi: 10.1083/jcb.147.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okuda T, Matsuda S, Nakatsugawa S, et al. Molecular cloning of macrophin, a human homologue of Drosophila kakapo with a close structural similarity to plectin and dystrophin. Biochem Biophys Res Commun. (1999);264:568–574. doi: 10.1006/bbrc.1999.1538. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Zhang J, Kraeft SK, et al. Molecular cloning and characterization of human trabeculin-alpha, a giant protein defining a new family of actin-binding proteins. J Biol Chem. (1999);274:33522–33530. doi: 10.1074/jbc.274.47.33522. [DOI] [PubMed] [Google Scholar]
- 18.Gong TW, Besirli CG, Lomax MI. MACF1 gene structure: a hybrid of plectin and dystrophin. Mamm Genome. (2001);12:852–861. doi: 10.1007/s00335-001-3037-3. [DOI] [PubMed] [Google Scholar]
- 19.Brown A, Bernier G, Mathieu M, Rossant J, Kothary R. The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat Genet. (1995);10:301–306. doi: 10.1038/ng0795-301. [DOI] [PubMed] [Google Scholar]
- 20.Leung CL, Zheng M, Prater SM, Liem RK. The BPAG1 locus: Alternative splicing produces multiple isoforms with distinct cytoskeletal linker domains, including predominant isoforms in neurons and muscles. J Cell Biol. (2001);154:691–697. doi: 10.1083/jcb.200012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CM, Chen HJ, Leung CL, Parry DA, Liem RK. Microtubule actin crosslinking factor 1b: a novel plakin that localizes to the Golgi complex. J Cell Sci. (2005);118:3727–3738. doi: 10.1242/jcs.02510. [DOI] [PubMed] [Google Scholar]
- 22.Goryunov D, He CZ, Lin CS, Leung CL, Liem RK. Nervous-tissue-specific elimination of microtubule-actin crosslinking factor 1a results in multiple developmental defects in the mouse brain. Mol Cell Neurosci. (2010);44:1–14. doi: 10.1016/j.mcn.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun D, Leung CL, Liem RK. Characterization of the microtubule binding domain of microtubule actin crosslinking factor (MACF): identification of a novel group of microtubule associated proteins. J Cell Sci. (2001);114:161–172. doi: 10.1242/jcs.114.1.161. [DOI] [PubMed] [Google Scholar]
- 24.Bernier G, Pool M, Kilcup M, Alfoldi J, De Repentigny Y, Kothary R. Acf7 (MACF) is an actin and microtubule linker protein whose expression predominates in neural, muscle, and lung development. Dev Dyn. (2000);219:216–225. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1041>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 25.Karakesisoglou I, Yang Y, Fuchs E. An epidermal plakin that integrates actin and microtubule networks at cellular junctions. J Cell Biol. (2000);149:195–208. doi: 10.1083/jcb.149.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Way M, Pope B, Weeds AG. Evidence for functional homology in the F-actin binding domains of gelsolin and alpha-actinin: implications for the requirements of severing and capping. J Cell Biol. (1992);119:835–842. doi: 10.1083/jcb.119.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winder SJ, Hemmings L, Maciver SK, et al. Utrophin actin binding domain: analysis of actin binding and cellular targeting. J Cell Sci. (1995);108(Pt 1):63–71. doi: 10.1242/jcs.108.1.63. [DOI] [PubMed] [Google Scholar]
- 28.Bandi S, Singh SM, Mallela KM. Interdomain linker determines primarily the structural stability of dystrophin and utrophin tandem calponin-homology domains rather than their actin-binding affinity. Biochemistry. (2015);54:5480–5488. doi: 10.1021/acs.biochem.5b00741. [DOI] [PubMed] [Google Scholar]
- 29.Jefferson JJ, Leung CL, Liem RK. Plakins: goliaths that link cell junctions and the cytoskeleton. Nat Rev Mol Cell Biol. (2004);5:542–553. doi: 10.1038/nrm1425. [DOI] [PubMed] [Google Scholar]
- 30.Jefferson JJ, Ciatto C, Shapiro L, Liem RK. Structural analysis of the plakin domain of bullous pemphigoid antigen1 (BPAG1) suggests that plakins are members of the spectrin superfamily. J Mol Biol. (2007);366:244–257. doi: 10.1016/j.jmb.2006.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sonnenberg A, Rojas AM, de Pereda JM. The structure of a tandem pair of spectrin repeats of plectin reveals a modular organization of the plakin domain. J Mol Biol. (2007);368:1379–1391. doi: 10.1016/j.jmb.2007.02.090. [DOI] [PubMed] [Google Scholar]
- 32.Rezniczek GA, de Pereda JM, Reipert S, Wiche G. Linking integrin α6β4-based cell adhesion to the intermediate filament cytoskeleton: direct interaction between the β4 subunit and plectin at multiple molecular sites. J Cell Biol. (1998);141:209–225. doi: 10.1083/jcb.141.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koster J, Geerts D, Favre B, Borradori L, Sonnenberg A. Analysis of the interactions between BP180, BP230, plectin and the integrin α6β4 important for hemidesmosome assembly. J Cell Sci. (2003);116:387–399. doi: 10.1242/jcs.00241. [DOI] [PubMed] [Google Scholar]
- 34.Hopkinson SB, Jones JC. The N terminus of the transmembrane protein BP180 interacts with the N-terminal domain of BP230, thereby mediating keratin cytoskeleton anchorage to the cell surface at the site of the hemidesmosome. Mol Biol Cell. (2000);11:277–286. doi: 10.1091/mbc.11.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Favre B, Fontao L, Koster J, et al. The hemidesmosomal protein bullous pemphigoid antigen 1 and the integrin β4 subunit bind to ERBIN. Molecular cloning of multiple alternative splice variants of ERBIN and analysis of their tissue expression. J Biol Chem. (2001);276:32427–32436. doi: 10.1074/jbc.M011005200. [DOI] [PubMed] [Google Scholar]
- 36.Yan Y, Winograd E, Viel A, Cronin T, Harrison SC, Branton D. Crystal structure of the repetitive segments of spectrin. Science. (1993);262:2027–2030. doi: 10.1126/science.8266097. [DOI] [PubMed] [Google Scholar]
- 37.Pascual J, Pfuhl M, Walther D, Saraste M, Nilges M. Solution structure of the spectrin repeat: a left-handed antiparallel triple-helical coiled-coil. J Mol Biol. (1997);273:740–751. doi: 10.1006/jmbi.1997.1344. [DOI] [PubMed] [Google Scholar]
- 38.Choi HJ, Park-Snyder S, Pascoe LT, Green KJ, Weis WI. Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure. Nat Struct Biol. (2002);9:612–620. doi: 10.1038/nsb818. [DOI] [PubMed] [Google Scholar]
- 39.Green KJ, Parry DA, Steinert PM, et al. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J Biol Chem. (1990);265:2603–2612. [PubMed] [Google Scholar]
- 40.Zaoui K, Benseddik K, Daou P, Salaun D, Badache A. ErbB2 receptor controls microtubule capture by recruiting ACF7 to the plasma membrane of migrating cells. Proc Natl Acad Sci U S A. (2010);107:18517–18522. doi: 10.1073/pnas.1000975107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margaron Y, Fradet N, Cote JF. ELMO recruits actin cross-linking family 7 (ACF7) at the cell membrane for microtubule capture and stabilization of cellular protrusions. J Biol Chem. (2013);288:1184–1199. doi: 10.1074/jbc.M112.431825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgo A, Proux-Gillardeaux V, Sotirakis E, et al. A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev Cell. (2012);23:166–180. doi: 10.1016/j.devcel.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 43.Sohda M, Misumi Y, Ogata S, et al. Trans-Golgi protein p230/golgin-245 is involved in phagophore formation. Biochem Biophys Res Commun. (2015);456:275–281. doi: 10.1016/j.bbrc.2014.11.071. [DOI] [PubMed] [Google Scholar]
- 44.Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat Genet. (1999);22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- 45.Kelly OG, Pinson KI, Skarnes WC. The Wnt co-receptors Lrp5 and Lrp6 are essential for gastrulation in mice. Development. (2004);131:2803–2815. doi: 10.1242/dev.01137. [DOI] [PubMed] [Google Scholar]
- 46.Gupta T, Marlow FL, Ferriola D, et al. Microtubule actin crosslinking factor 1 regulates the Balbiani body and animal-vegetal polarity of the zebrafish oocyte. PLoS Genet. (2010);6:e1001073. doi: 10.1371/journal.pgen.1001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prokop A, Uhler J, Roote J, Bate M. The kakapo mutation affects terminal arborization and central dendritic sprouting of Drosophila motorneurons. J Cell Biol. (1998);143:1283–1294. doi: 10.1083/jcb.143.5.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanchez-Soriano N, Travis M, Dajas-Bailador F, Goncalves-Pimentel C, Whitmarsh AJ, Prokop A. Mouse ACF7 and drosophila short stop modulate filopodia formation and microtubule organisation during neuronal growth. J Cell Sci. (2009);122:2534–2542. doi: 10.1242/jcs.046268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munemasa Y, Chang CS, Kwong JM, et al. The neuronal EGF-related gene Nell2 interacts with Macf1 and supports survival of retinal ganglion cells after optic nerve injury. PLoS One. (2012);7:e34810. doi: 10.1371/journal.pone.0034810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collard JF, Hinsenkamp M. Cellular processes involved in human epidermal cells exposed to extremely low frequency electric fields. Cell Signal. (2015);27:889–898. doi: 10.1016/j.cellsig.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Qian AR, Hu LF, Gao X, et al. Large gradient high magnetic field affects the association of MACF1 with actin and microtubule cytoskeleton. Bioelectromagnetics. (2009);30:545–555. doi: 10.1002/bem.20511. [DOI] [PubMed] [Google Scholar]
- 52.Hu LF, Su PH, Li RZ, et al. Knockdown of microtubule actin crosslinking factor 1 inhibits cell proliferation in MC3T3-E1 osteoblastic cells. BMB Rep. (2015);48:583–588. doi: 10.5483/BMBRep.2015.48.10.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fassett JT, Xu X, Kwak D, et al. Microtubule Actin Cross-linking Factor 1 regulates cardiomyocyte microtubule distribution and adaptation to hemodynamic overload. PLoS One. (2013);8:e73887. doi: 10.1371/journal.pone.0073887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang Y, Shi C, Yang J, et al. ACF7 regulates colonic permeability. Int J Mol Med. (2013);31:861–866. doi: 10.3892/ijmm.2013.1284. [DOI] [PubMed] [Google Scholar]
- 55.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. (2006);314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 56.Zhang J, Wu G, Miller CP, et al. Whole-genome sequencing identifies genetic alterations in pediatric lowgrade gliomas. Nat Genet. (2013);45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fleischer T, Frigessi A, Johnson KC, et al. Genomewide DNA methylation profiles in progression to in situ and invasive carcinoma of the breast with impact on gene transcription and prognosis. Genome Biol. (2014);15:435. doi: 10.1186/s13059-014-0435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Del Valle PR, Milani C, Brentani MM, et al. Transcriptional profile of fibroblasts obtained from the primary site, lymph node and bone marrow of breast cancer patients. Genet Mol Biol. (2014);37:480–489. doi: 10.1590/S1415-47572014000400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nat Commun. (2014);5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]